Abstract
Background:
Predictive biomarkers could allow more precise use of immune checkpoint inhibitors (ICI) in treating advanced cancers. Given the central role of HLA molecules in immunity, variation at the HLA loci may differentially impact response to ICI. The aim of this epidemiological study was to determine the effect of HLA-A*03 as a biomarker for predicting response to immunotherapy.
Methods:
We studied clinical outcomes (overall survival, progression free survival or objective response) of treatment for advanced cancer in eight cohorts, including 3,339 patients treated with various ICI agents and 10,867 treated with alternative therapies. We initially modelled the association of HLA amino acid variation, followed by detailed analysis of HLA-A*03 on clinical outcomes, with replication in additional cohorts.
Findings:
HLA-A*03 associated in an additive manner with reduced survival after immune checkpoint inhibition (ICI) treatment in a discovery cohort (hazard ratio, 1·54 per HLA-A*03 allele; 95% confidence interval [CI] 1·20-1·96, p<0·001), validation cohort (hazard ratio, 1·22 per HLA-A*03 allele; 95% CI 1·05-1·42, p=0·009) and clinical trial for bladder cancer (hazard ratio, 1·36 per HLA-A*03 allele; CI 1·005-1·851, p=0·05). The HLA-A*03 effect was observed across ICI agents and tumor types, but not in patients treated with alternative therapies. HLA-A*03+ patients had shorter progression-free survival in three clinical trials of nivolumab for renal cell cancer (RCC, hazard ratio, 1·31; 95% CI 1·0-1·71, p=0·04), but not in the control arm. Objective responses were observed in 0% (0/8) of HLA-A*03 homozygotes in the ICI arm. HLA-A*03 associated with shorter progression-free survival in patients receiving ICI in another randomized clinical trial for RCC (avelumab/axitinib; hazard ratio, 1·59 per HLA-A*03 allele; 95%CI 1·16-2·16, p=0·004), but not those receiving control therapy. Objective responses rate was 12·5% (1/8) among HLA-A*03 homozygotes in the ICI arm. HLA-A*03 associated with impaired outcome in meta-analysis of all 3,339 ICI treated patients at genome-wide significance (p=2·01x10−8, I2=0%, 95% CI 0%-0·76%).
Interpretation:
HLA-A*03 carriage should be considered in decisions regarding cancer therapy involving ICI.
Funding:
NIH, Merck KGaA, Darmstadt, Germany and Pfizer.
Introduction
Despite the success of PD-1, PD-L1, and CTLA-4 inhibitors in eliciting measurable shrinkage of tumors (objective response), slowing cancer progression, and/or improving overall survival in many cancer types, most patients who receive ICIs do not benefit. Identifying clinical or molecular features that selectively predict response to ICIs could improve patient outcomes through precision application of ICI, rationalize pathobiological targets to enhance efficacy, and help reduce inequality in ICI access through improved selection of patients who may benefit.
Identification and validation of clinically useful predictive biomarkers has remained elusive, though burgeoning understanding of tumor-immune interactions has fueled discovery of correlative biomarkers that provide important insights into biological mechanisms in vivo(1),(2). PD-L1 expression and tumor mutational burden(3) are examples of widely studied biomarkers that associate with response to ICI in some tumor types, yet remain complex to assay, and have considerable overlap between patients who do and do not respond to ICI, limiting clinical utility(4).
Variation of Human Leukocyte Antigens (HLA) has been a major focus of biomarker discovery efforts. The degree of heterozygosity at the locus was associated with improved survival in melanoma and non-small cell lung cancer (NSCLC) patients treated with ICI, but with inconsistent validation(5) (6),(7). Carriage of the HLA-B62 and -B44 supertypes were found to be associated with worse and improved survival, respectively, in patients treated with ICI in a discovery cohort, but HLA-B62 did not replicate in a second cohort (5). The association of the HLA-B44 supertype with survival was constrained to a subset of HLA-B44 supertype alleles applied to a specific subset of melanoma patients (5) and has not been observed in patients with NSCLC(6),(8). Disruption of antigen presentation through somatic loss of heterozygosity at HLA, or factors that diminish HLA expression are associated with reduced responses to ICI (9). Importantly, these effects concur with the central role of antigen presentation in the context of HLA for T-cell recognition of tumor cells and ICI response.
We hypothesized that responses to immune checkpoint inhibition (ICI) are modified by variation in HLA-I alleles, the functional unit of which is amino-acid level variation.
Methods
Patient cohorts and clinical endpoints
We studied three observational cohorts and five clinical trials comprising, in total, 3,339 patients with multiple tumor types treated with various ICI agents and 10,867 patients not treated with ICI, for overall survival, progression-free survival or objective response rate as clinical endpoints (Figure 1 and appendix page 5).
Figure 1:
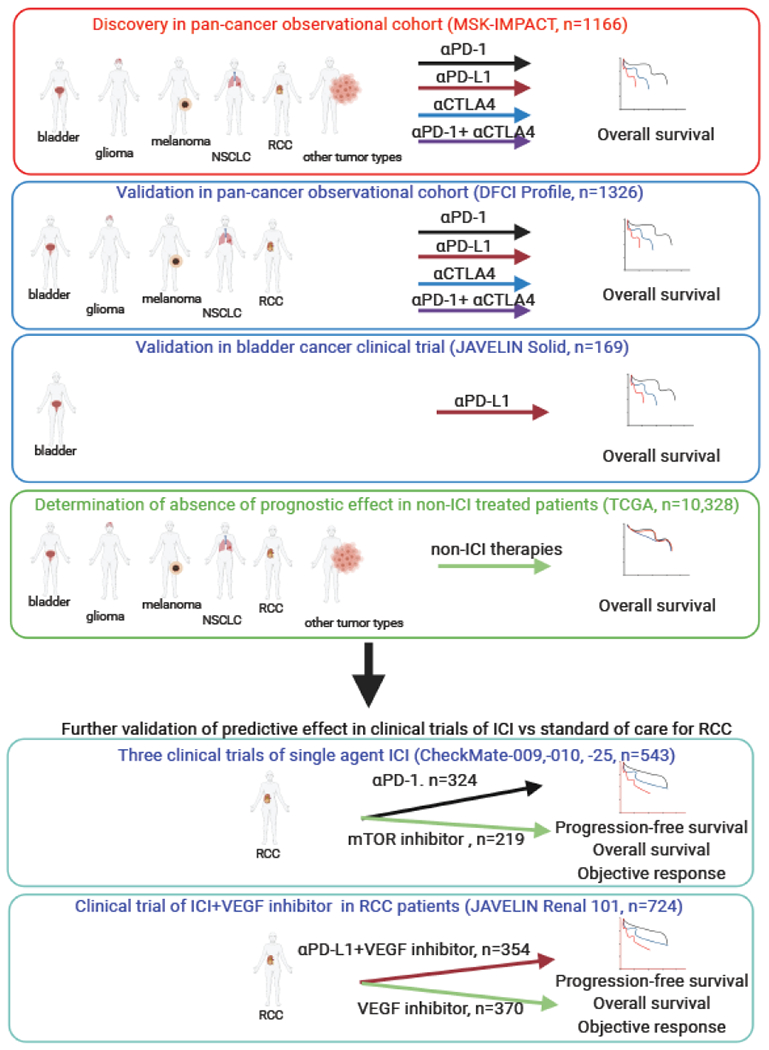
Schematic of discovery and validation of HLA-A*03 as a predictive biomarker in 3,339 patients treated with various ICI agents and 10,867 non-ICI treated patients with various cancers.
