Abstract
D-type cyclins (cyclin D1, D2, and D3, together cyclin D) are central drivers of the cell division cycle and well-described proto-oncoproteins. Rapid turnover of cyclin D is critical for its regulation, but the underlying mechanism has remained a matter of debate. Recently, AMBRA1 was identified as the major regulator of the stability of all three D-type cyclins. AMBRA1 serves as the substrate receptor for one of ∼40 CUL4-RING E3 ubiquitin ligase (CRL4) complexes to mediate the polyubiquitylation and subsequent degradation of cyclin D. Consequently, AMBRA1 regulates cell proliferation to impact tumor growth and the cellular response to cell cycle-targeted cancer therapies. Here we discuss the findings that implicate AMBRA1 as a core member of the cell cycle machinery.
Keywords: cell cycle, cancer, protein degradation, molecular biology
Introduction
Cell division is a complex multistep process that must be tightly regulated for proper development and tissue homeostasis, and its dysregulation lies at the heart of cancer. Initiating cell division requires the integration of multiple intra- and extracellular growth cues. Cyclin D couples these signals to the initiation of DNA replication, thereby acting as a key driver of the cell cycle (Choi and Anders, 2014).
In response to growth stimuli, cyclin D begins to accumulate early in the G1 phase of the cell cycle. Cyclin D binds and activates cyclin-dependent kinases 4 and 6 (CDK4/6), which, in turn, phosphorylate the retinoblastoma (RB) family of proteins, freeing the E2F transcription factors to turn on genes necessary for DNA replication and cell cycle progression. Precise regulation of cyclin D levels is, therefore, fundamental to proper cell division. Accordingly, loss of all D-type cyclins is embryonic lethal (Kozar et al., 2004), whereas too much cyclin D drives cancer (Sherr, 1996; Qie and Diehl, 2016). Although the rapid turnover of cyclin D is a well-established and critical facet of its regulation (Sherr, 1995; Qie and Diehl, 2020), we are continuing to uncover important details of the underlying mechanism, shedding new light on our understanding of the cell cycle.
Degradation of Cyclin D by the Ubiquitin-Proteasome System
Similar to other cyclins, cyclin D is regulated in a cell cycle-dependent manner, in part, through targeted degradation by the ubiquitin-proteasome system. Cyclin D has a short half-life throughout the cell cycle (Guo et al., 2002); however, phosphorylation of a C-terminal degron accelerates polyubiquitylation and degradation of cyclin D at the G1/S transition, and cyclin D levels remain low throughout S phase (Diehl et al., 1997; Guo et al., 2005). Stabilized phosphomutant cyclin D accelerates proliferation and promotes tumor growth in mouse models (Lin et al., 2007), indicating that the rapid turnover of cyclin D helps restrain CDK4/6 activity and maintain proper cell cycle control.
Nevertheless, the identity of the E3 ubiquitin ligase responsible for ubiquitylating cyclin D has remained a long-standing matter of debate. Previous studies have attributed the regulation of cyclin D to several SKP1-CUL1-F-box protein (SCF) E3 ubiquitin ligase complexes, which belong to the larger family of cullin-RING E3 ubiquitin ligases (CRLs) (Lee and Zhou, 2010). Each CRL is a multisubunit complex composed of a cullin scaffold, a RING-box protein that binds E2 enzymes, an adaptor protein, and a substrate receptor that specifies target substrates. A family of F-box proteins (69 in human) serves as the substrate receptors of SCFs. Multiple F-box proteins have been reported to target cyclin D1 (reviewed by Qie and Diehl, 2020), and distinct F-box proteins have been linked to cyclin D2 and cyclin D3 (Chen et al., 2012a, 2012b; Yoshida et al., 2021). Still other studies have implicated the anaphase promoting complex/cyclosome (APC/C) in cyclin D1 ubiquitylation (Agami and Bernards, 2000; Pawar et al., 2010). However, a 2012 study by Kanie and colleagues found that deletion of up to four F-box proteins purportedly targeting cyclin D1 (FBXO4, FBXW8, FBXO31, and SKP2) had no effect on cyclin D1 stability (Kanie et al., 2012). Inhibition of CUL1 or APC/C also failed to increase the half-life of cyclin D1. Although these experiments did not exclude the possibility that previously reported complexes may regulate cyclin D stability in some contexts, they strongly suggested the existence of another E3 ubiquitin ligase responsible for ubiquitylation and degradation of cyclin D.
