Abstract
Resistance represents a major challenge for antibody-based therapy for COVID-191-14. Here we engineered an immunoglobulin M (IgM) neutralizing antibody (IgM-14) to overcome the resistance encountered by immunoglobulin G (IgG)-based therapeutics. IgM-14 is over 230-fold more potent than its parental IgG-14 in neutralizing SARS-CoV-2. IgM-14 potently neutralizes the resistant virus raised by its corresponding IgG-14, three variants of concern—B.1.1.7 (Alpha, which first emerged in the UK), P.1 (Gamma, which first emerged in Brazil) and B.1.351 (Beta, which first emerged in South Africa)—and 21 other receptor-binding domain mutants, many of which are resistant to the IgG antibodies that have been authorized for emergency use. Although engineering IgG into IgM enhances antibody potency in general, selection of an optimal epitope is critical for identifying the most effective IgM that can overcome resistance. In mice, a single intranasal dose of IgM-14 at 0.044 mg per kg body weight confers prophylactic efficacy and a single dose at 0.4 mg per kg confers therapeutic efficacy against SARS-CoV-2. IgM-14, but not IgG-14, also confers potent therapeutic protection against the P.1 and B.1.351 variants. IgM-14 exhibits desirable pharmacokinetics and safety profiles when administered intranasally in rodents. Our results show that intranasal administration of an engineered IgM can improve efficacy, reduce resistance and simplify the prophylactic and therapeutic treatment of COVID-19.
The respiratory tract is the major target for SARS-CoV-2 infection5. High viral load in the respiratory tract correlates with severe disease in patients with COVID-196. So far, almost all neutralizing monoclonal antibodies (monoclonal antibodies) that have been tested in clinical trials are the IgG1 isotype and are administered through intravenous infusion. Circulating IgG antibodies lack efficient access to mucosal compartments7. After intravenous infusion, antibody levels in the lung are around 200–500 times lower than those in the serum8. Potent neutralizing IgG1 monoclonal antibodies need to be administered at high doses (up to 8 g); even so, the antiviral effect is small in the respiratory tract in patients treated with drug compared to those treated with placebo9. Emerging SARS-CoV-2 variants of concern (VOCs) are resistant to many potent neutralizing IgG1 monoclonal antibodies, including those that are in clinical trials and that have been authorized for emergency use2-4. Therefore, the development of new antibody therapies that can overcome these challenges is urgently needed.
IgM and IgA are mucosal antibodies that constitute the first line of defence against mucosal pathogens. Typically, IgM assembles into pentamers and IgA1 into dimers in the presence of the joining chain (J-chain), which facilitates efficient mucosal transcytosis of antibodies10,11. IgM and IgA1 can also be nebulized and reach airway tissues after inhalation12. Owing to avidity effects, multivalent antibodies can exhibit enhanced neutralization of SARS-CoV-213 and reduce the evasion of antibodies by the virus14. An IgM pentamer is naturally decavalent owing to the repetitive antigen-binding variable fragments (Fvs)11. These unique features make the intranasal delivery of IgM neutralizing monoclonal antibodies appealing for the prevention and treatment of COVID-19.
Engineering of neutralizing IgM and IgA1
To develop human IgM and IgA1 neutralizing monoclonal antibodies, we performed antibody engineering based on the CR3022 monoclonal antibody15 and five IgG1 monoclonal antibodies (CoV2-06, CoV2-09, CoV2-12, CoV2-14 and CoV2-16) that were previously isolated from a phage-displayed antibody library1. These six monoclonal antibodies recognize different epitopes on the receptor-binding domain (RBD) (Fig. 1a, Extended Data Fig. 1a, b). The Fv of IgG1 was engineered into human IgM or IgA1 scaffolds for co-expression with the J-chain. The engineered IgM is a pentamer and IgA1 is a dimer (Fig. 1b). After production, the monoclonal antibodies of different isotypes assembled correctly and exhibited a purity of greater than 95% (Fig. 1c, d). IgM and IgA1 bound to the RBD more strongly than did IgG1 (Fig. 1e, Extended Data Fig. 1c). The higher binding activities, as evidenced by smaller values of the half-maximal effective concentration (EC50), were consistent with the higher valencies of IgA1 and IgM (Fig. 1f). Neutralization of live SARS-CoV-2 at monoclonal antibody concentrations of 0.1 and 1 μg ml−1 showed stronger inhibition of virus infection by IgM and IgA1, as compared to IgG1, with IgM CoV2-14 being the most potent monoclonal antibody (Fig. 1g, Extended Data Fig. 1d). These results demonstrate successful engineering of IgM and IgA1 monoclonal antibodies. The enhancement in binding and neutralization for IgM and IgA1 antibodies derived from different IgG1 antibodies exhibited different patterns, as exemplified by CoV2-06 and CoV2-14 (Fig. 1h), which suggests that, in addition to valency, IgG1 epitopes affect the neutralizing potency of the corresponding IgM and IgA1 antibodies.
Fig. 1 ∣. Engineering of IgM and IgA1 neutralizing monoclonal antibodies.
a, The IgG1 epitope residues are shown as spheres on the RBD. b, Illustration of antibody engineering from IgG1 into IgM and IgA1. c, d, SDS–PAGE (left) and native PAGE (right) (c) and size-exclusion chromatography (d) analysis showing monoclonal antibody assembly and purity. The gel images were from one experiment. HC, heavy chain; LC, light chain. e, ELISA binding to the RBD by the indicated isotypes of CoV2-14. Data are mean of duplicate wells. f, Antibody valency versus EC50 for the indicated isotypes. g, Neutralization of SARS-CoV-2 by the indicated monoclonal antibodies at 0.1 μg ml−1. Data are mean ± s.d. of triplicates. h, The EC50 and per cent neutralization of the three isotypes for CoV2-06 and CoV2-14 are plotted to illustrate different correlation patterns between binding and neutralization.
Enhanced potency of IgM over IgG
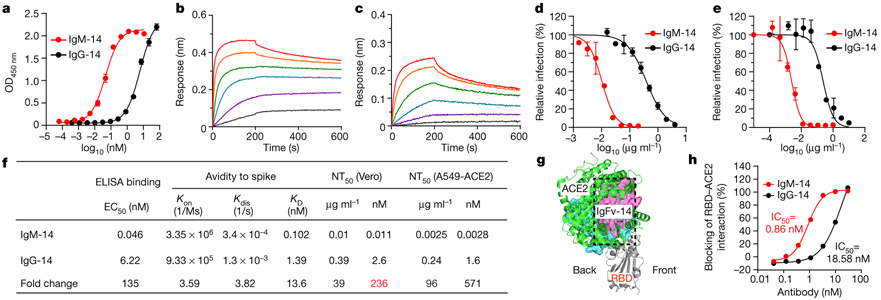
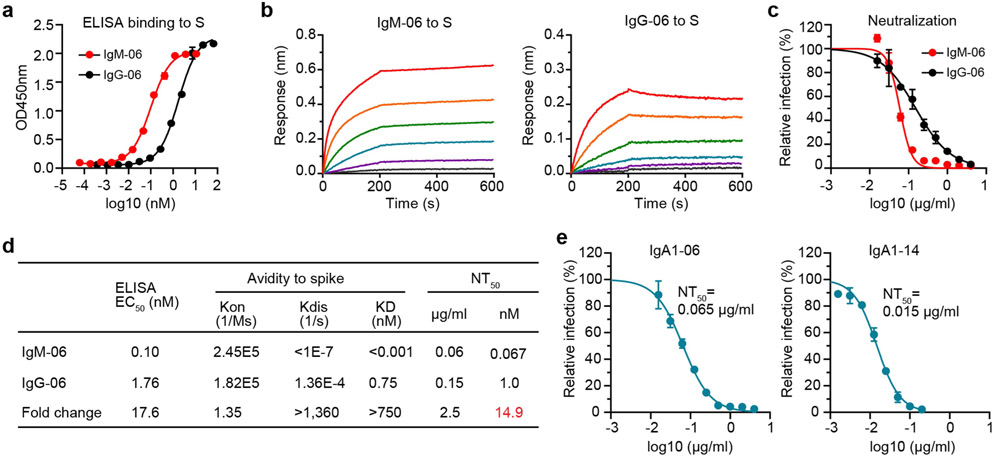
We focused on IgM CoV2-14 (IgM-14) for detailed characterization in parallel with IgG-14. To examine how epitopes affect the potency of engineered IgM antibodies, we also characterized the IgM and IgG1 pairs of CoV2-06 (IgM-06 and IgG-06). In an enzyme-linked immunosorbent assay (ELISA), IgM-14 showed much stronger binding to the spike protein than did IgG-14 (Fig. 2a). In kinetic binding, IgM-14 exhibited faster association with and slower dissociation from the spike protein than did IgG-14 (Fig. 2b, c). In neutralization titrations in Vero and human ACE2-overexpressing A549 (A549-ACE2) cells, IgM-14 markedly shifted the curves towards a higher potency relative to IgG-14 (Fig. 2d, e). Figure 2f summarizes the binding EC50, the avidity-related parameters (association Kon, dissociation Kdis and avidity KD), the half-maximal neutralization titre (NT50) and the fold changes of these values between IgM-14 and IgG-14. The change was 135-fold for EC50, 13.6-fold for KD, 39-fold for NT50 on a weight basis and 236-fold for NT50 on a molar basis. For IgM-06 and IgG-06, there was a 17.6-fold change in EC50 and a change of more than 750-fold in KD, but the change in NT50 was only 2.5-fold on a weight basis and 14.9-fold on a molar basis (Extended Data Fig. 2a-d). A substantial enhancement of neutralizing activity compared to the corresponding IgG1 was observed for IgA1-14 (26-fold) but not for IgA1-06 (2.3-fold) (Extended Data Fig. 2e). The contrast between CoV2-06 and CoV2-14 supports the conclusion that epitope selection is critical for identifying the most potent neutralizing IgM.