The discovery analysis was a test for potential effects of individual HLA class I amino acids on overall survival following ICI in the MSK-IMPACT cohort, consisting of 1166 patients with a range of five main tumor types and 25 smaller subgroups treated with various ICI agents (obtained from ‘cohort 2’ of supplementary data(5)). The initial validation cohort (DFCI Profile) was chosen with particular focus on a comparable study design to the discovery MSK-IMPACT cohort; namely, an observational cohort of patients in clinical care with overall survival as the outcome measure. The DFCI Profile cohort used here consisted of 1326 patients with advanced bladder cancer, high grade glioblastoma, stage 3 or 4 melanoma, NSCLC, or stage 3 or 4 RCC treated with ICI at the Dana Farber Cancer Institute (DFCI) between 2016 and 2019 (DFCI Profile, IRB number 11-104, 17-000, and 19-025). Patients were treated with therapies at their physician’s discretion. An additional validation cohort of 169 participants in the phase 1 clinical trial of avelumab for treatment of bladder cancer (JAVELIN Solid Tumor urothelial carcinoma cohort(10), ClinicalTrials.gov number, NCT01772004) was included. The primary endpoint in the discovery and validation cohorts was overall survival defined as the time from treatment start until death. Prospectively assessed progression-free survival outcomes were not available in these clinical cohorts. Data from The Cancer Genome Atlas (TCGA) were used to examine the prognostic effect of HLA variation on overall survival in non-ICI treated patients.
Data from four clinical trials of ICI in RCC were included as additional validation and to extend the interpretation of findings through comparison of effects of HLA-A*03 on progression-free survival (defined by RECIST v1·1) in patients randomized to receive ICI containing regimens vs. non-ICI containing regimens. In CheckMate-009(11) and -010(12), single-agent nivolumab was evaluated for treatment of clear cell RCC in patients previously treated with anti-angiogenic therapy, and compared with everolimus (an mTOR inhibitor) in CheckMate-025(13) (ClinicalTrials.gov NCT01358721, NCT01354431, NCT01668784). Biomarker analyses based on exome sequence analysis of participants in all three trials combined have been previously presented (15); and to gain power, we also analyzed the unified trial datasets. In JAVELIN Renal 101, patients with newly diagnosed advanced RCC were randomized to receive avelumab and axitinib (an anti-angiogenic agent targeting vascular endothelial growth factor [VEGF]), or sunitinib alone (an alternate anti-VEGF agent) as previously described (ClinicalTrials.gov number, NCT02684006)(13,14). For the CheckMate and JAVELIN Renal 101 RCC trials, biomarker analyses have been previously reported for a subset of patients(14,15), which are used herein. Investigator-assessed objective response rates (ORR) were available for analyses in all four trials.
HLA typing
HLA typing in MSK-IMPACT was obtained from prior reports(5). HLA typing in DFCI Profile was performed by HLA-imputation using SNP2HLA(16) with variants imputed with STITCH(17) from low-coverage sequence reads derived from targeted sequence of recurrently mutated genes in cancer. The accuracy of imputation at HLA-A from low-coverage tumor-sequencing was verified in 881 individuals and was 91·6+−8·8% at four-digit resolution as measured by correlation with imputed dosages from germline genome-wide genotyping, which is known to have very high accuracy. We note that the slightly lower imputation accuracy in this cohort may result in less precision in the estimates in these settings and hence bias towards the null. HLA typing in the JAVELIN Solid Tumor urothelial carcinoma cohort was performed using targeted next generation sequencing of DNA extracted from buffy coats or peripheral blood mononuclear cells (appendix page 1). For CheckMate-009, -010 and -025, and for JAVELIN Renal 101, HLA typing was obtained from tumor exomes(14,15). HLA amino acid frequency, and coding of expression levels and haplotypes is detailed in the appendix (page 1)
Statistical analysis
Analyses were performed in R v4·0·2 using the survival, survminer, meta, and foresttable packages. Survival analyses were performed to study the association between HLA derived measures and overall survival or progression-free survival as the outcome using cox proportional hazard models (coxph() function). Kaplan-Meier curves were drawn using the survfit() function and medians annotated on figures are derived from the surv_median() function.
Individuals with missing data were removed from analyses. Relevant variables available for each cohort, namely age, gender, tumor mutational burden (z-score transformed) and HLA-A expression were first tested in univariate analyses and used as covariates in multivariate analyses of HLA-A*03 and the outcome if significant. Analyses combining two or more tumor types were performed including tumor type as a stratifying variable in the cox proportional hazards model, which is more appropriate than treating tumor types as a covariate in the model because of the expected differences in hazard functions for each tumor type resulting in violation of the proportional hazards assumption if included as a covariate.
We explicitly initiated analyses in the discovery cohort and added analyses of validation cohorts in the order they are presented and did not have an a priori sample size. For analysis of an effect of HLA amino acid variation on overall survival in the MSK-IMPACT discovery cohort, each position of the two allotypes present in each subject was coded as 0 if the amino acid was not the most common or as 1 if the amino acid was the most common, and a score at each site was computed as the sum (0,1, or 2) for the two HLA-A allotypes, the two HLA-B allotypes, or the two HLA-C allotypes. We tested for an effect on overall survival adjusting for tumor mutation burden. A Bonferroni correction was applied to adjust for the total number of HLA-A, -B and -C residues tested. Conditional analyses were performed by adjusting for variation at HLA-A position 161.
An effect of HLA-A*03 on survival in MSK-IMPACT and DFCI Profile was tested by coding the number of HLA-A*03 alleles present as 0,1 or 2. Subgroup analyses of the effect of HLA-A*03 on overall survival were performed for the major tumor types in MSK-IMPACT having sufficient size (n≥75), and adjusting for covariates that were nominally associated with outcomes in each specific tumor type. We performed interaction analysis according to the checkpoint target of the ICI agent for each patient. Individual alleles of the HLA-A3 supertype (coded as 0,1,2), and the A3 supertype excluding HLA-A*03 (coded as presence or absence) were tested in the two cohorts for association with overall survival. Neither HLA-B44 supertype (defined in full, without exclusion of alleles) nor HLA-B62 associated with outcomes consistently in melanoma or any other disease in this study. Adjustment for z-score transformed tumor mutation burden, and/or HLA-A expression was performed where mentioned. Age in the MSK-IMPACT cohort was only available in the form of five ranges which were encoded as 1,2,3,4 and 5 (from the lowest range to the highest range), and tested as such in the models. In DFCI Profile, imputed HLA allelic dosages on the continuous scale were converted to allele count probabilities (0,1, or 2) by rounding the imputed dosage estimate. Ancestry was adjusted for using the top two principal components estimated from genome-wide genotyping data and scaled to allow comparison with 1000 Genomes populations. Additional covariates included were age, gender, line of therapy and whether concurrent chemotherapy was given. Outcome measures for the RCC randomized control trials (RCTs) included progression-free survival, overall survival and objective response. Comparisons of ORR between groups in the CheckMate and JAVELIN Renal 101 studies were performed using the chi-squared test. In the CheckMate RCC studies, MSKCC risk score was explored as a confounder. All p-values shown are two-sided. Additional analyses in JAVELIN Renal 101 of other immune factors that may be associated with HLA-A*03 were performed using supplementary data available in prior publications (14). Fixed effects meta-analyses were performed using the metagen() function implementing inverse-variance pooling of standardized effects sizes and standard errors.