An Unexpected Role for an Understudied E3 Ubiquitin Ligase
In independent studies (Chaikovsky et al., 2021; Simoneschi et al., 2021), our groups found that the E3 ubiquitin ligase in question was not the SCF complex, but rather CRL4, guided by the substrate receptor AMBRA1. AMBRA1 is a large mostly unstructured protein that has previously been associated with a variety of cellular functions, including autophagy, proliferation, and apoptosis (Cianfanelli et al., 2015a). AMBRA1 was first identified as DDB1 and CUL4-associated factor 3 (DCAF3), a member of the family of substrate receptors for CRL4 (Jin et al., 2006); however, only one protein, Elongin C, had previously been proposed to be ubiquitylated by CRL4AMBRA1 (Chen et al., 2018). We found that loss of AMBRA1 stabilizes all three D-type cyclins in every context examined, including malignant and untransformed human cell lines, developing mouse embryos, and mouse tumor models. Furthermore, AMBRA1 binds cyclin D, and these interactions are enhanced upon cyclin D phosphorylation. Recurrent mutations at and around the phosphodegron are found in all three D-type cyclins in human tumors, and these mutations disrupt AMBRA1 binding. Thus, AMBRA1 plays a prominent role in limiting the levels of D-type cyclins in cells.
Loss of AMBRA1 leads to many of the same cellular phenotypes as overexpression of cyclin D. AMBRA1-null cells exhibit higher levels of CDK4/6 activity and RB phosphorylation. In some cell lines, this leads to shorter G1 duration and faster basal proliferation rates. In fact, data from the Cancer Dependency Map show that, across hundreds of human cancer cell lines, the consequences of AMBRA1 loss on cell proliferation are highly correlated with loss of multiple members of the RB pathway. Using this data set to group genes into co-essential modules—indicative of related cellular functions—a recent study grouped AMBRA1 with known cell cycle genes (Wainberg et al., 2021). Of note, AMBRA1 did not map to the module encompassing autophagy genes. Furthermore, using mass spectrometry to compare the proteomes of wild-type and AMBRA1 knockout cells, we found relatively few proteins to be upregulated in the absence of AMBRA1, and D-type cyclins (and their binding partners CDK4 and p27) were the most dramatically upregulated. Therefore, AMBRA1's role in regulating cyclin D appears to be one of its most consequential functions across cell types.
AMBRA1 in Cancer
Cyclin D is a well-described proto-oncoprotein, so, it is perhaps unsurprising that AMBRA1 acts as a tumor suppressor. A previous study reported increased incidence of spontaneous tumors in mice lacking one copy of Ambra1 (Cianfanelli et al., 2015b). We expanded upon these findings using genetically engineered mouse models and xenograft models to show that loss of AMBRA1 accelerates tumor growth in multiple cancer types, including diffuse large B cell lymphoma and lung adenocarcinoma (LUAD) (Fig. 1). Data from The Cancer Genome Atlas indicate that low AMBRA1 expression correlates with worse overall survival in several cancer types. Interestingly, in the case of LUAD, this correlation is only observed in patients with KRAS-mutant tumors, suggesting that genetic context impacts the tumor suppressive function of AMBRA1. Whether such context specificity is related to the regulation of cyclin D or other potential functions of AMBRA1 remains unclear.
