Abstract
Clinical trials evaluating cardiac progenitor cells (CPC) demonstrated feasibility and safety, but no clear functional benefits. Therefore a deeper understanding of CPC biology is warranted to inform strategies capable to enhance their therapeutic potential. Here we have defined, using a label-free proteomic approach, the differential cytoplasmic and nuclear compartments of human CPC (hCPC). Global analysis of cytoplasmic repertoire in hCPC suggested an important hypoxia response capacity and active collagen metabolism. In addition, comparative analysis of the nuclear protein compartment identified a significant regulation of a small number of proteins in hCPC versus human mesenchymal stem cells (hMSC). Two proteins significantly upregulated in the hCPC nuclear compartment, IL1A and IMP3, showed also a parallel increase in mRNA expression in hCPC versus hMSC, and were studied further. IL1A, subjected to an important post-transcriptional regulation, was demonstrated to act as a dual-function cytokine with a plausible role in apoptosis regulation. The knockdown of the mRNA binding protein (IMP3) did not negatively impact hCPC viability, but reduced their proliferation and migration capacity. Analysis of a panel of putative candidate genes identified HMGA2 and PTPRF as IMP3 targets in hCPC. Therefore, they are potentially involved in hCPC proliferation/migration regulation.
Subject terms: Cell biology, Cardiology
Introduction
The adult mouse heart has low but intrinsic cardiomyocyte turnover1–3, mainly associated with the function of multipotent resident cardiac stem or progenitor cells (CSC/CPC) populations4,5. The eventual contribution of adult de-differentiating cardiomyocytes to heart homeostasis remains to be consolidated6. Several markers have been proposed to identify and purify CSC/CPC (reviewed in4) but the first5 and the more intensively studied has been the expression of c-Kit+ by CSC (reviewed in7), seeking to explode their expected potential for cardiovascular therapy8–12. In spite of the recent controversy13–15 several important cues have been provided during the last years, and main technical limitations at the origin of the conflict have been mostly clarified16–18. Results using these mouse lines (c-Kit-Cre/Rosa26-floxed-STOP-reporters)14,15 or dual-recombinase approaches13, concluded that cardiac c-Kit+ populations have a marginal cardiomyogenic capacity (0.01% of total cardiomyocytes). Torella et al. provided strong evidences indicating that KitCre alleles, both inducible and constitutive, promote an inefficient recombination of the several reporter constructs evaluated in the cardiac c-Kit+ cells, including a very low recombination (≤ 1%) of c-Kit+lowCD45-CD31-cells. Compelling experiments concluded that c-kit haploinsufficiency in all these genetic fate-mapping experiments is at the most probable origin of all discrepancies, provoking growth and clonogenesis, cardiosphere formation defects. All these deficits, including the cardiomyogenic differentiation capacity were completely rescued by BAC transfection, that harbors the complete cloned c-Kit locus, and, finally, Vicinanza et al. confirmed the described phenotypes in vivo, using cloned cardiac c-kit+lowCD45-CD31- cells17. Therefore, currently murine CSC/CPC population is defined as Sca‐1pos/c‐kitpos/CD31neg/CD45neg/Tryptaseneg 10,18, distinguishing them from cardiac c‐kitpos endothelial (CD31pos) and mast (CD45pos/Tryptasepos) cells; CSC/CPCs are a rare population in the adult heart19. The recent identification of c-Kit+ stem cells in adult vessels20 and the characterization of human cardiac atrial myxomas as the first-described CSC (c-Kit+CD45-CD31-)-related human heart disease21, clearly offer indirect support for ckit+ CSC.
Cardiosphere-derived cells (CDC) and c-kitpos CPC have been characterized and evaluated in pigs and humans. Results from transplantation studies in swine models of cardiac ischemic injury revealed a moderate but reproducible improvement in cardiac function22–27. Multiple lines of evidence from preclinical studies on the transplantation of human or swine CSC/CPC suggested that the mechanisms of action are mainly indirect (reviewed in28), resulting in durable benefits despite low engraftment and cell survival of the transplanted cells29,30.
Human c-kitpos CPC (hCPC) showed weak expression of classical embryonic pluripotency factors (OCT4, NANOG, SOX2 and ECAD). These data and cell membrane expression analyses, coupled with their demonstrated immunoregulatory capacity, indicated that hCPC could be a resident mesechymal stem cell (MSC)-like population31–35. Finally, analysis of the hCPC secretome revealed a strong angiogenic potential and highlighted CXCL6 as an important paracrine factor that signals mainly through CXCR236.
Based on promising preclinical results22,24, a phase I/IIa clinical trial was carried out, using allogeneic hCPC, for the treatment of pacients with large cardiac infarcts (EudraCT 2013–001,358-81;37). While the results demonstrated the feasibility and safety of the approach, no statistical significant functional benefits were demonstrated38. Given all of these data, a deeper understanding of hCPC biology and their behavior in response to acute or diffuse chronic damage might be critical for a better definition of the mechanism of action of these therapies, which might lead to improvements in the current strategies based on hCPC.
With this in mind, here have compared, by a proteomics approach, the differential cytoplasmic and nuclear compartments of hCPC, hMSC and fibroblasts. From this analysis, we focused on two overexpressed nuclear proteins in hCPC, IL1A and IMP3 (IGF2BP3). IL1A was demonstrated to be a dual-function cytokine with a plausible role in apoptosis regulation, and IMP3 regulated proliferation and migration of hCPC.
Results
Comparative analysis of the human CPC cytoplasmic compartment
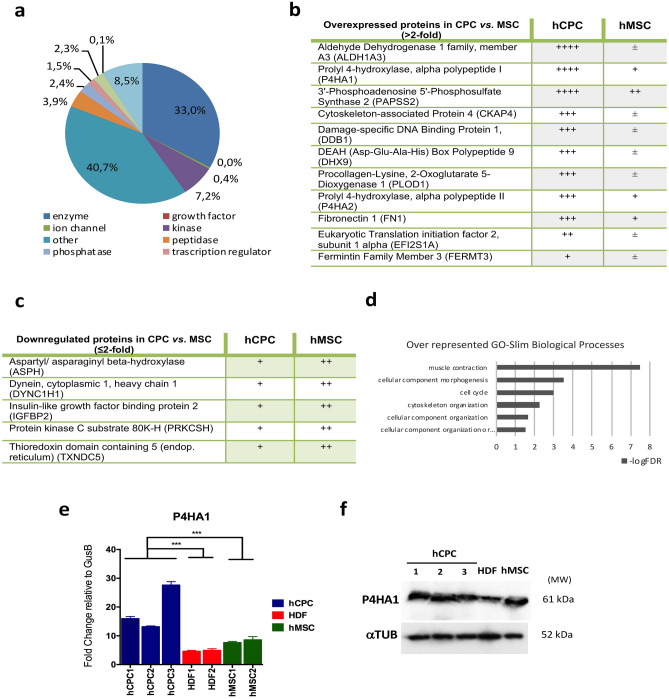
Whole label-free (LF) proteomic analyses of hCPC and hMSC34 yielded 1,260 and 1,176 cytoplasmic proteins, respectively; 95% of which could be mapped onto a GO category. Ingenuity Pathway Analysis (IPA) of the cytoplasmic hCPC subproteome is shown (Fig. 1a). Cytoplasmic fractions of hCPC and hMSC were obtained and analyzed first by LF proteomics; 748 and 707 cytoplasmic proteins were identified in hCPC and hMSC, respectively (Supplementary Fig. S1 online). Among the cytoplasmic proteins expressed more differentially in hCPC, we identified 11 upregulated proteins, including 3'-phosphoadenosine 5'-phosphosulfate synthase 2 (PAPSS2), procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 (PLOD1) and prolyl 4-hydroxylase, alpha polypeptide I/II, (P4HA1 and P4HA2) (Fig. 1b). We also identified 5 moderately downregulated proteins in hCPC, including aspartate beta-hydroxylase (ASPH) and insulin-like growth factor mRNA binding protein 2 (IGFBP2) (Fig. 1c). PANTHER GO-Slim analysis of biological processes using the upregulated hCPC cytoplasmatic proteins clearly indicated an over-representation of muscle contraction-associated proteins in hCPC (Fig. 1d) and PANTHER Pathway analysis showed an over-representation of cytoskeletal regulation by Rho GTPases (Supplementary Fig. S1 online).
Figure 1.
Comparative analysis of hCPC cytoplasmic compartment. (a) Ingenuity pathway analysis of the cytoplasmic hCPC subproteome obtained by whole label-free (LF) proteomics; (b), (c) Main overexpressed (b) or downexpressed (c) proteins in purified cytoplasmic fractions of hCPC compared with hMSC, analyzed by LF proteomics (n = 3); code: ++++ , indicates > 10 peptides; +++ , 5–10 peptides; ++, 2–4 peptides; + , 1 peptide and + / − , 0–1 peptides. (d) PANTHER GO-Slim Biological Processes analysis of overexpressed cytoplasmic proteins in hCPC. (e) Comparative RT-qPCR expression analysis of P4HA1 in the three independent isolates of hCPC (hCPC 1–3), two human fibroblasts (HDF1 and F3) and two hMSC isolates (MSC19 and MSC45). Assays were performed three times and data are expressed as mean ± SD; black lines summarize p-values (*** < 0.002) for hCPC vs. fibroblasts or hMSC (one-way ANOVA analysis of variance followed by the Bonferroni correction for multiple comparison). (f) Western blot analysis of P4HA1 in the three independent isolates of hCPC (hCPC 1–3), human dermal fibroblasts (HDF1) and an hMSC isolate (MSC19); tubulin was used as a loading control. 'Full-length blots/gels' are presented in Supplementary Figure S5 online.