Role of the funding sources
The funding sources had no involvement in the study design, data collection, analysis, interpretation, nor the decision to submit the paper for publication.
Results
Previous studies have indicated the importance of variation at the HLA class I locus in response to ICI(5,14). We identified 71, 76 and 69 polymorphic amino acid sites among HLA-A, -B and -C alleles, respectively, prevalent in the MSK-IMPACT discovery cohort. Testing each variant for an effect on overall survival in the 30 tumor types combined showed that variation at position 161 of HLA-A, where glutamic acid is the most common variant across all HLA-A allotypes, was the most strongly associated (HR 1·47 for each non-glutamic acid residue at HLA-A position 161, 95% CI 1·20-1·81, nominal p=2x10−4, Bonferroni corrected p=0·04, Figure 2A). Furthermore, after conditioning on variation at position 161 no other HLA-A residues associated with overall survival (Figure 2B). Variation at HLA-A position 161 distinguishes alleles of the HLA-A*03 lineage from all other lineages, as all known HLA-A*03 alleles encode aspartic acid (D) at position 161 whilst others encode glutamic acid (E) (or rarely another amino acid). Position 161 of HLA-A is part of the alpha helix adjacent to, but not within a pocket of the peptide binding groove.
Figure 2:
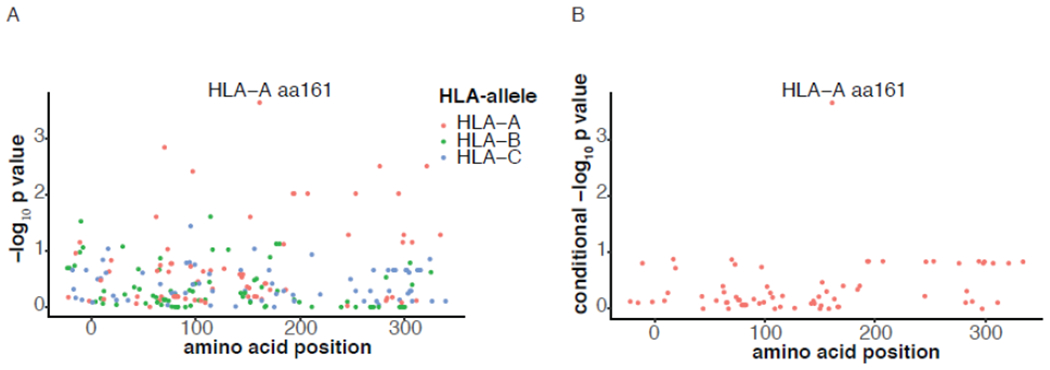
Amino acid variation at HLA-A position 161 and association with overall survival after ICI initiation in the discovery cohort (MSK-IMPACT n=1166). Panel A shows the −log10 p-value (unadjusted) for association at each amino acid site in HLA-A (red circles), HLA-B (green circles) and HLA-C (blue circles). HLA-A position 161 is marked. Panel B shows the −log10 p-value for association at HLA-A residues after conditioning on HLA-A position 161.
Carriage of one or two HLA-A*03 alleles associated with significantly shorter overall survival, stratifying for tumor type in MSK-IMPACT (hazard ratio, 1·54; 95% CI 1·20-1·96, p<0·001). Notably, this association is additive (Figure 3A; hazard ratio, 1·48 per HLA-A*03 allele; 95% CI 1·20-1·82, p<0·001), where individuals homozygous or heterozygous for HLA-A*03 have a 2·31-times (95%CI 1·23-4·33) or 1·47-times (95% 1·14-1·90) greater hazard of death, respectively, relative to non-carriers. HLA-A*03 associated with impaired overall survival in each of the five major tumor types in the MSK IMPACT cohort (bladder cancer, glioma, melanoma [adjusting for tumor mutational burden], NSCLC [adjusting for HLA-A expression] and RCC [adjusting for tumor mutational burden]), and in the remaining minor tumor types collectively (Figure 3B and appendix page 7). Larger cohorts of these minor tumor types are required to definitively demonstrate an effect in each of them. There was no evidence of variation in the effect of HLA-A*03 on overall survival for patients treated with ICI targeted against PD-1/PD-L1 or the combination of PD-1 and CTLA-4, demonstrating that the observed association of HLA-A*03 with outcome to ICI is, at least in this limited analysis, independent of target or ICI agent. The effect of HLA-A*03 on overall survival was independent of HLA-A zygosity (hazard ratio adjusting for HLA-A zygosity, 1·47 per HLA-A*03 allele; 95% CI 1·20-1·81, p<0·001, appendix page 3).
Figure 3:
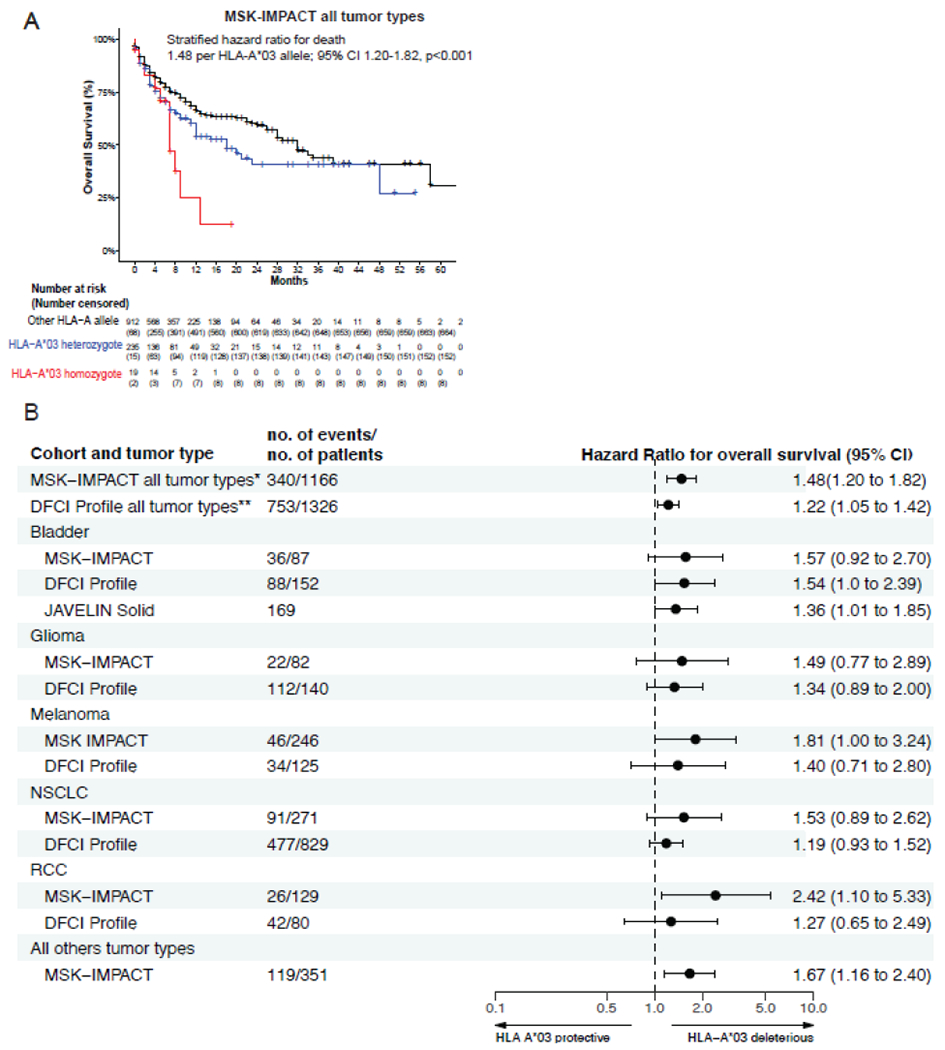
Effect of HLA-A*03 on overall survival in MSK-IMPACT across all tumor types, with statistical estimates stratified for tumor type (Panel A). Forest plot of HLA-A*03 effects on overall survival in MSK-IMPACT, DFCI Profile, and JAVELIN Solid Tumor urothelial carcinoma cohort for all tumor types combined (HR shown is stratified for tumor type), and for each tumor type subgroup individually (Panel B). Statistics in Panel B are adjusted for other covariables that impact the specific tumor type (appendix page 7): tumor mutational burden (RCC and Melanoma) and HLA-A expression (NSCLC). * denotes analysis was stratified for 30 tumor types; ** denotes analysis was stratified for 5 tumor types. HR=Hazard ratio per HLA-A*03 allele. CI=Confidence Interval.
