Abstract
Functional analyses of mitochondria have been hampered by few effective approaches to manipulate mitochondrial DNA (mtDNA) and a lack of existing animal models. Recently a TALE-derived base editor was shown to induce C-to-T (or G-to-A) sequence changes in mtDNA. We report here the FusX TALE Base Editor (FusXTBE) to facilitate broad-based access to TALE mitochondrial base editing technology. TALE Writer is a de novo in silico design tool to map potential mtDNA base editing sites. FusXTBE was demonstrated to function with comparable activity to the initial base editor in human cells in vitro. Zebrafish embryos were used as a pioneering in vivo test system, with FusXTBE inducing 90+% editing efficiency in mtDNA loci as an example of near-complete induction of mtDNA heteroplasmy in vivo. Gene editing specificity as precise as a single nucleotide was observed for a protein-coding gene. Nondestructive genotyping enables single-animal mtDNA analyses for downstream biological functional genomic applications. FusXTBE is a new gene editing toolkit for exploring important questions in mitochondrial biology and genetics.
Introduction
Mitochondria have critical roles in cellular homeostasis, metabolism, apoptosis, and aging.1 Approximately 80% of the adult-onset and 25% of the pediatric-onset mitochondrial disorders can be attributed to genetic lesions in the mitochondrial genome. Mutations in both nuclear DNA and mitochondrial DNA (mtDNA) are pathogenic.2,3 For example, a recurring 5 kb deletion underlies the Kearns–Sayre syndrome (KSS), and a single-point mutation in mtDNA is the basis for the mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes (MELAS) syndrome.4,5 The complex genetics, including penetrance and phenotypic heterogeneity observed in the case of mtDNA-associated disorders, can be explained by the homoplasmic or heteroplasmic condition of the cell.
A prototypical cell harbors around 100–100,000 copies of circular, double-stranded mtDNA molecules. The proportion of the mutant to wild-type mtDNA within a cell governs the progression of the mitochondrial disease, the severity of which is directly proportional to the number of mutant mtDNA molecules. Despite this clear association of genotype with the disease, there are no current treatments for patients with mitochondrial disease. Understanding the pathophysiology of mitochondrial disorders contributed by variations in the mtDNA has been impeded due to the lack of an effective toolkit to engineer the powerhouse of the cell. The lack of systems for the quantitative delivery of exogenous DNA or RNA to mitochondria also restricts approaches for mtDNA editing.
While gene-editing tools such as CRISPR and TALENs enable the rapid manipulation of the nuclear genome,6–10 there are few tools to manipulate/engineer vertebrate mtDNA mainly due to the absence of any common DNA repair mechanisms. DNA nucleases that create double-stranded breaks in the nuclear genome and subsequently recruiting repair machinery instead are proposed to induce the degradation of mtDNA.11–14 These inherent mitochondrial gene editing challenges have been circumvented by expanding on the existing protein-based gene editing delivery systems. Currently, few strategies take advantage of this feature to target and degrade pathogenic mtDNA via double-stranded breaks.
Gene editing tools, such as mitochondrially targeted zinc-finger nucleases (mtZFNs), and mitochondrially targeted transcription activator-like effector nucleases (mitoTALENs) have been reported to shift mtDNA heteroplasmy in models of mitochondrial disorders.11,12,15 Mitochondrially targeted nucleases selectively bind the mtDNA harboring deletions/edits and introduce double-stranded breaks, thereby triggering rapid degradation of mtDNA. In a previous study by our group, we developed an alternative methodology to introduce precise deletions in the mtDNA using a novel “block and nick” approach to model KSS in zebrafish.16 However, the utility of these nuclease-based gene editing systems cannot be extended to correct or model homoplasmic and single-nucleotide pathogenic mtDNA variations.
Recently, Mok et al. described a novel base editing system in the mitochondria using a bacterial cytidine deaminase toxin, DddAtox.17 The present system uses split halves of DddAtox fused to each of the TALE monomers programmed to bind a specific sequence on mtDNA. Mitochondrial DdCBE (DddA-derived cytosine base editor) introduces C-to-T base edits in the spacer region between the two TALE arms with optimal editing efficiency in vitro. This system has a strong sequential preference for the presence of T at the 5′ end of the C around the targeted editing site. Furthermore, Mok et al. show that to target mtDNA, DddAtox can be split into two halves at two different positions, each of the split-half molecules swapped with any one of the TALE monomers.
Using this as a critical study design constraint, one needs to test four different combinations of the DdCBE to interrogate a single target, a process that can be quite time-consuming if extended to edit multiple cytosine residues present on a given mtDNA molecule. The application of this editing system has been extended to in vivo in a study in which Lee et al. have demonstrated to introduce base edits in the mice mtDNA utilizing DddA-TALE fusion deaminases. C-to-T transition was detected in multiple tissues and was also observed to be germline transmitted in two F1 offsprings born to an edited F0 animal.18
Here we describe a universal and programmable vertebrate mtDNA engineering toolbox suitable for the establishment of new in vitro cell and in vivo animal models. We build upon the existing DdCBE system to develop a rapid, accessible, and efficient programmable FusX19 TALE base editor system with high editing efficiency in vitro and in vivo.
To predict all potential editable sites in mtDNA, we have designed a customizable script that also determines the target sequences amenable to premature termination codon (PTC) induction via C-to-T base editing.
A noninvasive method well adapted to zebrafish provides a unique advantage to the genotype and propagate injected embryos that harbor up to 90% edits in their mtDNA. The findings from this study provide insights in exploring the mitochondrial biology and genetics of disorders caused by mutations in mtDNA.
Materials and Methods
Generation of mitoFusX TALE Base Editor (FusXTBE)
For the synthesis of this next-generation human mitochondrial TALE base editor, the cassette containing the N-terminus half of the DddAtox (split at 1397 amino acid position), uracil glycosylase inhibitor (UGI), and 3′UTR of human ATP5B was amplified from the published DdCBE construct (Addgene plasmid no. 157843). The PCR product was digested with the restriction enzymes XbaI and EcoRI. Digested DNA fragments were then subsequently cloned in the pKTol2C-COX8A-mTALEN vector linearized with XbaI and EcoRI to obtain pKTol2C-FusXTBE-N. To generate pKTol2C-FusXTBE-C, the cassette containing the C terminus half of the DddAtox (split at 1397 amino acid position) and UGI was amplified from the DdCBE plasmid (Addgene plasmid no. 157844).
The PCR amplicon was digested with XbaI and BsU36I restriction enzyme and cloned in the pKTol2C-FusXTBE-N vector linearized with XbaI and Bsu36I to generate the pKTol2C-FusXTBE-C plasmid. Clones were confirmed by Sanger sequencing (GENEWIZ LLC). For zebrafish mitochondrial FusX TALE base editor constructs, individual cassettes including partial N- and C-terminus of TALE domain, along with Dddatox (split at 1397 amino acid position), and UGI were ordered as clonal fragments in pTwist Amp High Copy plasmids from Twist Bioscience.
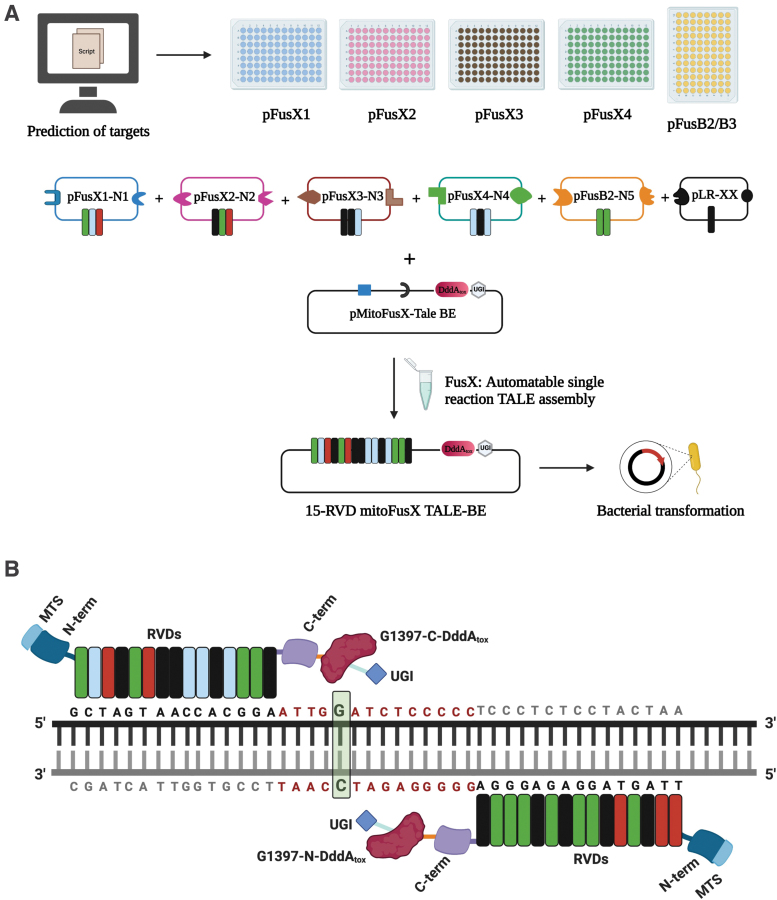
Each of the clonal fragments was then separately cloned in the BclI-PstI cut pkidh2GoldyTAL-lacZ-Fok1 vector backbone, generating plasmids pT3-FusXTBE-N and pT3-FusXTBE-C. The final FusX mitoTALE-Dddatox backbone plasmids will be made available via our Addgene repository. Repeat variable di-residues (RVDs) specific to prioritized mtDNA genetic loci (Supplementary Table S1) were then cloned in the receiving vector backbones using the one-step FusX19 assembly system (Fig. 1A). Each of the constructs was sequence-verified by Sanger sequencing.
FIG. 1.
Schematic for the assembly of mitochondrial TALE base editors by the FusX reaction with the FusXTBE architecture. (A) FusX library of all combinations of RVD dimers and trimers enables the mitochondrial TALE base editor to be built in a single Golden Gate cloning step. (B) Schematic of FusXTBE binding to the human mitochondrial DNA target, ND4. Repeat divariable residues are shown in different colors (green-guanine-NN; blue-cytosine-HD; red-thymine-NG; black-adenine-NI). DNA binding region for each of the TALE monomers is black in color, and the spacer region is depicted in red. Genomic target C is highlighted in green with bold font. Figure 1 was created using Biorender.com. FusXTBE, FusX TALE Base Editor; MTS, mitochondrial targeting sequence; RVD, repeat variable di-residues; UGI, uracil glycosylase inhibitor. Color images are available online.
FusX Extender series was built using the previously published FusX system where an additional RVD in all possible combinations was added to each of the pLR (last half-repeat) plasmids. This modified design enables us to synthesize TALEs binding to sequences 17 bp long on mitochondrial or nuclear genome.