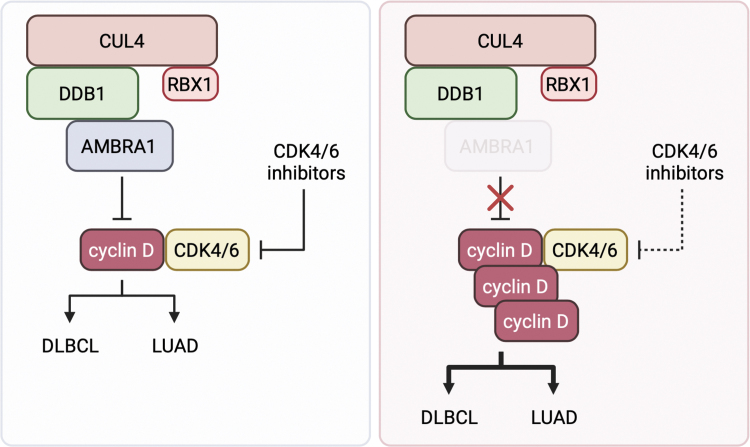
FIG. 1.
AMBRA1 in cancer. CRL4AMBRA1 ubiquitylates cyclin D for proteasomal degradation. Loss of AMBRA1 leads to high levels of cyclin D, which promotes the growth of multiple cancer types, such as DLBCL and LUAD, and decreases the sensitivity of cancer cells to CDK4/6 inhibitors. Created with BioRender.com. DLBCL, diffuse large B cell lymphoma; LUAD, lung adenocarcinoma.
AMBRA1 status may also play an important role in how cancer cells respond to treatment, as we found that loss of AMBRA1 decreases cellular sensitivity to small molecule inhibitors of CDK4/6 (Fig. 1). CDK4/6 inhibitors have been approved for the treatment of hormone receptor-positive breast cancer and are currently being tested in several other cancer types. However, inherent and acquired resistance present a major clinical challenge, and identifying mechanisms of resistance is an active area of investigation (Schoninger and Blain, 2020). In the absence of AMBRA1, high levels of cyclin D increase the levels of active p27-cyclin D-CDK4 trimers, which have been shown to be unable to bind the CDK4/6 inhibitor palbociclib (Guiley et al., 2019). In addition, accumulated cyclin D binds CDK2, forming “atypical” cyclin-CDK complexes that can phosphorylate RB and are impervious to CDK4/6 inhibitors. Thus, AMBRA1 loss, as well as other mechanisms that acutely upregulate cyclin D, may represent important mechanisms of resistance to CDK4/6 inhibition. At the same time, we observed that AMBRA1 loss leads to more potent resistance to CDK4/6 inhibitors than overexpression of stabilized cyclin D1 alone, suggesting that additional mechanisms in AMBRA1-null cells may contribute to CDK4/6 inhibitor resistance.
Revisiting the Model of Cyclin D Degradation
Although many phenotypic consequences of AMBRA1 loss match what we expect from cyclin D stabilization, revisiting the current model of cyclin D degradation is warranted in light of our recent findings (Fig. 2). First, in the current model of cyclin D degradation, cyclin D is exported to the cytoplasm before ubiquitylation and degradation. However, AMBRA1 works specifically with CUL4B to ubiquitylate cyclin D. Unlike CUL4A, CUL4B contains a nuclear localization signal at the N-terminus (Zou et al., 2009), suggesting that CRL4AMBRA1 ubiquitylates cyclin D in the nucleus. Accordingly, immunofluorescence experiments revealed that AMBRA1 localizes to the nucleus, and upon proteasome inhibition or AMBRA1 loss, cyclin D accumulates in the nucleus. In addition, inhibition of nuclear export with leptomycin B does not stabilize cyclin D, as would be expected if cyclin D is degraded solely in the cytoplasm. It remains possible that nuclear export of phosphorylated cyclin D provides a secondary mechanism to quickly evict cyclin D from the nucleus at the G1/S transition, but the evidence thus far suggests that AMBRA1-mediated ubiquitylation and degradation of cyclin D occurs in the nucleus.