Additional analyses ruled out the possibility that the HLA-A*03 association was confounded by several plausible characteristics intrinsic to HLA-A or the tumor itself. HLA-A alleles vary in expression level in a lineage-dependent manner(18), and HLA-A*03 has the lowest expression amongst common HLA-A alleles, but controlling for predicted HLA-A expression levels based on HLA-A genotypes did not weaken the effect of HLA-A*03 on overall survival (adjusted hazard ratio, 1·75; 95% CI 1·34-2·29 p<0·001). Spurious HLA allelic associations can arise from confounding by an allele with strong opposing effects at the locus; however, controlling for the two more common HLA-A lineages (HLA-A*01 allele frequency, 13·5%; HLA-A*02 allele frequency, 24·7%) did not ablate the HLA-A*03 effect on overall survival (adjusted hazard ratio, 1·47; 95% CI 1·19-1·83 p<0·001). HLA alleles can be grouped into supertypes according to their binding preferences for certain amino acid residues within anchor pockets of the groove, such that alleles of a given supertype bind overlapping peptide repertoires. However, other HLA-A alleles of the HLA-A3 supertype (HLA-A*11:01, -A*31:01, -A*33:01, -A*66:01, -A*68:01), all of which encode Eat position 161, did not individually or collectively associate with overall survival, nor did the HLA-A1, HLA-A2, or HLA-A24 supertypes. Finally, HLA-A*03 was not associated with mutation counts in the MSK-IMPACT cohort (tumor mutational Z-score −3·1x10−4 per A*03 allele; standard error 0·06, p=1·0); accordingly, adjusting for mutational burden did not attenuate the association between HLA-A*03 and shorter overall survival (adjusted hazard ratio, 1·38; 95% CI 1·20-1·82, p<0·001).
The HLA-A*03 effect on overall survival replicated in a second large observational cohort, the DFCI Profile cohort, consisting of 1326 patients treated with ICI for one of the five main cancer types also studied in the discovery cohort, namely bladder cancer, glioma, melanoma, NSCLC, or RCC. HLA-A*03 was significantly associated with impaired overall survival in the entire cohort (hazard ratio stratified by tumor type, 1·22 per HLA-A*03 allele; 95%CI 1·05-1·42, p=0·009), and for each tumor type, the effect estimate was comparable to that in MSK-IMPACT (Figure 3B). As in the MSK-IMPACT cohort, the effect of HLA-A*03 on overall survival did not differ by the target of the ICI agent (interaction p>0·1). Adjusting for somatic mutations that may have prognostic importance in specific tumor types (STK11 and KEAP1 amplification for NSCLC(19), and PBRM1 for RCC(15,20)) did not alter the effect of HLA-A*03 in these tumor types. A key consideration in genetic studies is confounding by ancestry/population structure. The frequency of individuals of Asian, African and European descent in this cohort was 4.5%, 4.5% and 91%, respectively, and the allele frequency of HLA-A*03 was 4.2%, 8.4% and 13.0%, respectively, consistent with expectations(21). In a multivariate model incorporating age, gender, line of therapy, concurrent chemotherapy, and the top two principal components reflecting ancestry, HLA-A*03 remains associated with survival (HR 1·21 95% CI 1·04-1·41, p=0.01).
Evaluation of 169 bladder cancer patients treated with avelumab in the phase 1 JAVELIN Solid Tumor clinical trial urothelial carcinoma cohort (10,22) also replicated the deleterious effect of HLA-A*03 on overall survival (hazard ratio, 1·36 per HLA-A*03 allele; CI 1·005-1·851, p=0·05).
HLA-A*03 was not associated with survival in patients treated with non-ICI therapies in The Cancer Genome Atlas (TCGA) overall or for any of the five main tumor types included in the subgroup analyses (appendix page 8). Taken together, these data suggest that HLA-A*03 may be a predictive biomarker of impaired survival outcomes in patients treated with ICIs irrespective of indication or agent, even though it is not in patients treated with alternative therapies in TCGA.
A substantial effect of HLA-A*03 on overall survival in MSK-IMPACT was observed in RCC patients treated with ICI (Figure 3B and 4A). No effect of HLA-A*03 on overall survival was observed in patients with RCC in TCGA (Figure 4B), highlighting the predictive nature of HLA-A*03 in the context of ICI specifically. We focused subsequent analyses of HLA-A*03 on randomized trials of ICI in RCC where comparison of the association in ICI and non-ICI treated patients ameliorates selection bias and permits simulation of biomarker performance at a potentially relevant decision branchpoint(13,23). We studied 543 participants (324 received nivolumab, 219 received everolimus) who had exome sequencing data available from CheckMate-009, -010, and -025 trials(11,13,24),(15). The treatment effect of nivolumab relative to everolimus interacted significantly with HLA-A*03 allele carriage in predicting progression-free survival (interaction p-value 0·04). Presence of one or more HLA-A*03 alleles was associated with shorter progression-free survival in patients treated with nivolumab in the three trials combined (hazard ratio, 1·31; 95% CI 1·01-1·71 p=0·04, Figure 4C). In contrast, there was no association between HLA-A*03 and progression-free survival in the everolimus arm of CheckMate-025 (hazard ratio, 0·89; 95% CI 0·64 - 1·23 p=0·47, Figure 4D). HLA-A*03 was not associated with MSKCC risk score(25), which predicts overall survival in patients with metastatic RCC, and adjusting for MSKCC risk score did not alter the association of HLA-A*03 and progression-free survival in the ICI arm. HLA-A*03 was not significantly associated with overall survival in nivolumab treated patients (hazard ratio, 1·22; 95% CI 0·91-1·62, p=0·18) nor those treated with everolimus (hazard ratio, 0·89; 95% CI 0·64-1·22, p=0·47). Objective response rates in patients who received single agent nivolumab in CheckMate-009, -010, and -025 were 25·3% (59/233) among HLA-A*03 non-carriers, 15·7% (13/83) in HLA-A*03 heterozygotes, and 0% (0/8) in HLA-A*03 homozygotes (Figure 4E, chi-squared p=0·06). Although 62·5% (5/8) HLA-A*03 homozygotes treated with nivolumab achieved stable disease, 80% (4/5) experienced disease progression within 6 months.