Fig. 2 ∣. Enhanced binding, neutralization and ACE2-blocking by IgM-14 over IgG-14.
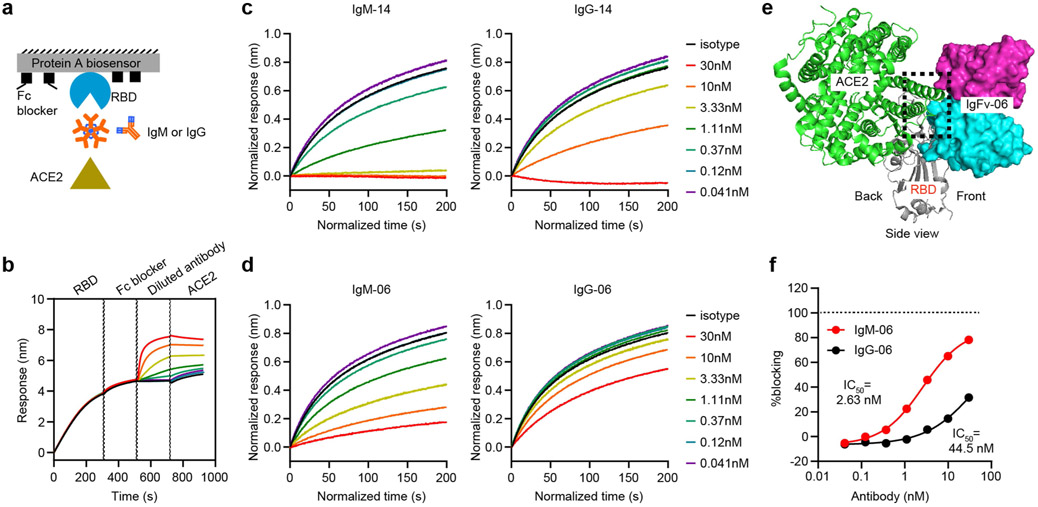
a, ELISA for binding to the spike protein. Data are mean of duplicate wells. OD450 nm, optical density at 450 nm. b, c, Binding kinetics of IgM-14 (b) and IgG-14 (c) to the spike protein. d, e, Neutralization of SARS-CoV-2 in Vero (d) and A549-ACE2 (e) cells. Data are mean of duplicate wells. f, Summary of binding EC50, association (Kon), dissociation (Kdis), avidity (KD) and neutralization NT50 values. The fold change of NT50 values between IgM-14 and IgG-14 is highlighted in red. g, Superposition of the IgFv-14–RBD and ACE2–RBD complexes. IgFv-14 is shown as a surface with the variable heavy chain (Vh) coloured in cyan and the variable light chain (Vl) in magenta. The RBD–ACE2 complex is shown as a cartoon with the RBD coloured in grey and ACE2 in green. The dashed box indicates steric clash. h, Blocking of RBD and ACE2 interaction. Data for each antibody concentration are from one biosensor.
To understand the structural mechanism of this observation, we performed antibody docking to simulate the Fv and RBD complex using Rosetta-based protocols16. The Fv of IgG-14 (IgFv-14) targets the back side of the RBD, whereas IgFv-06 targets the front side of the RBD (Extended Data Fig. 3a, b). Therefore, IgFv-14 has a larger steric clash with ACE2 (Fig. 2g). To support the structural analysis, we used a bio-layer interferometry (BLI)-based assay to assess the ACE2-blocking activities of IgM and IgG1 (Extended Data Fig. 4a-d). IgM-14 inhibited RBD–ACE2 interaction more strongly than did IgG-14 (Fig. 2h). By contrast, IgFv-06 has a smaller steric clash with ACE2 (Extended Data Fig. 4e). IgM-06 also blocked RBD–ACE2 interaction more strongly than did IgG-06, however, neither IgM-06 nor IgG-06 achieved full blocking even at the highest concentration tested (Extended Data Fig. 4f). These results demonstrate that epitope-dependent steric hindrance is an important mechanism for IgM-14 to exhibit potent neutralization.
Broad coverage of variants by IgM-14
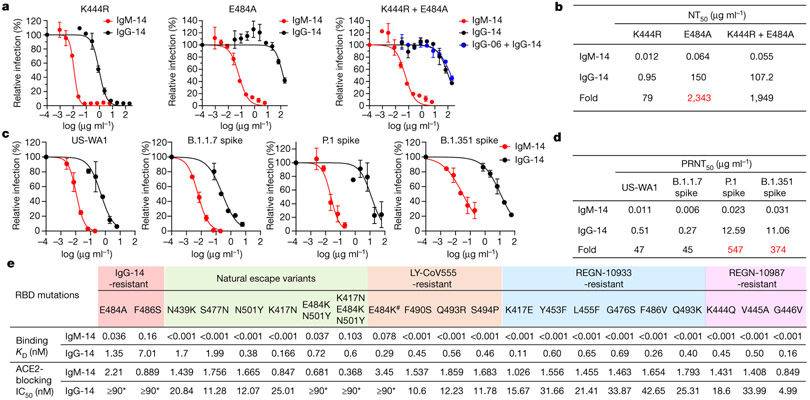
SARS-CoV-2 escapes from antibody neutralization by acquiring mutations in resistance-selection experiments and in natural circulation1-4,17,18. We previously identified neutralization-resistant RBD mutations K444R for IgG-06 and E484A for IgG-14 (ref. 1). To test whether IgM can neutralize these IgG escape mutants, we constructed three SARS-CoV-2 variants that contain K444R, E484A or both K444R and E484A mutations (K444R + E484A) (Extended Data Fig. 5a, b). IgG-14 effectively neutralized the K444R variant and marginally neutralized the E484A and K444R + E484A variants. Notably, IgM-14 potently neutralized all three variants including the K444R + E484A variant, which is resistant to the cocktail of IgG-06 + IgG-14 (Fig. 3a). The NT50 values of IgM-14 against the E484A and the K444R + E484A variants were 0.064 μg ml−1 and 0.055 μg ml−1, respectively (Fig. 3b), which are comparable to the NT50 (0.01 μg ml−1) value of IgM-14 against the wild-type virus (Fig. 2f). IgM-14 is 2,343-fold and 1,949-fold more potent than IgG-14 in neutralizing the E484A and K444R + E484A variants, respectively (Fig. 3b). Similarly, in A549-ACE2 cells, IgM-14 is 714-fold and 2,217-fold more potent than IgG-14 in neutralizing the E484A and K444R + E484A variants, respectively (Extended Data Fig. 5c, d). By contrast, IgM-06 only neutralized the E484A variant and not the K444R or K444R + E484A variants (Extended Data Fig. 5e, f). These data show that IgM-14 can effectively neutralize IgG-14 escape variants. The contrast between CoV2-06 and CoV2-14 further underscores that epitope selection is critical for identifying the IgM antibody that can overcome IgG escape mutations.