To validate the proteomic data, we compared P4HA1 and ASPH expression in hCPC, hMSC and fibroblasts by RT-qPCR analysis. The data confirmed P4HA1 overexpression in hCPC (Fig. 1e). This was also confirmed by western blotting, although the difference in hCPC P4HA1 expression was less pronounced when compared with human fibroblasts, and no differences were evident when compared with hMSC (Fig. 1f). When we analyzed the gene expression of ASPH, we found that it was not downregulated in hCPC (Supplementary Fig. S1 online), as inferred from the proteomic analysis (Fig. 1c), suggesting a post-transcriptional regulation in hCPC. Additionally, we discarded the possibility that the expression differences found would be associated with the cardiac origin of the hCPC analyzed. We compared their differential expression with total human heart samples confirming all results with the the sole exception of CDH5 (Supplementary Fig. S1 online).
Comparative analysis of the human CPC nuclear compartment
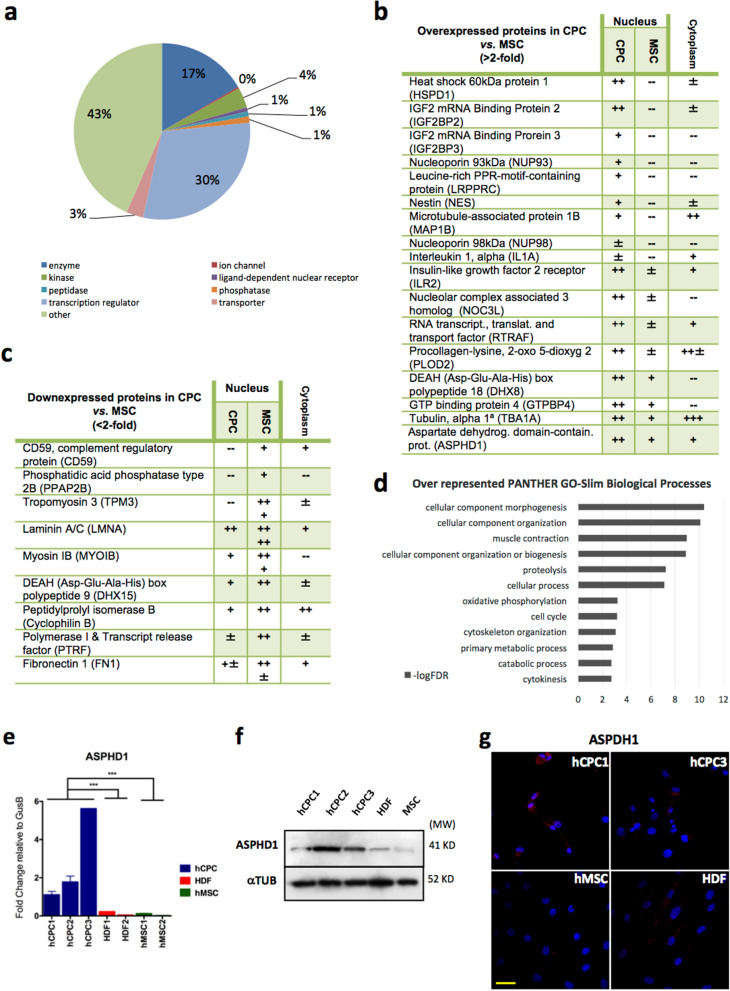
Analysis of the whole LF proteome in hCPC and hMSC yielded 446 and 514 nuclear proteins, respectively34; 95% of which could be included in a GO category. IPA analysis of the nuclear hCPC subproteome is shown (Fig. 2a). LF-proteomic analysis of purified nuclear fractions of hCPC and hMSC rendered 369 and 348 proteins, respectively (Supplementary Fig. S1 online). To confirm the proteins identified in the nuclear fraction, we compared with their representation in the cytoplasm (Fig. 2b,c). The comparative proteomic analysis of hCPC versus hMSC nuclear purified fractions revealed the potential differential expression of 27 proteins in hCPC (Fig. 2b,c). Of the more clearly overexpressed proteins in the hCPC nuclear compartment only IMP3 (also known as IGF2BP3), nestin and IL1A (Fig. 2b) also showed a parallel significant increase in mRNA expression in hCPC relative to hMSC. In addition, hCPC expressed lower nuclear levels of several proteins including Polymerase I and Transcript release factor (PTRF) (Fig. 2c); previous RNAseq studies34 confirmed all dentified proteins. PANTHER GO-Slim analysis of biological processes using the upregulated hCPC nuclear proteins indicated a strong involvement in cellular component morphogenesis and organization, as well as muscle contraction (Fig. 2d). PANTHER Pathway analysis also revealed an important representation of the ubiquitin proteasome pathway (Supplementary Fig. S2 online).
Figure 2.
Comparative analysis of hCPC nuclear compartment. (a) Ingenuity pathway analysis (IPA) of the nuclear hCPC subproteome obtained by whole label-free (LF) proteomics; (b), (c). Main overexpressed (b) or downregulated (c) proteins in purified nuclear fractions of hCPC compared with hMSC, analyzed by LF proteomics (n = 3); code: + + + , indicates 5–10 peptides; + + , 2–4 peptides; + , 1 peptide, + / − , 0–1 peptide and –-, no peptide detected. In parallel, comparative levels of the indicated proteins in cytoplasmic extracts are also indicated. (d) PANTHER GO-Slim Biological Processes analysis of overexpressed nuclear proteins in hCPC. (e) Comparative RT-qPCR expression analysis of ASPHD1 in the three independent isolates of hCPC (hCPC 1–3), two human fibroblasts (HDF1 and HDF2) and two hMSC isolates (MSC19 and MSC45). Assays were performed three times and data are expressed as mean ± SD; black lines summarize p-values (*** < 0.002) for hCPC vs. fibroblasts or hMSC (One-way ANOVA analysis of variance followed by the Bonferroni correction for multiple comparison). (f) Western blot analysis of ASPHD1 in the three independent isolates of hCPC (hCPC 1–3), human dermal fibroblasts (HDF1) and hMSC (MSC19) isolate; tubulin was used as a loading control.'Full-length blots/gels' are presented in Supplementary Figure S4 online. (g) Comparative immunofluorescence analysis of ASPHD1 (red) in hCPC (hCPC 1 and 3), hMSC (MSC19) and human dermal fibroblasts (HDF); nuclei were counterstained with DAPI (blue). Bar, 20 μm.
Aiming to validate the proteomics nuclear data we evaluated the expression of ASPHD1 and PTRF, which were up- and down-regulated, respectively, in hCPC versus hMSC, by proteomics. RT-qPCR analysis confirmed a significant differential expression of ASPHD1 in all hCPC isolates (hCPC1–3) in comparison with hMSC and fibroblasts (Fig. 2e). The preferential expression in hCPC was also confirmed by western blotting and by immunofluorescence (Fig. 2f,g). By contrast, PTRF downregulation in hCPC was not confirmed by RT-qPCR (Supplementary Fig. S2 online), suggesting again a relevant post-transcriptional regulation.
Because a very significant fraction of regulatory nuclear proteins is expressed at low levels, below the detection limits of proteomics, we validated by RT-qPCR several transcriptional factors found up- and down-regulated, by RNAseq in hCPC compared with hMSC34. GATA4, SOX17, WT1 and GATA2 were robustly overexpressed in hCPC in comparison with hMSC (Supplementary Fig. S2 online). The expression levels of TBX3 and MEF2C were also significantly higher in hCPC than in hMSC, but less pronounced. We also confirmed that HOXD8 and HOXA10 were barely expressed by hCPC in comparison with hMSC (Supplementary Fig. S2 online).
IL1A is a dual-function cytokine in hCPC
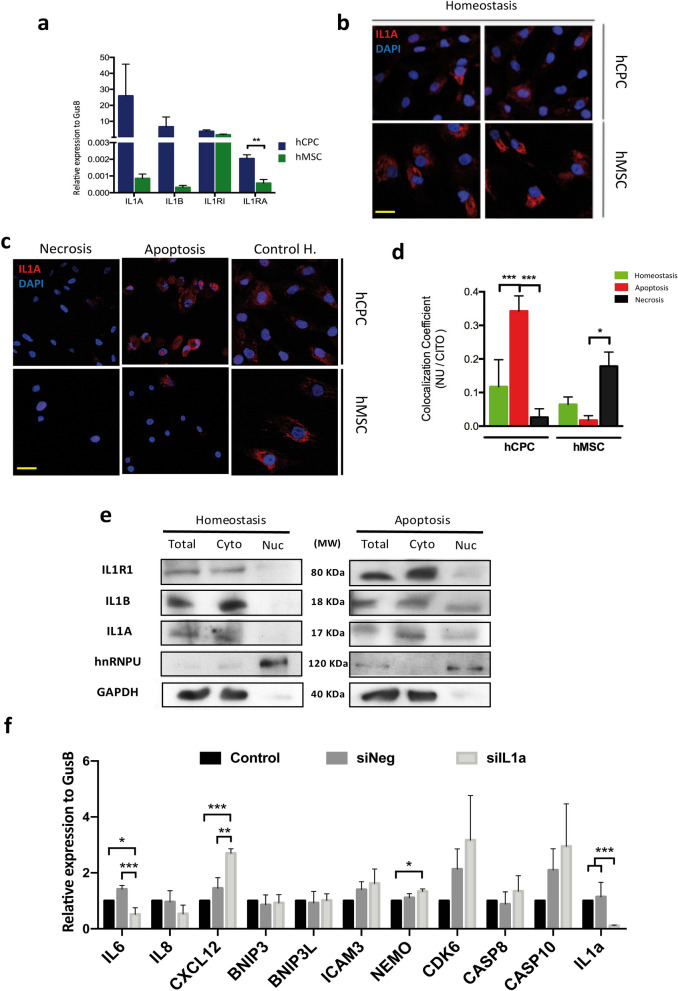
Two of the most differentially expressed genes in hCPC were IL1A (as mentioned above) and IL1B, whose overexpressions were also validated by RNAseq34. IL1A was found over-represented by comparative LF-proteomic analysis in the nuclear compartment of hCPC compared with hMSC (Fig. 2b). IL1A is produced as a precursor protein that yields a mature form and an N-terminal propeptide, containing a nuclear localization sequence that allows access to the nuclear compartment. In this way, IL1A is a well established “dual-function cytokine” that plays a role in the nucleus, independently of its classical extracellular mediated effects39–41.