TALE-based technology design tool for mitochondrial genome engineering
TALE Writer, a script for the efficient design of TALE-based technologies, was developed using Python 3.7.3. TALE Writer is hosted at* under a GNU General Public License v3.0. The script can be used to (I) design TALENs, (II) design C-to-T base editors against 5′-TC-3′ loci,17 and (III) design C-to-T base editors specifically for PTC induction.20 The input of the script is a sequence of interest, the minimum and maximum lengths of the TALE arms and the spacer, a target window, and, only for TALEN design, a target coordinate.
In general, the target window is defined as the window within the spacer that contains the target site and for which design permutations will be calculated. In other words, the parameter “target window” denotes the nucleotide positions that are not parsed within the protospacer in search for on-target editable motifs. The algorithm looks for the target sites at least “x” base pairs from the target sequence of either TALE repeat array (where x = length of the target window and is a user-defined parameter). The parameters herein utilized for base editor design were TALE arms and spacer region ranging between 14 and 16 bp, and a target window of 4 bp.
The output of the script consists of a table containing all the design permutations found for the defined target (for nuclease design) or for each target identified within the sequence of interest (for base editor design). The table includes the following: (1) the target index, (2) the local coordinate of the target, (3) the target motif (i.e., a specific nucleotide or a short sequence), (4) the design index, (5) the left TALE arm preceded by a 5′T, (6) the right TALE arm followed by a 3′A, and (7) the sequence of the spacer (in which the target site is capitalized and highlighted in blue). In addition, for base editor design, potential off-target edit sites are highlighted in red, and a column marks the designs that contain such sites with an exclamation mark. Moreover, the table can be saved as a .csv file.
For PTC induction, herein defined as a DNA edit resulting in the generation of a nonfunctional gene product due to the premature termination of translation,20 three types of edits were identified. First, the in-frame 5′-TGA-3′ motif, in which a G-to-A edit (equivalent to a C-to-T edit on the opposite strand) results in the stop codon UAA. Second, the 5′-TCAA-3′ motif (CAA in-frame), in which a C-to-T edit also results in the stop codon UAA. Lastly, the 5′-TCAG-3′ motif (CAG in-frame), in which a C-to-T edit results in the stop codon UAG. Noteworthy, PTC induction should be possible both in nuclear DNA and mtDNA through any of these edits. It is also worth mentioning that the availability of types of edits is currently limited by the strong 5′-TC-3′ context preference of the first generation of DdCBEs.
Transfection of the mammalian cell line with mitoFusX TALE Base Editor
HEK293T cells were cultured and maintained at 37°C with 5% CO2. The HEK293T cell line was obtained from ATCC and cultured in DMEM GlutaMAX media (Thermo Scientific) supplemented with 10% fetal bovine serum (Gibco) and 1 × penicillin/streptomycin solution (Pen/Strep) (Thermo Scientific). A total of 30,000 cells/well were seeded in six-well plate 18 h before the time of lipofection by Lipofectamine3000 (Thermo Scientific).
Cells were transfected with 500 ng of each TALE-BE monomer for each of the three target genes, MT-ND4, MT-ND2, and MT-CO1 (left arm- MT-ND4 pKTol2C-FusXTBE-C and right arm- MT-ND4 pKTol2C-FusXTBE-N); (left arm- MT-ND2 pKTol2C-FusXTBE-C and right arm- MT-ND2 pKTol2C-FusXTBE-N); (left arm- MT-CO1 pKTol2C-FusXTBE-C and right arm- MT-CO1 pKTol2C-FusXTBE-N), and 500 ng of pKTol2C-EGFP as a positive control for the lipofection. Media was changed 24 h post-transfection and cells were cultured for 72 h before collection for mitochondrial genotyping.
Genomic DNA isolation and mitochondrial genotyping
Following 3 days post-transfection, media was aspirated, and cells were washed with 1 × phosphate-buffered saline (Thermo Scientific). Total genomic DNA was extracted from cells using the DNeasy Blood and Tissue Kit (Qiagen). Primers (Supplementary Table S2) flanking the target loci were used to amplify the edited loci using MyTaq polymerase (Bioline). PCR amplicons were gel extracted and purified using the QIAquick Gel Extraction Kit (Qiagen). Purified samples were submitted to Genewiz (GENEWIZ LLC) for Sanger sequencing. Mitochondrial heteroplasmy was quantified using EditR software21 to predict the percentage of edits in the target loci. Raw Sanger sequencing files in .ab1 format were uploaded for each of the control and nontransfected samples.
Zebrafish handling and husbandry
Adult zebrafish and embryos were maintained according to the guidelines established by the Mayo Clinic Institutional Animal Care and Use Committee (IACUC number: A34513-13-R16) and the Children's Hospital of Philadelphia IACUC (number IAC 21-001154). No Institutional Review Board (IRB) approval was required for this study.
Zebrafish microinjection
Targets predicted by TALE Writer were prioritized to introduce edits in protein-coding and tRNA genes. Mitochondrially encoded cytochrome c oxidase I (mt-co1) (nonsynonymous substitution), mitochondrially encoded cytochrome c oxidase III (mt-co3) (premature termination induction), and mitochondrially encoded tRNA-leu (mt-tl1) were chosen as target sites in the zebrafish mtDNA locus. mt-co1 and mt-co3 code for subunits of complex IV of the mitochondrial respiratory chain, whereas mt-tl1 is involved in tRNA aminoacylation.
Sequence-verified mitoTALE-BE clones were linearized with PstI restriction enzyme. Linearized plasmid was used as a template to in vitro transcribe mRNA using the T3 mMESSAGE mMACHINE Transcription Kit (Thermo Scientific). mRNA was purified using the NEB Monarch RNA cleanup kit (NEB). Three nanoliters of 40 pg mRNA of each arm of the FusX mitoTALE-BE (left- pT3-FusXTBE-N and right- pT3-FusXTBE-C) was injected into single-cell zebrafish embryos. Injected embryos were then raised in embryo water at 29°C.
Noninvasive enzymatic zebrafish genotyping
To assess the in vivo editing efficiency of the mitoTALE BE, a method of genotyping was needed that enabled the rapid investigation of the functional readout of mtDNA edits. A noninvasive enzymatic method to extract nuclear DNA from zebrafish embryos was used with modifications.22
Larvae aged 3 days postfertilization (dpf) were rinsed thrice in fresh embryo media, followed by three washes in collection buffer (30 mM Tris-HCl, 1 × tricaine solution). Larvae were aspirated in 40 μL of genotyping buffer (1.5 μg/mL proteinase K, 30 mM Tris-HCl, 1 × tricaine solution) into a 96-well plate and were incubated in a shaker at 37°C for 20 min. After incubation, 15 μL of the lysis buffer (10 mM Tris-HCl, pH 8.0, 50 mM KCl, 0.3% Tween-20, 0.3% NP-40, and 1 mM EDTA) was added to the incubation mix. The solution was incubated at 98°C for 5 min to deactivate the proteinase K and release DNA from cells shed by the larvae upon agitation. This DNA was used as a template for the PCR amplification to genotype the larvae.
Each larva was then individually rescued from the 96-well plate and rinsed in 4-quadrant Petri dishes with fresh embryo water, before being placed in 24-well plates for incubation at 29°C.
Next-generation amplicon sequencing
DNA extracted from transfected and control cells for the in vitro experiments and from mutant and control embryos for the in vivo studies was used as a template for the amplicon sequencing. PCR was carried out using primers with Illumina adaptor sequences attached at the 5′ end (Supplementary Table S2). Amplicons were gel extracted and purified using the QIAquick Gel Extraction Kit (Qiagen). Purified amplicons (a total amount of 150 ng/amplicon) were submitted to Genewiz (GENEWIZ LLC) for next-generation amplicon sequencing (GeneWiz AmpliconEZ). Raw fasta files obtained after the sequencing run were analyzed using CasAnalzyer23,† to estimate the editing frequency and nucleotide changes in each allele.
mtDNA genotyping in zebrafish embryos
PCR amplification was performed for each of mt-co1, mt-co3, and mt-tl1 using the respective primers mentioned in Supplementary Table S2. PCR consisted of 4 μL of 5 × MyTaq Red PCR buffer, 1 μL of 10 μM forward primer, 1 μL of 10 μM reverse primer, 0.2 μL of MyTaq DNA polymerase, 8 μL of template, and 5.8 μL dH2O. Samples were run in a thermal cycler with the following PCR conditions: (1) 95°C, 3 min; (2) 95°C, 1 min; (3) 58°C, 30 s; (4) 72°C, 30 s; (5) go to step 2 34 × ; (6) 72°C, 5 min; and (7) 12°C, hold. PCR amplicons were run on 2% agarose gel and analyzed.
Eight microliters of the PCR amplicon was used as the substrate DNA for the restriction fragment length polymorphism (RFLP) assay in a total reaction volume of 20 μL (1 × buffer, 5 U of restriction enzyme, and remaining volume dH2O). Embryos injected with mt-co1, mt-co3, and mt-tl1 TALE BE RNAs were surveyed for loss of BstYI, gain of MseI, and loss of SacI restriction sites, respectively. PCR amplicons for the samples confirmed positive by RFLP were then gel extracted and purified using the QIAquick Gel Extraction Kit (Qiagen).
Purified amplicons were then submitted to Sanger sequencing (GENEWIZ LLC) to estimate the editing efficiency and any off-target edits in the protospacer region (protospacer region is the sequence between two bound TALE monomers). Chromatogram files were then analyzed using EditR software21 to calculate the heteroplasmy in the injected embryos. Embryos harboring base edits were then placed on the fish facility at 6 dpf to enable the investigation of potential longitudinal functional studies, including assessing any tissue-specific differential maintenance of heteroplasmy of the induced edits.
Relative mitochondrial copy number
To estimate the relative mitochondrial copy number between mutants and control animals, individual embryos were placed in PCR tubes and incubated in the DNA extraction buffer (50 mM Tris-HCl pH 8.5, 1 mM EDTA, 0.5% Tween-20, 200 μg/mL proteinase K) at 55°C overnight. Following incubation, lysate was centrifuged at 16,000 g for 2 min and the supernatant was transferred to the fresh tube. Each DNA sample was screened for the editing by RFLP and those displaying nucleotide changes in the protospacer region were prioritized for the mtDNA copy number analysis. Relative mtDNA abundance was measured using primers for mt-nd1 as the mitochondrial control and polg as the nuclear control (Supplementary Table S2). PCR was performed using the SensiFAST SYBR Lo-ROX Kit (Bioline).
Mitochondrial respiration analysis, ATP, and lactate measurement
To carry out biochemical assays, 20 embryos were collected per tube and washed two times with E3 buffer. After buffer removal, embryos were immediately frozen in liquid nitrogen, and then stored at −80°C until ready for analysis. For respiratory chain complex and citrate synthase assays, frozen zebrafish larvae were homogenized in a mitochondrial isolation buffer (250 mM sucrose, 20 mM Tris-HCl, 3 mM EDTA, pH 7.4) on ice with a motorized pestle, followed by three freeze/thaw cycles with liquid N2. The lysate was then subjected to differential centrifugation to obtain mitochondrial-enriched fractions.