Figure 4:
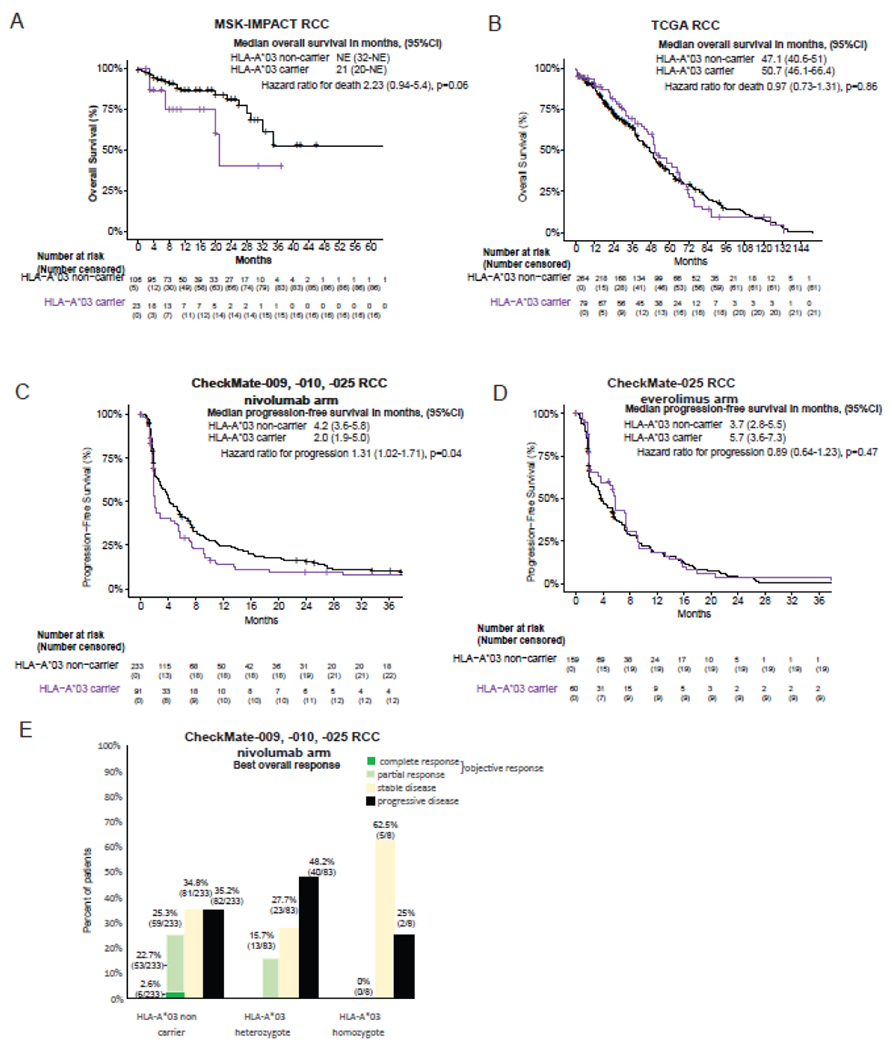
Effect of HLA-A*03 presence on outcomes of ICI therapy in renal cell cancer (RCC). The effect of HLA-A*03 presence on overall survival in RCC is shown for ICI-treated patients in MSK-IMPACT (Panel A) or patients treated with historical therapies in The Cancer Genome Atlas (TCGA) (Panel B). The effect on progression-free survival in the CheckMate-009, -010, and -025 randomized clinical trial is shown for the Nivolumab arm (Panel C) or Everolimus arm (Panel D). Panel E shows the numbers and percentage of patients treated with Nivolumab in CheckMate-009, -010, and -025 according to the best overall response they achieved (chi-squared p=0·06). A total of 11 HLA-A*03 non-carriers, 7 HLA-A*03 heterozygotes, and 1 HLA-A*03 homozygote had missing responses.
Amongst patients with objective response (i.e. complete or partial response) to nivolumab, progression-free survival was similar for HLA-A*03 non-carriers and heterozygotes. These data support HLA-A*03 as a potential predictive biomarker in ICI-treated patients with RCC, where HLA-A*03 homozygosity may predict absence of response to ICI and heterozygosity may predict reduced response (but similar durability of response amongst responders).
Current guidelines support use of ICI in combination with anti-angiogenic therapy as first-line therapy in metastatic RCC. In JAVELIN Renal 101, a phase 3 randomized controlled trial(23), avelumab with axitinib was associated with improved progression-free survival compared to sunitinib alone for first-line treatment of advanced RCC. Extensive biomarker discovery efforts suggested that HLA-A*03:01 carriage was associated with shorter progression-free survival in patients treated with avelumab plus axitinib, but not amongst patients treated with sunitinib(14). We re-examined trial outcomes with attention to HLA-A*03 allele count. The treatment effect of avelumab plus axitinib relative to sunitinib interacts significantly with HLA-A*03 allele count in predicting progression-free survival (interaction p-value 0·05). HLA-A*03 was associated in an additive manner with shorter progression-free survival amongst 354 patients randomized to avelumab plus axitinib (hazard ratio, 1·59 per HLA-A*03 allele; 95% CI 1·62-2·16, p=0·004), but not amongst the 370 patients treated with sunitinib (hazard ratio, 1·03 per HLA-A*03 allele; 95% CI 0·77-1·38, p=0·87) (Figure 5A, B). The effect was independent of PDL1 status. The magnitude of the effect was such that progression free survival benefit of ICI over standard therapy seen in HLA-A*03 non-carriers, was diminished in HLA-A*03 heterozygotes and reversed in HLA-A*03 homozygotes (appendix page 4). HLA-A*03 was also associated with lower objective response rates in Javelin Renal 101 (Figure 5C, chi-square p=0·01) of 63·8% (162/254) in HLA-A*03 non-carriers, 55·6% (40/72) in HLA-A*03 heterozygotes, and 12·5% (1/8) in HLA-A*03 homozygotes. These data support HLA-A*03 allele count as a predictive biomarker in this setting.
