Abstract
Peptide ligands presented by human leukocyte antigen (HLA) molecules on the cell surface represent the immunopeptidome that could be utilized for identification of antigenic peptides for immunotherapy and prevention of autoimmune diseases. Although T-cells are well-known key players in the destruction of pancreatic beta-cells in type 1 diabetes (T1D), increasing evidence points toward a role for B-cells in disease pathogenesis. However, as antigen presenting cells, little is known about the comprehensive immunopeptidome of B cells and their changes in the context of T1D. We performed HLA allele-specific quantitative immunopeptidomics using B lymphocytes derived from T1D patients and healthy controls. Hundreds of HLA-I and HLA-II immunopeptides were identified as differentially regulated in T1D per HLA allele for B cells sharing identical HLA alleles. The results were further validated using additional T1D and healthy B cells with partially overlapped HLA alleles. Differentially expressed immunopeptides were confirmed with targeted proteomics and for reactivity using known T-cell assays in the immune epitope database. Considering samples with identical HLA alleles are difficult to obtain for T1D and other similar HLA-restricted diseases, our work represents a viable approach to better understand HLA allele-specific antigen presentation and may facilitate identification of immunopeptides for therapeutic applications in autoimmune diseases. Data are available via ProteomeXchange with identifier PXD026184.
Keywords: Immunopeptidome, HLA ligandome, Type 1 Diabetes, B lymphocytes, HLA-I, HLA-II, HLA allele
Graphical Abstract
INTRODUCTION
The immune system plays critical roles in self-recognition and host defense through an intricate network of biological processes, and aberrant immune regulations are involved in a myriad of diseases, ranging from autoimmune diseases to cancers. Immunopeptidomics, the investigation of peptide ligands presented on the human leucocyte antigen (HLA) molecules on cell surfaces, has attracted a lot of attention in recent years owing to the clinical successes in cancer immunotherapy.1, 2 Together with the improvements in sample preparation methods for enriching HLA bound peptides and development of better algorithms for HLA peptide binding affinity prediction,3–5 advancements in high resolution mass spectrometry techniques for unbiased identification of peptide epitopes fueled the wide applications of immunopeptidomics to identify antigenic peptides for immunotherapy and as vaccines in prevention of cancer, autoimmune and infectious diseases.
HLA class I molecules are expressed nearly on all nucleated cells while HLA class II molecules are only found on antigen presenting cells such as B lymphocytes, macrophages, and dendritic cells, among others. Past immunopeptidomics works centered on immortal human cancer cell lines or human B cell lines transformed with Epstein-Barr virus (EBV). Using these model systems, the immunopeptide repertoire has been expanded, and a better understanding of the immune regulation is gained through the observation of differential HLA peptide presentation upon in vitro stimulation with immune modulating agents.3, 6, 7 Recently, with more streamlined sample preparation and more sensitive mass spectrometry-based proteomics platforms,3, 8 successful applications of immunopeptidomics have been achieved by direct analysis of clinically collected tumor tissues or T cells isolated from these tissues.3, 9–11 To improve prediction of allele-specific endogenous immunopeptides covering most of the human population, ground-breaking work was accomplished recently to broadly expand the HLA class I epitope database by using 95 HLA-I monoallelic cell lines.12
As an autoimmune disease, Type 1 Diabetes (T1D) is characterized by T cell mediated self-destruction of pancreatic beta cells and resulted insulin deficiency.13, 14 Genome-wide association studies have identified that HLA genes conferred the strongest risk, particularly HLA DR3/DR4 in populations of European descent.15 However, genetic variation among different racial and ethnic groups diminishes the predicting value of these genetic markers, and genetic predisposition cannot explain why T1D would selectively occur in individuals of the same families or more strikingly only in one of the homozygotic twins.16 The environmental factors that initiate and drive