For ATP and lactate assays, frozen 7 dpf zebrafish larvae were homogenized in ice-cold 0.5 M perchloric acid (PCA) by grinding them with motorized pestle followed by 2 s of sonication, and one freeze/thaw cycle in liquid nitrogen/room temperature water. Supernatant was then spun at 16,000 g for 15 min, and subsequently neutralized by ice-cold 1 M potassium carbonate. The supernatant after centrifuge at 16,000 g for 10 min was used. Spectrophotometric assays were performed at 37°C for lactate assay or at 30°C for respiratory complex and citrate synthase assays in 170 μL of final volume using a Tecan Infinite 200 PRO plate reader.
Complex I and complex II activities were estimated by the reduction of 2,6-dichlorobenzenone-indophenol sodium salt (2,6-DCPIP) at 600 nm (ɛ600 = 21 mM−1 cm−1). Complex I assay buffer contained 25 mM KH2PO4, pH 7.4, 5 mM MgCl2, 3 mg/mL bovine serum albumin (BSA), 25 μM ubiquinone Q1, 5 μM antimycin A, and mitochondrial-enriched fraction. After the addition of 100 μM NADH in the presence and absence of 5 μM rotenone, the rates were calculated after the subtracting rotenone-insensitive activities.
Complex II assays comprised 25 mM KH2PO4 pH 7.4, 5 mM MgCl2, 3 mg/mL BSA, 25 μM ubiquinone Q1, 5 μM antimycin A, 5 μM rotenone, and mitochondrial-enriched fraction. Reaction began with the addition of 20 mM succinate to the assay buffer. Oxidation of reduced cytochrome c at 550 nm (ɛ550 = 21 mM−1 cm−1) was used to investigate the activity of complex IV. The assay buffer contained 25 mM KH2PO4 pH 7.4, 5 mM MgCl2, 0.015% n-dodecyl-β-d-maltoside, 5 μM antimycin A, 5 μM rotenone, and mitochondrial-enriched fraction. Fifteen micromolar-reduced cytochrome c was used to start the reaction and the rates were calculated as a first-order constant. Fish numbers or protein concentration (estimated by Bradford assay)24 was used to normalize the enzyme activity.
ATP levels were investigated using a YMC-Pack ODS-A column (5 μm, 4.6 × 250 mm) preceded by a guard column at 50°C. Flow rate was set at 0.4 mL/min. Initially the mobile phase (A) was 100% mobile, 0.1 M NaH2PO4 buffer, pH 6.0, after which the concentration of methanol was linearly increased with mobile-phase solution (B) consisting of 0.1 M NaH2PO4 buffer, pH 6.0, containing 25% methanol to 20% over 10 min. The mobile phase B was increased to 100% for 5 min to aid in the washing of the column after each separation. UV absorbance was monitored at 260 nm with a Shimadzu SPD-M20A. Pertinent peak areas were integrated by the LabSolution software from Shimadzu. ATP levels were quantified using standard curves and normalized per fish or protein concentration.
A highly sensitive lactate oxidase (LOX)-based colorimetric assay was used to assess the lactate levels.25 (carboxymethylaminocarbonyl)-4,4′-bis(dimethylamino)diphenylamine sodium salt (DA-64) (FUJIFILM Wako Chemicals U.S.A Corporation), a dye that is a highly sensitive indicator, was used for the detection of hydrogen peroxide.26 The following reaction is catalyzed by LOX: l-lactate + O2 → pyruvate + H2O2.
Five microliters of neutralized PCA extract was added to 155 μL of lactate assay reaction mixture (0.2 mM DA-64, 1 mM EDTA, 0.1% Triton X-100, and 5 U/mL horseradish peroxidase in 100 mM HEPES, pH 7.4), mixed thoroughly, and then incubated at 37°C for 3 min. Absorbance was measured every 20 s at 727 nm for 15 min, after the addition of 10 μL of freshly prepared LOX (2 U/mL). Lactate concentrations were calculated using the standard curve obtained with sodium l-lactate and were normalized per fish or protein concentration.
Results
Assembly of FusXTBE plasmids
To assess the activity of FusXTBE plasmids in mammalian cells, we cloned the split halves of DddAtox (split at 1397 amino acid position) and UGI in an FusX-compatible, codon-optimized destination plasmid, pKTol2C-COX8A-mTALEN. This receiving plasmid contained the mitochondrial targeting sequence of human cytochrome c oxidase subunit 8a (COX8A) upstream of the N-terminal domain of the TALE cassette to facilitate the import of base editor inside mitochondria. The resulting mitochondrial targeting sequence (MTS)-TALE-DddAtox module was cloned upstream to 3′UTR sequence from the human ATP5B gene to increase the efficiency of mRNA sorting near the proximity of mammalian mitochondria.27
To quantify the editing efficiency in vertebrate zebrafish embryos, a cassette containing DddAtox was cloned at the 3′ end of the C-terminus of the TALE domain in the linearized pK-idh2GoldyTAL-lacZ-Fok1 destination vector. We identified a highly functional and sufficient mitochondrial targeting sequence from a reverse-engineered in vivo protein trap in a nuclearly encoded mitochondrial protein.16,28 This derived minimal MTS was highly active in zebrafish in vivo compared with the previously described MTS and was used to target an array of different proteins capable of interacting with the mtDNA genome.16
We were able to rapidly assemble and test different TALE constructs using the previously published FusX system.19 This method utilizes a library of dimer and trimer RVD (TALE repeat) modules that uses Golden Gate cloning to synthesize a TALE that targets any 15- and 16-mer DNA sequence of interest (Fig. 1A, B).
TALE Writer predicts potential base editing sites in the human and zebrafish mitochondrial genome
TALE Writer, a computational tool for the efficient design of TALE-based technologies, was developed utilizing Python 3.7.3 to rapidly identify potential target sites for base editing in the human and zebrafish mitochondrial genomes.
Based on specific design parameters from prior TALE-based nuclease and gene editor work (described in the Materials and Methods section), the script allows the identification of cytosines within 5′-TC-3′ loci that can potentially be targeted by FusXTBEs. Some of these edits can lead to the induction of PTCs, which can result in the generation of nonfunctional gene products due to the premature termination of translation.20 TALE Writer can specifically identify such targets and calculate all their possible FusXTBE design permutations. The calculation of these designs is contingent on user-specified parameters, such as the lengths of the TALE arms and the spacer region.
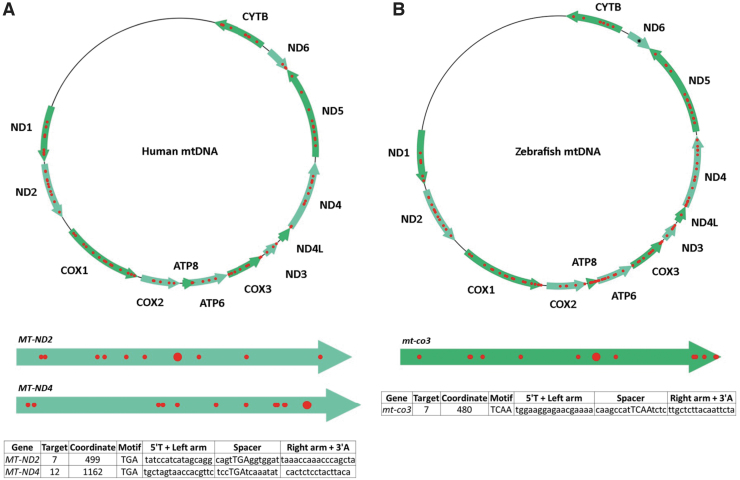
Following user-specified design parameters, TALE Writer identified a total of 1617 potential target sites for C-to-T base editing via FusXTBEs in the human mitochondrial genome, 94 of which were amenable to PTC induction (Fig. 2A). For example, multiple design permutations were found for the locus m.G11922, which coincides with an in-frame 5′-TGA-3′ motif in the forward strand of the human gene MT-ND4. In this case, a successful G-to-A edit (equivalent to a C-to-T edit on the opposite strand) leads to the stop codon UAA.
FIG. 2.
Targetable sites for PTC induction in human and zebrafish mtDNA. (A) Based on design parameters extrapolated from TALEN and initial mtDNA base editor work, and including assembly parameters for the FusX system, TALE Writer identified a total of 1617 potential target sites for C-to-T base editing in the human mitochondrial genome, 94 of which are amenable to PTC induction (red circles). The human mitochondrial genes MT-ND2 and MT-ND4 are represented below the human mtDNA map as light green arrows. The big red circles correspond to the targets chosen for this study. The table at the bottom includes the number of the targets within their respective genes, their local coordinates, their sequence motifs, and the sequences of their TALE arms and spacers. (B) Similarly, the zebrafish mitochondrial genome possesses a total of 1570 potential target sites for C-to-T base editing, 115 of which correspond to targets amenable to PTC induction (red circles). The zebrafish mitochondrial gene mt-co3 is represented below the zebrafish mtDNA map as a green arrow. The big red circle corresponds to the target chosen for this study. The table at the bottom includes the number of the target within mt-co3, its local coordinate, its sequence motif, and the sequences of its TALE arms and spacer. The single black asterisk in the zebrafish gene mt-nd6 represents a site for PTC induction made available by setting the maximum TALE arm and spacer lengths to 17 bp; such a FusXTBE format can be assembled via our FusX extender kit. mtDNA, mitochondrial DNA. PTC, premature termination codon. Color images are available online.
Similarly, a total of 1570 potential target sites for C-to-T base editing via FusXTBEs were found in the zebrafish mitochondrial genome, 115 of which coincided with loci amenable to PTC induction (Fig. 2B). For example, multiple design permutations were found for the locus m.C10215, which coincided with a 5′-TCAA-3′ motif in the forward strand of the zebrafish gene mt-co3. In this case, a successful C-to-T edit leads to the stop codon UAA.
Based on the originally selected design parameters, no targetable sites amenable to PTC induction in the zebrafish gene mt-nd6 were found (Fig. 2B). However, by expanding the allowable lengths of the TALE arms and the spacer region up to 17 bp, various design permutations for PTC induction were identified around a single target in this gene (Fig. 2B). Specifically, the zebrafish locus m.C14967, which coincides with an in-frame 5′-TGA-3′ motif in the heavy strand of the gene mt-nd6, can be targeted. A successful G-to-A edit in this site (equivalent to a C-to-T edit in the opposite strand) would result in the UAA stop codon. Our FusX Extender series system can enable the manufacturing of such a design permutation as a single-tube reaction.