Figure 5:
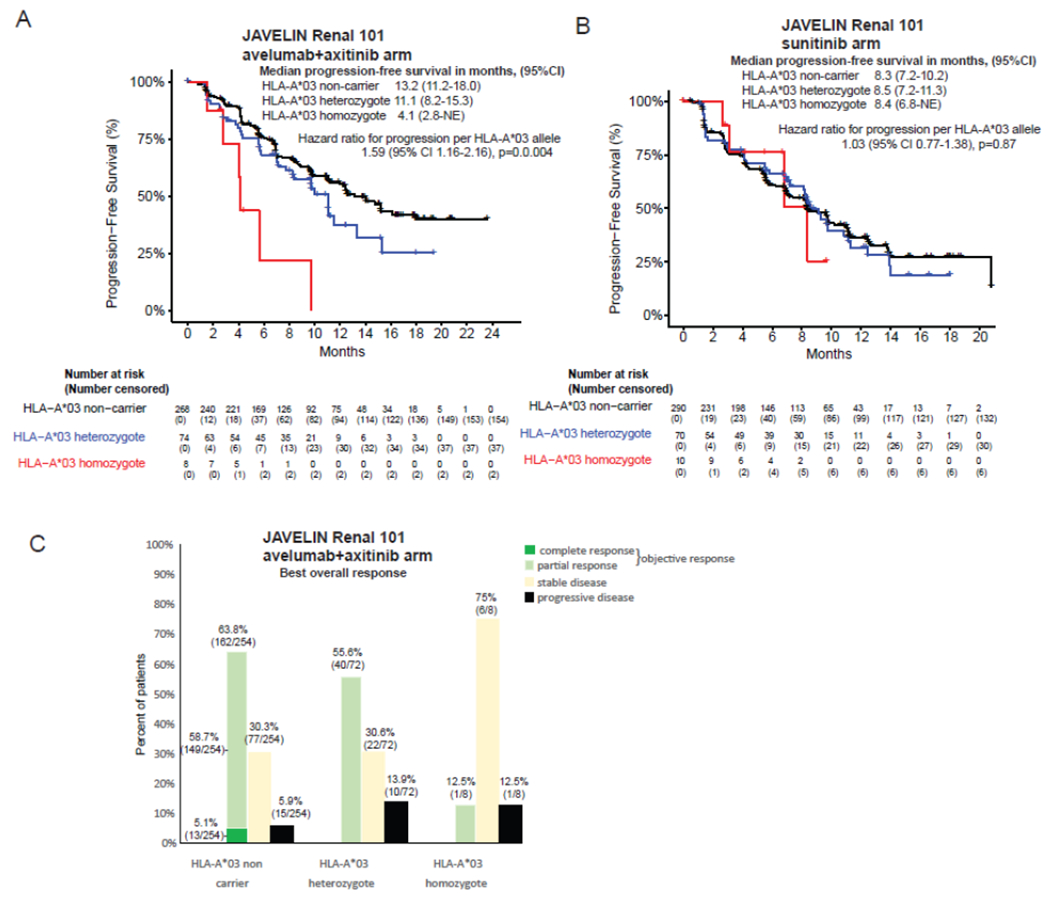
Effect of HLA-A*03 allele count on progression free survival in JAVELIN Renal 101 randomized controlled clinical trial in the avelumab and axitinib arm (Panel A), or the sunitinib arm (Panel B). Panel C shows the numbers and percentage of patients according to the best overall response they achieved (chi-squared p=0·01); a subset of 14 individuals had missing responses.
In a fixed effects meta-analysis including all 3,339 patients treated with ICI in the discovery and validation analyses, HLA-A*03 was associated with poor outcomes at a genome-wide significant level (p=2·01x10−8, Table 1) and there was no evidence of heterogeneity in effect (I2=0%, 95% confidence interval 0%-0·76%).
Table 1:
Summary of HLA-A*03 frequency and effects in ICI treated patients according to cohort, and estimate of significance and heterogeneity from fixed-effects meta-analysis.
| Cohort | HLA-A*03 allele frequency (%) | HLA-A*03 carrier frequency (%)* | Exposure variable | Outcome | Covariates | no. of events/no. of patients | Hazard Ratio | 95% Confidence interval lower bound | 95% Confidence interval upper bound |
|---|---|---|---|---|---|---|---|---|---|
| MSK-IMPACT (pan-cancer*) | 11·7% (273/2332) | 21·8% (254/1166) | HLA-A*03 allele count | Overall Survival | Stratified for 30 tumor types | 340/1166 | 1·48 | 1·20 | 1·82 |
| DFCI Profile (pan-cancer*) | 18·8% (311/2652) | 21·8% (289/1326) | HLA-A*03 allele count | Overall Survival | Stratified for 5 tumor types | 753/1326 | 1·22 | 1·05 | 1·42 |
| JAVELIN Solid Tumor urothelial carcinoma cohort (Urothelial) | 12·7% (43/338) | 21·3% (36/169) | HLA-A*03 allele count | Overall Survival | - | 128/169 | 1·36 | 1·01 | 1·85 |
| CheckMate-009,010,025, Nivolumab arm (RCC) | 15·3% (99/648) | 28·1% (91/324) | HLA-A*03 carrier status | Progression-free survival | - | 276/324 | 1·31 | 1·01 | 1·71 |
| JAVELIN Renal 101, avelumab+axitinib arm (RCC) | 12·7% (90/708) | 23·2% (82/354) | HLA-A*03 allele count | Progression-free survival | - | 157/354 | 1·59 | 1·16 | 2·16 |
|
Fixed effects meta-analysis: p=2x10−8
I2 heterogeneity=0% (95%CI 0%-0·76%) | |||||||||
Proportion of population carrying at least one allele
Discussion
Identifying biomarkers of ICI response could allow greater precision in therapy selection and management of patients with cancer. We found that HLA-A*03 allele count associates with poor overall survival and progression free survival in multiple tumor types. HLA-A*03 interacts with therapy arm in two large RCT’s of RCC and predicts progression free survival after immunotherapy but not alternative therapies. The magnitude of effect is such that HLA-A*03 homozygotes may have potential harm from ICI therapy.
The mechanism for the effect of HLA-A*03 is unclear. HLA-A alleles are expressed on normal and most neoplastic cells, and present antigens to T-cells. Since the mechanism by which ICI appears to work is through enhancing priming and effector T-cell function resulting in induction of largely novel T-cell responses, we speculate that HLA-A*03 specifically impairs this process. It is, however, notable that HLA-A*03 binds several inhibitory innate immune receptors including KIR3DL2, LILRB1 and LILRB2, which could impact efficacy of the innate response to tumor cells. Although HLA-A*03 is the lowest expressed HLA-A lineage, HLA-A expression level did not explain the effect of HLA-A*03 on overall survival, and even if there exists a threshold of expression level beneath which antigen presentation becomes biologically impaired, the presence of other class I alleles should serve to mitigate the effect of HLA-A*03. In addition, the specificity of the association to HLA-A*03 and not to other members of the HLA-A3 supertype, the consistency across tumor types, and the magnitude of effects in HLA-A*03 homozygous individuals suggest an active HLA-A*03-specific mechanism(s). HLA-A*03 is not known to associate with any common disease, and we did not identify any association between HLA-A*03 and intratumor lymphocyte profiles, gene expression in tumor tissue, or somatic mutation landscape prior to immunotherapy from available data(14). However, since HLA-A*03 appears to be predictive of ICI response (but is not prognostic), study of tumor biopsies following immunotherapy may be illuminating. Interestingly, in HIV infection, HLA-A*03 restricted CD8+ T-cells were more susceptible to regulatory T-cell mediated inhibition than other evaluated HLA-A alleles(26). Enhanced regulatory T-cell function has been linked to hyper-progression(27) after ICI, and ICI is known to have complex effects on these cells(28). Further elucidation of the mechanism behind HLA-A*03 predicting poor response is necessary and is likely to be complex, as HLA-A*03 may mediate effects through allotype-specific characteristics such as peptide specificity, expression level, differential affinity for innate immune receptors, and other factors.