To validate the proteomic analysis, we assessed the expression of IL1A and IL1B by RT-qPCR in hCPC and hMSC. The results clearly confirmed the overexpression of IL1A and IL1B in hCPC (6,788- and 1,409-fold, respectively, Fig. 3a). We also tested the expression of other members of the IL1 signaling pathway. A significant increase (3.22-fold change) was found for the expression of the natural antagonist of IL1R1 (IL1RA) (Fig. 3a). Other main members of the IL1 pathway, IL1 receptor (IL1RI), IL38 and the secondary IL1 receptor (IL1R2) were, or not differentially expressed (IL1RI) or not detected. We next evaluated whether IL1A could acts as a dual-function cytokine in hCPC, as has previously been described in other cell lineages42. We analyzed in hCPC the behavior of IL1A, IL1B and IL1RI expression in response to apoptosis- or necrosis-promoted by oxidative stress (see Methods). We found that IL1RI expression was equivalently and moderately reduced (~ 25%) by both treatments (preferential apoptosis or necrosis). IL1A and IL1B expression were also decreased by both treatments, but to a much greater extent; IL1A expression was more pronouncedly reduced (90–95% reduction) when necrosis was induced (Supplementary Fig. S3 online).
Figure 3.
Comparative functional evaluation of IL1A in hCPC. (a) Comparative RT-qPCR expression analysis of IL1A, IL1B, IL1R1 and IL1RA in hCPC1 compared with hMSC (MSC19). (b) Immunofluorescence analysis of IL1A expression (red) in hCPC1 compared with hMSC (MSC19), in homeostasis; nuclei were counterstained with DAPI (blue). Bar, 20 μm. (c) Comparative immunofluorescence analysis of IL1A expression (red) in hCPC1 and MSC19, in homeostasis or after induction apoptosis or necrosis; nuclei were counterstained with DAPI (blue). Bar, 20 μm. (d) Quantification by immunofluorescence of nuclear/cytoplasmic location of IL1A in hCPC1 and MSC19, comparing homeostasis and after apoptosis or necrosis induction; co-localization of IL1A with DAPI signal was compared with cytoplasmic pool; fluorescence intensity was measured using ImageJ software (NIH, Bethesda, MD). (e) Representative western blot analysis of IL1R1, IL1A and IL1B expression in purified cytoplasmic (Cyto) and nuclear (Nuc) fractions of hCPC1, in homeostasis (left) or after induction of apoptosis (right). GAPDH and hnRNPU were used as internal controls of cytoplasmic and nuclear fractions, respectively; lower contrast in these blots is caused by the higher intensity signal of these proteins. 'Full-length blots/gels' are presented in Supplementary Figure S5 online. (f) Target evaluation of IL1A in hCPC subjected to oxidative-mediated apoptosis. A panel of candidate genes, previously reported to be involved in apoptosis or immunoregulation, were evaluated by RT-qPCR in hCPC where IL1A was downregulated (siIL1A) in comparison with negative control hCPC (siNeg) and untransfected hCPC cells (Control). IL1A was confirmed to be significantly downregulated (> 90%). Assays were performed three times and data are expressed as mean ± SD; black lines summarize p-values (*** < 0.002; ** < 0.02; * < 0.05; one-way ANOVA analysis of variance followed by the Bonferroni correction for multiple comparison).
Immunofluorescence analysis of IL1A, with an antibody against the full size protein, revealed a highly preferential cytoplasmic location in basal conditions (homeostasis; Fig. 3b). However, while hCPC showed significantly higher levels of IL1A mRNA expression than hMSC (Fig. 3a), the latter showed higher levels of IL1A protein (Fig. 3b), suggesting an important lineage-specific post-transcriptional regulation. In homeostasis the results confirmed an almost exclusive cytoplasmic location for all proteins analyzed. The fraction of nuclear IL1A detected by proteomics (Fig. 2b) thus seems to be minor. Of note, IL1A behaved differently to the two stmuli in hCPC and hMSC, as evaluated by immunofluorescence. Upon induction of apoptosis, IL1A protein was significantly upregulated in hCPC, whereas it was clearly reduced in hMSC (Fig. 3c); induction of necrosis provoked a major loss of IL1A in both cell types (Fig. 3c). Quantification of nuclear versus cytoplasmic localization by immunofluorescence of IL1A in hCPC, comparing homeostasis with the induction of apoptosis or necrosis, revealed a significant increase in the nuclear location of IL1A (co-localization with DAPI signal) after the induction of apoptosis (Fig. 3d). The opposite was found in hMSC where co-localization of IL1A with the nuclear compartment was poorer upon apoptosis induction, although it was augmented after necrosis induction (Fig. 3d). All these results suggest a high variable cell-type linage behavior.
We therefore performed western blotting of subcellular compartments in hCPC subjected to apoptosis compared with homeostasis (Fig. 3e); in necrotic cells, expression of the three proteins was quite low and difficult to quantify because of the strong loss of cellular content. In agreement with the immunofluorescence study (Fig. 3c), a substantial fraction of IL1A and IL1B was found in the nuclear compartment (Fig. 3e) of apoptotic cells whereas the subcellular localization of IL1RI was barely unchanged by the induction of apoptosis. Densitometric analysis of the representative western experiment shown (Fig. 3e) yielded an important nuclear/cytoplasmic ratio increment for IL1A and IL1B, in apoptotic cells compared with cells in homostasis, and a modest variation on IL1RI (Supplementary Fig. S3 online).
Furthermore we analyzed whether IL1A could be regulating transcription during early apoptosis. For that we have compared expression level of several genes involved in apoptosis or inflammation in hCPC cells knockdown (~ 20%) for IL1A (hCPC-siIL1A) in comparison with control hCPC cells (hCPC-siNeg) or negative-control tranfected cells (Supplementary Fig. S3 online); all cell populations were exposed to H2O2 during a short period (500 μM; 5 h) (Fig. 3f). Results indicated that although several genes involved in apoptosis (BNIP3, BNIP3L and ICAM3), are non-affected by IL1A knockdown, CDK6, Casp8, Casp10 showed a clear trend for overexpression in IL1A knockdown hCPC-siIL1A although the differences did not result statitistically significant. CXCL12 and NEMO were clearly overexpressed in for hCPC-siIL1A compared with control hCPC-siNeg. Finally, expression of IL6 was also clearly downregulated in hCPC-siIL1A. Altogether, these data reinforced that idea that IL1A could be acting as dual-cytokine in hCPC with a potential role in the transcriptional regulation in apoptosis.
Given the known immunoregulatory capacity of hCPC32–34 and their definition as an MSC-like cell subpopulation33, it is possible that IL1A could play a role in hCPC in homeostasis. We thus evaluated whether IL1A could contribute to the immunoregulatory capacity of hCPC. Thus, we co-cultivated phytohemagglutinin-stimulated human CD3 T cells with control hCPC, hCPC siIL1A, or hCPC siNeg. All cell populations (hCPC, hCPC siIL1A and hCPC siNeg) demonstrated similar immunoregulatory capacity at the higher cell doses analyzed (1:10–1:20), which was lost when lower doses were evaluated (1:40). Therefore no significant changes in immunoregulatory capacity were found (Supplementary Fig. S3 online), indicating that IL1A seems not to have a relevant role in the T cell immunoregulatory capacity of hCPC.
Functional evaluation of IMP3 in hCPC in homeostasis and in response to oxidative damage
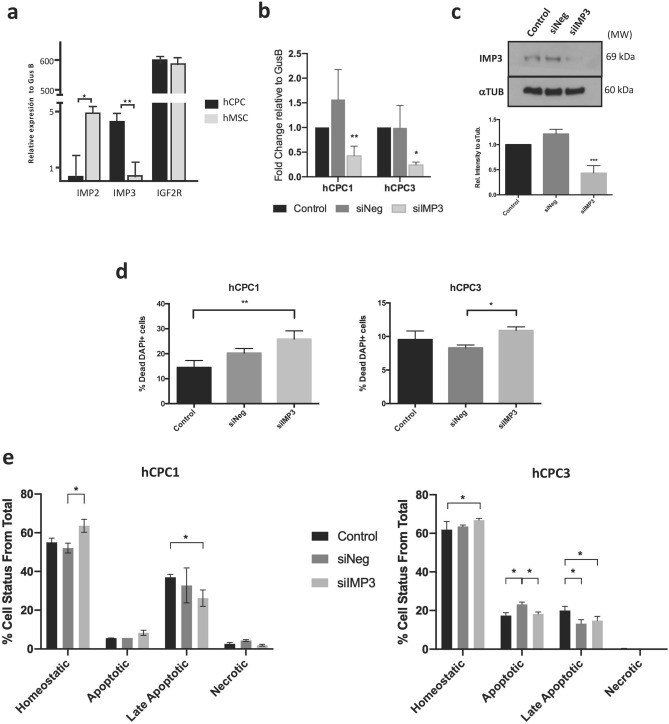
IGF2 is the predominant form of IGF in humans43 and it binds to insulin-like growth factor 1 receptor (IGF1R), insulin-like growth factor 2 receptor (IGF2R; CD222) and the insulin receptor A isoform (IR-A). It seemed interesting that in addition to IGF2R, two additional members of the IGF2 pathway (insulin-like growth factor mRNA binding proteins 2 and 3; IMP2/IMP3) were identified as over-represented in the hCPC nuclear subproteome by LF-proteomics (Fig. 2b). RT-qPCR analysis validated the high levels of IMP3 expression in hCPC versus hMSC (> 40-fold overexpression), but the opposite was observed for IMP2 (Fig. 4a).
Figure 4.
