Abstract
Acyl modifications vary greatly in terms of elemental composition and site of protein modification. Developing methods to identify acyl modifications more confidently can help to assess the scope of these modifications in large proteomic datasets. We analyze the utility of acyl-lysine immonium ions for identifying the modifications in proteomic datasets. We demonstrate that the cyclized immonium ion is a strong indicator of acyl-lysine presence when its rank or relative abundance compared to other ions within a spectrum is considered. Utilizing a stepped collision energy method in a shotgun experiment highlights the immonium ion strongly. By implementing an analysis that accounted for features within each MS2 spectrum, the method clearly identified peptides with short chain acyl-lysine modifications from complex lysates. Immonium ions can also be used to validate novel acyl-modifications; in this study we report the first examples of 3-hydroxylpimelyl-lysine modifications and validate them using immonium ions. Overall these results solidify the use of the immonium ion as a marker for acyl-lysine modifications in complex proteomic datasets.
1. Introduction
As proteomic technology advances, the depth and breadth of proteomic data that can be analyzed has increased concurrently. This increased scope of proteomic information has resulted in a plethora of data on post-translational modifications (PTMs), which can affect protein function and modulate protein activity in a cell without the energetic burden of synthesizing new proteins.[1–5] These modifications fall into different classes based on their physicochemical properties, such as phosphorylation, glycosylation, oxidation and many more. One class of modification that has been observed across biological systems is lysine acylation.[6–8] Acetylation has been long known as an epigenetic regulator, modulating protein expression through the modified lysine side chains on histone tails.[9–11] Later, it was discovered that acetyl and other acyl modifications not only impact histone function, but are also ubiquitous in mammalian metabolic pathways as well as in other eukaryotic and prokaryotic systems.[6,9] In eukaryotic organisms, these modifications have been shown to correlate with aging as well as metabolic state.[7,8] In prokaryotes, these modifications impact the activity of enzymes in metabolic pathways.[12,13] Acylation even plays a role in viruses, as viral proteins have been shown to require acetylation to function.[14]
Acyl-modifications have proven to be challenging to identify on a proteome-wide scale for a number of reasons. These modifications tend to have low stoichiometries relative to unmodified proteoforms; e.g., mitochondrial acylation stoichiometries are around 0.02%.[15] This low abundance makes it difficult to consistently identify modified peptides in complex samples using untargeted proteomic methods, and enrichment strategies during sample preparation are often necessary.[16] A second challenge is that acyl modifications come in many different varieties. Chain length, elemental composition, and degree of unsaturation differ between different types of acylations.[4] For example, an acetyl group and a propionyl group differ by one carbon. Although it is often unclear whether these modifications assume different functions, delineating their presence is important. However, creating such a list requires knowing a priori which acyl compositions might be present. Acylation can occur spontaneously; that is, reactive intermediates such as acetyl phosphate or an acyl-CoA can modify certain primary amines even without enzymatic catalysis.[17] Such processes suggest that a given residue could be tagged by many acyl compositions, even on a single peptide. Not including all possible acyl modifications during database searching in proteomic workflows may miss critical information about a given protein, particularly when quantification is sought. Broadening the number of modifications considered, however, can increase the false discovery rate (FDR) or, if FDR is treated properly, reduce the sensitivity for identifying peptides. All of these challenges add to the difficulty in characterizing the full range of acyl modifications.
Physicochemical differences in PTMs have meant that different experimental strategies may need to be considered if each modification is to be detected and localized optimally. For example, peptide phosphates are often labile to collision induced dissociation (CID) and the associated mass shift may not be apparent in MS2 spectra or it may migrate to another residue.[18] To overcome this limitation, phosphoproteomic studies have adopted strategies that utilize electron transfer dissociation (ETD) as an alternative MS2 dissociation method.[18,19] Glycosylated peptides also present challenges for MS fragmentation given the fragility and complexity of their structures. One strategy for identifying these modifications uses collision energy stepping in conjunction with detecting the characteristic oxonium ions associated with specific glycans.[20] Oxonium ions[21–23] are low mass ions resulting from fragmentation of oligosaccharides and glycopeptides.
Similarly, immonium ions can serve as diagnostic markers for specific acyl-lysine PTMs.[24,25] Immonium ions are internal product ions resulting from two-bond cleavages that retain a single amino acid side-chain. These ions, common in peptide tandem mass spectra, can often verify the presence of certain amino acids.[26,27] It has been shown that, in addition to the canonical amino acids, immonium ions can be generated for acyl-lysine residues. The immonium ion for acetyl-lysine is observed at m/z 143.1179; however, a related ion observed at m/z 126.0913 that originates from cyclization has shown special utility.[24,28] Other acyl-lysines present unique immonium ions.[25] These diagnostic indicators are typically used for post-identification validation, because their presence and intensity depend heavily on sequence context and instrument parameters. Here, we posit a means to overcome limitations in the use of immonium ions by optimizing collision energies and by defining comparative criteria. This strategy can be extended to identifying novel acyl PTMs in large proteomic data sets.
2. Experimental Section
2.1. Reagents and Materials
Synthetic peptides (lyophilized, >95% purity) were obtained from Genscript, Inc. and reconstituted in water. The sequences, which originate from Syntrophus aciditrophicus, are: KSTPEAMAK, FKDEIPVVIK, STDPKGPSVR, with the lysine residues indicated in bold containing an acetyl-, butyryl-, or crotonyl-modification on the ε-amine.
2.2. Preparation and Digestion of Acyl-Bovine Serum Albumin
Acetylated-bovine serum albumin (BSA, Promega Product #R3961) was diluted in 100 mM ammonium bicarbonate. Butyrylated-BSA was prepared in a process adapted from Baez, et al.[29] Butyric anhydride (~25 μmol) was added to 100 μL of a 1 mg/ml solution of BSA (Sigma Aldrich, Product #A8022) in 100 mM ammonium bicarbonate. The solution was incubated for 20 minutes at 4°C, after which the solution pH was adjusted to ~8 using ammonium hydroxide. The incubation/pH adjustment process was repeated two more times. To reverse adventitious O-acylation, hydroxylamine hydrochloride was added to 50% (w/v) of the final concentration (e.g., 55 mg added to 110 μL) and the pH was readjusted with NH4OH to ~8. The solution was incubated at room temperature overnight. Butyrylated-BSA was then buffer exchanged into 100 mM ammonium bicarbonate using 10kD MWCO Amicon spin filters (Millipore).