Engineering base edits in vitro using FusXTBE
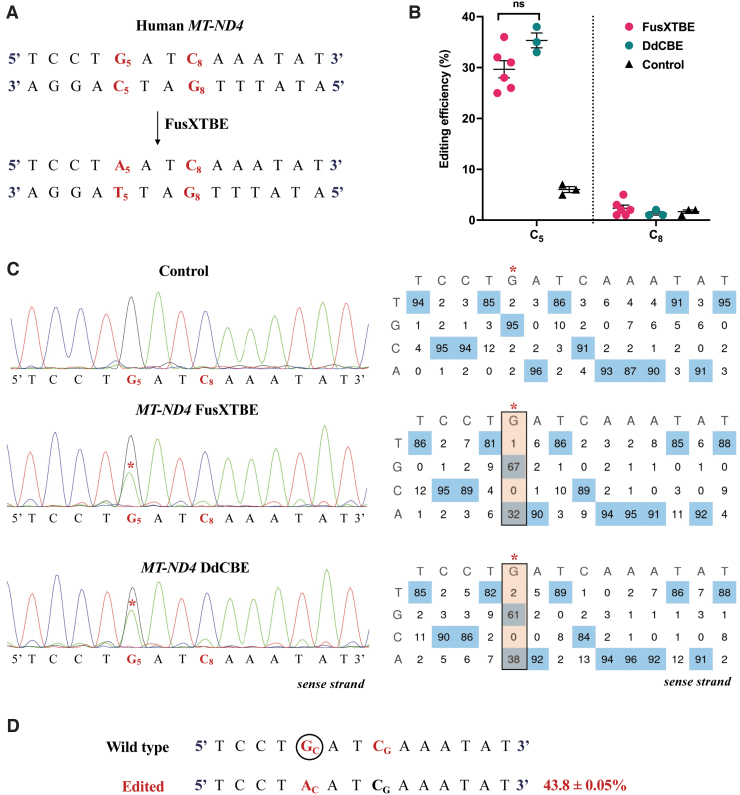
To assess the activity of the modular mitoFusXTBE system in vitro, we used the computational tool TALE Writer, to predict the target sites in the human mtDNA locus that are amenable to PTC induction. Out of the 94 potential predicted target sites, we chose to edit the cytosine on the reverse strand at the m.G11922 position of MT-ND4 gene (Fig. 3A). We transfected HEK293T cells with each of the monomers of the FusX compatible TALE base editor with G1397-C-DddAtox (left) and G1397-N-DddAtox (right) and measured the editing efficiencies at 3 days post-transfection. PCR across the target locus and subsequent Sanger sequencing revealed editing efficiency at C5 in the range of 30% ± 4% (Fig. 3B, C). No other edits were observed in the spacer region between the two TALE binding sites, despite favorable C1 and C8 present upstream and downstream to the PTC target.
FIG. 3.
Demonstrating the editing activity of FusXTBE system in vitro for mtDNA. (A) Potential target sites favorable for base editing in the protospacer region of the targeted human MT-ND4 locus. (B) Editing efficiency of the mitochondrial FusXTBE and DdCBE systems in HEK293T cells for the C5 and C8 positions, assessed by Sanger sequencing. Data are represented from independent experiments. Red and green colored data points represent editing efficiency by FusXTBE and DdCBE systems, respectively. Black colored data points represent the wild-type heteroplasmy in the target locus in the nontransfected cells. Error bars are represented as standard error of the mean. No significant difference was observed in the editing efficiencies between the FusXTBE and DdCBE systems (p > 0.05 as determined by Student's t-test). (C) Representative chromatogram of the control (nontransfected) and cells transfected with FusXTBE and DdCBE plasmids. Asterisk (*) denotes the site of edit (C5 position) with corresponding editing percentage (C-to-T or G-to-A). Chromatograms and editing table plot were obtained using EditR. (D) Editing frequency of MT-ND4 alleles estimated by deep sequencing of the target amplicon. Edited cytosine (antisense strand) residue (or guanine on the sense strand) is highlighted in red and circled. Frequency reflects the mean percentage ± standard deviation from independent biological replicates (N = 6 for FusXTBE and N = 3 for DdCBE). A combination of left arm- pKTol2C-FusXTBE-C and right arm- pKTol2C-FusXTBE-N was used to obtain the edits. Color images are available online.
To gain a better understanding of the effect of the programmable FusX-TALE modular domain on the editing activity, we compared the FusXTBE system with the original DdCBE construct reported,17 targeting the same mtDNA locus. Cells transfected with the original DdCBE system exhibited editing efficiencies in the range of 35% ± 3% across a series of experiments (Fig. 3B, C). These findings were corroborated by NGS amplicon sequencing of the MT-ND4 locus, in which 43% of the alleles harbored single edit at the C5 position (Fig. 3D).
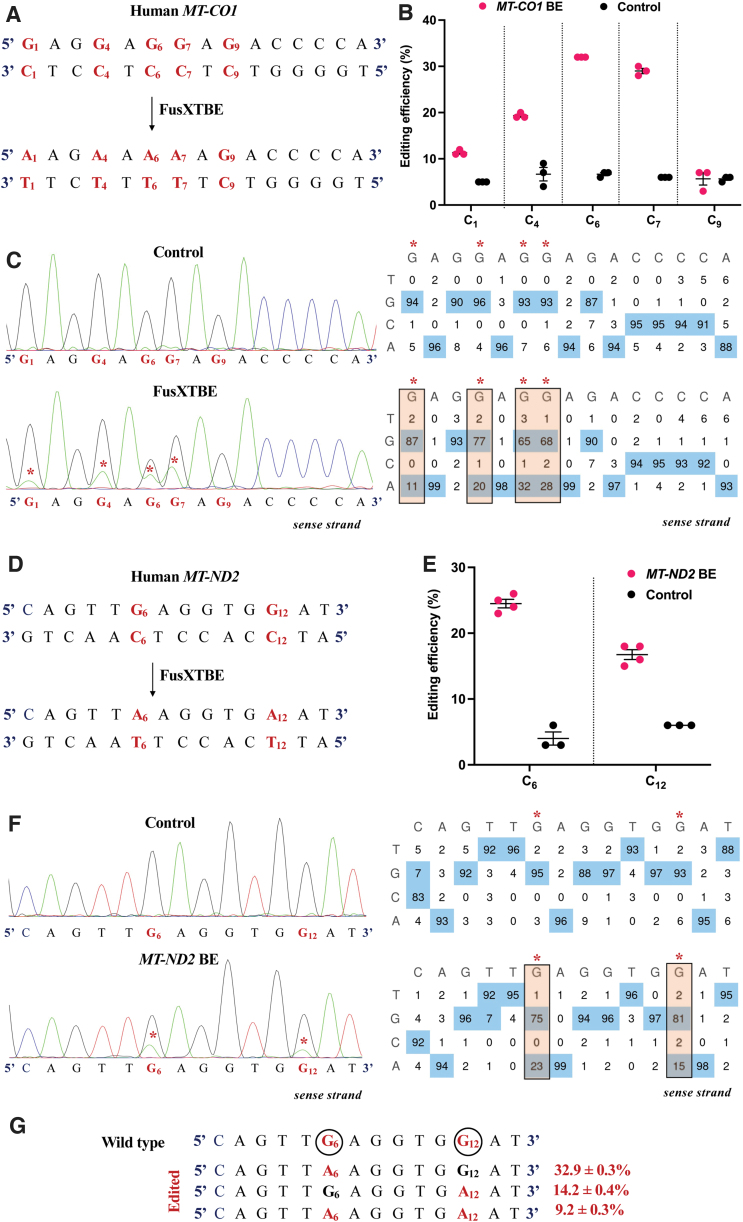
We also interrogated the activity of FusXTBE at two other genetic loci MT-ND2 and MT-CO1. Mitochondrial heteroplasmy assessment of the MT-CO1 locus revealed multiple C-to-T transitions (m.G6574, m.G6577, m.G6579, m.G6580, and m.G6582) in the protospacer region in the range between 11% and 32% (Fig. 4A). Maximum editing efficiency in the range of 29–32% was observed at the C6 and C7 position, where C6 was preceded by 5’TC (Fig. 4B, C).
FIG. 4.
Engineering base edits in MT-CO1 and MT-ND2 genes in vitro using the FusXTBE system. (A) Sequence of the potential editing sites in the MT-CO1 protospacer region. (B) Editing efficiency for the 5′TCs present in the protospacer region plotted as individual data points for each of the target nucleotides in the transfected and control HEK293T cells. Error bars are represented as standard error of the mean. (C) Representative chromatogram of the control and cells transfected with MT-CO1 FusXTBE plasmid. Asterisk (*) denotes the site of edit with corresponding editing percentage (C-to-T or G-to-A). (D) Protospacer region in the MT-ND2 locus highlighting the edited nucleotides. (E) Editing efficiency assessed by Sanger sequencing of mitochondrial FusXTBE for the each of the 5′TCs (C6 and C12) in the MT-ND2 target locus. Error bars are represented as standard error of the mean. (F) Representative chromatogram of the control (nontransfected) and cells transfected with MT-ND2 FusXTBE plasmid DNA. Asterisk (*) denotes the sites of edit (C6 and C12 positions). (G) Editing frequency of MT-ND2 alleles obtained by deep sequencing. Edited cytosine (antisense strand) residue (or guanine on the sense strand) is highlighted in red and circled. Frequency reflects the mean percentage ± standard deviation from independent biological replicates [N = 3 for MT-CO1, N = 4 for MT-ND2 (C6), and N = 3 for MT-ND2 (C12)]. Each point represents data from independent experiments. Chromatograms and editing table plot were obtained using EditR. The combination of left arm- pKTol2C-FusXTBE-C and right arm- pKTol2C-FusXTBE-N was used to obtain the edits. Color images are available online.
Protospacer in the target locus of MT-ND2 had two Cs (C6- m.G4969 and C12- m.G4975) on the antisense strand amenable to C-to-T transitions (Fig. 4D), which were edited at the frequency of 24.5% ± 1.3% and 16.8% ± 1.7%, respectively (Fig. 4E, F). Majority of the edited alleles, that is, 32.9% ± 0.3% harbored single edit at C6 position, followed by alleles with edits at C12 position. Only 9.2% of the edited alleles carried C-to-T edits at both C6 and C12 positions (Fig. 4G). C-to-T transition at C6 position (G on the sense strand) leads to an introduction of stop codon [p.(Trp161*)]. A bystander edit at m.C4846 in the MT-ND2 gene locus was also observed post-NGS analysis.
The results from the prioritized human mtDNA loci indicate that the FusXTBE system has a comparable editing efficiency in vitro when bound to the mtDNA and established this tool for potential additional applications, leveraging the informatic base mapping and rapid assembly system.