Functional evaluation of IMP3 in hCPC in homeostasis and in response to oxidative damage. (a) Comparative RT-qPCR expression analysis of IGF2R, IMP2 and IMP3 in hCPC1 and MSC19. (b), (c) Confirmation of downregulation of IMP3 in hCPC (1,3) transfected with sIMP3 compared with a negative control (siNeg) and untransfected control cells (control), by RT-qPCR relative to the expression of GusB (b) and western blot (c); bottom panel corresponds to the quantification, relative to tubulin, on the representative western blot (upper panel); 'full-length blots/gels' are presented in Supplementary Figure S5 on line. All samples were analyzed 48 h post-transfection. (d) Evaluation of cell viability in hCPC (1,3) transfected with sIMP3 compared with a negative control (siNeg) and untransfected control cells (control), by DAPI staining, evaluated 48 h post-transfection. (e) Analysis of the effects of IMP3 downregulation on hCPC (1,3) response to oxidative damage induced by H2O2. hCPC control, siIMP3- or siNeg-transfected cells were exposed to H2O2 (500 μM) for 48 h; then cultures were stained with AnnexinV/propidium iodide (Anex.V/P.I.) and homeostatic viable (Anex.V − /PI −), apoptotic (Anex.V + /PI −), late apoptotic (Anex.V + / PI +) or necrotic (Anex.V −/PI +) cells were quantified by cytometry. Assays were performed three times and data expressed as mean ± SD; black lines summarize p-values (*** < 0.002, ** < 0.02, * < 0.05; one-way ANOVA analysis of variance followed by the Bonferroni correction for multiple comparison).
IMP3 belongs to a family of mRNA-binding proteins that bind to multiple mRNAs in mammalian cells, including IGF244. Based on previous literature43, we hypothesized that high levels of IMP3 would lead to a decrease in the autocrine bioavailability of IGF2, reducing the potential signaling through IGF2R and triggering senescence/apoptosis. We first analyzed the impact of IMP3 knockdown in two independent hCPC isolates. Cells transfected with siIMP3 showed significantly reduced levels of IMP3 when compared with negative control or non-transfected control cells, analyzed both by RT-qPCR and western blotting (Fig. 4b,c). IMP3 silencing did not affect negativelly hCPC viability, 48 h post-transfection; in fact, IMP3-silenced cells showed a moderate increase in viability (Fig. 4d). We then analyzed the effects of IMP3 silencing on the response of hCPC to oxidative stress (500 μM H2O2, during 48 h) and evaluating apoptotic and necrotic cells with the Annexin V/propidium iodide (Supplementary Fig. S4 online). Neither of the two hCPC populations tested showed any remarkable difference in the percentages of homeostatic, apoptotic, late apoptotic or necrotic cells (Fig. 4e). Thus, IMP3 does not seem to play a critical role in the regulation of hCPC response to oxidative stress-mediated apoptosis.
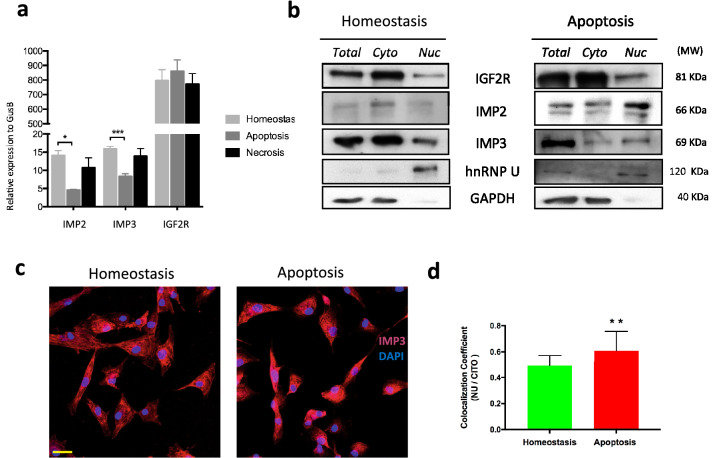
Although IMP3 seems not to be essential for apoptotic responses we investigated IMP3 regulation in hCPC damage responses. We first studied the impact of apoptosis or necrosis induction on the transcriptional activity of IMP3, IMP2 and IGF2R, and their subcellular localization. Neither apopotosis nor necrosis affected IGF2R expression; however, the induction of apoptosis (but not necrosis) promoted a significant decrease of IMP3 and IMP2 transcription in hCPC (Fig. 5a). Western blotting of hCPC showed that IGF2R and IMP3 were expressed at similar levels whereas IMP2 was expressed at apparently lower levels (Fig. 5b). Analysis of nuclear and cytoplasmic fractions by western blotting confirmed that a substantial fraction of IMP3, and to a lesser extent IMP2, was found in the nuclear fraction in homeostasis (Fig. 5b, left panel). After induction of apoptosis, both IMP2 and IMP3 showed an increased presence in the nuclear compartment with respect to the cytoplasmic compartment, whereas IGF2R was unchanged (Fig. 5b; right panel); densitometric analysis of the representative western blot shown (Fig. 5b) yield an increase in nuclear IMP2 and IMP3 of 8.5-fold and 13-fold, respectively, in apoptotic cells compared with cells in homeostasis (Supplementary Fig. S4 online). These results were confirmed by immunofluorescence (Fig. 5c,d). We analyzed the nuclear versus cytoplasmic localization of the IMP3 fluorescent signal and we confirmed that, upon induction of apoptosis, the nuclear pool of IMP3 singnificantly increases (co-localization coefficient referred to DAPI signal) (Fig. 5d).
Figure 5.
Evaluation of IMP3 expression and subcellular localization after apoptosis in hCPC. (a) Comparative RT-qPCR expression analysis of IMP2, IMP3 and IGF2R in hCPC1 in homeostasis and after the induction apoptosis and necrosis. Assays were performed three times and data are expressed as mean ± SD; black lines summarize p-values (** < 0.02; * < 0.05; one-way ANOVA analysis of variance followed by the Bonferroni correction for multiple comparison). (b) Representative western blot analyses of IGF2R, IMP2 and IMP3 expression in purified cytoplasmic (Cyto) and nuclear (Nuc) fractions of hCPC1 in homeostasis (left panel) or subjected to apoptosis (right panel). GAPDH and hnRNPU were used as internal controls of cytoplasmic and nuclear fractions, respectively; lower contrast in these blots is caused by the higher intensity signal of these proteins. 'Full-length blots/gels' are presented in Supplementary Figure S5 online. (c) Comparative immunofluorescence analysis of IMP3 expression (red) in hCPC1, in homeostasis or after induction apoptosis; nuclei were counterstained with DAPI (blue); Bar, 20 μm. (d) Quantification by immunofluorescence of nuclear/cytoplasmic location of IMP3 in hCPC1, comparing homeostasis and after apoptosis induction; co-localization coefficient of IMP3 with DAPI signal was compared with cytoplasmic pool; fluorescence intensity was measured using ImageJ software (NIH, Bethesda, MD). Assays were performed three times and data are expressed as mean ± SD; black lines summarize p-values (** < 0.02; t-test analysis for data with paired standard desviation).
Thus, apoptosis induction in hCPC triggers a significant decrease in IMP2 and IMP3 transcription, concomitant with an enrichement of both proteins in the nuclear compartment. These results suggest a non-essential role of IMP3 in gene expression regulation upon induction of oxidative stress-mediated apoptosis.
IMP3 regulates proliferation and migration of hCPC
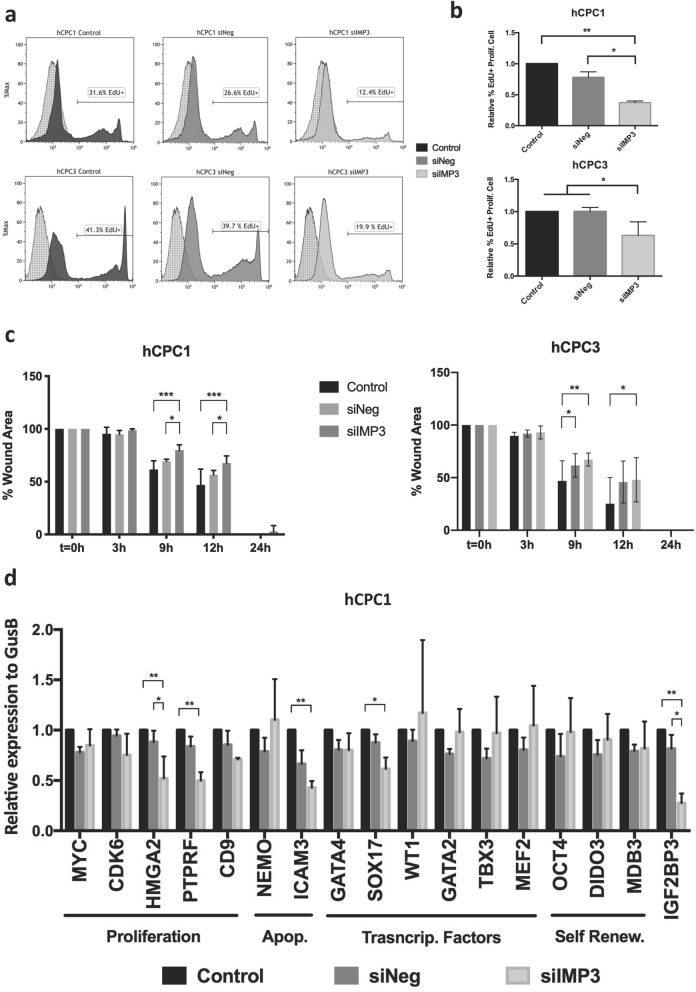
Analysis of proliferation in IMP3-silenced cells and controls estimated by EdU (5-ethynyl-2’-deoxyuridine) incorporation during 12 h and 48 h post-transfection revealed that the knockdown of IMP3 significantly reduced proliferation in hCPC1 cells (about twofold) and a similar effect was found in hCPC3 cells (Fig. 6a,b). Thus, IMP3 seems not to be relevant for survival, but is likely involved in hCPC proliferation regulation.
Figure 6.