Acylated-BSA preparations were heated to 95°C for 10 minutes, disulfide-reduced with 20 mM dithiothreitol (DTT) for 1 hour at 60°C, and alkylated with 50 mM iodoacetamide for 45 minutes at room temperature in the dark. Excess iodoacetamide was quenched with DTT and the samples were digested overnight with endoproteinase GluC (1:100) at room temperature (New England Biolabs, Product #P8100S). Digested peptides were dried in a vacuum concentrator, acidified with 0.1% acetic acid, desalted with STAGE tips assembled from 3M Empore C18 Solid Phase Extraction Disks[30] and dried again. Peptides were reconstituted in LC-MS injection buffer (3% acetonitrile, 0.1% formic acid) and quantified by Pierce Quantitative Fluorometric Peptide Assay (Thermo Scientific, Product #23290).
2.3. Peptide Preparation from S. aciditrophicus Cells
Cells were harvested from tricultures of Syntrophus aciditrophicus, Methanosaeta concilli, and Methanospirillum hungatei grown with benzoate as the carbon source. Peptides were prepared from cell pellets using enhanced filter-aided sample preparation (eFASP) as described by Erde, et al.[31,32] Briefly, cells were lysed in 4.0% (v/v) ammonium lauryl sulfate, 0.1% (w/v) sodium deoxycholate, and 5 mM tris(2-carboxyethyl)phosphine in 100 mM ammonium bicarbonate. The lysate was exchanged into a buffer containing 8 M urea, 0.1% (w/v) sodium deoxycholate and 0.1% (w/v) n-octyl glucoside using a 10 kDa Microcon ultrafiltration unit (Millipore). Within the ultrafiltration unit, proteins were alkylated in 17 mM iodoacetamide and digested overnight at 37°C with trypsin in a buffer containing 0.1% (w/v) sodium deoxycholate, 0.1% (w/v) n-octyl glucoside and 100 mM ammonium bicarbonate. Peptides were desalted with STAGE tips, as described earlier.
2.4. Mass Spectrometry (LC-MS/MS) Analysis
Processed peptides were measured by reversed phase liquid chromatography-tandem mass spectrometry (LC-MS/MS) on an EASY nLC1000 (Thermo Scientific) coupled to a quadrupole orbitrap mass spectrometer (Q-Exactive, Thermo Scientific). Peptides (100 ng of acyl-BSA) were loaded onto an Acclaim PepMap100 C18 trap column (Thermo Scientific, Product #16–494-6, 75 μm x 2 cm, 100 Å) and separated on an Acclaim PepMap RSLC C18 analytical column (Thermo Scientific, Product #03–251-873, 75 μm x 25 cm, 100 Å). Buffer A (0.1% formic acid) and buffer B (0.1% formic acid in 100% acetonitrile) were employed in the 300 nL/min gradient: 3–35% B in 30 min, 35–50% B in 5 min, and 50–80% B in 2 min.
Synthetic, acylated peptide standards (1 fmol of each) were spiked into 100 ng of a HeLa tryptic digest standard (Thermo Scientific, #PI88329). For the HeLa digest, 100 ng was loaded onto the column, whereas 200 ng of the triculture digest (containing S. aciditrophicus and two other organisms) was loaded. HeLa and triculture analyses used the gradient 3–20% B in 62 min, 20–30% B in 31 min, 30–50% in 5 min, and 50–80% in 2 min.
The mass spectrometer was operated in a data-dependent acquisition mode with an m/z 300–1800 MS scan acquired at 70,000 resolution using an automatic gain control (AGC) target of 1E6 (maximum fill: 100 ms). Collision induced dissociation MS/MS spectra were acquired at 17,500 resolution, AGC (maximum fill: 80 ms) of 1E5, and a normalized collision energy of 27 (unless otherwise indicated) on the top 10 abundant precursor ions. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD018758.
2.5. Proteomic Data Analysis
2.5.1. Acyl-BSA Data
RAW files were converted into MGF format and peak lists were submitted to Mascot (version 2.5; Matrix Science) and searched against the BSA sequence supplemented with protein sequences of common contaminants. GluC was specified as the cleavage enzyme with up to 6 missed cleavages considered, and a precursor mass tolerance of 10 ppm and product mass error of 0.02 Da. Cysteine carbamidomethylation (+57.021464), methionine oxidation (+15.994915), and the respective acyl-lysine modification, acetyl (+42.010565) or butyryl (+70.041865) were set as variable modifications. Peptide spectral matches (PSMs) were filtered to 1% false discovery rate using the target-decoy strategy.
2.5.2. Acyl-modifications in HeLa and S. aciditrophicus
All HeLa cell data was analyzed using Mascot (version 2.5). Files were searched against the UniProt Human database (as of January 23, 2019) supplemented with common laboratory contaminants and the three spiked synthetic peptide sequences. The S. aciditrophicus triculture RAW files were processed through ProteomeDiscoverer (version 1.4), using Mascot for the database search. Files were searched against UniProt S. aciditrophicus, Methanosaeta concilli, and Methanospirillum hungatei sequence databases that were concatenated and supplemented with contaminant sequences (as of July 8, 2019). The search parameters for both datasets were: enzyme specificity, trypsin; maximum number of missed cleavages, 2; precursor mass tolerance, 10 ppm; product mass error allowed, 0.02 Da; variable modifications included cysteine carbamidomethylation, methionine oxidation, lysine acetylation, lysine butyrylation, and lysine crotonylation (+68.026215). PSMs were filtered to 1% false discovery rate using the target-decoy strategy.
2.5.3. Immonium Ion Analysis for Acylated BSA
From the Mascot search results (DAT files), a more stringent secondary filter was applied to the data to increase our confidence in PSMs identified. PSMs with an ion score >25 were considered for immonium ion analysis and an in-house Python script was utilized to extract the corresponding MS/MS spectra containing the immonium ion of interest for further characterization. For all datasets, the mass tolerance was set to 10 ppm for the immonium ion of interest. Similar analysis was performed on the Svinkina et al.[33] data set (MassIVE MSV000079068).
3. Results
3.1. The m/z 126 Immonium Ion is Present in Great Abundance in an Acetyl-Lysine Data Set
Previous reports have indicated that the m/z 126 ion (126.0913) (hereafter called the “126 ion”) is diagnostic for acetyl-lysine.[24] To determine the prevalence of the 126 ion in PSMs, we investigated peptides from GluC-digested acetylated BSA, focused on comparing matches with and without acetyl-lysine, as indicated by the MASCOT search. Of those identified as acetylated (335/571), 97.3% displayed the 126 ion in MS/MS spectra (Table 1). However, the 126 ion was also present in 65.7% of non-acetylated spectra. There are several possible ways that non-acetyl-lysine containing peptides can yield an ion with the exact mass of the 126 ion, including “a-ion” type products that subsequently lose NH3 from sequences containing Gly-Ile, Gly-Leu, or Ala-Val and the reversed sequences. To verify that the observation of 126 ions from non-acetylated peptides also occurs with complex lysates, we performed the same analyses on a published dataset that utilized immunoprecipitation to enrich acetylated peptides from Jurkat E6–1 cells.[33] From this dataset, 96% of acetylated PSMs presented a 126 ion, but 73.9% of the non-acetylated PSMs also displayed a 126 product (Table 1). That the 126 ion appears in acetylated PSMs agrees with previous work detailing its sensitivity as an acetyl-lysine marker, but its presence in non-acetylated PSMs raises questions about its specificity.[24] This difference may reflect the higher sensitivity of current mass spectrometers as compared to those used previously.[24] It may also reflect the previous study’s metrics for determining false positives, which relied on the presence of specific dipeptide cleavages that may have been absent in spectra with limited sequence information.[24]
Table 1.