Fig. 3 ∣. Broader coverage of escape variants by IgM-14 over IgG-14.
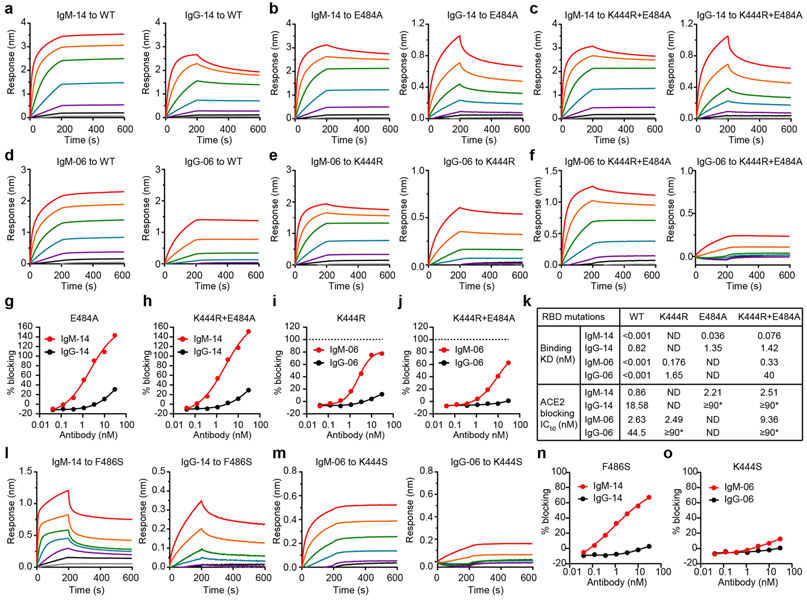
a, Neutralization of the K444R (IgG-06-resistant), E484A (IgG-14-resistant) and K444R + E484A (IgG-06 + IgG-14-resistant) SARS-CoV-2 variants. b, Summary of NT50 values against the indicated SARS-CoV-2 variants. c, PRNT assay using the US-WA1 strain and the recombinant B.1.1.7, P.1 and B.1.351 variants. d, Summary of PRNT50 values against the indicated viruses. Data are mean of duplicate wells for a, c. The fold changes in NT50 and PRNT50 values between IgM-14 and IgG-14 against the indicated resistant viruses are highlighted in red. e, Summary of binding KD and ACE2-blocking IC50 values to a panel of 21 RBD mutants. The hash symbol indicates that E484K is an escape mutation for both LY-CoV555 and REGN-10933. Asterisks indicate that the half-maximal blocking was not achieved at the highest monoclonal antibody concentration (30 nM) and the IC50 values are defined as ≥90 nM.
To assess the neutralizing activities of IgM-14 and IgG-14 against the recently emerged SARS-CoV-2 VOCs, we constructed recombinant viruses based on the US-WA1 strain and replaced its full spike gene with that of the B.1.1.7, P.1 or B.1.351 variants. In the plaque-reduction neutralization test (PRNT), IgG-14 potently neutralized the B.1.1.7 variant but weakly neutralized the P.1 and B.1.351 variants (Fig. 3c, d). These results are consistent with E484 being a critical epitope residue for IgG-14, as both the P.1 and B.1.351 variants contain the E484K neutralization-resistant mutation. Notably, IgM-14 potently neutralized all three variants (Fig. 3c). The half-maximal concentration values in PRNTs (PRNT50 values) of IgM-14 against the B.1.1.7, P.1 and B.1.351 variants were 0.006, 0.023 and 0.031 μg ml−1, respectively, which are comparable to the PRNT50 (0.011 μg ml−1) value of IgM-14 against the US-WA1 strain. IgM-14 is 45-fold, 547-fold and 374-fold more potent than IgG-14 in neutralizing the B.1.1.7, P.1 and B.1.351 variants, respectively (Fig. 3d). These data show that IgM-14, but not IgG-14, can tolerate the RBD mutations in VOCs.
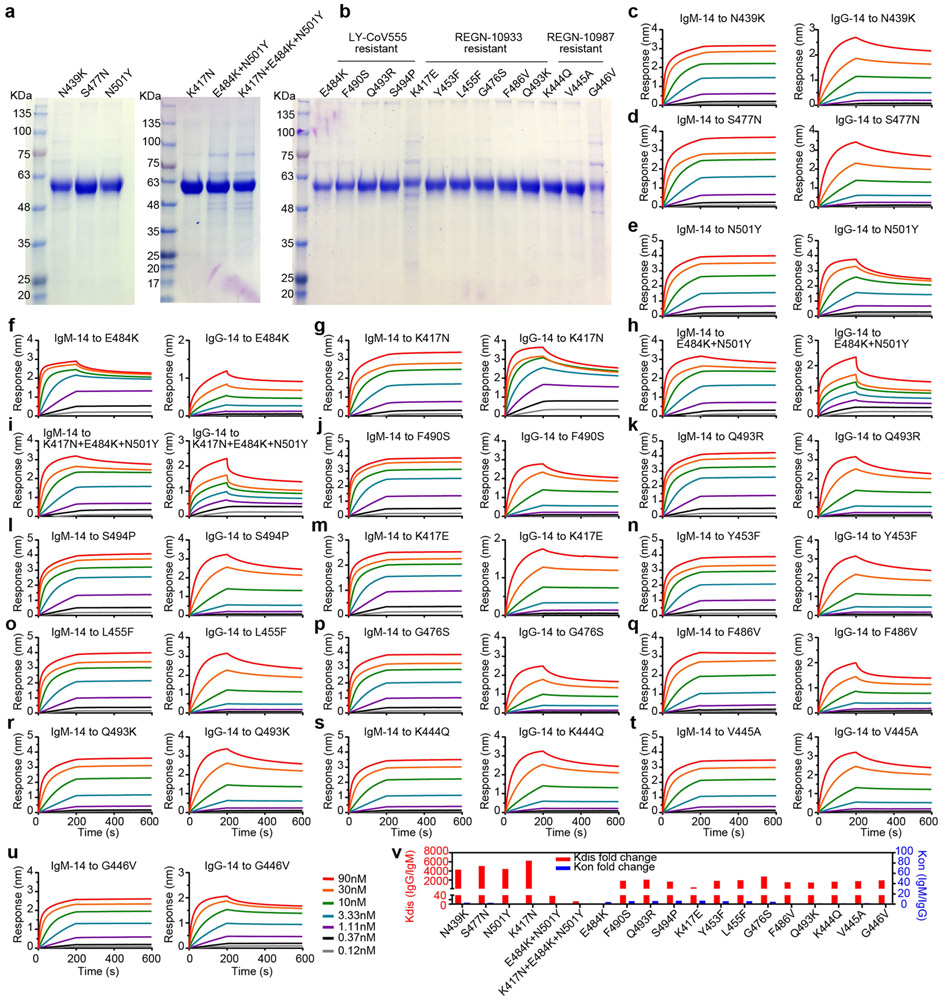
We used the wild-type, K444R, E484A and K444R + E484A RBD proteins to assess the binding and ACE2-blocking activities of IgM-14 versus IgG-14 and IgM-06 versus IgG-06 (Extended Data Fig. 6a-j). Consistent with the neutralization results, IgM-14 and IgM-06 bound to these RBD proteins and blocked ACE2 interactions more effectively than did their parental IgG1 antibodies (Extended Data Fig. 6k). Similar results were also observed using the F486S (another IgG-14-resistant mutation) and K444S (IgG-06-resistant) RBD proteins (Extended Data Fig. 6I-o).
We generated another 19 RBD mutants to characterize the binding and ACE2-blocking activities of IgM-14 and IgG-14 (Extended Data Figs. 7a-u, 8a-s). The N439K and S477N are resistant mutations for several neutralizing IgG1 monoclonal antibodies17,18. The K417N, N501Y, E484K + N501Y and K417N + E484K + N501Y mutations represent the B.1.351 variant2. The remaining 13 mutations are associated with resistance to three IgG1 monoclonal antibodies that have been authorized for emergency use19,20. Figure 3e summarizes the KD and half-maximal inhibitory concentration (IC50) values. Across all RBD mutants, IgM-14 exhibited much higher binding to RBD and higher ACE2-blocking activity compared to IgG-14. This superior activity is probably attributable to avidity effects, which result in much slower dissociation kinetics from the RBD mutants for IgM-14 than that for IgG-14 (Extended Data Fig. 7v, Supplementary Table 1). A good correlation between IC50 and NT50 values was observed for the VOCs (Extended Data Fig. 8t). Globally, these mutations emerged with varying frequencies in the 373,387 viral sequences analysed (Extended Data Fig. 8u). These mutations affect RBD functionality to various degrees (Extended Data Fig. 8v), indicating that RBD mutational effects on antibody binding and virus fitness together determine the neutralizing activities of monoclonal antibodies against these mutants. Together, these data demonstrate that IgM-14 is superior to IgG-14 in covering viral escape mutations.