IMP3 regulates proliferation and migration of hCPC. (a), (b) Analysis of the effect of IMP3 downregulation on hCPC proliferation rate, estimated by EdU (5-ethynyl-2’-deoxyuridine) incorporation during 12 h. hCPC (1,3) transfected with sIMP3 were compared with a negative control (siNeg) and untransfected control cells (Control), were evaluated 48 h post-transfection by flow cytometry. (a) Representative histograms. (b) Percentage of EdU + cells. (c) Evaluation of the effect of IMP3 downregulation (siIMP3) on the wound-healing capacity of two independent hCPC isolates (1,3) compared with a siNeg and control non-transfected cells 48 h post-transfection. Evolution of the wounded area was monitored during 24 h. (d) Target evaluation of IMP3 in hCPC. A panel of candidate genes, previously reported to be regulated by IMP3 in heterologous models or described as preferentially expressed in hCPC, were evaluated by RT-qPCR; genes are related to proliferation (MYC, CDK6, HMGA2, PTPRF, and CD9), apoptosis (NEMO and ICAM3), transcriptional factors (GATA4, SOX17, WT1 and GATA2) and self-renewal (OCT4, DIDO3 and MBD3). IMP3 was confirmed to be significantly downregulated (> 70%). IMP3 knockdown hCPC cells (siIMP3) were compared with a siNeg and non-transfected control cells. Assays were performed three times and data expressed as mean ± SD; black lines summarize p-values (*** < 0.002, ** < 0.02, * < 0.05; one-way ANOVA analysis of variance followed by the Bonferroni correction for multiple comparison).
We then evaluated the potential implication of IMP3 in cell motility, as previously proposed45 using wound-healing assays. Monolayers of hCPC cells silenced or not for IMP3 were compared in their capacity to repair a wound during 24 h, as described34. As shown in Fig. 6c, both IMP3-silenced hCPC isolates demonstrated a statistically significant delay (at 9–12 h) in wound healing, albeit with different kinetics (Fig. 6c).
Finally, we analyzed a panel of candidate target genes previously reported to be regulated by IMP3 in heterologous models, or described as preferentially expressed in hCPC34,36. We used RT-qPCR to compare the levels of gene expression in hCPC silenced for IMP3 knockdown (hCPC-siIMP3) in comparison with control hCPC cells or negative-control tranfected cells (hCPC-siNeg). Figure 6d shows the results obtained with isolate hCPC1, which showed more robust IMP3 silencing.
Concerning genes involved in proliferation, we found that PTRF (also known as Cavin1 or Cavin-1) and HMGA2 (high-mobility group AT-hook 2) were significantly downregulated in hCPC- siIMP3 cells (~ 50%). c-MYC, CDK6 and CD9 were moderately but not significantly downregulated (< 20%). In relation to genes involved in apoptosis, we found that ICAM3 expression was significantly reduced (~ 60%) in IMP3-silenced cells, but unexpectedly not NEMO (inhibitor of nuclear factor kappa B kinase subunit gamma).
We also tested the consequence of IMP3 silencing for the expression of a small panel of transcriptional factors previously (Supplementary Fig. S2 online) defined in hCPC. All them, except SOX17 expression (~ 40% reduction), did not modify the expression in hCPC-siIMP3. These results suggest that IMP3 might have a modest role in regulating hCPC fate-genes by regulation of SOX17. We additionally analyzed the potential impact of IMP3 knockdown on several genes associated in other cell types with self-renewal, such as OCT4, DIDO3 and MBD335,37,46. The expression of all three genes was unaffected by IMP3 silencing (Fig. 6d). These results suggest that IMP3 seems not to be mainly involved in the regulation of the undifferentiated state of hCPC. A similar analysis using hCPC3 yielded essentially identical but non-significant results.
Finally, because nuclear IL1A also regulates cell proliferation and migration39,40, we sought to evaluate the potential involvement of IMP3 in the regulation of the IL1 pathway. IMP3-silenced hCPC and controls were evaluated by RT-qPCR 48 h post-transfection. Compared with control hCPC, IMP3-silencing failed to affect IL1A expression but enhanced IL1B expression (Supplementary Fig. S4 online). IL1RA and IL1R1 were also unaffected by IMP3-silencing (Supplementary Fig. S4 online), indicating that IMP3 is likely not involved in the regulation of IL1 pathway in hCPC.
Discussion
Cardiosphere-derived cells (CDC) and c-kitpos hCPC have been previously characterized and evaluated in preclinical studies, demonstrating modest therapeutic efficacy in acute ischemia models22–25. Based on previous studies, hCPC were defined as an hMSC-like population with confirmed immunoregulatory capacity33,34,36. Considering the promising, but not statistically significant, results of the clinical studies based on these cells37,38 a more detailed description of hCPC populations might lead to a better understanding of their mechanism of action and, ultimately, the development of more effective treatments.
Analysis of the most relevant cytoplasmic proteins over-represented in hCPC suggests that these cells might be well suited to mount an effective response to hypoxia, demonstrating also an active collagen metabolism. P4HA1 and P4HA2 are both overexpressed (P4HA1 > P4HA2) in the cytoplasmic compartment of hCPC, being both activated by hypoxia47. We also found PLOD1 and PLOD2 to be significantly overexpressed. P4HA1 and P4HA2 are required for collagen deposition, whereas PLOD2 is required for extracellular matrix stiffening and collagen fiber alignment. Furthermore, DHX9, an ATP-dependent helicase of double-stranded RNA and DNA-RNA complexes48, is highly upregulated during activation of quiescent cells to collagen-producing cells49. In the context of different adult stem cell compartments, CKAP4 and DHX9, both overexpressed in the hCPC cytoplasm, have been related to differentiation regulation50. CKAP4 is a nucleoplasmic shuttle protein that acts as a high-affinity receptor for antiproliferative factor (APF)51. DHX9 has been also proposed as a RISC-loading factor52. Concerning the proteins that were found moderately downregulated in hCPC cytoplasm, ASPH and TXNDC5 are also implicated in proliferation and cell motility regulation53,54. Altogether, hCPC might demonstrate an effective in response to hypoxia associated-damage showing an active remodeling of the extracellular matrix.
Regarding the proteins preferentially expressed in the hCPC nuclear compartment, we confirmed high levels of expression of several cardiogenic transcriptional factors such as GATA4, SOX17, WT1, GATA2 and TBX3 (Supplementary Fig. S2 online), with GATA4 and SOX17 more differentially expressed in comparison with hMSC. In addition, comparative proteomics analysis of enriched nuclear fraction yielded a panel of proteins more represented in hCPC than in hMSC. Among them, ASPDH1 that was also confirmed by RT-qPCR is poorly characterized. Finally, among the proteins under-represented in the hCPC nuclear compartment, it is noteworthy that levels of PTRF/cavin-1 have been directly associated with cell senescence. PTRF has been demonstrated to mediate in transcription pausing and termination, and the final dissociation of the transcription complex55.
Among the nuclear over-represented proteins in hCPC, IL1A and IMP3 were selected for further analysis. IL1A is a pro-inflammatory cytokine with multiple immune-regulatory functions. It is mainly expressed as a cell-associated form and not actively secreted in healthy tissue, but its membrane-associated form is critically involved in cell senescence56. IL1A is one of the four (IL1A, IL33, HMGB1 and S100) “dual-function cytokines” described in mesenchymal cells. These cytokines play a role in the nucleus independently of their extracellular-mediated effects, as a classical cytokines, and have been also called “damage-associated molecular pattern” molecules or alarmins42. Unlike IL1B, processed IL1A has a nuclear localization sequence and is trafficked to the nucleus, regulating cell proliferation and migration40,41. For example, in acute lymphocytic leukemia T cells, overexpression of the IL1A nuclear propeptide has been demonstrated to promote proliferation and reduce apoptosis, by NFkB and SP1 up-regulation57. Analyses of hCPC in homeostasis demonstrated a strong post-transcriptional regulation of IL1A mRNA and a highly preferential cytoplasmic location of IL1A. We found that IL1A is not related with the immunoregulation capacity of hCPC but, upon induction of apoptosis, IL1A was clearly upregulated and a substantial nuclear fraction was found; this behavior was not paralleled by hMSC. We also found a similar intracellular pattern for IL1B, although less pronounced. In this sense, IL1A knockdown in hCPC, follow by a short period of oxidative stress-associated apoptosis, demonstrated significant alterations in several apoptosis or inflammatory genes. Overall these results suggested that IL1A, and probably IL1B, could have dual-cytokine profile in hCPC, playing a role in the regulation of response to apoptosis.
IMP3 is an mRNA binding protein that, among other functions, regulates IGF2 expression44. In the context of cancer, there are numerous examples of the critical role of IMP3 favoring chemoresistance, aggressiveness and metastasis58,59. In neural and pancreatic cancer cells, IMP2- and IMP3-bound transcripts are localized in cytoplasmic RNA granules that accumulate in dendrites or membrane protrusions, where they are preferentially translated60. In pancreatic ductal adenocarcinoma cells, IMP3 modulates miRNA-mRNA interactions61. However, IMP3 binding could result both in an enhanced expression of the target mRNA62 or its destabilization63. Although different pathways and targets have been associated with the overexpression of IMP3 in cancer, few studies have addressed the role of IMP3 in healthy developmental processes; i.e. muscle growth is regulated by IMP3 levels, controlled by let-7b64 and adult megakaryocyte development is also under the control of IMP3, by regulating P-TEFb45.
hCPC in homeostasis show a clear overepresentation of IMP3, but not IMP2, in the nuclear compartment and induction of apoptosis provoked an enrichment in the nuclear compartment. IMP3 knockdown reduced hCPC proliferation and migration capacity, although it had no obvious impact on viability. IMP3 has been found to promote cell migration in glioma by increasing the levels of p65 protein (RELA; subunit of NF-κB heterodimer), but without modifying transcript levels65. In glioblastomas IMP3 also promotes cell proliferation, migration and invasion by inducing epithelial-mesenchymal transition58.