Distribution of PSMs containing m/z 126 and 129 immonium ions from acetylated BSA (27 NCE) and from Jurkat E6-1 cells (25 NCE) (Svinkina et al.)[33]
| PSMs with 126 ion |
PSMs without 126 ion |
PSMs with 129 ion |
PSMs without 129 ion |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Acetyl-BSA | Non-acetyl | 155 | (65.7%) | 81 | (34.3%) | 212 | (89.8%) | 24 | (10.2%) |
| Acetyl | 326 | (97.3%) | 9 | (2.7%) | 304 | (90.7%) | 31 | (9.3%) | |
| Svinkina et al. | Non-acetyl | 4458 | (73.9%) | 1577 | (26.1%) | 5806 | (96.2%) | 229 | (3.8%) |
| Acetyl | 4415 | (96%) | 185 | (4%) | 4255 | (92.5%) | 345 | (7.5%) | |
3.2. Using the 126:129 Ion Abundance Ratio as an Acetyl-Lysine Indicator
Given the ubiquity of the 126 ion in MS2 spectra, a more specific diagnostic ion metric is needed to distinguish between acetylated and non-acetylated PSMs. Other low mass ions that may indicate the presence of an unmodified lysine include the m/z 101 (101.1079) immonium ion and a diagnostic ion at m/z 129 (129.1023).[26] The m/z 101 ion was rarely observed within lysine-containing PSMs and, when present, did not clearly differentiate acetylated from non-acetylated peptides (Supplemental Table S1). The m/z 129 diagnostic ion (hereafter called the “129 ion”), was present more often and proved to better indicate the presence of unmodified lysine (Figure 1). Similar to the low specificity of the 126 ion for acetyl-lysine containing peptides, the 129 ion was not very specific for unmodified lysine-containing peptides; 89.8% and 90.7% of non-acetyl and acetyl PSMs, respectively, contained the 129 ion (Table 1). The prevalence of the 129 ion in acetyl and non-acetyl PSMs was also noted by Svinkina, et al.[33]
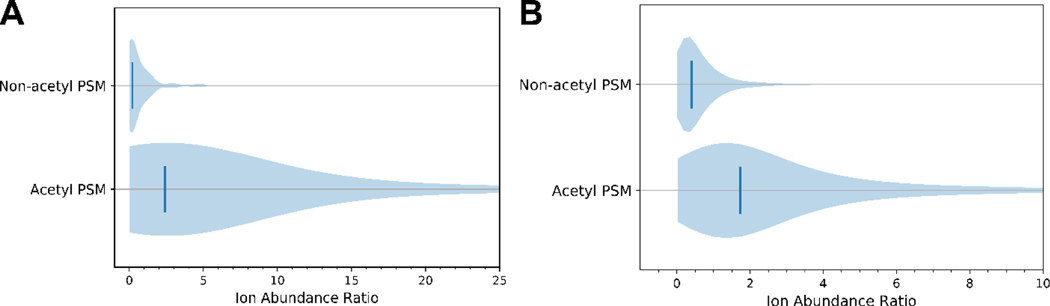
Figure 1. Ion abundance ratios [126]/[129] for acetylated and non-acetylated spectra.
A) Violin plots of the [126]/[129] abundance ratio for all PSMs in acetylated-BSA. Vertical lines denote median ion abundance ratios 0.2 and 2.4 for non-acetyl and acetyl PSMs, respectively. B) Violin plots of the [126]/[129] ion abundance ratio for all PSMs in the Jurkat E6–1 cell dataset of Svinkina et al.[33] Vertical lines denote the median ion abundance ratios of 0.4 and 1.7 for non-acetyl and acetyl PSMs, respectively.
Given the presence of 126 and 129 ions in both acetylated and non-acetylated PSMs, we considered whether their abundance ratio could yield an improved diagnostic for acetylation. The abundance of the 129 ion was compared to that of the 126 ion. Only 13.6% of non-acetylated PSMs had a 126 ion of greater abundance than the 129 ion, while 72.8% of acetylated PSMs contained 126 ions at abundances exceeding the 129 ions. Peak intensity ratios for acetylated and non-acetylated peptide spectra clearly differ, with acetyl PSMs having a [126]/[129] ratio greater than 1 (Figure 1A). In a large majority of cases where the [126]/[129] ratio is less than 1, the PSMs were identified as possessing an additional unmodified lysine residue (94%). Employing the [126]/[129] ratio to discriminate between non-acetylated and acetylated PSMs greatly increases the specificity as compared to using the 126 ion alone. Similar trends can be ascertained from the data published by Svinkina, et al.[33], with only 17.8% of non-acetylated PSMs and 71.8% of acetylated PSMs displaying the 126 ion at greater abundance than 129 (Figure 1B). This data suggests that the trends identified here are generalizable to large datasets and can increase the reliability for assigning spectra with lysine acetylation.
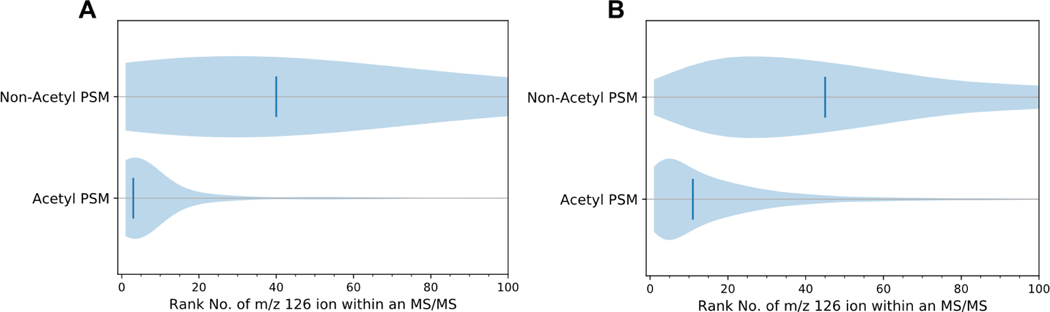
For acetylated peptide PSMs, the 126 ion tended to be more abundant than other ions in the MS2 spectrum. The ion abundances within each spectrum containing a 126 ion were ranked; approximately 80% of acetyl PSMs had m/z 126 as one of the 10 most abundant ions, as compared to 16% of non-acetyl PSMs (Figure 2A). The trend is also true in the re-mined Jurkat dataset (Figure 2B). [33]
Figure 2. Abundance rank m/z 126 in MS/MS spectra of non-acetylated and acetylated peptides.