The allele frequency of HLA-A*03 varies widely amongst global populations: ~13-16% in individuals of European, Indian, Native American, or Middle-Eastern descent, ~7% in those of Hispanic descent, ~5% in those of African descent, and 0-2% in those of Chinese, Philippino, Japanese, Korean, Vietnamese or other South-East Asian descent(21). This study had limited power to definitively determine whether the HLA-A*03 effect may vary by checkpoint target because most patients received anti-PD-1 therapies in conjunction with CTLA-4. This study was also limited in being able to probe the effects of HLA-A*03 in randomized settings outside of RCC. Whilst we find that HLA-A*03 is associated with poor outcomes in ICI treated patients across many cancer types with similar effect sizes, additional validation is desirable for cancer types not well represented in the observational cohorts. Similarly, additional validation of the potential for harm from ICI in HLA-A*03 homozygotes may be required through meta-analysis across multiple studies, as robust subgroup analyses of HLA-A*03 homozygotes are generally not feasible in any single study. Given the frequency distributions of HLA-A*03, our findings may have relevance to a substantial population of patients treated globally with ICI for a variety of indications, but further validation, ideally in trial settings in different populations with different tumor types and varying therapies, is needed.
HLA-A*03 has several properties desirable of a predictive biomarker. HLA-A*03 allele count is significantly and independently predictive of clinically relevant outcomes of ICI treatment. HLA genotyping is feasible, reproducible, and standardized in clinical laboratories. Use of HLA alleles as predictive biomarkers has become standard clinical practice in other settings; for example, prior to abacavir(29) or carbamazepine(30) therapy. Interpretation of the potential effect of HLA-A*03 is aided by its univariate association in most tumor types (melanoma and NSCLC being the exception, where adjusting for confounding factors was necessary). Given the common frequency of HLA-A*03 carriage (20-30% in the cohort or trial populations studied here), the magnitude of effect (such that even heterozygotes have markedly poorer responses to ICI) and the rare objective responses accrued by HLA-A*03 homozygotes, formal HLA-A*03 testing may be advisable to inform treatment decisions in patients eligible for ICI.
Supplementary Material
Research in context.
Evidence before this study
Although immune checkpoint inhibition (ICI) is used to treat many cancers, for most cancer types fewer than 50% of patients respond to therapy. We performed a systematic search of MEDLINE for relevant articles published between 2010 and December 2020, not restricted to the English language using the following search terms: “cancer”, “immunotherapy”, “immune checkpoint inhibitor (ICI)”, “HLA”, “MHC”, “biomarker”, “predictive”. We reviewed search results by title and abstract to identify relevant studies, and hand searched the reference list of these and two recent reviews on biomarkers of cancer immunotherapy outcomes by Keenan et al. in Nature Medicine 2019, and Van Ellen and Choueiri in Nature Reviews Clinical Oncology 2020. Existing evidence is limited to association studies in retrospective cohorts of HLA zygosity or other features. There are no robust data on the association of specific HLA alleles and lack of response to immunotherapy.
Added value of this study
This study is the largest epidemiological analysis of variation at HLA class I alleles and outcomes of immunotherapy in patients with cancer. By studying patients receiving immunotherapy or other therapies in clinical and trial settings for distinct cancers, we systematically show that HLA-A*03 associates with poor clinical outcomes. These findings support HLA-A*03 as a biomarker for predicting response to immunotherapy.
Implications of all the available evidence
HLA-A*03 is associated with impaired outcomes in patients with different types of cancer treated with immunotherapy. Caution in treating patients who are HLA-A*03 carriers with ICI should be considered when alternative therapeutic options exist. Clinical trials that stratify responses to HLA-A*03 genotype are necessary. Patients with renal cell cancer who are HLA-A*03 homozygotes may not benefit from ICI, and potentially do worse than if they were treated with other therapies.
Declaration of Interests
Dr. Braun reports non-financial support from Bristol Myers Squibb, personal fees from LM Education / Exchange Servies, personal fees from Octane Global, personal fees from Defined Health, personal fees from Dedham Group, personal fees from Adept Field Solutions, personal fees from Slingshot Insights, personal fees from Blueprint Parnterships, personal fees from Charles River Associates, personal fees from Trinity Group, personal fees from Insight Strategy, personal fees from Exelixis, personal fees from MDedge, personal fees from Schlesinger Associates, outside the submitted work; . Dr. Chin reports employment at EMD Serono during the conduct of the study. Dr. Wind-Rotolo reports employment and stock ownership at BMS. Dr. Mu reports employment, compensation and stock ownedship in Pfizer. Dr. Robbins reports employment from Pfizer during the conduct of the study. Dr. Choueiri reports grants, personal fees and non-financial support from Pfizer, grants, personal fees and non-financial support from Exelixis, grants, personal fees and non-financial support from Bristol Myers Squibb (BMS), grants, personal fees and non-financial support from Merck, grants, personal fees and non-financial support from Roche/Genentech, grants, personal fees and non-financial support from Novartis, grants, personal fees and non-financial support from Lilly, grants, personal fees and non-financial support from EMD Serono, grants from Dana-Farber/Harvard Cancer Center Kidney SPORE, outside the submitted work; Dr. Gulley reports being party to a cooperative research and development agreement with EMD Serono, during the conduct of the study.
Role of the funding source
This project has been funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research, under Contract No. HHSN261200800001E. This Research was supported in part by the Intramural Research Program of the NIH, Frederick National Lab, Center for Cancer Research. The results published here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga, the DFCI Oncology Data Retrieval System (OncDRS) and DFCI/BWH Data Sharing Group; who had no role in the analysis or decision to publish these findings. D.A.B. was supported by the DF/HCC Kidney Cancer SPORE Career Enhancement Program (P50CA101942-15), DOD CDMRP (KC170216, KC190130), and the DOD Academy of Kidney Cancer Investigators (KC190128T). TKC is supported in part by the Dana-Farber/Harvard Cancer Center Kidney SPORE and Program, the Kohlberg Chair at Harvard Medical School and the Trust Family, Michael Brigham, and Loker Pinard Funds for Kidney Cancer Research at DFCI. This research was financially supported by Merck KGaA, Darmstadt, Germany and Pfizer. Authors were not precluded from accessing data in the study, and they accept responsibility to submit for publication
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data sharing statement: Data (deidentified) are available to bona fide researchers via the original reports from which data was accessed, or from the authors subject to the restrictions relevant to each cohort. Code used is available by writing to the corresponding author.