IgM-14 delivered intranasally targets the airways
We evaluated the feasibility of IgM-14 for intranasal administration by tracking antibody bio-distribution in mice (Fig. 4a). After a single intranasal dose, IgM-14 (labelled with Alexa Fluor 750) was enriched in the nasal cavity and lasted for at least 96 h in whole-body imaging (Fig. 4b, Extended Data Fig. 9a). Ex vivo organ imaging showed that IgM-14 enriched in the nasal cavity and lung at various time points and was still evident in the nasal cavity at 168 h. The blood and other organs had minimal antibody exposure (Fig. 4c, d, Extended Data Fig. 9b, c). These results indicate that intranasally administered IgM-14 mainly targets the respiratory tract, with long-term retention in the nasal cavity and lung. Nasal epithelium is at first the dominant site for SARS-CoV-2 respiratory tract infection, followed by aspiration of virus into the lung21. Therefore, intranasal administration can efficiently load IgM-14 to the airways, which should confer protection against respiratory infection.
Fig. 4 ∣. Intranasally delivered IgM-14 confers protection against SARS-CoV-2 VOCs.
a, Experimental design for the evaluation of antibody bio-distribution. b, Representative whole-body images. c, Representative ex vivo images. Bl, blood (20 μl); Br, brain; H, heart; K, kidney; Lu, lung; Lv, liver; N, nasal cavity; S, spleen. d, Quantification of fluorescence signals. Data are mean ± s.d. of four mice. Dashed line shows the average autofluorescence of organs. e, Experimental design of dose-range evaluations of IgM-14 against a mouse-adapted SARS-CoV-2 (CMA4 strain). For prophylactic treatment, IgM-14 was evaluated at two sets of dose ranges to determine the minimal effective dose. f, g, Viral loads in the lungs of CMA4-infected mice after prophylactic (f) or therapeutic (g) treatment with IgM-14. n = 10 biologically independent mice for all groups, except for the 1.2 mg kg−1 group in g (n = 15). h, i, Viral loads in the lungs of mice infected with the P.1 (h) and B.1.351 (i) variant, after therapeutic treatment as indicated. n = 5 biologically independent mice for all groups. The solid lines in f–i indicate median viral loads in the lung. An ordinary one-way ANOVA with Sidak’s multiple comparisons was used in the statistical analysis for f, g. A two-sided Mann–Whitney test was used in the statistical analysis for h, i.
In vivo protection against VOCs by IgM-14
We evaluated the protective efficacy of IgM-14 using intranasal administration with a mouse-adapted virus that contains an N501Y mutation26, thus representing the RBD of the B.1.1.7 variant. First, we tested the three isotypes of CoV2-06 and CoV2-14 at a prophylactic dose of 3.5 mg per kg body weight (hereafter, mg kg−1) (Extended Data Fig. 10a). Peak lung viral loads on day 2 after infection were reduced to undetectable levels in all four mice of the IgG-06 and IgM-06 groups and in three of the four mice of the IgA1-06, IgG-14, IgA1-14 and IgM-14 groups (Extended Data Fig. 10b). We then focused on IgM-14 and performed dose range evaluations. Five dose levels (3.5, 1.2, 0.4, 0.13, and 0.044 mg kg−1) were tested for prophylactic treatment and three dose levels (3.5, 1.2 and 0.4 mg kg−1) were tested for therapeutic treatment (Fig. 4e). For prophylactic treatment, lung viral loads were reduced to undetectable levels in 90% (9/10), 70% (7/10), 90% (9/10) and 60% (6/10) of mice in the 3.5, 1.2, 0.4 and 0.13 mg kg−1 groups, respectively. Even with the 0.044 mg kg−1 dose, the median viral load (5.56-log) was significantly reduced by fivefold as compared to the isotype group (6.29-log; Fig. 4f). For therapeutic treatment, the median viral loads (excluding the mice with undetectable virus) were reduced by 13,667-fold, 13,667-fold and 56-fold in the 3.5, 1.2, and 0.4 mg kg−1 groups, respectively (Fig. 4g). Quantification of viral RNA in lung samples confirmed the anti-viral effects of IgM-14 (Extended Data Fig. 10c). IgM-14 showed more potent efficacy than IgG-14 at a therapeutic dose of 1.2 mg kg−1 (Extended Data Fig. 10d). No mutations were observed in the RBD of recovered virus after treatment with IgM-14 (Extended Data Fig. 10e). These data show that IgM-14 confers protection with an effective dose as low as 0.044 mg kg−1 for prophylactic treatment and 0.4 mg kg−1 for therapeutic treatment.
We also compared the therapeutic efficacy of IgM-14 and IgG-14 against the P.1 and B.1.351 VOCs. For the P.1 variant, IgM-14 reduced lung viral loads by 16,429-fold and 144-fold in the 3.5 and 1.2 mg kg−1 groups, respectively. In comparison, IgG-14 had a marginal anti-viral effect, reducing viral loads by around fourfold at either dose (Fig. 4h). Similarly, IgM-14 exhibited a significantly higher potency than IgG-14 against the B.1.351 variant (Fig. 4i). Together, our data show that intranasal administration of IgM-14 confers highly efficacious respiratory protection and that IgM-14 is superior to IgG-14 for protection against the tested VOCs.
Preclinical pharmacokinetics and safety
To further evaluate the translational potential of IgM-14, we conducted an intranasal pharmacokinetic study in mice. A single intranasal administration of 5 mg kg−1 of IgM-14 resulted in low levels of antibody in the blood (around 100 ng ml−1) that persisted for several hours (Extended Data Fig. 10f). In patients with COVID-19, the positive rate of viral RNA is high (93%) in bronchoalveolar lavage samples but very low (less than 1%) in blood samples22. Therefore, intranasally administered IgM-14 is better focused on targeting the site of virus replication in the respiratory tract, compared to intravenously infused IgG1. To further assess the tolerability of IgM-14, a pilot safety study was conducted in rats. Groups of rats were dosed intranasally with IgM-14 twice daily for five consecutive days. All rats survived to the end of the study with no change in body weight (Extended Data Fig. 10g).
Discussion
In this study, we have shown that six engineered IgM antibodies exhibit higher binding and neutralizing activities than their parental IgG1 antibodies, indicating a general mechanism of gaining potency through avidity. However, epitope selection is required to identify the most appropriate engineered IgM. Epitope-dependent steric hindrance is one mechanism for IgM to exhibit enhanced neutralization compared to IgG1. IgM may also engage more spike proteins than can an IgG1, and such a mechanism is related to antibody binding angles23. Epitope selection is also critical for identifying the IgM that can overcome resistance. The critical epitope residues for IgG-14, but not for IgG-06, make direct contact with ACE2. Investigating resistance for other monoclonal antibody pairs may provide additional insights into the precise mechanism. Nevertheless, the finding that IgM-14 has a broader coverage of variants than IgG-14 points to the potential for leveraging IgM to combat other dynamically evolving pathogens.
Intranasally delivered IgM mainly targets the airways, which allows dose-sparing and offers highly effective protection. Previously, intrarectal delivery of an IgM has been shown to reduce viral rectal infection in macaques24, and intranasal delivery of IgG antibodies has been studied. Together, these data indicate that mucosal delivery is a promising alternative to intravenous infusion for treating mucosal viral infections. Notably, intranasal dosing of IgM simplifies administration and also provides broader coverage and better protection against variants.
Traditionally, IgM antibodies have been considered difficult to produce. With recent advances in manufacturing, engineered IgM antibodies—including IgM-14—can be produced at high titres (greater than 1 g l−1) with good purity and stability. Several engineered IgM antibodies have successfully entered human clinical trials in oncology25. IgM-14 holds potential for development as an effective and flexible therapy for outpatients and high-risk populations.