A) Violin plots ranking the 126 ion abundance to all other ions in a spectrum for all acetyl-BSA PSMs. Vertical lines denote median rankings 40 and 3 for non-acetyl and acetyl peptides, respectively. B) Violin plots ranking the 126 ion abundance to all other ions present in a spectrum for all Jurkat E6-1 PSMs (dataset of Svinkina et al.[33]) Vertical lines denote median rankings 45 and 11 for non-acetylated and acetylated PSMs, respectively.
Butyrylated BSA was also analyzed in order to determine if a similar trend applied to other short-chain acylations. Data was limited, likely due to the low efficiency of butyrylation in aqueous solution. Nevertheless, the trends mirrored those for acetyl BSA. The presence of the m/z 126 analogue for butyryl-lysine (m/z 154.1232) in a spectrum corresponded to a butyrylated PSM 87.1% of the time (Table S2, Supplemental Figure S2), while 96.4% of butyrylated peptide spectra presented a 154 ion, indicating that it is both specific and sensitive.
3.3. Sequence Dependence of Immonium Ion Formation
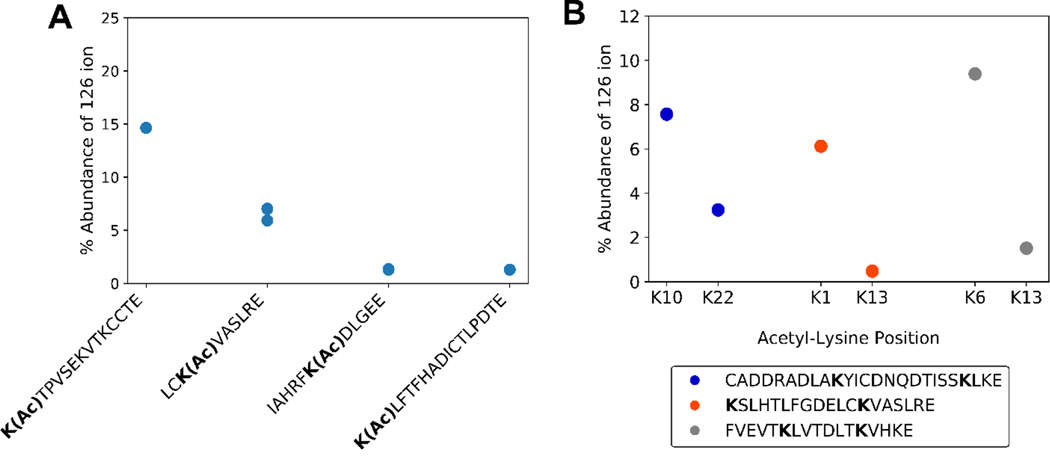
Efficient formation of immonium ions can depend on a modified residue’s location in a particular peptide sequence. Sequence-specific fragmentation is a well elucidated phenomenon, as certain local residues can affect CID fragmentation; for example, when a proline or glycine residue is near.[34–38] Some examples of this sequence dependence are shown in Figure 3. Generally an acetyl-lysine at the N-terminal position will yield the strongest signal for the 126 ion[24] (Figure 3A). This observation can be rationalized by recognizing that the first step in forming immonium ions requires an N-terminal amine. Hence, an internal lysine would require two fragmentation events, while an N-terminal lysine requires only one. Sequence composition is also important. For example, two different peptides with acetylation at the K1 position yield very different relative abundances for the 126 ions (Figure 3A). Multiple lysine peptides that are acetylated closer to the N-terminus tend to yield more abundant 126 ions (Figure 3B). Thus, there can be a localization bias when using the cyclized immonium ion to identify and/or validate acyl modifications.
Figure 3. Dependence of 126 ion abundance on sequence context within fragmentation spectra.
A) 126 ion abundance versus total product ion abundance for acetyl-BSA peptides. B) 126 ion abundance versus total product ion abundance for isobaric acetyl-BSA peptides. A single PSM of the same precursor charge state was selected for each acetyl-lysine position.
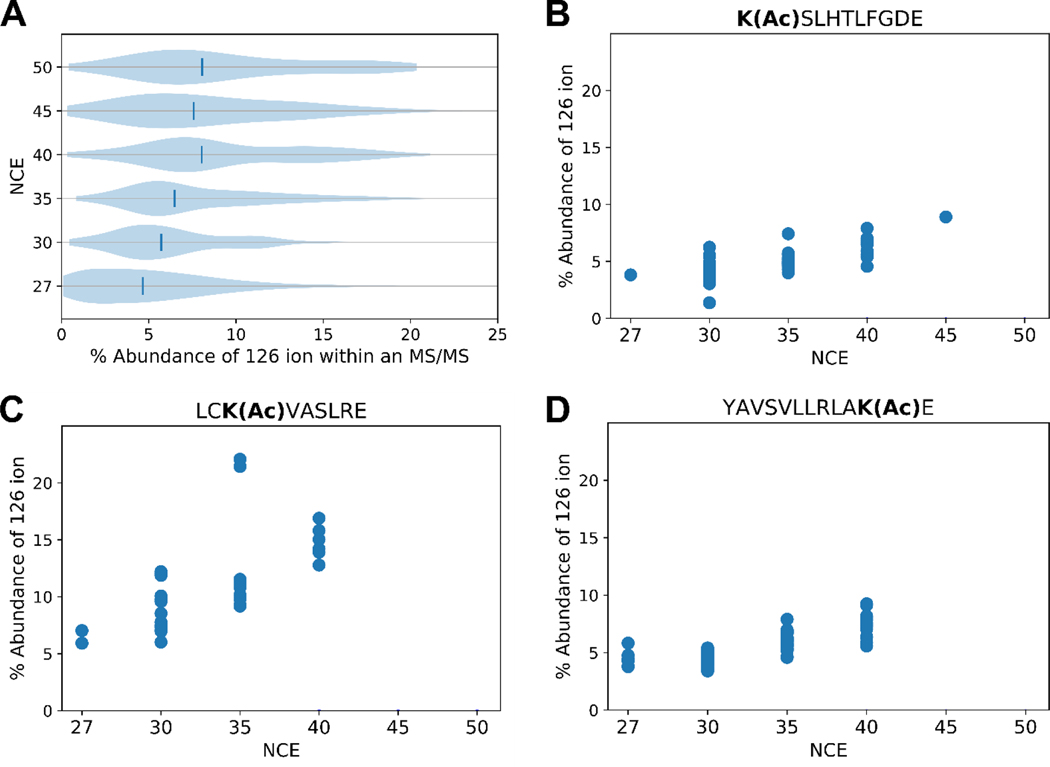
Normalized collision energy (NCE) was increased in an attempt to maximize the 126 ion’s signal. Global analysis with this optimization indicates, indeed, that the elevated NCE increases the relative abundance of the 126 ion from the acetylated BSA sample (Figure 4A). As per the violin plots, the 126 ion relative abundance increases with increasing NCE. Tandem mass spectra for individual peptides were examined to ensure that this trend applied to modifications at varied sequence locations. Figure 4B-D shows examples for different modified sequences. All of these peptides show that increased collisional energy increases the relative abundance of the immonium ion, congruent with the global analysis of Figure 4A.