Finally, we analyzed a panel of candidate target genes whose expression could be affected by the downregulation of IMP3 in hCPC. cMYC, CD44 and CDK6 were demonstrated previously to be targeted by IMP3 in mixed lineage leukemia, enhancing the half-life of the transcripts62. Silencing of IMP3 (hCPC-siIMP3 cells), however, resulted in a moderate and non-significant downregulation of these targets in hCPC. By contrast, silencing of IMP3 led to a significant downregulation of HMGA2 and PTPRF. HMGA2 is considered as an architectural transcription factor that is involved in growth regulation and tumorigenesis46. Interestingly, it has been demonstrated that IMP3 ribonucleoprotein complexes contain HMGA2 mRNA, preventing miRNA-directed mRNA decay during tumor progression66. In addition, it has been recently demonstrated that HMGA2 controls both, proliferation and migration / metastasis, in colon cancer67; analyses of other HMGA2 candidate genes associated with cell migration (ARF6, ARHGEF4) rendered negative results. It is also worth noting that HMGA2 mRNA is significantly overexpressed (17.8 fold) in hCPC versus hMSC34. By contrast, high PTRF expression levels correlate with an increased senescence in human fibroblasts55. Therefore, these data suggest that PTRF and HMGA2 are regulated by IMP3 and, consequently, could be involved in hCPC proliferation/migration regulation.
In conclusion, we have compared, using a label-free proteomic approach, the differential cytoplasmic and nuclear compartments of human CPC (hCPC) versus human mesenchymal stem cells (hMSC) and fibroblasts. Globally, hCPC, with a clear cardiogenic transcriptional factor profile, are well suited to mount an effective response to hypoxia with active collagen metabolism. IL1A, characterized as a dual-function cytokine, seems to play a role in the regulation of the hCPC response to apoptosis caused by oxidative stress. Finally, IMP3 was demonstrated to be involved in hCPC proliferation and migration.
Methods
Ethical approval
Human CPC were obtained from human right atria appendage from adult donors, with no relevant cardiac pathology, and subjected to cardiac surgery with extracorporeal circulation; during the procedure, this tissue is normally discarded during cannulation. Human CPC were isolated from human myocardial samples by c-kit immunoselection, as described32. Procedures were approved by the hospital ethical committees (Hospital 12 de Octubre and Hospital Universitario Gregorio Marañón, Madrid, Spain) with the corresponding patient informed consents. All methods were carried out in accordance with relevant guidelines and regulations (R.D. 9/2014 and Orden SSI/2057/2014, which transpose the European Commission Directive 2012/39/UE). hCPC1-hCPC3 isolates have been previously characterized34,36.
Cells and culture conditions
hCPC were maintained and expanded as previously indicated34,36, essentially under equivalent conditions to those used in the CAREMI clinical trial (EudraCT 2013-001358-81). See Supplementary "Methods" section online for details. All cells were expanded and manipulated (induction of oxidative damage and transfections) in an atmosphere of 3% O2/5% CO2, which mimics physiologic conditions and reduces the senescence evolution of the cultures63. Human bone marrow-derived MSC were obtained from the Inbiobank Stem Cell Bank (www.inbiobank.org) under specific regulations (R.D. 1301/2006). Human fibroblasts were purchased from the American Type Culture Collection (Manassas, VA; cat# CRL-2097), ScienceCell Research Laboratories (San Diego, CA; cat# 6300) and PromoCell (Heidelberg, Gemany; cat# C-12375 and C-12360). hMSC and fibroblasts were maintained and expanded under optimal conditions, previously described34,36, also in a 3% O2/ 5%CO2 atmosphere. A more detailed description can be found in expanded methods (Supplementary "Methods" section online details).
Label-free proteomics analysis
hCPC3 protein levels were compared with those of hMSC19, essentially as previously described34. Cells were expanded to P7- P8, recovered and, after several washes in PBS, pellets (5–8 × 107 cells) were collected. Subcellular cytoplasmic and nuclear protein fractions (see Supplementary "Methods" section online) were obtained using the Qproteome Cell Compartment Kit (Qiagen, Barcelona, Spain). Samples (~ 500 μg) were digested using an in-gel digestion protocol, as described20. Tryptic peptides were dissolved in 0.1% formic acid (FA) and loaded on a liquid chromatography-mass spectrometry (LC–MS/MS) system for online desalting on C18 cartridges and further analysis by LC–MS/MS, using a reverse-phase nanocolumn (75 μm inner diameter × 50 μm, 3 μm-particle size, Acclaim PepMap 100 C18; Thermo Fisher Scientific, San Jose, CA) in a continuous (0–30%) acetonitrile gradient consisting of B (90% acetonitrile, 0.5% formic acid), in 180 min, 30–43% in 5 min and 43–90% in 2 min. A ~ 200 nL/min flow rate was used to elute peptides from the nanocolumn to an emitter nanospray needle for real time ionization and peptide fragmentation onto an ion trap-orbitrap hybrid mass spectrometer (Orbitrap Elite, Thermo Fisher). Bioinformatic identification and analyses methods are described in Supplementary "Methods" section online. Relative representation of the different proteins identificated was estimated by peptide-counting; three replicas were analyzed for each comparison. When indicated pathway analysis with PANTHER software68 was carried out.
RT-qPCR analyses
Total mRNA was isolated as described33. cDNA first strands were synthesized from total RNA (1 μg) with the SuperScript III First-Strand Synthesis System (Invitrogen). Genes of interest (see Supplementary "Methods" section online) were evaluated by quantitative RT-qPCR in a Mastercycler Ep-Realplex platform (Eppendorf, Hamburg, Germany), using Power SYBR Green reagents (Applied Biosystems, Foster City, CA). Cycle conditions were 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Quantified gene expression values were normalized against those of GUSB or GAPDH. Supplementary Methods Table 1 section online includes the list of all primer sequences used.
Western blotting
Cells were harvested in RIPA (radioimmunoprecipitation assay) lysis buffer, and equal amounts of lysate were separated by 10% SDS-PAGE. When indicated, cytoplasmic or nuclear fractions were obtained using the NE-PER Nuclear and Cytoplasmic Extraction kit (Thermo Fisher Scientific). Proteins were transferred to PVDF membranes using the iBlot Dry Blotting System (Invitrogen). After incubation with primary and secondary antibodies, signals were developed using an ECL kit (GE Healthcare, Uppsala, Sweden). Supplementary Methods Table 2 section online includes the list of all primary and secondary antibodies used.
Immunofluorescence
Immunofluorescence protocols have been previously described in detail. Cells were fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton-X100 (5 min, room temperature), blocked with blocking buffer (PBS with 5% BSA; 30 min, room temperature) and then incubated with primary antibodies in PBS/1% BSA (overnight, 4 °C). Slides were washed three times in PBS/1% bovine serum albumin (BSA) and incubated in PBS/1% BSA with appropriate secondary antibodies (1 h, room temperature). Washed cells (three times in PBS/1% BSA) were mounted in Prolong DAPI mounting medium (Invitrogen). Images were captured with a Zeiss LSM 700 or a Leica TCS SP5 confocal microscope. Supplementary Methods Table 2 section online includes the list of all primary and secondary antibodies used. Fluorescence intensity was measured using ImageJ software (NIH, Bethesda, MD) (https://imagej.net/software/imagej/).
Gene silencing assays
hCPC were transfected in Opti-MEN medium (Gibco, Invitrogen) with 10 nM of small interfering RNA (siRNA) against IMP3 (siIMP3), IL1A (siIL1A) or an siRNA negative control (all provided by Origene Technologies, Rockville, MD) using Lipofectamine 2000 Reagent (Invitrogen, Thermo Fisher Scientific). Cells were maintained overnight with the transfection mix. RT-qPCR or western blotting within 24–48 h of transfection checked silencing efficiency. Functional effects were tested 48 h post-transfection, as maximum inhibition efficiency was confirmed at this time point.
Viability, proliferation and apoptosis assays
To study cell viability, hCPC were detached with trypsin–EDTA 48 h post-transfection, labeled with DAPI (1/1000; Sigma-Aldrich) and quantified by flow cytometry on a FACS Canto 3L flow cytometer (BD Biosciences, San Jose, CA). For proliferation assays, 5-ethynyl-2’-deoxyuridine (EdU; 10 μM) was added to hCPC cultures 12 h prior to analysis. Proliferating cells were detected with the Click-iT Flow-Cytometry Kit (Thermo Fisher Scientific). For apoptosis analysis, cells were exposed to H2O2 (500 μM, during 5 h), then collected (including detached cells) and labeled at 4ºC for 15 min with AnnexinV-FITC (diluted 1:10) in the binding buffer provided by the manufacturer (ApoScreen® Annexin V Apoptosis Kit-FITC; Southern Biotech, Birmingham, AL). Labeled cells were washed with PBS/0.01% BSA and resuspended in 390 μL of binding buffer. Propidium iodide (50 μg/ml, Beckman Couler, Nyon, Switzerland) was added (1:40 dilution) for dual-staining and cells were analyzed by flow cytometry. DAPI and AnexinV/PI positive-cells were quantified on a FACS Canto 3L flow cytometer (BD Biosciences). When indicated, necrosis of hCPC was induced by a short heat treatment (10 min, 60ºC) of attached monolayers.
Immunoregulation evaluation
Human peripheral mononuclear cells (MNC) were labeled with 1 µM carboxyfluorescein succinimidyl ester (CFSE CellTrace Cell Proliferation Kit; Molecular Probes, Invitrogen) and stimulated with 10 μg/mL phytohemagglutinin (Sigma-Aldrich) over three days, as described69. hCPC1 cells were used for the evaluation of hCPC immunoregulatory capacity on T cells. Cells were plated in triplicate at different cell densities (15 × 103, 30 × 103 or 60 × 103) in 24-well plates and incubated at 37 °C for 16 h in an atmosphere of 3% O2/5%CO2. hCPC were transfected during 6 h with siIL1A or siNeg Control (10 nM) using 1 μL of Lipofectamine 2000 per well, in a final volume of 500 μL of Opti-MEN. The medium was then replenished with 100 μL of fresh DMEM-complete medium. CFSE-labeled MNC (6 × 105) in 900 μL of RPMI-complete medium was added to the plates at different hCPC/MNC ratios (1:10, 1:20, 1:40) and incubated for 3 additional days. A comparative evaluation of viable (DAPI) CD3 + proliferative (CFSE +) cells was carried out by flow cytometry (Fortessa, BD Bioscience). Data were analyzed with ModFit LT (Verity Software House, Topsham, ME).