In summary, we show that engineered IgM is a promising drug modality with potent neutralization, broad coverage of variants, desirable pharmacokinetics and safety profiles, and effective respiratory protection. We envision that engineered IgM administered in an intranasal manner can serve as a therapeutic platform for COVID-19 as well as for other respiratory viral diseases.
Methods
Data reporting
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Cells and viruses
Vero (ATCC, CCL-81) and Vero-E6 (ATCC, CRL-1586) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Expi293F cells (Gibco, 100044202) were maintained in Expi293 expression medium without FBS. ExpiCHO-S cells (Thermo Fisher Scientific, A29127) were maintained in ExpiCHO expression medium without FBS. CHO cells (Horizon Discovery) were maintained in Ex-Cell Advanced CHO fed-batch medium (Millipore-Sigma, 14366C) supplemented with 4 mM L-glutamine. The human ACE2-overepressing A549 (A549-ACE2) cells were generated in-house using lentivirus transduction. All cells have been tested negative for mycoplasma contamination. The SARS-CoV-2 (US-WA1 strain) mNeonGreen (mNG) reporter virus was generated in a previous study using a SARS-CoV-2 infectious clone27. The SARS-CoV-2 variants with the K444R, the E484A or the K444R + E484A mutations were also generated based on this infectious clone. A mouse-adapted SARS-CoV-2 (CMA4 strain) used for in vivo studies was recombinantly generated. In brief, a previously used mouse-adapted SARS-CoV-2 (CMA3 strain)1 was passed in BALB/c mice for 20 passages. To generate the CMA4 strain, the adaptive mutations were identified and reintroduced into the backbone of the CMA3 strain. The generation and characterization of these mouse-adapted viruses are reported in a separate preprint26. The recombinant SARS-CoV-2 viruses with spike mutations from the B.1.1.7, P.1 and B.1.351 lineages were prepared on the genetic background of an infectious cDNA clone derived from clinical strain USA-WA1/2020 (ref. 27) using a previously described protocol28. The spike mutations for the B.1.1.7 variant included 69–70 deletion, Y145 deletion, N501Y, A570D, D614G, P681H, T716I, S982A and D1118H. The spike mutations for the P.1 variant included L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I and V1176F. The spike mutations for the B.1.351 variant included D80A, D215G, 242–244 deletion, K417N, E484K, N501Y, D614G and A701V. The recombinant viruses were sequenced to confirm the presence of the engineered mutations and to rule out undesired mutations.
Engineering and production of IgM and IgA1 antibodies
The Vh and Vl regions of five human anti-SARS-CoV-2 IgG1 antibodies—CoV2-06, CoV2-09, CoV2-12, CoV2-14, and CoV2-16—were incorporated into expression vectors encoding IgM and IgA1 constant regions and a human J-chain according to standard cloning protocols. The original isolated CoV2-06, CoV2-09, CoV2-12, CoV2-14, and CoV2-16 IgG1 antibodies had lambda light chains. For initial in vitro characterization, the light chains for these antibodies were constructed as lambda-Vl-kappa-Cl hybrid light chains and the antibodies were transiently expressed in Expi293 cells. For subsequent in vitro and in vivo experiments, complete lambda light chains were used and the antibodies were expressed in Expi293F, ExpiCHO-S or CHO cells. The control antibody-binding domain from anti-SARS-CoV antibody CR3022 was similarly constructed. The IgG1 and IgA1 antibody constructs were purified by affinity chromatography using protein A and Capture Select LC-lambda resins (Thermo Fisher Scientific), respectively. The IgM antibody constructs were purified by mixed-mode chromatography and anion-exchange chromatography25. The degree of assembly and purity were assessed by SDS–PAGE and native PAGE. For SDS–PAGE, all antibodies were run on 10% Mini-PROTEAN TGX gels (Bio-Rad). The gels were stained with Coomassie blue R-250 (Bio-Rad). For native PAGE, IgG1 and IgA1 antibodies were run on native 4–12% Bis-Tris gels (Life Technologies), whereas IgM antibodies were run on native 3–12% Bis-Tris gels (Life Technologies) by referring to a previously described electrophoretic method29. The gels were stained with Colloidal Blue Stain (Life Technologies). For gel source data, see Supplementary Fig. 1. The IgG1, IgA1 and IgM antibodies were also assessed by size-exclusion chromatography and the data were analysed using the UNICORN v.7.0 software.
Recombinant proteins
The spike protein (S1 + S2 ECD, His tag) (40589-V08B1) and the human ACE2 protein (10108-H08H) were purchased from Sino Biological. The His-tagged RBD protein (CoV2-RBD–His) of SARS-CoV-2 spike protein was purchased from ATUM (65639.1.a). The Fc-tagged wild type RBD protein and seven RBD proteins that contain amino acid mutations, including K444R, E484A, K444R + E484A, K444S and F486S, were generated in a previous study1. The expression constructs of 19 Fc-tagged RBD proteins that contain amino acid mutations, including N439K, S477N, N501Y, K417N, E484K + N501Y, K417N + E484K + N501Y, E484K, F490S, Q493R, S494P, K417E, Y453F, L455F, G476S, F486V, Q493K, K444Q, V445A and G446V, were generated by overlap PCR using specific primers (Supplementary Table 2) in this study. The proteins were produced from Expi293F cells and purified using the CaptivA protein A affinity resin (Repligen, CA-PRI-0100).
ELISA binding assay
ELISA titration of monoclonal antibody binding to the RBD of the SARS-CoV-2 spike protein was performed as follows. The 96-well white polystyrene ELISA plates (Pierce 15042) were coated with 100 μl per well of 0.5 μg ml−1 recombinant His-tagged RBD protein overnight at 4 °C. Plates were then washed five times with phosphate-buffered saline (PBS) with 0.05% Tween and blocked with 2% bovine serum albumin (BSA)-PBS. After blocking, 100-μl serial dilutions of monoclonal antibodies were added to the wells and incubated at room temperature for 2 h. The plates were then washed 10 times and incubated with HRP-conjugated mouse anti-human kappa (Southern Biotech, 9230-05, 1:6,000 diluted in 2% BSA-PBS) for 30 min. After 10 final washes using 0.05% PBS-Tween, the plates were read using Super Signal chemilumnescent substrate (Thermo Fisher Scientific, 37070). Luminescent data were collected on an EnVision plate reader (Perkin Elmer) and analysed with GraphPad Prism 8.
For ELISA titration of monoclonal antibody binding to SARS-CoV-2 spike protein, high-binding ELISA plates were coated with recombinant spike protein (1 μg ml−1) at 4°C overnight and blocked with 5% skimmed milk at 37 °C for 2 h. Antibodies were serially diluted in 1% skimmed milk and added at a volume of 100 μl per well for incubation at 37 °C for 2 h. The HRP-conjugated F(ab’)2 fragment goat anti-human IgA + IgG + IgM (H+L) antibody (Jackson ImmunoResearch, 109-036-064) was diluted 1:5,000 and added at a volume of 100 μl per well for incubation at 37 °C for 1 h. The plates were washed around three to five times with PBST (0.05% Tween-20) between incubation steps. TMB (3,3′,5,5′-tetr amethylbenzidine) substrate was added at 100 μl per well for colour development. The reaction was stopped by adding 50 μl per well 2M H2SO4. The OD450 nm was read by a SpectraMax microplate reader and analysed with GraphPad Prism 8.
Antibody avidity
The measurement of antibody avidity (apparent affinity) to the spike protein and the wild-type or mutant RBD proteins were performed on the ForteBio Octet RED96 system. The His-tagged spike protein (15 μg ml−1) was captured on the Ni-NTA biosensor and the Fc-tagged RBD proteins (20 μg ml−1) were captured on the protein A biosensor. Following 10 s of baseline run in kinetics buffer, the sensors were dipped in threefold serially diluted antibodies (0.12 nM to 90 nM) for 200 s to record association kinetics. Then, the sensors were dipped into kinetics buffer for 400 s to record dissociation kinetics. For avidity measurement with Fc-tagged RBD proteins, the biosensors were blocked with a control Fc protein (150 μg ml−1) for 200 s to occupy the free protein A. ForteBio Octet Data Analysis software was used to fit the KD data using the global fitting method.