Figure 4. Abundance of the 126 ion versus total product ion abundance in MS2 spectra of acetyl PSMs.
A) Each violin plot shows data obtained at a given NCE that reveals for each acetyl-PSM the percentage of total ion signal due to m/z 126. Vertical lines indicate the median relative abundances of the 126 ion. B-D) Percentage of total signal due to the 126 ion versus NCE for acetylated peptides selected for their varied acetyl-lysine position and sequence context. Some peptides were not identified at higher NCEs.
3.4. Stepped Collisional Activation Mitigates a Loss of Sequence Identifications
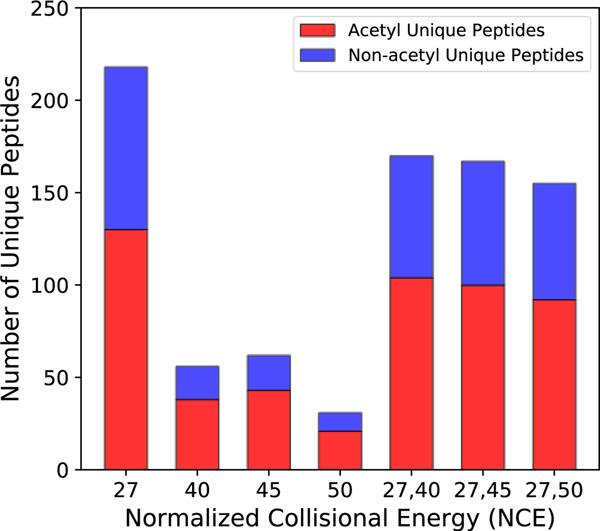
Increasing collision energy increases the intensities of low mass peaks, such as immonium ions, but in doing so, information critical to identifying the peptide sequence may be lost. At an NCE of 27, typical for most proteomic CID fragmentation, over 200 unique peptides could be identified from acetylated-BSA LC-MS/MS runs. At 30 NCE, the number of identified peptides dropped moderately to just below 100. However, at NCE 35 and 40, the number falls dramatically to just over 20 (Figure 5). While higher collision energies highlight the 126 ion in MS2 spectra, the benefit comes at the cost of poor sequence information, making it less likely that peptides can be matched to the correct sequence confidently.
Figure 5. Unique peptides identified with different values of fixed and stepped NCE.
Acetylated (red) and non-acetylated (blue) peptides.
To recover this information, a stepped collision energy (CE) approach was taken to enhance the intensity of the immonium ion peaks while retaining enough high mass information to identify the peptide sequence. In the stepped CE strategy, precursor ions are fragmented at low and high NCE, and the product ions from each CE are pooled for detection. Figure 5 shows how stepped CE recovers the sequence information lost by using excessive collision energy. Several stepped collision energy combinations (NCEs of 27/40, 27/45, and 27/50) were tested to optimally balance maximal signal for the 126 ion with retention of sequence-related information. A stepped NCE method of 27/40 was found to be optimum, recovering 80% of unique peptides relative to the single energy method at 27 NCE (Figure 5). There was still some loss of peptides identified due to the complexity of the spectra produced by the stepped method, as the higher collisional energy results in greater neutral loss and low mass product ions. Applying this approach, we rescued the number of peptides identified while maintaining a strong immonium ion, such that 95.4% of acetylated PSMs presented a 126 ion within the 10 most abundant ions (compared to 80% from 27 NCE). Butyrylation (m/z 154.1232) showed similar trends with respect to stepped NCE (Supplementary Figure S3).
3.5. Immonium Ions Identify Acyl Modifications in a Complex Lysate Using a Stepped Collision Energy
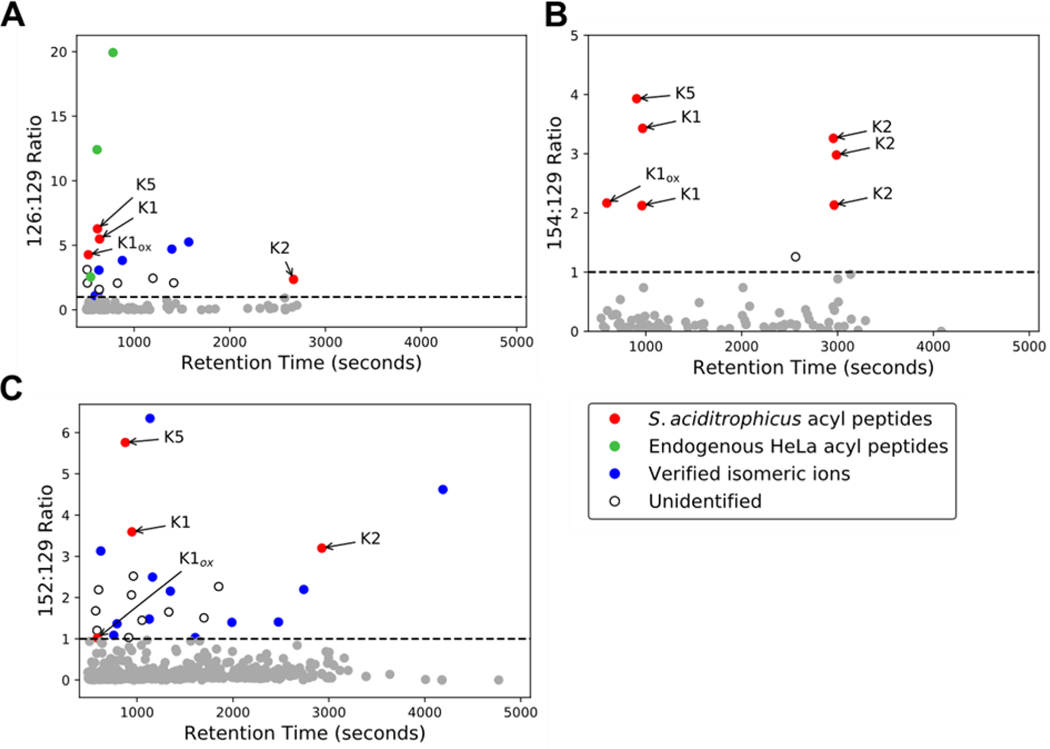
A question that remained with the stepped collision energy method was whether it would readily reveal immonium ions for acetyl-lysine and other acyl-lysines (e.g., butyryl- and crotonyl-lysine [m/z 152.107]) in a complex biological sample. To investigate this question, synthetic acylated peptides were spiked into a trypsinized HeLa lysate from which the respective immonium ion: m/z 129 abundance ratios were monitored. The spiked peptides (KSTPEAMAK, FKDEIPVVIK, STDPKGPSVR) differ in the type and location of acylation. In additional to acetyl-lysine, we chose butyryl- and crotonyl-lysine because they are relevant to Syntrophus aciditrophicus, a syntrophic bacterium for which we expect extensive protein acylation due to high cellular concentrations acyl-CoA metabolites.[17] Our lab identified these sequences as highly modified in the course of S. aciditrophicus proteomic studies.[39] Plotted are the cyclized immonium: 129 ion relative abundance ratios for each MS2 spectrum (Figure 6). Acylated peptides are clearly identified in the population of PSMs with [immonium]/[129] ratios exceeding 1. Isomers of the 126 [24] and 152 ions are also present (Supplemental Figure S4) and some peptides containing these sequences were identified (Figure 6). The quality of some MS2 spectra with abundance ratios exceeding 1 was insufficient to ascribe a sequence. These instances are labeled as unidentified in Figure 6.