Wound healing assay
For migration (scratch) assays, hCPC cells were cultured to confluence and starved in serum-free medium (24 h). The cell monolayer was then scraped with a pipette tip (t = 0 h) and cultures were monitored (t = 6–24 h) to evaluate their wound healing capacity. Images were acquired and migration rates were measured using ImageJ software (NIH, Bethesda, MD).
Statistics
Assays were performed three times and data expressed as mean ± SD; black lines summarize p-values (*** < 0.002, ** < 0.02, * < 0.05) for hCPC versus fibroblasts or hMSC (one-way analysis of variance followed by the Bonferroni correction for multiple comparison).
Supplementary Information
Acknowledgements
This study was initiated by European Commission funding (HEALTH-2009_242038) and by grants to AB from the Spanish Ministry of Science and Innovation RTI2018-097604-B-I00 (AEI/FEDER, UE) and SAF2015-70882-R. The Research Program of the Comunidad Autónoma de Madrid (S2017/BMD-3692) and the Instituto de Salud Carlos III (RETICS-RTI2018-097604-B-I00) to AB also funded parts of the work. We also wish to thank to K McCreath for editorial work.
Abbreviations
- B-CPC
Murine cardiac progenitor cells Bmi1high+
- c-KIT, CD177
Receptor for stem cell factor
- hCPC
Human cardiac progenitor cells
- DAPI
4',6-Diamidino-2-phenylindol
- EdU
5-Ethynyl-2’-deoxyuridine
- hMSC
Human mesenchymal stem cells
- HDF
Human dermal fibroblasts
- H2O2
Hydrogen peroxide
- IPA
Ingenuity Pathway Analysis
- IL1A
Interleukin 1A, interleukin 1α
- IL1B
Interleukin 1B, interleukin 1β
- IMP2
IMP U3 Small Nucleolar Ribonucleoprotein 2, IGF2BP2
- IMP3
IMP U3 Small Nucleolar Ribonucleoprotein 3, IGF2BP3
- iTRAQ
Isobaric tags for relative and absolute quantitation
- LF
Label-free
- Log FC
Log2 fold-change
- RNAseq
RNA sequencing
- RT-qPCR
Quantitative polymerase chain reaction coupled to reverse transcription
- siRNA
Small interfering RNA
Author contributions
G.A. and S.A. performed the main block of experiments, collected data and performed data analysis. J.A.L. and J.V. were the main responsible of the proteomic study. J.L.T. was mainly involved in the supervision and analysis of the proteomic data. R.M.Y. was involved in the evaluation of the putative role of IL1A in the immunoregulation capacity of CPC. A.B. conceived the project, designed the global strategy and supervised research. A.B. wrote and edited the manuscript with the collaboration of C.M. and G.A.. All authors read and approved the final manuscript.
Data availability
Mass spectrometry proteomics data are deposited in Peptide Atlas (http://www.peptideatlas.org/repository/) and are accessible through the PASS00827 accession number. All transcriptomic data related to this study are deposited in Gene Expression Omnibus (GEO) database repository (www.ncbi.nlm.nih.gov/geo/) and are accessible through the GSE84070 accession number.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Guillermo Albericio and Susana Aguilar.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-03956-8.
References
- 1.Bergmann O, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergmann O, et al. Dynamics of cell generation and turnover in the human heart. Cell. 2015;161:1566–1575. doi: 10.1016/j.cell.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 3.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin-Puig S, Wang Z, Chien KR. Lives of a heart cell: tracing the origins of cardiac progenitors. Cell Stem Cell. 2008;2:320–331. doi: 10.1016/j.stem.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Beltrami AP, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 6.Nakada Y, et al. Hypoxia induces heart regeneration in adult mice. Nature. 2017;541:222–227. doi: 10.1038/nature20173. [DOI] [PubMed] [Google Scholar]
- 7.Hesse M, Fleischmann BK, Kotlikoff MI. The role of c-kit expressing cells in heart repair at the neonatal and adult stage. Stem Cells. 2014;32:1701–1712. doi: 10.1002/stem.1696. [DOI] [PubMed] [Google Scholar]
- 8.Burgess KA, et al. Functionalised peptide hydrogel for the delivery of cardiac progenitor cells. Mater. Sci. Eng. C Mater. Biol. Appl. 2021;119:111539. doi: 10.1016/j.msec.2020.111539. [DOI] [PubMed] [Google Scholar]
- 9.Amini H, Rezaie JA, Rahbarghazi R, Nouri M. Cardiac progenitor cells application in cardiovascular disease. J. Cardiovasc. Thorac. Res. 2017;9:127–132. doi: 10.15171/jcvtr.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellison GM, et al. Adult c-kitpos cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell. 2013;154:827–842. doi: 10.1016/j.cell.2013.07.039. [DOI] [PubMed] [Google Scholar]
- 11.Mohsin S, et al. Human cardiac progenitor cells engineered with Pim-I kinase enhance myocardial repair. J. Am. Coll. Cardiol. 2012;60:1278–1287. doi: 10.1016/j.jacc.2012.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madonna R, Rokosh G, de Caterina R, Bolli R. Hepatocyte growth factor/Met gene transfer in cardiac stem cells–potential for cardiac repair. Basic Res. Cardiol. 2010;105:443–452. doi: 10.1007/s00395-010-0102-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He L, et al. Enhancing the precision of genetic lineage tracing using dual recombinases. Nat. Med. 2017;23:1488–1498. doi: 10.1038/nm.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sultana N, et al. Resident c-kit+ cells in the heart are not cardiac stem cells. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nat. Commun. 2015;6:8701. doi: 10.1038/ncomms9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Berlo JH, et al. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature. 2014;509:337–341. doi: 10.1038/nature13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aquila I, et al. c-kit Haploinsufficiency impairs adult cardiac stem cell growth, myogenicity and myocardial regeneration. Cell Death Dis. 2019;10:436. doi: 10.1038/s41419-019-1655-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vicinanza C, et al. Kit cre knock-in mice fail to fate-map cardiac stem cells. Nature. 2018;555:E1–E5. doi: 10.1038/nature25771. [DOI] [PubMed] [Google Scholar]
- 18.Vicinanza C, et al. Adult cardiac stem cells are multipotent and robustly myogenic: c-kit expression is necessary but not sufficient for their identification. Cell Death Differ. 2017;24:2101–2116. doi: 10.1038/cdd.2017.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis-McDougall FC, et al. Aged-senescent cells contribute to impaired heart regeneration. Aging Cell. 2019;18:e12931. doi: 10.1111/acel.12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng J, et al. Single-cell gene profiling and lineage tracing analyses revealed novel mechanisms of endothelial repair by progenitors. Cell Mol Life Sci. 2020;77:5299–5320. doi: 10.1007/s00018-020-03480-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scalise M, et al. Atrial myxomas arise from multipotent cardiac stem cells. Eur. Heart J. 2020;41:4332–4345. doi: 10.1093/eurheartj/ehaa156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crisostomo V, et al. Dose-dependent improvement of cardiac function in a swine model of acute myocardial infarction after intracoronary administration of allogeneic heart-derived cells. Stem Cell Res. Ther. 2019;10:152–169. doi: 10.1186/s13287-019-1237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanazawa H, et al. Durable benefits of cellular postconditioning: long-term effects of allogeneic cardiosphere-derived cells infused after reperfusion in pigs with acute myocardial infarction. Am. Heart Assoc. 2016;5(2):e002796. doi: 10.1161/JAHA.115.002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crisostomo V, et al. Delayed administration of allogeneic cardiac stem cell therapy for acute myocardial infarction could ameliorate adverse remodeling: experimental study in swine. J. Transl. Med. 2015;13:156–172. doi: 10.1186/s12967-015-0512-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keith MC, et al. Safety of intracoronary infusion of 20 million C-kitpositive human cardiac stem cells in pigs. PLoS ONE. 2015;10:e0124227. doi: 10.1371/journal.pone.0124227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koudstaal S, et al. Sustained delivery of insulin-like growth factor-1/hepatocyte growth factor stimulates endogenous cardiac repair in the chronic infarcted pig heart. J. Cardiovasc. Transl. Res. 2014;7:232–241. doi: 10.1007/s12265-013-9518-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellison GM, et al. Endogenous cardiac stem cell activation by insulin-like growth factor-1/hepatocyte growth factor intracoronary injection fosters survival and regeneration of the infarcted pig heart. J. Am. Coll. Cardiol. 2011;58:977–986. doi: 10.1016/j.jacc.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 28.Madonna R, et al. ESC Working Group on Cellular Biology of the Heart: position paper for Cardiovascular Research: tissue engineering strategies combined with cell therapies for cardiac repair in ischaemic heart disease and heart failure. Cardiovasc. Res. 2019;115:488–500. doi: 10.1093/cvr/cvz010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malliaras K, et al. Stimulation of endogenous cardioblasts by exogenous cell therapy after myocardial infarction. EMBO Mol. Med. 2014;6:760–777. doi: 10.1002/emmm.201303626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pavo N, et al. Long-acting beneficial effect of percutaneously intramyocardially delivered secretome of apoptotic peripheral blood cells on porcine chronic ischemic left ventricular dysfunction. Biomaterials. 2014;35:3541–3550. doi: 10.1016/j.biomaterials.2013.12.071. [DOI] [PubMed] [Google Scholar]
- 31.Sebastião MJ, et al. Human cardiac stem cells inhibit lymphocyte proliferation through paracrine mechanisms that correlate with indoleamine 2,3-dioxygenase induction and activity. Stem Cell Res. Ther. 2018;9:290–307. doi: 10.1186/s13287-018-1010-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lauden L, et al. Allogenicity of human cardiac stem/progenitor cells orchestrated by programmed death ligand 1. Circ. Res. 2013;112:451–464. doi: 10.1161/CIRCRESAHA.112.276501. [DOI] [PubMed] [Google Scholar]