Neutralization assays
The neutralization of SARS-CoV-2 viruses with wild-type RBD, K444R RBD, E484A RBD and K444R + E484A RBD was performed using the mNG reporter viruses. In brief, a total of 1.5 × 104 Vero cells or A549-ACE2 cells were plated into each well of a black transparent flat-bottom 96-well plate (Greiner Bio-One, 655090). The next day, antibodies (serial dilutions) were mixed with an equal volume of SARS-CoV-2-mNG virus (multiplicity of infection = 0.5). After 1 h incubation at 37 °C, the antibody–virus complexes were inoculated with Vero cells. At 20 h after infection, nuclei were stained by the addition of Hoechst 33342 (Thermo Fisher Scientific) to a final concentration of 10 nM. Fluorescent images were acquired using a Cytation 7 multimode reader (BioTek). Total cells (in blue) and mNG-positive cells (in green) were counted, and the infection rate was calculated. The relative infection rates were calculated by normalizing the infection rate of each well to that of control wells (no antibody treatment).
The neutralization of the SARS-CoV-2 US-WA1 strain and the SARS-CoV-2 viruses containing the spike mutations from the B.1.1.7, P.1 and B.1.351 lineages were performed using the PRNT. In brief, antibodies were serially diluted in culture medium and incubated with 100 PFU of wild-type or mutant viruses at 37 °C for 1 h, after which the antibody–virus mixtures were inoculated onto Vero E6 cell monolayer in six-well plates. After 1 h of infection at 37 °C, 2 ml of 2% SeaPlaque agar (Lonza) in DMEM containing 2% FBS and 1% penicillin–streptomycin was added to the cells. After 2 d of incubation, 2 ml of 2% SeaPlaque agar in DMEM containing 2% FBS, 1% penicillin–streptomycin and 0.01% neutral red (Sigma-Aldrich) were added on top of the first layer. After another 16 h of incubation at 37 °C, plaque numbers were counted. The relative infection was obtained by the plaque counts from the antibody-treated groups to the untreated groups. The relative infection versus the concentration of the antibody (in log10 scale) was plotted. PRNT50 titres were calculated using a nonlinear regression model. All SARS-CoV-2 manipulations were conducted at the Biosafety Level-3 facility with approval from the Institutional Biosafety Committee at the University of Texas Medical Branch.
Antibody structure modelling and molecular docking
The three-dimensional (3D) structures of antibody-RBD complexes were built using the Rosetta-based computational protocols16. In brief, the 3D structures of the antibody Fv region were first predicted from their amino acid sequences by the antibody function of Rosetta. Then, the structure of the Fv region was docked to the published RBD structure (Protein Data Bank (PDB) code: 6M0J) by using the SnugDock function of Rosetta. The binding interface of the antibody Fv relative to the RBD was positioned using PyMol software. The antibody epitope residues were previously identified using an alanine-scanning RBD mutant library1 and were incorporated into the analysis to inform docking studies. All the modelling and docking tasks were performed by querying the online server (https://rosie.graylab.jhu.edu/). The Fv–RBD structures with the lowest interface energy and having good coverage of epitope residues in the docking were selected for further analysis. To visualize the binding model of the antibody relative to the ACE2, the Fv–RBD structures were aligned with the ACE2–RBD complex structure (PDB: 6M0J) based on the RBD using PyMol software.
Antibody blocking of RBD and ACE2 interaction
The wild-type RBD or the mutant RBD proteins (4 μg ml−1) were captured on the protein A biosensor for 300 s. The sensors were then blocked by a control Fc protein (150 μg ml−1) for 200 s to occupy the free protein A on the sensor. The serially diluted antibodies (0.041 nM to 30 nM) were incubated with the sensors for 200 s to allow antibody and RBD binding. The irrelevant isotype antibodies (30 nM) were used as controls. After 10 s of baseline run in kinetics buffer, the sensors were dipped in the ACE2 solution (8 μg ml−1) for 200 s to record the response signal. For analysis of the IC50, the ACE2 response values were normalized to the starting points. The blocking percentages at each concentration were calculated as: ((normalized ACE2 response of isotype antibody – normalized ACE2 response of tested antibody)/normalized ACE2 response of isotype antibody) × 100. The dose-blocking curves were plotted and the blocking IC50 values were calculated by nonlinear fit using GraphPad Prism 8.
Bioinformatics analysis of RBD mutations in circulating virus
As of 8 February 2021, 373,387 human SARS-CoV-2 genomic sequences were analysed for the occurring frequencies of individual RBD mutations in the global circulating virus variants. The database of the COVID-19 Viral Genome Analysis Pipeline was queried and the Tracking Mutations tool (https://cov.lanl.gov/content/sequence/TRACK_MUT/trackmut.html) was used to obtain the numbers of virus variants that contain amino acid mutations at individual RBD sites. The frequencies of individual RBD mutations were calculated by dividing the numbers of virus variants by a total of 373,387 virus sequences. The frequencies were expressed as number of variants per 10,000 sequences.
Tracking antibody bio-distribution in mice
Antibodies were conjugated with Alexa Fluor 750 dye for in vivo imaging studies. In brief, 1.78 mg ml−1 solution of IgM-14 in PBS (pH 7.4) was reacted with Alexa Fluor 750 succinimidyl ester (AF750-NHS,Thermo Fisher Scientific) in the presence of 3% DMSO and 10% sodium biocarbonate buffer (v/v, pH 8.3) using the molar ratio of 1:10 protein to fluorescent probe at room temperature for 1 h. Unreacted dye was removed by dialysis, the labelled antibody was washed in PBS and concentrated with an Amicon ultra centrifugal filter unit (MWKO 10 kDa). All procedures were performed under dimmed light. The CD-1 mice (6–8 weeks, female, Charles River Laboratories) were anaesthetized by inhalation of 2% isoflurane and placed in a supine position. The mice were administered intranasally with Alexa Fluor 750-labelled IgM-14 to both nostrils of the mice using a fine pipet tip (40 μl total) to achieve the final antibody dose of 1.2 mg kg−1. The mice were imaged at predetermined time points after administration (fluorescence ex = 740 nm, em = 790 nm, auto-exposure setting, n = 4 mice in each group) using an IVIS Lumina XRMS Imager (Perkin Elmer). At the time of euthanasia, 20 μl of blood, the heart, lung, liver, spleen, kidney, brain and nasal cavity samples were excised and imaged. Regions of interest (ROIs) were drawn and average radiant efficiency ((p s−1 cm−2 sr−1)/(μW cm−2)) was measured. This parameter represents the sum of the radiance from each pixel inside the ROI divided by the number of pixels. All images were processed using Living Image software (Perkin Elmer) and the same fluorescence threshold was applied for group comparison.
Mouse infection and antibody protection
The animal study was carried out in accordance with the recommendations for care and use of animals by the Office of Laboratory Animal Welfare, National Institutes of Health. The Institutional Animal Care and Use Committee (IACUC) of the University of Texas Medical Branch approved the animal studies under protocol 1802011. Ten-to-twelve-week-old female BALB/c mice were purchased from Charles River Laboratories and maintained in Sealsafe HEPA-filtered air in/out units. A previously described mouse infection model was used to evaluate antibody protections1. Animals were anaesthetized with isoflurane and infected intranasally with 104 PFU of mouse-adapted SARS-CoV-2 (CMA4 strain; N501Y)26, the P.1 variant or the B.1.351 variant in 50 μl of PBS. Antibodies were intranasally delivered at 6 h before or 6 h after viral infection. Two days after infection, lung samples of infected mice were collected and homogenized in 1 ml PBS using the MagNA Lyser (Roche Diagnostics). The homogenates were clarified by centrifugation at 15,000 rpm for 5 min. The supernatants were collected for measuring infectious virus titres by plaque assay as mentioned in the neutralization assay. Quantitative PCR with reverse transcription (qRT–PCR) assay was also used for measuring viral RNA (nucleocapsid gene) titres in the lung. In brief, the clarified tissue homogenates were mixed with a fivefold excess of TRIzol LS Reagent (Thermo Fisher Scientific, 10296010). Total RNA was extracted according to the manufacturer’s instructions. The extracted RNA was finally dissolved in 40 μl nuclease-free water. Two microlitres of RNA samples were used for qRT–PCR assays using the iTaq SYBR Green one-step kit (Bio-Rad) on the QuantStudio Real-Time PCR systems with fast 96-well module (Thermo Fisher Scientific). The quantification of viral RNA was determined by a standard curve method using an RNA standard (in vitro transcribed 3,839 bp RNA at the nucleotide positions from 26,044 to 29,883 of SARS-CoV-2 genome) and the primers 2019-nCoV_N2-F (5′-TTACAAACATTGGCCGCAAA-3′) and 2019-nCoV_N2-R (5′-GCGCGACAT TCCGAAGAA-3′).