FusXTBE introduces base edits in vivo
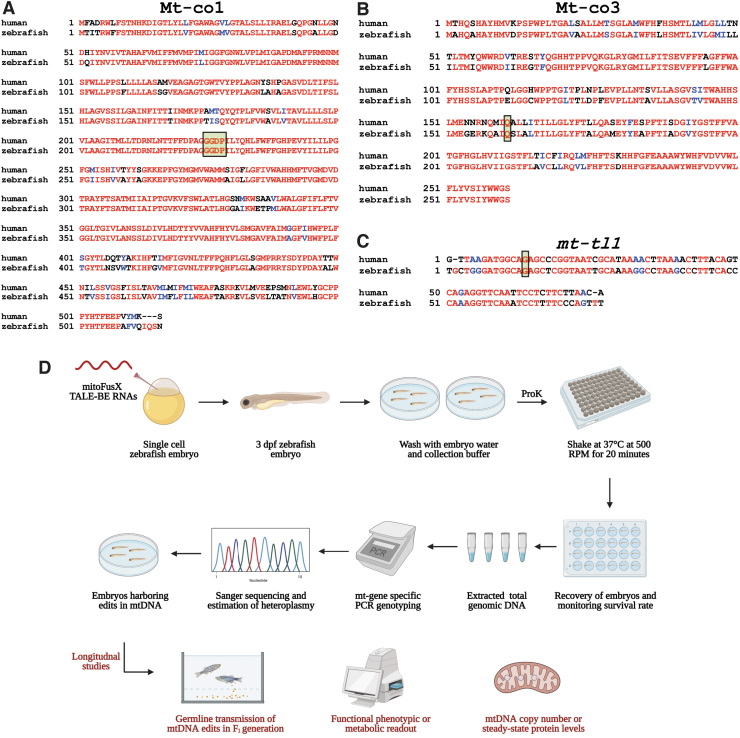
We used TALE Writer to predict potential sites amenable to base editing in the protein-coding and tRNA genes in zebrafish mtDNA. Out of the 1570 potential sites, we prioritized three targets: two protein-coding genes, mt-co1 and mt-co3, and the mt-tRNA-leu gene (or mt-tl1). Amino acid sequences of the zebrafish Mt-Co1 (Fig. 5A) and Mt-Co3 (Fig. 5B) proteins displayed >80% similarity to their human orthologs indicating a high level of sequence conservation between these vertebrate species. Zebrafish mt-tl1 displayed ∼69% sequence similarity with the human ortholog at the nucleotide level (Fig. 5C).
FIG. 5.
Schematic to prioritize base editing targets and noninvasively genotype mtDNA from zebrafish embryos. (A, B) Targets identified in the mitochondrial protein-coding genes, mt-co1 and mt-co3, were prioritized to test the editing efficiency of FusXTBE in vivo. Amino acid alignments are shown for the human MT-CO3 and MT-CO1 proteins with zebrafish Mt-co1 and Mt-co3 proteins. Edited amino acids are shaded and boxed. (C) The mitochondrial-encoded tRNA gene, mt-tl1, was prioritized to introduce base edits. Sequence alignment of human MT-TL1 and zebrafish ortholog mt-tl1 is shown. Target nucleotide is shaded and boxed. Red indicates sequences fully conserved between the two species. Blue indicates similar and black indicates positions with no similar or identical relationship. (D) Longitudinal study design assay for potential functional phenotype of the engineered edits in the zebrafish mtDNA locus. Zebrafish embryos are gently shaken in the genotyping buffer supplemented with proteinase K. Following recovery of the embryos, DNA is isolated from the shed cells, and mitochondrial genotyping is conducted for the target locus. Positive embryos can then be screened for germ line transmission or different functional and molecular studies. For Figure 4A–C, sequences were aligned using T-COFFEE multiple sequence alignment webserver and color-coded using BoxShade server. Figure 4D was created using Biorender.com. Color images are available online.
The zebrafish-applicable version of the FusXTBE specific to each of the target loci was assembled using the FusX assembly protocol. The expression of the mitochondrially targeted TALE base editor system (left- pT3-FusXTBE-N and right- pT3-FusXTBE-C) as in vitro synthesized mRNA was injected into the single-cell zebrafish embryos (Fig. 5D). More than 70% survival was observed for the embryos injected with mt-co1, mt-co3, and mt-tl1 BE RNAs over a range of experiments.
Three dpf injected embryos were subjected to noninvasive genotyping to facilitate downstream longitudinal analyses during the future course of studies (Fig. 5D). Post-genotyping, survival was monitored for 2 days, and mortality in the range between 15% and 20% was observed across the three target-specific genotyping experiments. Subsequently, DNA was extracted from the shed cells in the solution and was used as a template for downstream assays such as PCR and RFLP. Base edits in any of the target sites either created a loss or gain of restriction enzyme site, facilitating the scoring for embryos harboring variations in the mtDNA locus.
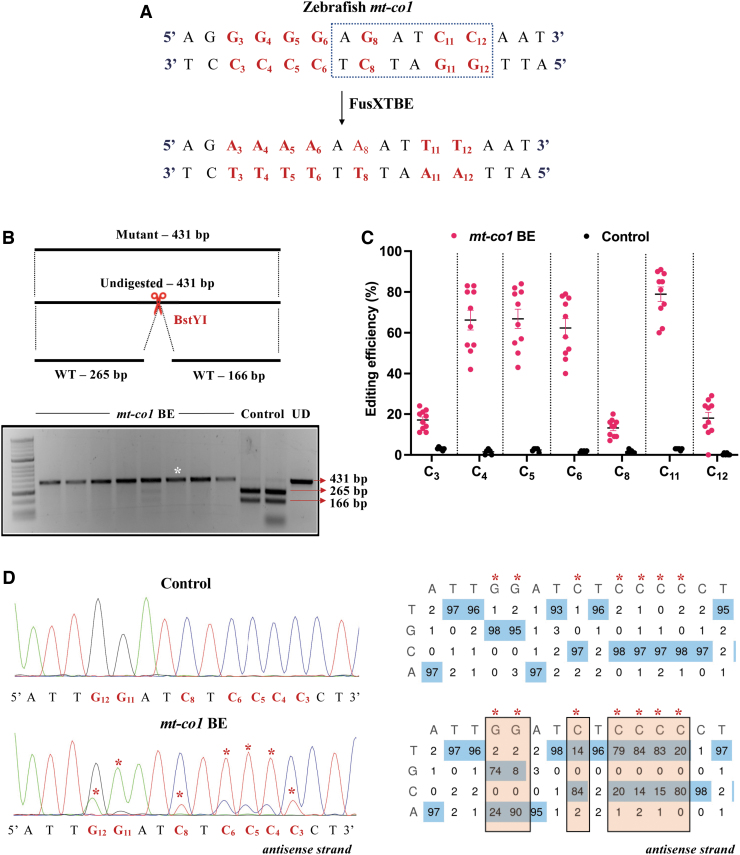
mt-co1 BE RNA was designed to edit the cytosine (C8) on the reverse strand at the m.C7106 of zebrafish mtDNA (Fig. 6A). m.G7106A edit on the forward strand, or the C-to-T transition on the reverse strand, leads to the loss of a restriction site for the enzyme BstYI. Amplification of the target locus from the extracted DNA followed by mitochondrial genotyping revealed the presence of undigested band (431 bp) in the case of injected embryos compared with the digested products (265 and 166 bp observed) in the noninjected control embryos (Fig. 6B).
FIG. 6.
Introducing base edits in the zebrafish mt-co1 gene using the FusXTBE system. (A) Protospacer region in the mt-co1 locus highlighting the potential editing sites. Restriction site for the enzyme BstY1 is marked by dotted rectangle. (B) Screening of embryos harboring mutations by RFLP. C-to-T edits in the protospacer region leads to loss of recognition site for BstYI restriction enzyme. PCR amplicons from control embryos show the presence of two wild-type digested products. PCR amplicons from the injected embryos indicate the presence of an undigested band marked by white asterisk (*). Each lane represents an individual embryo. (C) Editing efficiency for the favorable 5′TC sites present in the protospacer region, both in mt-co1 BE RNA and control zebrafish embryos. Embryos harboring edits and control embryos are highlighted by red and black color circles, respectively. Each data point represents an individual zebrafish embryo. Data are represented from independent experiments (Injected: N = 120 embryos and were screened and N = 10 embryos [RFLP positive] were genotyped by Sanger sequencing; Control = 60 embryos were screened and N = 4 embryos were genotyped by Sanger sequencing for which data have been included in the graph). (D) Representative chromatogram of the control and mutant embryos. Asterisk (*) denotes the site of edit with corresponding editing percentage (C-to-T or G-to-A). Chromatograms and editing table plot were obtained using EditR. The combination of left arm- pT3-FusXTBE-N and right arm- pT3-FusXTBE-C was used to obtain the edits. RFLP, restriction fragment length polymorphism. Color images are available online.
Sanger sequencing files (.ab1) were then assessed using EditR software21 to quantify the level of editing in the injected embryos. This analysis demonstrated that FusXTBE was highly efficient in vivo compared with in vitro, showing editing efficiencies as high as up to 90% (Fig. 6C). More than 70% of the injected embryos exhibited mutant mtDNA heteroplasmy. Analysis of the protospacer region revealed that the editing happened in both strands in the pyrimidine stretch present in the protospacer region, with efficiency in the range of 60–80% (Fig. 6C, D) in individual zebrafish embryos.
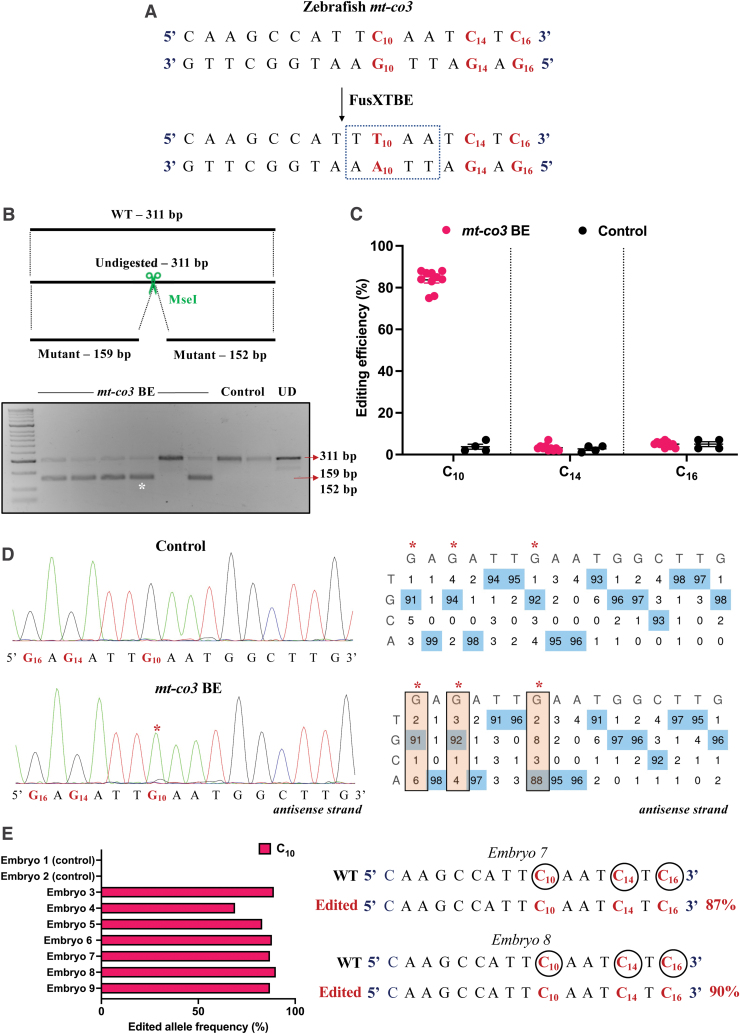
To leverage the potential of this construct in studying loss-of-function studies for mitochondrial genes, we chose to test the system against the site m.C10215 (C10) present in the mt-co3 gene (Fig. 7A). A C-to-T edit at this position creates a new PTC, UAA, leading to a truncated protein of 160 amino acids [p.(Gln161*)], as determined by in silico analyses. Embryos were scored based on the gain of restriction site for the enzyme MseI. RFLP analyses from the injected animals revealed the presence of digested products at the expected size of 159 and 152 bp along with the 311 bp undigested WT band (Fig. 7B).