- 33.Moscoso I, et al. Podocalyxin-like protein 1 is a relevant marker for human c-kit+ cardiac stem cells. J. Tissue Eng. Regen. Med. 2016;10:580–590. doi: 10.1002/term.1795. [DOI] [PubMed] [Google Scholar]
- 34.Torán JL, et al. Definition of a cell surface signature for human cardiac progenitor cells after comprehensive comparative transcriptomic and proteomic characterization. Sci. Rep. 2019;9:4647–4663. doi: 10.1038/s41598-019-39571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomes-Alves P, et al. Exploring analytical proteomics platforms toward the definition of human cardiac stem cells receptome. Proteomics. 2015;15:1332–1337. doi: 10.1002/pmic.201400318. [DOI] [PubMed] [Google Scholar]
- 36.Torán JL, et al. CXCL6 is an important paracrine factor in the pro-angiogenic human cardiac progenitor-like cell secretome. Sci. Rep. 2017;7:12490–12504. doi: 10.1038/s41598-017-11976-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanz-Ruiz R, et al. Rationale and design of a clinical trial to evaluate the safety and efficacy of intracoronary infusion of allogeneic human cardiac stem cells in patients with acute myocardial infarction and left ventricular dysfunction: the randomized multicenter double-blind controlled CAREMI trial (cardiac stem cells in patients with acute myocardial infarction) Circ. Res. 2017;121:71–80. doi: 10.1161/CIRCRESAHA.117.310651. [DOI] [PubMed] [Google Scholar]
- 38.Fernández-Avilés F, et al. Safety and Efficacy of Intracoronary Infusion of Allogeneic Human Cardiac Stem Cells in Patients With ST-Segment Elevation Myocardial Infarction and Left Ventricular Dysfunction. Circ. Res. 2018;123:579–589. doi: 10.1161/CIRCRESAHA.118.312823. [DOI] [PubMed] [Google Scholar]
- 39.Cohen I, et al. Differential release of chromatin-bound IL-1alpha discriminates between necrotic and apoptotic cell death by the ability to induce sterile inflammation. Proc. Natl. Acad. Sci. USA. 2010;107:2574–2579. doi: 10.1073/pnas.0915018107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luheshi NM, Rothwell NJ, Brough D. Dual functionality of interleukin-1 family cytokines: implications for anti-interleukin-1 therapy. Br. J. Pharmacol. 2009;157:1318–1329. doi: 10.1111/j.1476-5381.2009.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luheshi NM, McColl BW, Brough D. Nuclear retention of IL-1 alpha by necrotic cells: a mechanism to dampen sterile inflammation. Eur. J. Immunol. 2009;39:2973–2980. doi: 10.1002/eji.200939712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bertheloot D, Latz E. HMGB1, IL-1α, IL-33 and S100 proteins: dual-function alarmins. Cell Mol. Immunol. 2017;14:43–64. doi: 10.1038/cmi.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D'Amario D, et al. Insulin-like growth factor-1 receptor identifies a pool of human cardiac stem cells with superior therapeutic potential for myocardial regeneration. Circ. Res. 2011;108:1467–1481. doi: 10.1161/CIRCRESAHA.111.240648. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Lin S, et al. Let-7b Regulates Myoblast Proliferation by Inhibiting IGF2BP3 Expression in Dwarf and Normal Chicken. Front Physiol. 2017;8:477–488. doi: 10.3389/fphys.2017.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhargava S, et al. IGF2 mRNA binding protein 3 (IMP3) promotes glioma cell migration by enhancing the translation of RELA/p65. Oncotarget. 2017;8:40469–40485. doi: 10.18632/oncotarget.17118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valiente-Alandi I, Albo-Castellanos C, Herrero D, Sanchez I, Bernad A. Bmi1 (+) cardiac progenitor cells contribute to myocardial repair following acute injury. Stem Cell. Res. Ther. 2016;7:100–111. doi: 10.1186/s13287-016-0355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilkes DM, Bajpai S, Chaturvedi P, Wirtz D, Semenza GL. Hypoxia-inducible factor 1 (HIF-1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. J. Biol. Chem. 2013;288:10819–10829. doi: 10.1074/jbc.M112.442939. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Jain A, Bacolla A, Del Mundo IM, Zhao J, Wang G, Vasquez KM. DHX9 helicase is involved in preventing genomic instability induced by alternatively structured DNA in human cells. Nucl. Acids Res. 2013;41:10345–10357. doi: 10.1093/nar/gkt804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manojlovic Z, Stefanovic B. A novel role of RNA helicase A in regulation of translation of type I collagen mRNAs. RNA. 2012;18:321–334. doi: 10.1261/rna.030288.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leone S, Bär D, Slabber CF, Dalcher D, Santoro R. The RNA helicase DHX9 establishes nucleolar heterochromatin, and this activity is required for embryonic stem cell differentiation. EMBO Rep. 2017;18:1248–1262. doi: 10.15252/embr.201744330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zacharias DA, Mullen M, Planey SL. Antiproliferative factor-induced changes in phosphorylation and palmitoylation of cytoskeleton-associated protein-4 regulate its nuclear translocation and DNA binding. Int. J. Cell Biol. 2012;201:150918–150923. doi: 10.1155/2012/150918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu Q, Yuan YA. Structural insights into RISC assembly facilitated by dsRNA-binding domains of human RNA helicase A (DHX9) Nucl. Acids Res. 2013;41:3457–3470. doi: 10.1093/nar/gkt042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao WF, Liu JW, Huang DS. MiR-200a inhibits cell proliferation and EMT by down-regulating the ASPH expression levels and affecting ERK and PI3K/Akt pathways in human hepatoma cells. Am. J. Transl. Res. 2018;10:1117–1130. [PMC free article] [PubMed] [Google Scholar]
- 54.Ogawa K, et al. Aspartate β-hydroxylase promotes pancreatic ductal adenocarcinoma metastasis through activation of SRC signaling pathway. J. Hematol. Oncol. 2019;12:144–160. doi: 10.1186/s13045-019-0837-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bai L, et al. Regulation of cellular senescence by the essential caveolar component PTRF/ Cavin-1. Cell Res. 2011;21:1088–1101. doi: 10.1038/cr.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Orjalo AV, Bhaumik D, Gengler BK, Scott GK, Campisi J. Cell surface-bound IL-1α is an upstream regulator of the senescence-associated IL-6/IL-8 cytokine network. Proc. Natl. Acad. Sci. USA. 2009;106:17031–17036. doi: 10.1073/pnas.0905299106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, et al. Propiece IL-1α facilitates the growth of acute T-lymphocytic leukemia cells through the activation of NF-κB and SP1. Oncotarget. 2017;8:15677–15688. doi: 10.18632/oncotarget.14934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu C, Ma H, Qi G, Chen F, Chu J. Insulin-like growth factor II mRNA-binding protein 3 promotes cell proliferation, migration and invasion in human glioblastoma. OncoTargets Ther. 2019;12:3661–3670. doi: 10.2147/OTT.S200901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Er LM, et al. Insulin-like growth factor II mRNA binding protein 3 regulates proliferation, invasion and migration of neuroendocrine cancer cells. Int. J. Clin. Exp. Pathol. 2017;10:10269–10275. [PMC free article] [PubMed] [Google Scholar]
- 60.Taniuchi K, Furihata M, Hanazaki K, Saito M, Saibara T. IGF2BP3-mediated translation in cell protrusions promotes cell invasiveness and metastasis of pancreatic cancer. Oncotarget. 2014;5:6832–6845. doi: 10.18632/oncotarget.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ennajdaoui H, et al. IGF2BP3 modulates the interaction of invasion-associated transcripts with RISC. Cell Rep. 2016;15:1876–1883. doi: 10.1016/j.celrep.2016.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palanichamy JK, et al. RNA-binding protein IGF2BP3 targeting of oncogenic transcripts promotes hematopoietic progenitor proliferation. J. Clin. Invest. 2016;126:1495–1511. doi: 10.1172/JCI80046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mizutani R, et al. Oncofetal protein IGF2BP3 facilitates the activity of proto-oncogene protein eIF4E through the destabilization of EIF4E-BP2 mRNA. Oncogene. 2016;35:3495–3502. doi: 10.1038/onc.2015.410. [DOI] [PubMed] [Google Scholar]
- 64.Elagib KE, et al. Neonatal expression of RNA-binding protein IGF2BP3 regulates the human fetal-adult megakaryocyte transition. J. Clin. Invest. 2017;127:2365–2377. doi: 10.1172/JCI88936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brants JR, Ayoubi TA, Chada K, Marchal K, Van de Ven WJ, Petit MM. Differential regulation of the insulin-like growth factor II mRNA-binding protein genes by architectural transcription factor HMGA2. FEBS Lett. 2004;569:277–283. doi: 10.1016/j.febslet.2004.05.075. [DOI] [PubMed] [Google Scholar]
- 66.Jønson L, Christiansen J, Hansen TVO, Vikeså J, Yamamoto Y, Nielsen FC. IMP3 RNP safe houses prevent miRNA-directed HMGA2 mRNA decay in cancer and development. Cell Rep. 2014;7:539–551. doi: 10.1016/j.celrep.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 67.Xiangpeng X, et al. MicroRNA-204-3p represses colon cancer cells proliferation, migration, and invasion by targeting HMGA2. J. Cell Physiol. 2020;235:1330–1338. doi: 10.1002/jcp.29050. [DOI] [PubMed] [Google Scholar]
- 68.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis A, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucl. Acids Res. 2010;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quah BJ, Warren HS, Parish CR. Monitoring lymphocyte proliferation in vitro and in vivo with the intracellular fluorescent dye carboxyfluorescein diacetate succinimidyl ester. Nat. Protoc. 2007;2:2049–2056. doi: 10.1038/nprot.2007.296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry proteomics data are deposited in Peptide Atlas (http://www.peptideatlas.org/repository/) and are accessible through the PASS00827 accession number. All transcriptomic data related to this study are deposited in Gene Expression Omnibus (GEO) database repository (www.ncbi.nlm.nih.gov/geo/) and are accessible through the GSE84070 accession number.