References
- 1.Keenan TE, Burke KP, Van Allen EM. Genomic correlates of response to immune checkpoint blockade. Vol. 25, Nature Medicine. Nature Publishing Group; 2019. p. 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Allen EM, Choueiri TK. Dissecting the immunogenomic biology of cancer for biomarker development. Nature Reviews Clinical Oncology. Nature Research; 2020. p. 1–2. [DOI] [PubMed] [Google Scholar]
- 3.Osipov A, Jin Lim S, Popovic A, Azad NS, Laheru DA, Zheng L, et al. CLINICAL CANCER RESEARCH | PRECISION MEDICINE AND IMAGING Tumor Mutational Burden, Toxicity, and Response of Immune Checkpoint Inhibitors Targeting PD(L)1, CTLA-4, and Combination: A Meta-regression Analysis. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu S, Stein JE, Rimm DL, Wang DW, Bell JM, Johnson DB, et al. Comparison of Biomarker Modalities for Predicting Response to PD-1/PD-L1 Checkpoint Blockade: A Systematic Review and Meta-analysis. Vol. 5, JAMA Oncology. American Medical Association; 2019. p. 1195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chowell AD, Morris LGT, Grigg CM, Weber JK, Robert M, Makarov V, et al. Patient HLA class I genotype influences response to immune checkpoint blockade therapy. Science [Internet]. 2017;4572(December):1–14. Available from: 10.1126/science.aao4572 [DOI] [Google Scholar]
- 6.Abed A, Calapre L, Lo J, Correia S, Bowyer S, Chopra A, et al. Prognostic value of HLA-I homozygosity in patients with non-small cell lung cancer treated with single agent immunotherapy. Journal for ImmunoTherapy of Cancer. 2020. November 23;8(2):1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Negrao MV, Lam VK, Reuben A, Rubin ML, Landry LL, Roarty EB, et al. PD-L1 Expression, Tumor Mutational Burden, and Cancer Gene Mutations Are Stronger Predictors of Benefit from Immune Checkpoint Blockade than HLA Class I Genotype in Non-Small Cell Lung Cancer. Journal of Thoracic Oncology. 2019. June 1;14(6):1021–31. [DOI] [PubMed] [Google Scholar]
- 8.Cummings AL, Gukasyan J, Lu HY, Grogan T, Sunga G, Fares CM, et al. Mutational landscape influences immunotherapy outcomes among patients with non-small-cell lung cancer with human leukocyte antigen supertype B44. Nature Cancer. 2020. December1;1(12):1167–75. [DOI] [PubMed] [Google Scholar]
- 9.Sade-Feldman M, Jiao YJ, Chen JH, Rooney MS, Barzily-Rokni M, Eliane JP, et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nature Communications. 2017. December 1;8(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel MR, Ellerton J, Infante JR, Agrawal M, Gordon M, Aljumaily R, et al. Avelumab in metastatic urothelial carcinoma after platinum failure (JAVELIN Solid Tumor): pooled results from two expansion cohorts of an open-label, phase 1 trial. The Lancet Oncology. 2018. January 1;19(1):51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choueiri TK, Fishman MN, Escudier B, McDermott DF, Drake CG, Kluger H, et al. Immunomodulatory activity of nivolumab in metastatic renal cell carcinoma. In: Clinical Cancer Research. American Association for Cancer Research Inc.; 2016. p. 5461–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motzer RJ, Rini BI, McDermott DF, Redman BG, Kuzel TM, Harrison MR, et al. Nivolumab for metastatic renal cell carcinoma: Results of a randomized phase II trial. Journal of Clinical Oncology. 2015. May 1;33(13):1430–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. New England Journal of Medicine. 2015. November 5;373(19):1803–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motzer RJ, Robbins PB, Powles T, Albiges L, Haanen JB, Larkin J, et al. Avelumab plus axitinib versus sunitinib in advanced renal cell carcinoma: biomarker analysis of the phase 3 JAVELIN Renal 101 trial. Nature Medicine. 2020;26(11):1733–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braun DA, Hou Y, Bakouny Z, Ficial M, Sant’ Angelo M, Forman J, et al. Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nature Medicine. 2020;26(6):909–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia X, Han B, Onengut-Gumuscu S, Chen W-M, Concannon PJ, Rich SS, et al. Imputing Amino Acid Polymorphisms in Human Leukocyte Antigens. Tang J, editor. PLoS ONE. 2013. June 6;8(6):e64683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies RW, Flint J, Myers S, Mott R. Rapid genotype imputation from sequence without reference panels. Nature Genetics. 2016. August 1;48(8):965–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramsuran V, Kulkarni S, O’huigin C, Yuki Y, Augusto DG, Gao X, et al. Epigenetic regulation of differential HLA-A allelic expression levels. Human molecular genetics. 2015. August 1;24(15):4268–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discovery. 2018. July 1;8(7):822–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao D, Margolis CA, Gao W, Voss MH, Li W, Martini DJ, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018. February 16;359(6377):801–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gragert L, Madbouly A, Freeman J, Maiers M. Six-locus high resolution HLA haplotype frequencies derived from mixed-resolution DNA typing for the entire US donor registry. Human Immunology. 2013. October 1;74(10):1313–20. [DOI] [PubMed] [Google Scholar]
- 22.Apolo AB, Infante JR, Balmanoukian A, Patel MR, Wang D, Kelly K, et al. JOURNAL OF CLINICAL ONCOLOGY Avelumab, an Anti-Programmed Death-Ligand 1 Antibody, In Patients With Refractory Metastatic Urothelial Carcinoma: Results From a Multicenter, Phase Ib Study. J Clin Oncol. 2017;35:2117–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Motzer RJ, Penkov K, Haanen J, Rini B, Albiges L, Campbell MT, et al. Avelumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. New England Journal of Medicine. 2019. March 21;380(12):1103–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Motzer RJ, Rini Bl, McDermott DF, Redman BG, Kuzel TM, Harrison MR, et al. Nivolumab for metastatic renal cell carcinoma: Results of a randomized phase II trial. Journal of Clinical Oncology. 2015. May 1;33(13):1430–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Motzer RJ, Mazumdar M, Bacik J, Berg W, Amsterdam A, Ferrara J. Survival and prognostic stratification of 670 patients with advanced renal cell carcinoma. Journal of Clinical Oncology. 1999. September;17(8):2530–40. [DOI] [PubMed] [Google Scholar]
- 26.Elahi S, Dinges WL, Lejarcegui N, Laing KJ, Collier AC, Koelle DM, et al. Protective HIV-specific CD8+ T cells evade T reg cell suppression. Nature Medicine. 2011. August 17;17(8):989–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamada T, Togashi Y, Tay C, Ha D, Sasaki A, Nakamura Y, et al. PD-1+ regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proceedings of the National Academy of Sciences of the United States of America. 2019. May 14;116(20):9999–10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Read S, Greenwald R, Izcue A, Robinson N, Mandelbrot D, Francisco L, et al. Blockade of CTLA-4 on CD4 + CD25 + Regulatory T Cells Abrogates Their Function In Vivo . The Journal of Immunology. 2006. October 1;177(7):4376–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alfirevic A, Pirmohamed M, Marinovic B, Harcourt-Smith L, Jorgensen AL, Cooper TE. Genetic testing for prevention of severe drug-induced skin rash. Vol. 2019, Cochrane Database of Systematic Reviews. John Wiley and Sons Ltd; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen P, Lin J-J, Lu C-S, Ong C-T, Hsieh PF, Yang C-C, et al. Carbamazepine-Induced Toxic Effects and HLA-B*1502 Screening in Taiwan. New England Journal of Medicine. 2011. March 24;364(12):1126–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