FIG. 7.
Engineering precise base edits in vivo in the zebrafish mt-co3 gene. (A) Sequence of the potential editing sites in the mt-co3 protospacer region. The resulting gain of the restriction site for the enzyme MseI postediting event is marked by a dotted rectangle. (B) Screening of embryos harboring mutations by RFLP. C-to-T edits in the protospacer region lead to gain of recognition site for the MseI restriction enzyme. PCR amplicons from the control embryos show the presence of uncut product (311 bp). PCR amplicons from the injected embryos indicate the presence of expected 159 and 152 bp digested bands marked by a white asterisk (*). Each lane represents an individual embryo. (C) Editing efficiency for the target C10 present in the protospacer region in both groups of zebrafish embryos. Red and black color circles denote mutant and control embryos, respectively. Each data point represents an individual zebrafish embryo. Data are represented from independent experiments (Injected: N = 140 embryos and were screened and N = 10 embryos [RFLP positive] were genotyped by Sanger sequencing; Control = 60 embryos were screened and N = 4 embryos were genotyped by Sanger sequencing for which data have been included in the graph). (D) Representative chromatogram of the control and injected embryos. Asterisk (*) denotes the site of edit with corresponding editing percentage (C-to-T or G-to-A). Chromatograms and editing table plot were obtained using EditR. The combination of left arm- pT3-FusXTBE-N and right arm- pT3-FusXTBE-C was used to obtain the edits. (E) Percentage of reads with C-to-T transitions estimated by deep sequencing of target amplicon. Representative data from embryo 8 show the editing frequency of cytosine residues (C10, C14, and C16) in the protospacer region (highlighted in red and circled). The combination of left arm- pT3-FusXTBE-N and right arm- pT3-FusXTBE-C was used to obtain the edits. Color images are available online.
RFLP analysis was confirmed by Sanger sequencing, in which RFLP-positive embryos displayed 85% mutant heteroplasmy (Fig. 7C, D). We did not observe any other C-to-T edits in the protospacer region (C14 and C16). Deep sequencing of the target amplicon was consistent with the findings from the Sanger sequencing. In the individual embryos, editing frequency of the alleles as high as up to 80–90% was observed with precise edit of the introduction of [p.(Gln161*)] in the mt-co3 gene (Fig. 7E and Supplementary Fig. S1).
However, we did also observe two bystander edits at m.C10253 and m.C10179, which lies outside the TALE binding window. Bystander C-to-T edit at m.C10179 position leads to an amino acid change from histidine to tyrosine. Edit at m.C10253 lies outside the truncated protein region and leads to a synonymous amino acid substitution.
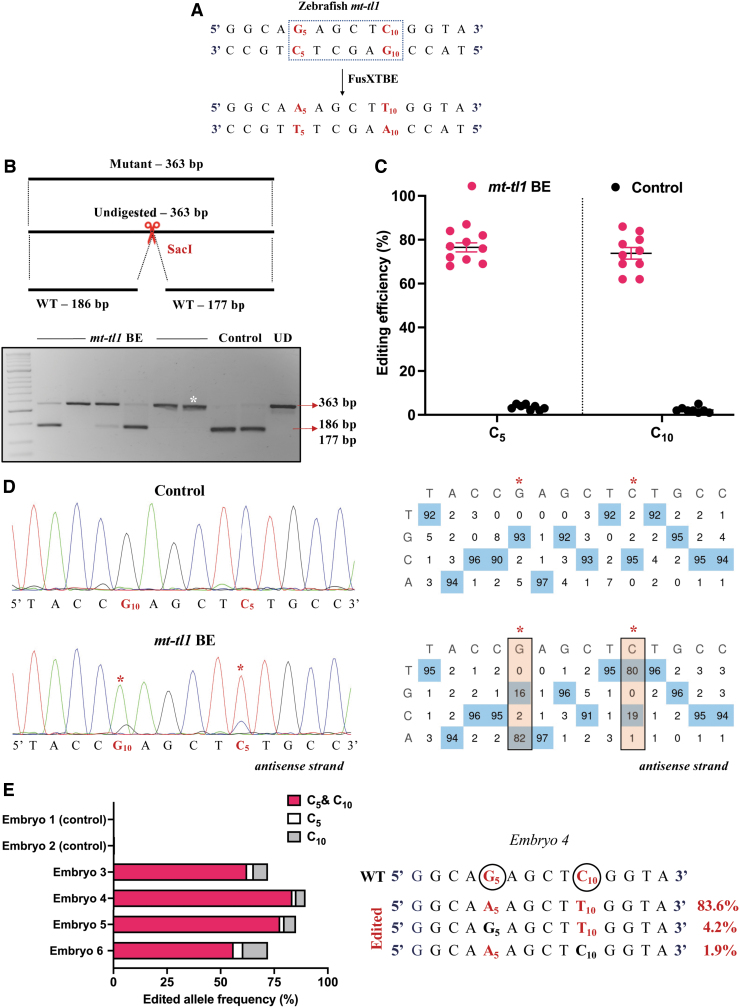
To confirm that our results were not specific to just protein-coding genes, we decided to test our system against the zebrafish mitochondrial tRNA gene, mt-tl1 (Fig. 8A). We chose a site that was conserved across the human and zebrafish genomes. A C-to-T transition at the target site m.G3739 (C5 on the reverse strand) created a loss of restriction site for the enzyme SacI (Fig. 8B).
FIG. 8.
Modeling mutations in the zebrafish mt-tl1 gene. (A) Protospacer region highlighting the putative cytosine targets in the mt-tl1 locus. Restriction site for the enzyme SacI is marked by a dotted rectangle. (B) Screening of embryos harboring mutations by RFLP. C-to-T editing event in the target locus leads to loss of restriction site for the SacI restriction enzyme. The presence of undigested WT bands of expected size, 186 and 177 bp (marked by red arrow), is observed in control embryos. PCR amplicons from the injected embryos show the undigested band, indicated by the loss of restriction site postediting (marked by white asterisk*). Each lane represents an individual embryo. (C) Editing efficiency for the favorable 5′TC sites present in the protospacer region, both in mutant and control zebrafish embryos. Control embryos and embryos harboring desired C-to-T edits are highlighted by black and red color circles, respectively. Each data point represents an individual zebrafish embryo. Data are represented from independent experiments (Injected: N = 150 embryos and were screened and N = 10 embryos [RFLP positive] were genotyped by Sanger sequencing; Control = 60 embryos were screened and N = 7 embryos were genotyped by Sanger sequencing for which data have been included in the graph). (D) Representative chromatogram of the control and mutant embryos indicating the percentage of editing at each cytosine residue in the protospacer region. Asterisk (*) denotes the site of edit with corresponding editing percentage (C-to-T or G-to-A). Chromatograms and editing table plot were obtained using EditR. (E) Percentage of edited alleles with C-to-T edits at different cytosine residues obtained from deep sequencing. Representative data from embryo 4 show the editing frequency of cytosine (antisense strand) residue (or guanine on the sense strand), which is highlighted in red and circled. The combination of left arm- pT3-FusXTBE-N and right arm- pT3-FusXTBE-C was used to obtain the edits. Color images are available online.
Individual embryos that displayed the loss of restriction site corresponding to the uncut band at 363 bp (Fig. 8B) harbored 60–70% mtDNA mutant heteroplasmy levels for the target locus (Fig. 8C, D). We also observed another edit at m.C3744 (C10) in the protospacer region with comparable mutant heteroplasmy. This observation cannot be deemed as off-target since any C preceded by T at the 5′ end is observed to be amenable to base editing by the DdCBE system as reported previously.17
NGS sequencing revealed that the edited alleles had C-to-T transitions both at C5 and C10 positions predominantly (Fig. 8E and Supplementary Fig. S2). Precise C5 edit was observed to be the least favorable event in the embryos injected with mt-tl1 BE RNA. The observations from the deep sequencing of amplicon analysis also revealed two bystander edits at positions m.G3621 and m.C3581. These edits were observed to be in the noncoding region of the mtDNA in the mitochondrially encoded 16S RNA gene, which lies outside the DNA-binding region of the TALE BE molecule.
Introduction of base edits in mtDNA affects the mitochondrial function in vivo
The ability of the FusXTBE system to introduce C-to-T transitions in vivo was successfully demonstrated by the molecular genotyping studies. Gaining confidence from these observations, we were interested to assess the impact of the mutant mtDNA heteroplasmy on the mitochondrial function and mtDNA content. mt-co3 encodes for a protein cytochrome c oxidase III that forms an important subunit of the mitochondrial respiratory chain complex IV, which plays an critical role in the reduction of oxygen molecule to water during the oxidative phosphorylation. Mutations in this gene have been observed to be associated with clinical phenotypes such as complex IV deficiency, lactic acidosis, encephalopathy, exercise intolerance, and myopathy.29
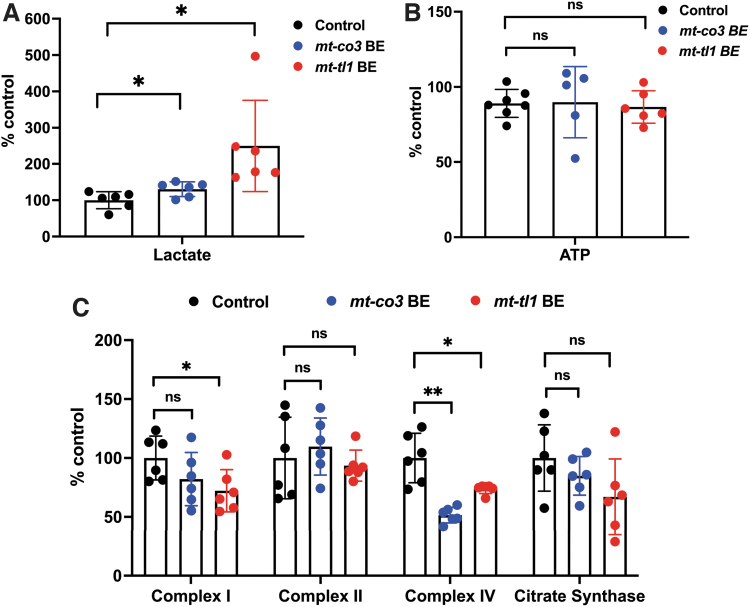
We used 7 dpf injected larvae to characterize the respiratory chain activity in the enriched mitochondrial fraction obtained from homogenization of whole larvae in mitochondrial isolation buffer (Fig. 9A–C). Complex I activity appeared to be slightly reduced, but the reduction was insignificant in the mt-co3 BE larvae, whereas complex II enzyme activity was maintained across the control and injected animals. Interestingly, we observed the complex IV activity to be significantly reduced to 51.5% in the embryos injected with mt-co3 BE RNA, supporting a specific downregulation of target enzyme activity of the mitochondrial electron transport chain. No difference in the citrate synthase enzyme activity was observed across both genotypes (Fig. 9C).
FIG. 9.