Figure 6. Immonium ion abundance ratio reveals low abundance synthetic acyl-peptides in a HeLa lysate background.
Scatterplots of the immonium ion abundance ratio versus retention time for A) lysine acetylation ([126]/[129]), B) lysine butyrylation ([154/[129]], and C) lysine crotonylation ([152]/[129]), respectively. Grey dots represent spectra with an abundance ratio less than one. K1, K2, and K5 correspond to the synthetic S. aciditrophicus sequences KSTPEAMAK, FKDEIPVVIK, and STDPKGPSVR, respectively, where the bolded residues are acylated. Verified isomeric ions and unidentified ions are described in the text.
3.6. Using Immonium Ions to Identify the Presence of Novel PTMs
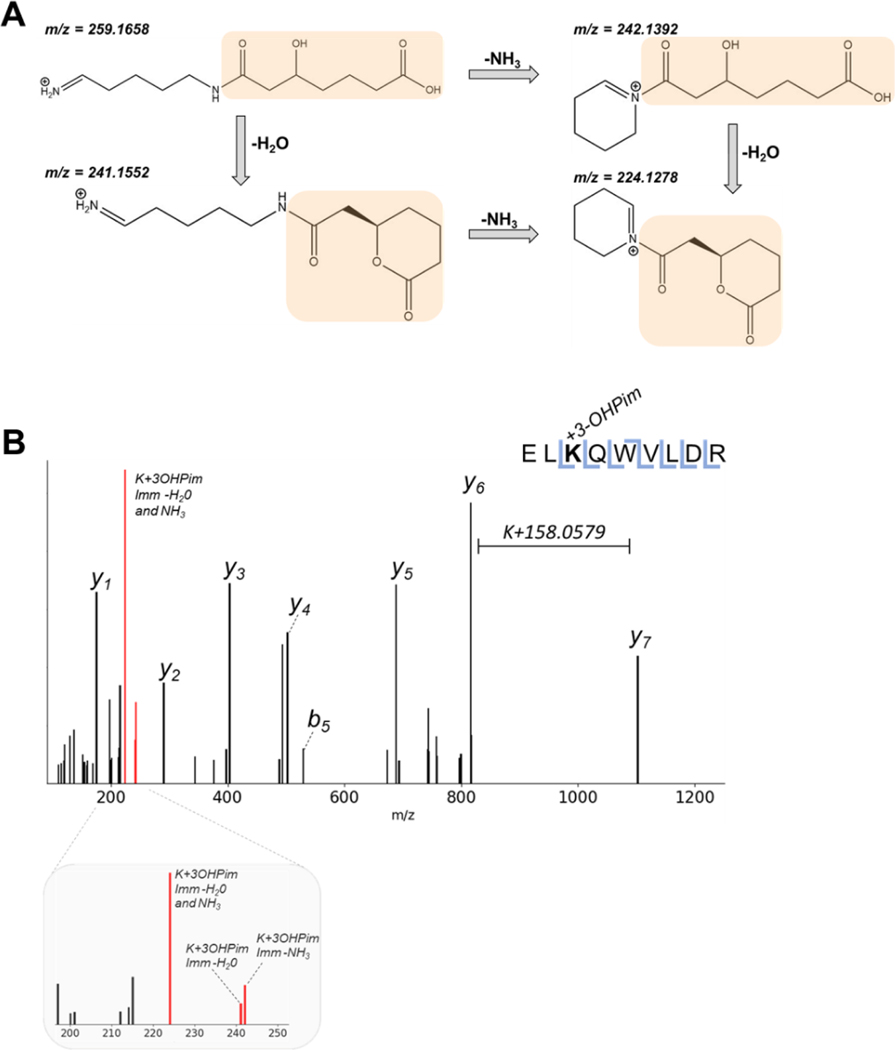
S. aciditrophicus has unique metabolic pathways that allow for the formation of novel protein PTMs. Given that its short chain and aromatic fatty acid metabolism generates a variety of reactive acyl-CoA metabolites, we expect the S. aciditrophicus proteome to display unique acyl-lysine modifications. These acyl-CoA intermediates can spontaneously acylate lysine side chain amines under physiological conditions.[40] One acyl-CoA intermediate within the benzoate degradation pathway is 3-hydroxypimelyl-CoA.[41] Interestingly, we noted that some mass spectra from the benzoate-cultivated S. aciditrophicus proteome presented mass shifts of K +158.0579 Da. To determine the validity of this novel modification, immonium-related ions were examined to verify that the shift was associated with a novel lysine PTM on a benzoate-CoA ligase peptide, rather than a misidentification.[42] Based on the immonium ion structure, one expects certain facile neutral losses (Figure 7) to be present, an NH3 neutral loss (similar to that of the acetyl-lysine 126 ion), an H2O loss, and the loss of both NH3 and H2O. Tandem mass spectra from two peptides containing the putative modification, benzoate-CoA ligase (RS03815/RS03820) and acetyl-CoA transferase (RS12490), presented ions corresponding to neutral loss of NH3, H2O, and NH3+H2O from the predicted immonium ion (Figure 7, Supplemental Figure S5); i.e., 242.1392, 241.1552, and 224.1278 Da, respectively. These immonium-related ions validated the presence of a hydroxypimelyl-lysine, the first observation of this PTM. Although only two 3-hydroxypimelyl-lysine peptides were identified in this analysis, we have found many other 3-hydroxylpimelyl modifications from other S. aciditrophicus sample preparations that will be addressed in future studies.