the disease progression still remain elusive.17 Recently, hybrid insulin peptides and peptides produced from translational errors of insulin gene are discovered, and diabetogenic T cells recognizing these neoantigens are capable of killing human pancreatic beta cells.18–20 Although T cells are well-known key players in the process of pancreatic beta-cell destruction in T1D, increasing evidence points toward a role for B cells in disease pathogenesis.21, 22 In this respect, B cells have been established as critical antigen-presenting cells to diabetogenic T cells in T1D. 23, 24 In addition, anti-insulin B cells are known to express higher surface levels of antigen presenting MHC-II molecules in nonobese diabetic (NOD) mice.25 Recent studies also highlight that HLA immunopeptides presented by B cells have the ability to trigger the T-cell-mediated beta-cell-specific autoimmunity.26, 27 And importantly, depletion of B cells using anti-CD20 antibody can prevent or delay the development of T1D in NOD mice and in humans.28–30
Therefore, to get a better understanding of the pathogenic mechanism of T1D, it is imperative to comprehensively profile the repertoire of the immunopeptides related to this disease and investigate the differences in peptide presentation between T1D and healthy individuals sharing the same HLA alleles. To this end, we selected T1D and healthy B cell lines derived from the same family that have exactly matching HLA-I and HLA-II alleles. Through sequential enrichment of HLA class I and II immunopeptides and LC-MS/MS based proteomics analyses, we quantitatively compared the changes of immunopeptidome in an allele-specific manner. The HLA allele-specific-immunopeptide changes were further verified using T1D and healthy B cells originating from different families with partially overlapped HLA alleles. Some HLA-II immunopeptides identified are associated with the immunoregulatory functions which might have potential roles in T1D pathogenesis.
EXPERIMENTAL PROCEDURES
Cell Culture and HLA-I and HLA-II Antibody Production
The Epstein-Barr Virus-transformed B-Lymphocytes (GM02675, GM02676, GM02678, GM02679, GM02812, GM02765, GM02741 and GM03027) were purchased from Coriell Institute for Medical Research (CIMR, Camden, New Jersey). The clinical data such as age, gender, and race were included in supplemental Table 1. Cells were maintained in RPMI-1640 Medium (ATCC® 30–2001™) supplemented with fetal bovine serum (FBS) (Thermo Scientific, 16000044) as suggested by CIMR. When cells were grown to the required amount, we collected them by centrifugation at 200 g for 10 min, washed twice with ice cold PBS (Gibco, 10010023), and stored as dry cell pellets at −20 °C until use. The W6/32 (HLA-I) and IVA12 (HLA-II) monoclonal antibodies were produced from HB95 (ATCC® HB-95™) and HB145 (ATCC® HB-145™) cells that were purchased from American Type Culture Collection (ATCC) and grown in DMEM (ATCC® 30–2002™) medium supplemented with FBS to a final concentration of 10%. The W6/32 and IVA12 monoclonal antibodies were purified from the growth medium using protein A-Sepharose 4B conjugate (Invitrogen, 101042). The purified antibodies were stored at −20 °C for future use.
DNA Extraction and HLA Allele Typing
DNA was extracted from B lymphocyte cell lines using the QIAamp DNA Mini Kit (Qiagen, 51304). The extracted DNA samples were genotyped at the high-resolution HLA typing facility of the Children’s Hospital Oakland Research Institute (Oakland, CA, USA) and at the HLA core facility of the Barbara Davis Center at the University of Colorado Anschutz Medical Campus. All the cells contain 4 digital data for 6 loci: A, B, C, DRB, DPA/B, and DQA/B as shown in the supplemental Table 1.
Generation of Antibody-Cross-linked Protein A-Sepharose Conjugate Beads
Following a previous cross-linking approach,31 purified HLA-I and HLA-II antibodies were separately incubated with protein A-sepharose beads at a ratio of 5 mg of antibodies per 1 mL volume of slurry for 2 h at 4 °C. Chemical cross-linking was performed by addition of 20 mM dimethyl pimelimidate dihydrochloride (DMP, Thermo Scientific, 21667) in 0.05 M borate buffer, pH 9 (Thermo Scientific, 28384) for 30 min at 4 room temperature. The reaction was quenched by incubation with 0.2 M ethanolamine pH 8 (Sigma-Aldrich, 411000) for 2 h at room temperature. Antibody-cross-linked protein A beads were stored at 4 °C until use.