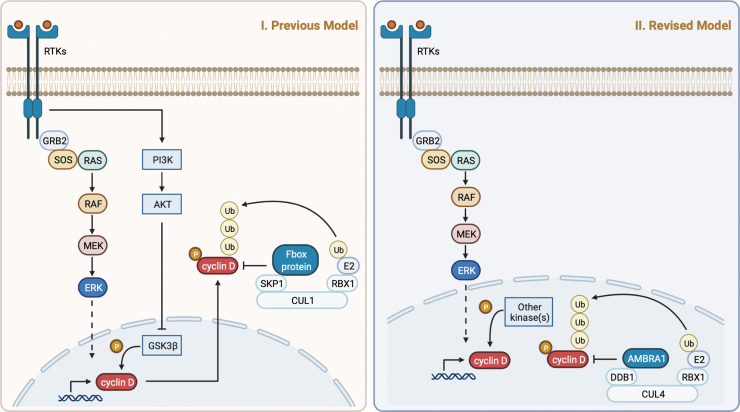
FIG. 2.
A potential new model of cyclin D ubiquitylation. Schematics of the current (left) and revised (right) models of cyclin D ubiquitylation. Identification of CRL4AMBRA1 as the main regulator of cyclin D stability suggests that cyclin D ubiquitylation occurs in the nucleus and may occur throughout the cell cycle. The identity of the major cyclin D kinase(s) warrants further investigation. Created with BioRender.com.
In the current model, cyclin D ubiquitylation and degradation occurs primarily at the G1/S transition, ensuring low levels of cyclin D in S phase. However, constant growth factor signaling, which drives transcription of CCND genes, is required throughout G1 to maintain cyclin D expression (Matsushime et al., 1991). Moreover, a recent study using live imaging of fluorescently tagged cyclin D1 confirmed that cyclin D1 is actively degraded in all cell cycle phases (Min et al., 2020). In cells lacking AMBRA1, steady-state levels of cyclin D1 are elevated in all cell cycle phases. Hence, CRL4AMBRA1 may ubiquitylate cyclin D in all phases, with phosphorylation of cyclin D enhancing the rate of ubiquitylation at the G1/S transition, when cyclin D levels must remain low to avoid DNA replication defects (Pagano et al., 1994; Maiani et al., 2021). Further studies are needed to clarify whether AMBRA1 regulates cyclin D in a cell cycle-dependent manner.
Finally, given the role of phosphorylation in regulating cyclin D degradation, identifying the major kinase(s) responsible is a key area of interest. Previous studies have implicated glycogen synthase kinase-3β (GSK-3β) as the major cyclin D kinase (e.g., Diehl et al., 1998). However, other groups have found no effect of GSK-3β activity on cyclin D levels (e.g., Yang et al., 2006; Alao, 2007). Although it is possible that multiple kinases phosphorylate cyclin D in different contexts, the conflicting data highlight the need for a systematic characterization of cyclin D kinase(s). Hopefully, insights gleaned from the identification of CRL4AMBRA1 as the major E3 ubiquitin ligase targeting cyclin D for degradation will aid this endeavor.
Conclusion
Decades after cyclin D was first discovered, we have answered a long-standing question about its regulation, demonstrating that CRL4AMBRA1 serves as a major E3 ubiquitin ligase targeting cyclin D for degradation. AMBRA1 is thus a critical cell cycle gene with a direct role in regulating the RB pathway. Going forward, it would be interesting to understand how AMBRA1 itself is regulated, both with relation to the cell cycle, as well as in light of the multiple functions that have previously been ascribed to this protein. Multiple AMBRA1 isoforms are annotated in the human genome; however, it remains to be determined whether multiple protein isoforms exist in cells, and, if so, how their functions differ. Finally, the impact of AMBRA1 in tumor growth appears to be context specific. Understanding the contexts in which AMBRA1 is a potent tumor suppressor may provide new insight into AMBRA1 function, the relationship between the RB pathway and tumor growth, and the response of tumors to certain therapeutics, such as CDK4/6 inhibitors.
Author Contributions
A.C.C. and D.S. wrote the article with input from J.S. and M.P. D.S. generated the figures.