Figure 7. Novel modification (3-hydroxypimelylation) on benzoate-CoA ligase in S. aciditrophicus with its diagnostic ion.
A) Immonium ion for 3-hydroxypimelylation (K+158.0579 Da) (highlighted in orange) undergoes facile neutral losses. B) MS/MS spectrum showing mass shift corresponding to 3-hydroxypimelylation and its low mass region enlarged to highlight immonium-related ions (colored in red).
4. Discussion
Immonium and immonium-related ions are excellent proxies for the presence of acetylation.[24,43] Despite their long use in global analyses, current instrumentation and methods have not been optimized for their use. While previous reports found that the specificity for acetylation of the m/z 126 ion was superior to that of m/z 143,[24] the former is often observed in mass spectra lacking any acetylation, as is confirmed in our analysis. Hence, the diagnostic ion’s presence alone is not sufficient to claim modification with high confidence, or to restrict data acquisition or analysis to only acetylated peptides. Incorporating abundance ratios provides a nuanced way to utilize this marker. The [126]/[129] ratio is meaningful when identifying candidate PSMs that contain acetyl-lysine residues. Although some sequence contexts make it an imperfect criterion, ions likely containing acetylated lysine can be identified by the [126]/[129] ratio in MS2 spectra prior to database searching. This can help to narrow candidate spectra prior to sequence assignment or help to identify modified targets in the case of limited sequence information. This criterion can be also used in conjunction with other evidence for more rigorous identification. Likewise, incorporating a stepped collisional energy CID experiment may increase confidence in acyl-lysine assignments. Akin to oxonium ion analysis, using a stepped NCE strategy highlights the diagnostic ions of acyl-lysine while also optimizing sequence-related information in a data-dependent acquisition (DDA) method. Stepped NCE is particularly important for modifications present in the middle of a peptide, because generating these immonium ions require more collision energy.
While using immonium ions is appropriate for CID, transferring that use to other activation methods requires consideration of the mechanisms by which those methods fragment peptides. Another common fragmentation approach in the analysis of peptides and PTMs is electron transfer dissociation (ETD),[44–46] and interest in ultraviolet (UV) photodissociation (PD) is growing as well.[47] CID fragmentation yields many products, including the neutral losses that form immonium ions. ETD mostly cleaves along the Cα-N bond of the peptide backbone,[44] thereby making immonium ion formation unlikely. However, the production of immonium ions by ETD and PD has not been widely explored to date and requires further examination.
Future applications of stepped NCE with immonium ion abundance analyses can take two possible routes. One is to expose and verify novel PTMs. As the list of putative acyl-modifications grows,[4,48,49] it becomes critical to obtain additional constraints (metrics) to validate the presence of increasingly complex modifications. Many acyl-modified peptides can be isobaric with di- or tri-peptide related ions or be indistinguishable from non-acylated ions on low resolution instruments.[42] For example, acetyl and propionyl modifications differ by a single methyl group, as do propionyl and butyryl modifications. The presence of a methylated residue or an amino acid that differs from another by a methyl group (Ser/Thr, Asn/Gln, Ala/Val, etc) may make it impossible to distinguish the correct acyl modification.[50] Similarly, carbamylation has a 43.00582 Da mass shift, whereas acetylation combined with deamidation produce a 42.99458 Da shift. Depending on the mass of the ion and the resolving power of the instrument, these PTMs could be indistinguishable in the event of poor fragmentation and unincorporated diagnostic ions.
Stepped NCE can also be employed in conjunction with data-independent acquisition (DIA) approaches. Currently, DIA is greatly limited by the complexity of spectral deconvolution, particularly by fragment ion interference when co-eluting peptides have very similar masses.[51] Diagnostic ions, however, can increase the confidence that an acyl-lysine precursor is present in the convoluted spectra; thereby providing more information for elegant algorithms to consider when identifying and assigning PTMs. Furthermore, experiments targeting acyl modifications can exploit immonium and immonium-related ions in parallel reaction monitoring (PRM) or product ion scanning.
Increased understanding of acyl modifications has suggested some biological functions for these modifications.[52] Given the reactive nature of acyl-CoA species[17] and the new depths proteomics can reach, it should be expected that more acyl modifications will be found and may show biological significance in metabolic pathways across organisms. As datasets grow in size and complexity, confident assignments are essential. Properly incorporating immonium ions into the assignment of acyl-modifications will increase confidence in established PTM identifications and support/validate assignment of those yet to be discovered.
Supplementary Material
Statement of Significance.
Acyl-lysine modifications come in a variety of elemental compositions. There is increasing evidence that these modifications can have a functional effect on proteins and are present in species across all domains of life. Here we describe a new method that can allow for more confident identification of acyl modifications in proteomes by utilizing their immonium ions. We also report the first observation of a 3-hydroxypimelyl lysine.
Acknowledgements
Funding from the Department of Energy Office of Science (BER) contract DE-FC-02-02ER63421 (to J.A.L.; UCLA/DOE Institute for Genomics and Proteomics), NIH Ruth L. Kirschstein National Research Service Award GM007185, and NSF Graduate Research Fellowship (to J.Y.F.; DGE-1650604) is acknowledged. J.M.M. was supported by a UCLA Molecular Biology Institute Whitcome Fellowship. We would also like to thank Drs. Michael J. McInerny (University of Oklahoma) and Robert P. Gunsalus (UCLA) for the S. aciditrophicus samples used in the study.
Footnotes
Conflict of Interest Statement
The authors declare no conflict of interest.