Purification of HLA-I and HLA-II Complexes and Immunopeptides
HLA immunopeptides were purified as described below following an established protocol.31 Briefly, cell lysis was performed with PBS containing 0.25% sodium deoxycholate (Sigma-Aldrich, 30970), 1 mM EDTA (Invitrogen, AM9260G), 1:200 Protease Inhibitors Mixture (Sigma-Aldrich, P8340), 1 mM Phenylmethylsulfonylfluoride (Sigma-Aldrich, P7626), and 1% octyl-beta-D glucopyranoside (Sigma-Aldrich, O8001) at a concentration of 1 × 108 cells/mL at 4 °C for 1 h. Lysates were cleared by centrifugation of 14,000 rpm at 4 °C for 30 min. For sequential HLA-I and -II complexes purification, lysates were first added to the HLA-I column containing W6/32 antibody covalently bound to protein-A Sepharose beads and incubated for 1 h at room temperature. Subsequently, flow-through fractions were collected by gravity flow through the HLA-I column. Collected fractions were then loaded into the HLA-II purification column containing IVA12 antibody cross-linked protein-A Sepharose beads. To remove the contaminates, the HLA-I and -II columns were washed as follows: 10 mL of 150 mM NaCl (Fisher Scientific, S271–500), 20 mM Tris–HCl (Invitrogen, 15568025), 10 mL of 400 mM NaCl, 20 mM Tris–HCl, and finally with 10 mL of 20 mM Tris–HCl. HLA molecule-immunopeptide complexes were eluted from the antibodies by adding 1 mL of 0.1 M acetic acid (Sigma, A6283). To further separate the immunopeptides from HLA molecules, elutes were further purified by Sep-Pak tC18 96-well plate (Waters, 186002319). Wells were prewashed first with 1 mL methanol and then twice with 1 mL 0.1% TFA. Samples were loaded onto the wells and washed twice with 1 mL 0.1% TFA. Thereafter, we eluted the HLA peptides with 200 μL of 30% ACN in 0.1% TFA. Both HLA-I and -II peptide elutes were dried by vacuum centrifugation and the dried peptides were stored at −20 °C until MS analysis.
LC-MS/MS Analysis
Dried HLA-I and HLA-II peptide samples were re-suspended in 20 μL of 0.1% FA before MS analysis. For each peptide sample 10 μL was injected and separated using the nanoflow UHPLC Ultimat3000 system (Thermo Fisher Scientific). The chromatographic separation of peptides was performed on a 50 cm × 75 μm column packed with 3 μm C18 particles (Thermo Fisher Scientific, ES903). For HLA-I peptides, elution of peptides used the following gradient at a flow rate of 250 nL per min using a mix of 0.1% FA (buffer A) and 0.1% FA in 100% ACN (buffer B): 0–5 min (4% B); 5–85 min (5–28% B); 85–100 min (28–48% B); 100–100.1 min (48–70% B); 100.1–110 min (70% B); 110–110.01 min (70–4% B) and 110.01–130 min (4% B). The gradient for HLA-II peptides: 0–3 min (4% B); 3–46 min (4–20% B); 46–95 min (20–50% B); 95–95.01 min (50–70% B); 95.01–105 min (70% B), 105–105.01 min (70–4% B) and 105.01–125 min (4% B). Eluted peptides were ionized by nano-electrospray and analyzed using a QExactive HF mass spectrometer (Thermo Fischer Scientific). The data were acquired with a data-dependent “top15” method with a 1.4 m/z precursor isolation window. The fifteen most abundant precursor ions were fragmented by higher-energy collision dissociation (HCD) at normalized collision energy of 27. The mass spectrometer scan range was set to 300 to 1650 m/z with a resolution of 60,000 and an AGC target value of 3 × 106 ions. For MS/MS, AGC target values of 1 × 105 were used with a maximum injection time of 120 ms at mass resolution of 15,000. No fragmentation was performed: 1) for HLA-I peptides, precursor ion charge states of five and above; and 2) for HLA-II peptides, assigned precursor ion charge states of one, and from six and above. The peptide match option was disabled. The dynamic exclusion of precursor ions from further selection was set for 20 s.