Disclosure Statement
J.S. has received research funding from Stemcentrx/Abbvie, Pfizer, and Revolution Medicines. M.P. is a cofounder of Coho Therapeutics. M.P. is also a consultant for, a member of the scientific advisory board of, and has financial interests in Coho Therapeutics, CullGen, Kymera Therapeutics, Santi Therapeutics, and SEED Therapeutics. The authors declare no other competing interests.
Funding Information
This work was funded by grants from the National Institutes of Health: A.C.C., F99CA245471; J.S. R01CA228413 and 1R35CA231997; M.P., R01CA76584 and R35GM136250. M.P. is an investigator with the Howard Hughes Medical Institute.
References
- Agami, R., and Bernards, R. (2000). Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell 102, 55–66. [DOI] [PubMed] [Google Scholar]
- Alao, J.P. (2007). The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention. Mol Cancer 6, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaikovsky, A.C., Li, C., Jeng, E.E., Loebell, S., Lee, M.C., Murray, C.W., et al. (2021). The AMBRA1 E3 ligase adaptor regulates the stability of cyclin D. Nature 592, 794–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B.B., Glasser, J.R., Coon, T.A., and Mallampalli, R.K. (2012a). F-box protein FBXL2 exerts human lung tumor suppressor-like activity by ubiquitin-mediated degradation of cyclin D3 resulting in cell cycle arrest. Oncogene 31, 2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B.B., Glasser, J.R., Coon, T.A., Zou, C., Miller, H.L., Fenton, M., et al. (2012b). F-box protein FBXL2 targets cyclin D2 for ubiquitination and degradation to inhibit leukemic cell proliferation. Blood 119, 3132–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S.-H., Jang, G.M., Hüttenhain, R., Gordon, D.E., Du, D., Newton, B.W., et al. (2018). CRL4AMBRA1 targets Elongin C for ubiquitination and degradation to modulate CRL5 signaling. EMBO J 37, e97508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y.J., and Anders, L. (2014). Signaling through cyclin D-dependent kinases. Oncogene 33, 1890–1903. [DOI] [PubMed] [Google Scholar]
- Cianfanelli, V., Fuoco, C., Lorente, M., Salazar, M., Quondamatteo, F., Gherardini, P.F., et al. (2015b). AMBRA1 links autophagy to cell proliferation and tumorigenesis by promoting c-Myc dephosphorylation and degradation. Nat Cell Biol 17, 20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfanelli, V., Zio, D.D., Bartolomeo, S.D., Nazio, F., Strappazzon, F., and Cecconi, F. (2015a). Ambra1 at a glance. J Cell Sci 128, 2003–2008. [DOI] [PubMed] [Google Scholar]
- Diehl, J.A., Cheng, M., Roussel, M.F., and Sherr, C.J. (1998). Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev 12, 3499–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl, J.A., Zindy, F., and Sherr, C.J. (1997). Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev 11, 957–972. [DOI] [PubMed] [Google Scholar]
- Guiley, K.Z., Stevenson, J.W., Lou, K., Barkovich, K.J., Kumarasamy, V., Wijeratne, T.U., et al. (2019). p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition. Science 366, eaaw2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y., Harwalkar, J., Stacey, D.W., and Hitomi, M. (2005). Destabilization of cyclin D1 message plays a critical role in cell cycle exit upon mitogen withdrawal. Oncogene 24, 1032–1042. [DOI] [PubMed] [Google Scholar]
- Guo, Y., Stacey, D.W., and Hitomi, M. (2002). Post-transcriptional regulation of cyclin D1 expression during G2 phase. Oncogene 21, 7545–7556. [DOI] [PubMed] [Google Scholar]
- Jin, J., Arias, E.E., Chen, J., Harper, J.W., and Walter, J.C. (2006). A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell 23, 709–721. [DOI] [PubMed] [Google Scholar]