Functional consequence of base edits in mtDNA in zebrafish larvae. (A-B) Zebrafish larvae injected with mt-co3 and mt-tl1 BE RNAs raised to 7 dpf were collected and homogenized in 0.5 M perchloric acid and used for lactate (A) and ATP (B) assays. (A) Lactate concentrations in the samples were quantified (20 larvae each replicate: N = 6). Larvae injected with mt-co3 and mt-tl1 RNA displayed significant elevated lactate levels compared with the control (p < 0.05). Each data represent one biological replicate and error bars are represented as standard deviation. (B) No significant difference was observed in the steady-state ATP levels of larvae injected with the BE RNAs for mt-co3 and mt-tl1 genes compared with control larvae (p > 0.05) (N = 5 for mt-co3 and N = 6 for mt-tl1). ATP levels were maintained for both the groups. (C) For the mitochondrial respiratory chain complex activities, 20 larvae each tube were homogenized in the mitochondrial isolation buffer (250 mM sucrose, 20 mM Tris-HCl, 3 mM EDTA, pH 7.4) followed by differential centrifugation (to enrich mitochondrial fraction), and an assay was performed (N = 6 biological replicates of 20 pooled 7 dpf zebrafish larvae). Slight decrease in the complex I enzyme activity was observed for both mt-co3 and mt-tl1 BE groups compared with the control larvae, whereas activity for the complex II was maintained across all the three groups. For complex IV, a significant decrease in the activity was observed for both sets of larvae injected with mt-co3 and mt-tl1 BE RNA compared with the control. No difference in the citrate synthase activity was observed between the injected and control larvae (*p < 0.05; **p < 0.01). p-Values were determined by Student's t-test. Error bars represent the standard deviation. dpf, days postfertilization. Color images are available online.
Lactate was also observed to be significantly elevated by ∼30% in the mt-co3 BE group (Fig. 9A), with ATP levels being maintained in the injected animals (Fig. 9B). For the embryos injected with mt-tl1, predominantly edited alleles harbored mutations at both C5 and C10 positions. To explore the functional consequence of the altered tl1 sequence on the mitochondrial function, injected larvae were subjected to the same enzyme activity assays as previously described. Introduction of the C-to-T transitions did influence the mitochondrial function in the tl1 BE-injected animals. Lactate levels were observed to be significantly increased by 2.5-fold compared with the control larvae (Fig. 9A). This mimics the clinical phenotype observed in patients presenting with pathogenic variations in MT-TL1 gene.
Complex I and complex IV activities were reduced to 72.2% and 73.8%, respectively (Fig. 9C). Complex II activity and steady-state ATP levels (Fig. 9B) were maintained across both the experimental and control groups. We also interrogated the relative levels of copies of mitochondrial genome in the cell for both mutant and control animals, but did not detect any evidence showing that base edits modulate mtDNA copy number (Supplementary Fig. S3). Relative mtDNA copy number was maintained across all genotypes.
Discussion
mtDNA engineering had witnessed significant advancements in the past decade due to the advent of mitochondrially targeted nucleases that leveraged the potential of the absence of double-stranded break repair in shifting the heteroplasmy from mutant to wild-type mtDNA levels.11,12,15 However, the utility of this toolkit could not address treating mutant mtDNA homoplasmy or introducing precise edits in the mitochondrial genome.
Mok et al. recently described a bacterial toxin, cytosine deaminase, that catalyzes the conversion of C•G to T•A in the mtDNA.17 This obligate two-component system utilizes two separate nontoxic halves of DddAtox fused to TALE monomers to introduce single-nucleotide changes in the mtDNA in vitro. This system was not readily accessible due to the nature of the TALE construction approach used and did not include any corresponding informatic-based genomic analyses for exploring the potential gene editing outcomes. We built upon this initial architecture and extended the application of mtDNA base editing to model different mutations in mitochondrial genes implicated with clinical manifestations and to leverage it as a tool to understand basic mitochondrial biology.
As described previously, investigating a single target would require a user to test four different combinations of the DdCBE system (left-G1397-N and right-G1397C, left-G1397-C and right-G1397N, left-G1333-N and right-G1333C, or left-G1333-C and right-G1333N).17 This system does open an avenue to model or treat single-nucleotide pathogenic variations in animal and cellular models. However, traditional methods of TALE assembly either by conventional Golden Gate cloning or via expensive gene blocks impede the progress that the community can make in establishing mtDNA disease animal models.
To circumvent these challenges, we integrated our rapid one-step Golden Gate FusX TALE assembly system for use with the DdCBE base editor. We created the mitochondrial FusX TALE base editor (FusXTBE) architecture, where split halves of DddAtox and UGI were fused with a modular FusX19 compatible with the MTS-TALE module. The FusX system consists of 340 plasmids, which are preassembled trimers (spanning across six libraries), to generate 14.5- to 16.5-mer TALE repeats in a single Golden Gate reaction with a 3-day turnaround time. This toolbox offers an additional advantage of being compatible with mitochondrially and nuclearly targeted TALE scaffolds.
The FusX system can be automated and integrated to a high-throughput robotic liquid handling device, thereby scaling up the number of TALE base editors that can be assembled with minimal human error. This system is available at Addgene and has been successfully utilized for nuclear and mitochondrial gene editing applications, both in vitro and in vivo, with high efficiency.16,19
The DdCBE system is known to edit C in a 5′TC sequence-specific manner on any of the strands. To aid in interrogating different potential base editing sites, we developed TALE Writer, a customizable Python-based script. TALE Writer not only allowed predicting 5′TC sites across the human and zebrafish mitochondrial genomes, contingent on user-specified parameters, it also provided a list of all the possible combinations of TALE arms and spacer regions specific for each identified site.
While we identified >1500 such potential 5′TC sites in each of the two species, we were further interested in modeling loss-of-function studies for mitochondrially encoded protein-coding genes. Loss of function or translational defects due to the creation of premature stop codons in a protein-coding gene will allow us to elucidate its role in mitochondrial disease biology. TALE Writer identified >90 such sites in each of the interrogated genomes and can be used to model clinically significant loss-of-function mtDNA variations in the protein-coding genes, both in cellular and animal models (such as zebrafish).
Regarding the computation of the script, the time between the moments a user provides an input sequence to obtaining an output with all the possible design permutations for each of the targets identified within that sequence is usually negligible. All the targets used in this study were identified by the algorithm and were experimentally validated in different in vitro and in vivo model systems.
As a quantitative test of the FusX TALE modular cassette to DddAtox as a base editor, we compared our system with the previously published DdCBE system for the same mtDNA locus.17 It was notable from our findings that FusXTBE demonstrated no significant difference in activity in this in vitro assay (Fig. 3B). This served as a reference test system for expanding applications, including additional work in vivo. For example, the modular linker present in FusXTBE between the RVD domain and DddAtox enables changes in the potential editing window in the protospacer region. FusXTBE also offers an opportunity to drive heteroplasmy levels well beyond 30% for other mtDNA targets in mammalian cell lines.
Multisystemic clinical manifestations and varying mtDNA heteroplasmy from tissue to tissue in the case of mitochondrial disorders cannot be replicated by cells in a dish. Therefore, we explored the opportunity of testing the FusXTBE system in vivo. Zebrafish is a suitable animal model for studying mitochondrial disorders due to the 65% sequence conservation with human mtDNA as well as the shared mitochondrial-specific genetic code, strand-specific nucleotide frequencies, and fully syntenic gene order.30,31 Microinjection of FusXTBE RNAs in single-cell zebrafish embryos not only enhances the cellular delivery but also gives a rapid turnaround time to genotype-injected embryos within 3 dpf.
The poor correlation of phenotype to genotype and the stochastic mode of transmission make it challenging to study the biology of the mtDNA-encoded disorders. To overcome this technical hurdle, we adopted a noninvasive enzymatic genotyping approach22 to screen for embryos harboring desired edits. This approach allowed us to subject the animals to functional assessment or to monitor the germ line transmission of the edits in the F1 generation.
This assay involves the extraction of DNA from the cells that are shed after an event of modest shaking, ensuring optimal survival. This assay was originally described earlier for the nuclear genotyping,22 and with some modifications, we have adapted this assay to genotype mtDNA from zebrafish embryos. The high survival rate of injected embryos, that is, 80%, and successful amplification of the mtDNA RFLP, coupled with rapid TALE assembly, accelerates the process to model various pathogenic mutations in vivo.
One of the rationales to prioritize targets in zebrafish was to look for genes involved in clinically associated symptoms and those that are indispensable for mitochondrial function. Mutations in MT-CO1 and MT-CO3 are associated with conditions such as Leber optic atrophy, cytochrome c oxidase deficiency.32,33 MELAS disease is primarily associated with mutations in the gene MT-TL1 and is presented with symptoms such as mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes.4
Initially, we chose a pyrimidine-rich target on the zebrafish mt-co1 gene to identify the editing window within the protospacer region. Genotyping analyses revealed that ∼70% of the injected embryos displayed editing frequencies of up to 90%. Interestingly, we also observed several C-to-T edits having a 5′TCC sequence context, which may be due to the accessibility of the DddAtox molecule to the unstacked pyrimidine strand during the editing event.
Gaining insights from these observations, we chose a target in the mt-co3 locus to introduce a PTC upon a successful C-to-T edit at the m.C10215 position, thereby truncating the amino acid sequence, instead of the usual glutamine at that location. Sequence specificity for such targets is of utmost importance to avoid any nonspecific amino acid substitutions either upstream or downstream of the target nucleotide. Consistent with the high editing efficiency percentage observed for the previous target, frequencies of C•G to T•A transitions ranged between 75% and 88% for the injected individual embryos.
Such high percentages of precise edits in vivo generated with FusXTBE in zebrafish mtDNA allow us to understand the functional consequence of the truncation of mitochondrial Co-3 protein in the context of mitochondrial biology. Introduction of the premature stop codon in the mt-co3 gene appeared to significantly downregulate the activity of complex IV of the mitochondrial respiratory chain in addition to elevation of lactate concentration. These differences between mutant and control larvae indicated towards mitochondrial dysfunction in the edited larvae. Moreover, these assays were performed on the larvae, and it is possible that the difference could be enriched in specific extensive energy requiring organs such as muscle, brain, and heart in adult animals.
It will be interesting to see if such high percentage of edits are able to withstand the oocyte selection of mutant mtDNA as observed previously with low heteroplasmy of mtDNA large-scale deletion in zebrafish.16 Recently, a study describing mice editing with a custom-designed DdCBE demonstrated base edits in mice F0 and F1 embryos although with maximum mtDNA editing efficiency up to 57% in F0 generation for one of the loci. Two pups born to the mutant F0 female mouse harboring edits exhibited edits at a frequency that ranged between 6% and 26%.18
Our analysis documented C•G to T•A transitions in another target, mt-tl1, an ortholog of human gene MT-TL1 that is implicated in the pathogenicity of the MELAS. Approximately 70% of MELAS cases are attributed to the m.A3243G mutation in the human MT-TL1 gene.34 However, modeling this exact variation was not possible utilizing the DddAtox system due to the lack of a 5′TC in the target locus. We adopted a sequence similarity and secondary structure alignment informatic analyses to prioritize targets in the zebrafish locus (data not shown). One of the targets identified in the zebrafish mt-tl1 gene was upstream to the conserved nucleotide to m.A3243 of the human ortholog.