Parallel Reaction Monitoring (PRM) Analysis
PRM analysis was performed using QExactive HF mass spectrometer equipped with EASY-nano LC 1000 (Thermo Scientific). Enriched immunopeptide samples spiked with stable isotope labeled peptides were loaded onto a trap column (Acclaim PepMap 100, 2 cm × 75 μm, Thermo fisher) and PepMap RSLC C18 50 cm × 75 μm analytical column (Thermo Fisher). Peptides were separated using the low pH mobile phases (A: 0.1% FA in water; B: 0.1% FA in ACN) with a flow rate of 300 nL per min and a linear gradient of 3–7% buffer B in 0–3 min, 30% in 103 min, 40% in 106 min, and an increase to 75% in 111 min with a hold for 6 min before returning to the initial condition of 3% buffer B. Peptide samples were analyzed using the PRM method based on scheduled inclusion lists of HLA-I and HLA-II peptides. The PRM scan events collected using an Orbitrap mass resolution of 30,000, an AGC value of 1 × 106, and maximum injection time of 250 ms with an isolation width of 1 m/z. Fragmentation was performed with a normalized collision energy of 28.
Proteomic Data Analysis
We employed MaxQuant32 (version 1.6.10.43) to search and sequence the peak lists against the UniProt database (Human 20412 entries, 2018). The enzyme specificity was set as unspecific. N-terminal acetylation and methionine oxidation were used as variable modifications. Immunopeptidomics analysis was performed applying a false discovery rate (FDR) of 0.01 for peptide identifications and no protein FDR. Possible sequence matches were restricted to 8 to 25 amino acids. We enabled the “match between runs” option, which allows matching of identifications across different replicates in a time window of 0.7 min and an initial alignment time window of 20 min. Other parameters for MaxQuant were used as default. PRM data was processed using Skyline33 (version 20.2.0.343).
Statistical Analysis
The peptide quantification results obtained after MaxQuant analysis were further processed by Perseus34 (version 1.5.3.1) for statistical analyses and visualization. Briefly, the peptides with reverse hits, potential contaminants and missing value were removed, then the peptide intensities were log2–transformed and width adjustment normalization was performed. The t-test with both sides and 250 randomizations was adapted for significance analysis with S0 of 0.1 and FDR cutoff 0.05 or 0.01.
Bioinformatic Analyses
Binding affinities of immunopeptides to HLA-I and HLA-II molecules were predicted according to NetMHCpan 4.1 and NetMHCIIpan 4.04, respectively. For each B cell line, we applied a threshold of top 2% HLA-I and top 10% HLA-II ranked immunopeptide sequences to annotate their binding affinities specific to the HLA alleles. Next, Gibbscluster-2.035 was employed with the default options for aligning and clustering of the immunopeptide sequences of each B cell line. The recommended default configurations were loaded by choosing HLA class I ligands of length 8–13 and HLA class II ligands to output the clusters of HLA-I and HLA-II immunopeptides, respectively. These default parameters output 1–5 clusters with a motif length of 9 amino acids. We selected 4 and 1 cluster(s) that showed highest KLD (Kullbatch Leibler Distance) score for HLA-I and HLA-II, respectively in case of 2675 (T1D) and 2676 (HC) B cells. For better comparisons with 2675 (T1D) and 2676 (HC), we chose the same number of cluster(s) regardless of KDL score in case of additional six B cells. The motifs plotted for each specific HLA alleles were then compared and verified with the naturally presented ligand motifs using Motif viewer, which is available on NetMHCpan 4.1 server for HLA-I and on NetMHCIIpan 4.0 server for HLA-II. For additional validation of HLA-II peptides, selected HLA-II peptides were also analyzed using the web version of MixMHC2pred5, %Rank_best score was recorded, which indicates the percent of random peptides that would have a score higher than the input peptide among peptides of sizes 12–25 amino acids, with best score about 0 and worst score 100.