Pharmacokinetic and safety studies
For the pharmacokinetic studies, female BALB/c mice between the ages of 6 and 8 weeks were obtained from Charles River Laboratories. Five groups of mice (three per group) were used. Dosing of IgM-14 was done via the intranasal route according to an approved IACUC protocol. In brief, mice were lightly anaesthetized with isoflurane and 50 μl of IgM-14 at 2 mg ml−1 (total dose 5 mg kg−1) was slowly instilled with a pipette, with the volume split between the two nares. Survival and terminal bleeds were collected by either retro-orbital or cardiac puncture for sample collection at 15 min, 30 min, 1 h, 2 h, 4 h, 8 h, 24 h, 48 h, 72 h and 96 h. Approximately 200 μl of blood was processed for plasma. An ELISA assay was used to determine IgM-14 concentrations in mouse plasma. In brief, Pierce 96-well ELISA plates (Thermo Fisher, 15042) were coated with 100 μl of SARS-CoV-2 spike protein RBD (319-591, C-term His tag, LakePharma,46438) at 1 μg ml−1 in 1× PBS overnight at room temperature. After blocking with 3% BSA in PBS at room temperature for 2 h and washing in PBS + Tween-20, samples prepared in PBS containing 3% BSA and 1% diluted mouse plasma were added and incubated for 2h at room temperature. After washing, mouse anti-human lambda-HRP (SouthernBiotech, 9180-05, 1:2,000 diluted) was added and the plates were incubated for 1 h at room temperature. Following washing and addition of SuperSignal ELISA Pico Chemiluminescent Substrate (Thermo Fisher Scientific, 37069), plates were read on a Perkin Elmer EnVision 2104 Multilabel MicroPlate Reader and data were analysed by applying a four-parameter logistic fit to standards using GraphPad Prism. Sample concentrations were interpolated from the standard curve. The lower limit of quantification (LLOQ) for the assay was 0.02 μg ml−1.
For the safety study, groups of rats (four per group) were dosed intranasally with IgM-14 or the vehicle control twice daily for five consecutive days. The top dose was 4 mg per kg per day (20 mg total over the 5-day period). All rats survived to the end of the study. Clinical observations, body weight changes, macroscopic observations, clinical pathology observations or organ weight effects were monitored. After the completion of dosing, the nasal cavities of each rat were also removed, sectioned and examined histologically.
Statistical analysis
All statistical analysis was performed using GraphPad Prism 8 and the statistic tests are described in the indicated figure legends. Nonlinear regression curve fitting was performed to calculate the EC50, IC50 and NT50 values.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Extended Data
Extended Data Fig. 1 ∣. The epitopes of IgG1 monoclonal antibodies used for engineering and additional characterizations of IgM and IgA1.
a, The RBD is shown as a cartoon with the core region coloured in grey and the receptor-binding motif (RBM) coloured in red. b, Summary of the binding region on the RBD, key epitope residues and cross-reactivity to SARS-CoV of the six monoclonal antibodies. These epitope residues were mapped using an alanine-scanning RBD mutant library in a previous study1. c, ELISA binding to RBD by IgG1, IgA1 and IgM isotypes of the indicated monoclonal antibodies. Data are mean of duplicate wells. d, Neutralization of live SARS-CoV-2 by IgG1, IgA1 and IgM monoclonal antibodies at 1 μg ml−1. Data are mean ± s.d. of triplicates.
Extended Data Fig. 2 ∣. Binding and neutralization characterizations of IgM-06, IgG-06, IgA1-06 and IgA1-14.
a, ELISA binding to the spike protein (S) by IgM-06 and IgG-06. Data are mean of duplicate wells. b, Binding kinetics of IgM-06 and IgG-06 to the spike protein. c, Neutralization of SARS-CoV-2 by IgM-06 and IgG-06. Data are mean of duplicate wells. d, Summary of binding EC50, association (Kon), dissociation (Kdis), avidity (KD) and neutralization NT50 values of IgM-06 and IgG-06. e, Neutralization of SARS-CoV-2 by IgA1-06 and IgA1-14. Data are mean of duplicate wells.
Extended Data Fig. 3 ∣. Structural docking of the Fv–RBD complex.
a, b, Docking of IgFv-14–RBD (a) and IgFv-06–RBD (b) complex structures. The RBD is shown as a cartoon and coloured in grey. The Fv is shown as a surface with Vh coloured in cyan and Vl coloured in magenta. Antibody epitope residues are shown as sticks and coloured in blue.
Extended Data Fig. 4 ∣. Antibody blocking of RBD and ACE2 interaction as measured by a BLI assay.
a, Schematic diagram showing a BLI assay for IgM and IgG blocking of RBD and ACE2 interaction. b, A representative binding response curve of the BLI assay. The vertical dashed lines indicate the separation of each binding phase. c, d, The normalized ACE2 response curves after blocking by IgM-14 and IgG-14 (c) and by IgM-06 and IgG-06 (d). e, Superposition of IgFv-06–RBD and ACE2–RBD complexes. The IgFv-06 is shown as a surface with Vh coloured in cyan and Vl coloured in magenta. The RBD–ACE2 complex is shown as a cartoon with RBD coloured in grey and ACE2 coloured in green. The dashed box indicates steric clash. f, IgM-06 and IgG-06 blocking of RBD and ACE2 interaction. The horizontal dashed line indicates 100% blocking.
Extended Data Fig. 5 ∣. Construction of SARS-CoV-2 escaping variants and additional neutralization characterizations.
a, Schematic diagram showing the construction of the indicated mNeonGreen SARS-CoV-2 viruses using an infectious clone method. b, Plaque morphologies of mNeonGreen SARS-CoV-2 viruses with wild-type RBD or the indicated RBD mutations. c, Neutralization of SARS-CoV-2 viruses with K444R (IgG-06-resistant), E484A (IgG-14-resistant) and K444R + E484A (IgG-06 + IgG-14-resistant) mutations by IgM-14 and IgG-14 on A549-ACE2 cells. Data are mean of duplicate wells. d, Summary of the NT50 values against indicated mutant viruses and the fold changes of NT50 values between IgM-14 and IgG-14. e, Neutralization of indicated SARS-CoV-2 mutant viruses by IgM-06 and IgG-06 on Vero cells. Data are mean of duplicate wells. f, Summary of the NT50 values against indicated mutant viruses and the fold changes of NT50 values between IgM-06 and IgG-06. NA, not available.
Extended Data Fig. 6 ∣. Binding kinetics and ACE2-blocking activities of IgM-14, IgG-14, IgM-06 and IgG-06 against selected RBD mutants.
a–c, The binding kinetics of IgM-14 and IgG-14 to wild-type RBD (a), E484A RBD (b) and K444R + E484A RBD (c). d–f, The binding kinetics of IgM-06 and IgG-06 to wild-type RBD (d), K444R RBD (e) and K444R+E484A RBD (f). g, h, IgM-14 and IgG-14 blocking of E484A RBD (g) and K444R + E484A RBD (h) interaction with ACE2. i, j, IgM-06 and IgG-06 blocking of K444R RBD (i) and K444R + E484A RBD (j) interaction with ACE2. k, Summary of the binding avidities (KD) and ACE2-blocking activities (IC50) to indicated RBD proteins by IgM-06 and IgG-06. ND, not determined. *Half-maximal blocking was not achieved at the highest monoclonal antibody concentration (30 nM) and the IC50 values are defined as ≥90 nM. l, m, The binding kinetics of IgM-14 and IgG-14 to F486S RBD (l) and IgM-06 and IgG-06 to K444S RBD (m). n, o, IgM-14 and IgG-14 blocking of F486S RBD interaction with ACE2 (n) and IgM-06 and IgG-06 blocking of K444S RBD interaction with ACE2 (o).
Extended Data Fig. 7 ∣. Binding kinetics of IgM-14 and IgG-14 to the 19 RBD mutants.
a, SDS–PAGE images of six RBD proteins with mutations that represent natural escape variants in circulation. b, SDS–PAGE image of thirteen RBD proteins with mutations associated with neutralization-resistance to LY-CoV555, REGN-10933 and REGN-10987. The SDS–PAGE gel images were from one experiment. c–u, Binding kinetics of IgM-14 (left) and IgG-14 (right) to the indicated RBD mutants. v, Fold changes of Kdis and Kon between binding of IgM-14 and IgG-14 to the 19 RBD mutants. The ratios were calculated as indicated in the y axis.