- Kanie, T., Onoyama, I., Matsumoto, A., Yamada, M., Nakatsumi, H., Tateishi, Y., et al. (2012). Genetic reevaluation of the role of F-box proteins in cyclin D1 degradation. Mol Cell Biol 32, 590–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozar, K., Ciemerych, M.A., Rebel, V.I., Shigematsu, H., Zagozdzon, A., Sicinska, E., et al. (2004). Mouse development and cell proliferation in the absence of D-Cyclins. Cell 118, 477–491. [DOI] [PubMed] [Google Scholar]
- Lee, J., and Zhou, P. (2010). Cullins and cancer. Genes Cancer 1, 690–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, D.I., Lessie, M.D., Gladden, A.B., Bassing, C.H., Wagner, K.U., and Diehl, J.A. (2007). Disruption of cyclin D1 nuclear export and proteolysis accelerates mammary carcinogenesis. Oncogene 27, 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiani, E., Milletti, G., Nazio, F., Holdgaard, S.G., Bartkova, J., Rizza, S., et al. (2021). AMBRA1 regulates cyclin D to guard S-phase entry and genomic integrity. Nature 592, 799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushime, H., Roussel, M.F., Ashmun, R.A., and Sherr, C.J. (1991). Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell 65, 701–713. [DOI] [PubMed] [Google Scholar]
- Min, M., Rong, Y., Tian, C., and Spencer, S. (2020). Temporal integration of mitogen history in mother cells controls proliferation of daughter cells. Science 368, 1261-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano, M., Theodoras, A.M., Tam, S.W., and Draetta, G.F. (1994). Cyclin D1-mediated inhibition of repair and replicative DNA synthesis in human fibroblasts. Genes Dev 8, 1627–1639. [DOI] [PubMed] [Google Scholar]
- Pawar, S.A., Sarkar, T.R., Balamurugan, K., Sharan, S., Wang, J., Zhang, Y., et al. (2010). C/EBPδ targets cyclin D1 for proteasome-mediated degradation via induction of CDC27/APC3 expression. Proc Natl Acad Sci U S A 107, 9210–9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qie, S., and Alan Diehl, J. (2020). Cyclin D degradation by E3 ligases in cancer progression and treatment. Semin Cancer Biol 67, 159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qie, S., and Diehl, J.A. (2016). Cyclin D1, cancer progression, and opportunities in cancer treatment. J Mol Med 94, 1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoninger, S.F., and Blain, S.W. (2020). The ongoing search for biomarkers of CDK4/6 inhibitor responsiveness in breast cancer. Mol Cancer Ther 19, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr, C.J. (1995). D-type cyclins. Trends Biochem Sci 20, 187–190. [DOI] [PubMed] [Google Scholar]
- Sherr, C.J. (1996). Cancer cell cycles. Science 274, 1672–1677. [DOI] [PubMed] [Google Scholar]
- Simoneschi, D., Rona, G., Zhou, N., Jeong, Y.-T., Jiang, S., Milletti, G., et al. (2021). CRL4 AMBRA1 is a master regulator of D-type cyclins. Nature 592, 789–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainberg, M., Kamber, R.A., Balsubramani, A., Meyers, R.M., Sinnott-Armstrong, N., Hornburg, D., et al. (2021). A genome-wide atlas of co-essential modules assigns function to uncharacterized genes. Nat Genet 53, 638–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, K., Guo, Y., Stacey, W.C., Harwalkar, J., Fretthold, J., Hitomi, M., et al. (2006). Glycogen synthase kinase 3 has a limited role in cell cycle regulation of cyclin D1 levels. BMC Cell Biol 7, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, A., Choi, J., Jin, H.R., Li, Y., Bajpai, S., Qie, S., et al. (2021). Fbxl8 suppresses lymphoma growth and hematopoietic transformation through degradation of cyclin D3. Oncogene 40, 292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, Y., Mi, J., Cui, J., Lu, D., Zhang, X., Guo, C., et al. (2009). Characterization of nuclear localization signal in the N terminus of CUL4B and its essential role in cyclin E degradation and cell cycle progression. J Biol Chem 284, 33320–33332. [DOI] [PMC free article] [PubMed] [Google Scholar]