In 70% of the injected embryos, we observed C-to-T edits at m.G3744 (C10) in the range between 69% and 82%. We did notice another edit with similar percentage at m.G3739 (C5) within the protospacer window. This edit at C10 could be masked by using our FusX Extender series to design 17-mer TALE repeats that could bind to this nucleotide and make it inaccessible for cytidine deaminase activity. This improved FusXTBE architecture could aid in eliminating nonspecific edits near the targeted cytosine residue.
The two edits in the protospacer region appeared to affect complex I and complex IV of the mitochondrial electron transport chain, but not complex II, which does not contain any mtDNA-encoded subunit. These findings were consistent with the clinical phenotypes strengthened further by elevated levels of lactate in whole larvae. It needs to be appreciated that the effects that appear to be in the range of 25–30% appear to be milder, but that decrease is at the whole organism level.
Mitochondrial heteroplasmy varies from tissue to tissue and the decrease may be underrepresented by those tissues with normal mitochondrial function compared with the ones with impaired bioenergetics. Tissue-specific effects of this innovational FusXTBE are underway. However, the functional studies carried out in vivo do underscore the ability of FusXTBE to create novel or already known clinical pathogenic variations in animal models, which is critical to understand disease mechanism and also useful to perform preclinical drug screens to identify potential future therapies.
In summary, our study reports the first incidence of in vivo editing of mtDNA in zebrafish using a programmable FusX compatible TALE base editor. Rapid one-tube FusXTBE system with TALE Writer enables the facile design of TALE targets against potential base editing (5′TC) sites in human and mitochondrial genomes. The FusXTBE system demonstrated upto 90% C•G to T•A transitions in individual zebrafish embryos for both protein-coding and tRNA genes. Studies measuring the germ line transmission of high mutant mtDNA heteroplasmy load and genotype to phenotype correlation will shed more light on the underlying mitochondrial disease biology of mtDNA variations.
Conclusion
Mitochondrial medicine was galvanized by the recent discovery of the DdCBE system enabling single-nucleotide edits in the mitochondrial genome. In this study, we build upon this paradigm to extend the base editing in vivo using the rapid one-tube Golden Gate TALE assembly system, FusXTBE. Our programmable system also provides an adaptable algorithm to predict potential base editing sites in the human and zebrafish genome based on known editing guidelines, facilitating the design of TALE targets, and accelerating turnaround time.
The FusXTBE system has demonstrated editing efficiencies of upto 90% in the mtDNA locus in vivo, offering unique insights into using this automatable and high-throughput toolbox to model or correct pathogenic mutations in the mitochondrial genome in cellular and animal models. The findings presented in this study open an avenue to explore the biology of the mitochondrial genome and its implications in mitochondrial disorders.
Supplementary Material
Acknowledgments
The authors thank Dr. David Liu, Harvard University, for kindly providing the DdCBE plasmids through Addgene. They also acknowledge Dr. Eric Schon, Columbia University, for providing valuable input during the preparation of this article. The authors also thank Brandon Simone, Kavini Nanayakkara, and the Mayo Clinic Zebrafish Facility staff.
Authors' Contributions
The idea was conceived by K.J.C. and S.C.E. The article was written by A.S., S.R.C., and S.C.E. Experiments were executed by A.S., B.K., S.R.C., S.R.H., N.D.M., B.L.K., and C.S. with experimental guidance from E.N.O., K.J.C., and S.C.E. Data analysis was completed by A.S., B.K., S.R.C., and E.N.O. FusX Extender series was assembled by Z.W.J., R.P.C., and K.J.C. The article was reviewed and edited by A.S., B.K., S.R.C., S.R.H., N.D.M., C.S., E.N.O., and S.C.E.
Data Availability
Plasmids are available on request. The FusX assembly kit is available through Addgene (Kit no. 1000000063).
Author Disclosure Statement
The Mayo Clinic has an issued patent (US20180002707A1) on the FUSX TALE assembly system used here.
Funding Information
This work was funded by grants from the NIH U01-Somatic cell gene editing consortium (SCGE) AI 142773 and GM063904, Mayo Foundation for Medical Education and Research, a generous gift from the Marriott Foundation, and The Children's Hospital of Philadelphia (CHOP) Mitochondrial Medicine Frontier program.
Supplementary Material
References
- 1. Wallace DC. Mitochondrial genetic medicine. Nat Genet. 2018;50:1642–1649. DOI: 10.1038/s41588-018-0264-z. [DOI] [PubMed] [Google Scholar]
- 2. Thorburn DR. Mitochondrial disorders: Prevalence, myths and advances. J Inherit Metab Dis. 2004;27:349–362. DOI: 10.1023/B:BOLI.0000031098.41409.55. [DOI] [PubMed] [Google Scholar]
- 3. Skladal D, Sudmeier C, Konstantopoulou V, et al. The clinical spectrum of mitochondrial disease in 75 pediatric patients. Clin Pediatr (Phila). 2003;42:703–710. DOI: 10.1177/000992280304200806. [DOI] [PubMed] [Google Scholar]
- 4. Ciafaloni E, Ricci E, Shanske S, et al. MELAS: Clinical features, biochemistry, and molecular genetics. Ann Neurol. 1992;31:391–398. DOI: 10.1002/ana.410310408. [DOI] [PubMed] [Google Scholar]
- 5. Zeviani M, Moraes CT, DiMauro S, et al. Deletions of mitochondrial DNA in Kearns-Sayre syndrome. Neurology. 1988;38:1339–1346. DOI: 10.1212/wnl.38.9.1339. [DOI] [PubMed] [Google Scholar]
- 6. Bedell VM, Wang Y, Campbell JM, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. DOI: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. DOI: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. DOI: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miller JC, Tan S, Qiao G, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. DOI: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 10. Sander JD, Cade L, Khayter C, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697–698. DOI: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bacman SR, Williams SL, Pinto M, et al. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat Med. 2013;19:1111–1113. DOI: 10.1038/nm.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gammage PA, Viscomi C, Simard ML, et al. Genome editing in mitochondria corrects a pathogenic mtDNA mutation in vivo. Nat Med. 2018;24:1691–1695. DOI: 10.1038/s41591-018-0165-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kukat A, Kukat C, Brocher J, et al. Generation of rho0 cells utilizing a mitochondrially targeted restriction endonuclease and comparative analyses. Nucleic Acids Res. 2008;36:e44. DOI: 10.1093/nar/gkn124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peeva V, Blei D, Trombly G, et al. Linear mitochondrial DNA is rapidly degraded by components of the replication machinery. Nat Commun. 2018;9:1727. DOI: 10.1038/s41467-018-04131-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bacman SR, Kauppila JHK, Pereira CV, et al. MitoTALEN reduces mutant mtDNA load and restores tRNA(Ala) levels in a mouse model of heteroplasmic mtDNA mutation. Nat Med. 2018;24:1696–1700. DOI: 10.1038/s41591-018-0166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Campbell JM, Perales Clemente E, Ata H, et al. Engineering targeted deletions in the mitochondrial genome. bioRxiv. 2018:287342. DOI: 10.1101/287342. [DOI] [Google Scholar]
- 17. Mok BY, de Moraes MH, Zeng J, et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature. 2020;583:631–637. DOI: 10.1038/s41586-020-2477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee H, Lee S, Baek G, et al. Mitochondrial DNA editing in mice with DddA-TALE fusion deaminases. Nat Commun. 2021;12:1190. DOI: 10.1038/s41467-021-21464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma AC, McNulty MS, Poshusta TL, et al. FusX: A rapid one-step transcription activator-like effector assembly system for genome science. Hum Gene Ther. 2016;27:451–463. DOI: 10.1089/hum.2015.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lueck JD, Yoon JS, Perales-Puchalt A, et al. Engineered transfer RNAs for suppression of premature termination codons. Nat Commun. 2019;10:822. DOI: 10.1038/s41467-019-08329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kluesner MG, Nedveck DA, Lahr WS, et al. EditR: A method to quantify base editing from Sanger sequencing. CRISPR J. 2018;1:239–250. DOI: 10.1089/crispr.2018.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang X, Zhang Z, Zhao Q, et al. Rapid and efficient live Zebrafish embryo genotyping. Zebrafish. 2020;17:56–58. DOI: 10.1089/zeb.2019.1796. [DOI] [PubMed] [Google Scholar]
- 23. Park J, Lim K, Kim JS et al. Cas-analyzer: An online tool for assessing genome editing results using NGS data. Bioinformatics. 2017;33:286–288. DOI: 10.1093/bioinformatics/btw561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. DOI: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 25. Guha S, Mathew ND, Konkwo C, et al. Combinatorial glucose, nicotinic acid and N-acetylcysteine therapy has synergistic effect in preclinical C. elegans and zebrafish models of mitochondrial complex I disease. Hum Mol Genet. 2021;30:536–551. DOI: 10.1093/hmg/ddab059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takagi K, Tatsumi Y, Kitaichi K, et al. A sensitive colorimetric assay for polyamines in erythrocytes using oat seedling polyamine oxidase. Clin Chim Acta. 2004;340:219–227. DOI: 10.1016/j.cccn.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 27. Sylvestre J, Margeot A, Jacq C, et al. The role of the 3’ untranslated region in mRNA sorting to the vicinity of mitochondria is conserved from yeast to human cells. Mol Biol Cell. 2003;14:3848–3856. DOI: 10.1091/mbc.e03-02-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ichino N, Serres MR, Urban RM, et al. Building the vertebrate codex using the gene breaking protein trap library. eLife. 2020;9. DOI: 10.7554/eLife.54572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hanna MG, Nelson IP, Rahman S, et al. Cytochrome c oxidase deficiency associated with the first stop-codon point mutation in human mtDNA. Am J Hum Genet. 1998;63:29–36. DOI: 10.1086/301910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Broughton RE, Milam JE, Roe BA. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 2001;11:1958–1967. DOI: 10.1101/gr.156801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sabharwal A, Campbell JM, WareJoncas Z, et al. A primer genetic toolkit for exploring mitochondrial biology and disease using zebrafish. bioRxiv. 2019:542084. DOI: 10.1101/542084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koilkonda RD, Guy J. Leber's hereditary optic neuropathy-gene therapy: from benchtop to bedside. J Ophthalmol. 2011;2011:179412. DOI: 10.1155/2011/179412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rak M, Benit P, Chretien D, et al. Mitochondrial cytochrome c oxidase deficiency. Clin Sci (Lond). 2016;130:393–407. DOI: 10.1042/CS20150707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Niedermayr K, Polzl G, Scholl-Burgi S, et al. Mitochondrial DNA mutation “m.3243A>G”-Heterogeneous clinical picture for cardiologists (“m.3243A>G”: A phenotypic chameleon). Congenit Heart Dis. 2018;13:671–677. DOI: 10.1111/chd.12634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plasmids are available on request. The FusX assembly kit is available through Addgene (Kit no. 1000000063).