Extended Data Fig. 8 ∣. Antibody blocking of interactions between ACE2 and the 19 RBD mutants, the frequency of RBD mutations and mutational effects on RBD functionality.
a–s, The dose-dependent blocking of the interactions between ACE2 and the indicated RBD mutants. t, The correlation between ACE2 blocking IC50 and neutralization NT50 values. Two-tailed Spearman correlation was used in the statistical analysis. u, The frequency of SARS-CoV-2 circulating variants with indicated RBD mutations. The Tracking Mutations tool was used for analysis as described in the methods. v, The effects of indicated RBD mutations on RBD binding affinity to ACE2 and RBD protein expression. The online web source on the sequence-to-phenotype maps of the RBD of SARS-CoV-2 (https://jbloomlab.github.io/SARS-CoV-2-RBD_DMS/) was referred to for analysis. The y axis indicates log10 scale changes to wild-type RBD. Positive values indicate an improving effect and negative values indicate a decreasing effect by the mutations.
Extended Data Fig. 9 ∣. Bio-distribution of AF750-labelled IgM-14 after intransal delivery in mice.
a, Whole-body imaging of four mice at different time points after a single intranasal dose of IgM-14. b, Ex vivo imaging of blood (20 μl and different organs. c, Quantification of average radiant efficiency of blood and indicated organs. Data are mean ± s.d. of four independent mice.
Extended Data Fig. 10 ∣. Additional in vivo characterizations of antibody efficacy, pharmacokinetics and safety.
a, Experimental design of prophylactic evaluation of the indicated monoclonal antibodies. n = 4 independent mice for all groups. b, Virus PFU titres in the lung samples of mice prophylactically treated with the indicated monoclonal antibodies. c, Virus RNA (N gene) titres in the lung samples of mice prophylactically treated with IgM-14. The cut-off for the qRT–PCR method, shown as dotted line, is defined as mean + 2 standard deviations of corresponding RNA copies in the qRT–PCR using lung samples from five uninfected mice. d, Viral loads (PFU titres) in the lung samples of mice therapeutically treated with IgM-14 or IgG-14 at the indicated doses. The lines of median lung viral loads are shown for each group. n = 10 biologically independent mice for all groups except that n = 5 for IgM-14 group. A two-sided Mann–Whitney test was used in the statistical analysis for b, d. An ordinary one-way ANOVA with Sidak’s multiple comparisons was used in the statistical analysis for c. e, Sequencing analysis of viruses recovered from lung samples of the ten most outlier mice. A representative chromatogram representing the amino acids 483–489 of the RBD is shown to indicate that no mutations of the critical residues E484 and F486 were observed. f, The plasma concentrations of IgM-14 after a single intranasal dose of 5 mg kg−1 in BALB/c mice. Data are mean ± s.d. of three independent mice.The values lower than LLOQ (0.02 μg ml−1) were defined as 0.01. g, Body weight changes of rats after intranasal administrations of 2 mg kg−1 per dose of IgM-14 or the vehicle control. Data are mean ± s.d. of four independent rats. The arrows indicate dosing twice daily for five consecutive days. Statistical differences between IgM-14 and vehicle groups were analysed by a two-sided multiple t-test. ns, P ≥ 0.05.
Supplementary Material
Acknowledgements
We thank W. Strohl for his scientific advice; J. Tang, N. Vora and Y. Ng for their technical support; and J. Jarecki and G. Salazar for their editorial support. This work was supported in part by a Welch Foundation grant AU-0042-20030616 and Cancer Prevention and Research Institute of Texas (CPRIT) grants RP150551 and RP190561 (Z.A.); NIH grants AI134907 and UL1TR001439, awards from the Sealy Smith Foundation, Kleberg Foundation, John S. Dunn Foundation, Amon G. Carter Foundation, Gillson Longenbaugh Foundation and Summerfield Robert Foundation (P.-Y.S.); and NIH grant R15CA182769 and CPRIT RP150656 (X.L.).
Footnotes
Competing interests The University of Texas System has filed a patent on the SARS-CoV-2 antibodies and the reverse genetic system and reporter SARS-CoV-2. The University of Texas System and IGM Biosciences have filed a joint patent on the SARS-CoV-2 IgM antibodies. The antibodies are being developed by IGM Biosciences for prophylactic and therapeutic treatment of COVID-19. P.R.H., D.C.N., D.P., S.R., Y-A.C., K.B.C., E.J.H., B.A.K. and S.F.C. are employees of IGM Biosciences. The other authors declare no competing interests.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-021-03673-2.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-021-03673-2.
Data availability
Data associated with figures are available from the corresponding authors upon reasonable request. The COVID-19 Viral Genome Analysis Pipeline Tracking Mutations tool is available at https://cov.lanl.gov/content/sequence/TRACK_MUT/trackmut.html. The published structure of the RBD bound to ACE2 is available at the PDB with accession code 6M0J. Source data are provided with this paper.
References
- 1.Ku Z et al. Molecular determinants and mechanism for antibody cocktail preventing SARS-CoV-2 escape. Nat. Commun 12, 469 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wibmer CK et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med 27, 622–625 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Wang P et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593, 130–135 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Starr TN et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science 371, 850–854 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wölfel R et al. Virological assessment of hospitalized patients with COVID-2019. Nature 581, 465–469 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Pujadas E et al. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir. Med 8, e70 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwasaki A Exploiting mucosal immunity for antiviral vaccines. Annu. Rev. Immunol 34, 575–608 (2016). [DOI] [PubMed] [Google Scholar]
- 8.DeFrancesco L COVID-19 antibodies on trial. Nat. Biotechnol 38, 1242–1252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinreich DM et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with covid-19. N. Engl. J. Med 384, 238–251 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar N, Arthur CP, Ciferri C & Matsumoto ML Structure of the secretory immunoglobulin A core. Science 367, 1008–1014 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Li Y et al. Structural insights into immunoglobulin M. Science 367, 1014–1017 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Vonarburg C et al. Topical application of nebulized human IgG, IgA and IgAM in the lungs of rats and non-human primates. Respir. Res 20, 99 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z et al. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med 13, eabf1555 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galimidi RP et al. Intra-spike crosslinking overcomes antibody evasion by HIV-1. Cell 160, 433–446 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ter Meulen J et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 3, e237 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weitzner BD et al. Modeling and docking of antibody structures with Rosetta. Nat. Protocols 12, 401–416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomson EC et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 184, 1171–1187 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 29, 477–488 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.FDA. Fact sheet for health care providers. Emergency use authorization (EUA) of casirivimab and imdevimab, https://www.fda.gov/media/143892/download (2020). [Google Scholar]
- 20.FDA. Fact sheet for health care providers. Emergency use authorization (EUA) of bamlanivimab, https://www.fda.gov/media/143603/download (2020). [Google Scholar]
- 21.Hou YJ et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182, 429–446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W et al. Detection of SARS-CoV-2 in different types of clinical specimens. J. Am. Med. Assoc 323, 1843–1844 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barnes CO et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong S et al. Anti-HIV IgM protects against mucosal SHIV transmission. AIDS 32, F5–F13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keyt BA, Baliga R, Sinclair AM, Carroll SF & Peterson MS Structure, function, and therapeutic use of IgM antibodies. Antibodies 9, 53 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muruato A et al. Mouse-adapted SARS-CoV-2 protects animals from lethal SARS-CoV-2 challenge. Preprint at 10.1101/2021.05.03.442357 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie X et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 27, 841–848 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie X et al. Engineering SARS-CoV-2 using a reverse genetic system. Nat. Protocols 16, 1761–1784 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vorauer-Uhl K, Wallner J, Lhota G, Katinger H & Kunert R IgM characterization directly performed in crude culture supernatants by a new simple electrophoretic method. J. Immunol. Methods 359, 21–27 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with figures are available from the corresponding authors upon reasonable request. The COVID-19 Viral Genome Analysis Pipeline Tracking Mutations tool is available at https://cov.lanl.gov/content/sequence/TRACK_MUT/trackmut.html. The published structure of the RBD bound to ACE2 is available at the PDB with accession code 6M0J. Source data are provided with this paper.