References
- [1].Walsh G, Jefferis R, Nat. Biotechnol 2006, 24, 1241. [DOI] [PubMed] [Google Scholar]
- [2].Moremen KW, Tiemeyer M, Nairn AV, Nat. Rev. Mol. Cell Biol 2012, 13, 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Guccione E, Richard S, Nat. Rev. Mol. Cell Biol 2019, 20, 642. [DOI] [PubMed] [Google Scholar]
- [4].Sabari BR, Zhang D, Allis CD, Zhao Y, Nat. Rev. Mol. Cell Biol 2017, 18, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Humphrey SJ, James DE, Mann M, Tr. Endocrinol. Metab 2015, 26, 676. [DOI] [PubMed] [Google Scholar]
- [6].VanDrisse CM, Escalante-Semerena JC, Annu. Rev. Microbiol 2019, 73, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sack MCN, Finkel T, Cold Spring Harb. Perspect. Biol 2012, 4, DOI 10.1101/cshperspect.a013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Carrico C, Meyer JG, He W, Gibson BW, Verdin E, Cell Metab. 2018, 27, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Verdin E, Ott M, Nat. Rev. Mol. Cell Biol 2015, 16, 258. [DOI] [PubMed] [Google Scholar]
- [10].Turner BM, BioEssays 2000, 22, 836. [DOI] [PubMed] [Google Scholar]
- [11].Turner BM, Cell. Mol. Life Sci 1998, 54, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Crosby HA, Heiniger EK, Harwood CS, Escalante-Semerena JC, Mol. Microbiol 2010, 76, 874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gardner JG, Grundy FJ, Henkin TM, Escalante-Semerena JC, J. Bacteriol 2006, 188, 5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ott M, Schnölzer M, Garnica J, Fischle W, Emiliani S, Rackwitz HR, Verdin E, Curr. Biol 1999, 9, 1489. [DOI] [PubMed] [Google Scholar]
- [15].Hansen BK, Gupta R, Baldus L, Lyon D, Narita T, Lammers M, Choudhary C, Weinert BT, Nat. Commun 2019, 10, DOI 10.1038/s41467-019-09024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li Y, Silva JC, Skinner ME, Lombard DB, Methods Mol. Biol 2013, 1077, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Trub AG, Hirschey MD, Trends Biochem. Sci 2018, 43, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Boersema PJ, Mohammed S, Heck AJR, J. Mass Spectrom 2009, 44, 861. [DOI] [PubMed] [Google Scholar]
- [19].Sweet SMM, Bailey CM, Cunningham DL, Heath JK, Cooper HJ, Mol. Cell. Proteomics 2009, 8, 904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cao Q, Zhao X, Zhao Q, Lv X, Ma C, Li X, Zhao Y, Peng B, Ying W, Qian X, Anal. Chem 2014, 86, 6804. [DOI] [PubMed] [Google Scholar]
- [21].Halim A, Westerlind U, Pett C, Schorlemer M, Rüetschi U, Brinkmalm G, Sihlbom C, Lengqvist J, Larson G, Nilsson J, J. Proteome Res 2014, 13, 6024. [DOI] [PubMed] [Google Scholar]
- [22].Carr SA, Huddleston MJ, Bean MF, Protein Sci. 1993, 2, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zaia J, Mass Spectrom. Rev 2004, 23, 161. [DOI] [PubMed] [Google Scholar]
- [24].Trelle MB, Jensen ON, Anal. Chem 2008, 80, 3422. [DOI] [PubMed] [Google Scholar]
- [25].Zolg DP, Wilhelm M, Schmidt T, Médard G, Zerweck J, Knaute T, Wenschuh H, Reimer U, Schnatbaum K, Kuster B, Mol. Cell. Proteomics 2018, 17, 1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Falick AM, Hines WM, Medzihradszky KF, Baldwin MA, Gibson BW, J. Am. Soc. Mass Spectrom 1993, 4, 882. [DOI] [PubMed] [Google Scholar]
- [27].Hung CW, Schlosser A, Wei J, Lehmann WD, Anal. Bioanal. Chem 2007, 389, 1003. [DOI] [PubMed] [Google Scholar]
- [28].Yalcin T, Harrison AG, J. Mass Spectrom 1996, 31, 1237. [DOI] [PubMed] [Google Scholar]
- [29].Baeza J, Smallegan MJ, Denu JM, ACS Chem. Biol 2015, 10, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rappsilber J, Mann M, Ishihama Y, Nat. Protoc 2007, 2, 1896. [DOI] [PubMed] [Google Scholar]
- [31].Erde J, Loo RRO, Loo JA, in Methods Mol. Biol, vol. 1550, Humana Press Inc., 2017, pp. 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Erde J, Loo RRO, Loo JA, J. Proteome Res 2014, 13, 1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Svinkina T, Gu H, Silva JC, Mertins P, Qiao J, Fereshetian S, Jaffe JD, Kuhn E, Udeshi ND, Carr SA, Mol. Cell. Proteomics 2015, 14, 2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].van Dongen WD, Ruijters HFM, Luinge H-J, Heerma W, Haverkamp J, J. Mass Spectrom 1996, 31, 1156. [DOI] [PubMed] [Google Scholar]
- [35].Kapp EA, Schütz F, Reid GE, Eddes JS, Moritz RL, O’Hair RAJ, Speed TP, Simpson RJ, Anal. Chem 2003, 75, 6251. [DOI] [PubMed] [Google Scholar]
- [36].Loo JA, Edmonds CG, Smith RD, Anal. Chem 1993, 65, 425. [DOI] [PubMed] [Google Scholar]
- [37].Breci LA, Tabb DL, Yates JR, Wysocki VH, Anal. Chem 2003, 75, 1963. [DOI] [PubMed] [Google Scholar]
- [38].Huang Y, Triscari JM, Tseng GC, Pasa-Tolic L, Lipton MS, Smith RD, Wysocki VH, Anal. Chem 2005, 77, 5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Nguyen H, McInerney M, Gunsalus R, Loo JA, Loo RRO, in Present. 64th ASMS Conf. Mass Spectrom. Allied Top., 2016. [Google Scholar]
- [40].Wagner GR, Payne RM, J. Biol. Chem 2013, 288, 29036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Elshahed MS, Bhupathiraju VK, Wofford NQ, Nanny MA, McInerney MJ, Appl. Environ. Microbiol 2001, 67, 1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kim MS, Zhong J, Pandey A, Proteomics 2016, 16, 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nakayasu ES, Wu S, Sydor MA, Shukla AK, Weitz KK, Moore RJ, Hixson KK, Kim J-S, Petyuk VA, Monroe ME, Pasa-Tolic L, Qian W-J, Smith RD, Adkins JN, Ansong C, Int. J. Proteomics 2014, 2014, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF, Proc. Natl. Acad. Sci. U. S. A 2004, 101, 9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JEP, Shabanowitz J, Hunt DF, Biochim. Biophys. Acta - Proteins Proteomics 2006, 1764, 1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JEP, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF, Proc. Natl. Acad. Sci. U. S. A 2007, 104, 2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Halim MA, MacAleese L, Lemoine J, Antoine R, Dugourd P, Girod M, J. Am. Soc. Mass Spectrom 2018, 29, 270. [DOI] [PubMed] [Google Scholar]
- [48].Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, Liu W, Kim S, Lee S, Perez-Neut M, Ding J, Czyz D, Hu R, Ye Z, He M, Zheng YG, Shuman HA, Dai L, Ren B, Roeder RG, Becker L, Zhao Y, Nature 2019, 574, 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Huang H, Zhang D, Wang Y, Perez-Neut M, Han Z, Zheng YG, Hao Q, Zhao Y, Nat. Commun 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lee S, Tan M, Dai L, Kwon OK, Yang JS, Zhao Y, Chen Y, J. Proteome Res 2013, 12, 1007. [DOI] [PubMed] [Google Scholar]
- [51].Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R, Mol. Cell. Proteomics 2012, 11, O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Xiong Y, Guan KL, J. Cell Biol 2012, 198, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.