Proteomaps and IEDB Analyses of Cellular Functions of HLA Antigens
Gene ontology analysis was performed using DAVID36. The quantitative composition of differentially regulated HLA-I and HLA-II antigens were constructed with Proteomaps37 with a focus on cellular protein functions based on the KEGG Pathways gene classification.10 The functionally related antigens were arranged in common regions and antigens with polygon structures represented building blocks of these regions with the size of polygon weighted by the number of immunopeptides identified for each antigen. On the other hand, immune-regulatory functions of candidate HLA-I and HLA-II antigens based on the T-cell assay or tetramer assay were in silico verified using the downloaded immune epitope database (IEDB)38 (IEDB epitope_full_v3 on 12.2020).
RESULTS
Enrichment of HLA Class I and II Immunopeptides of T1D and Healthy B Cells
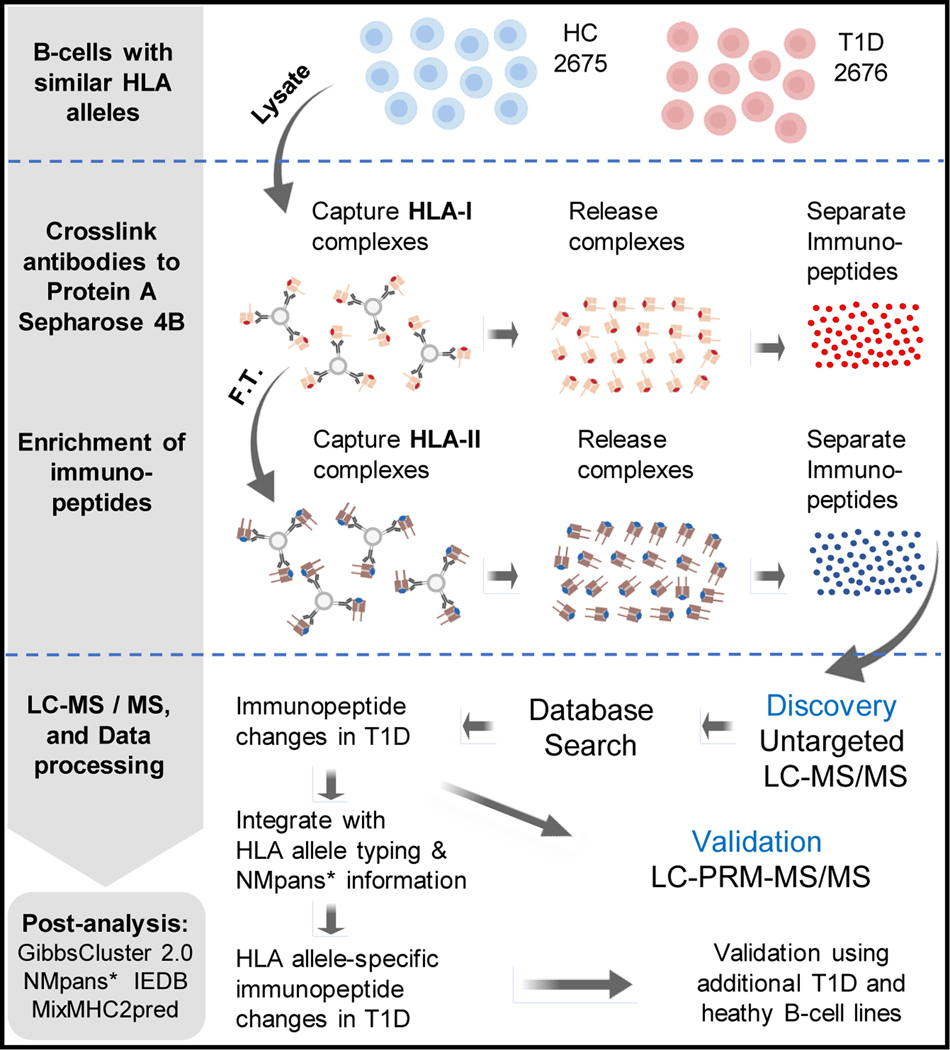
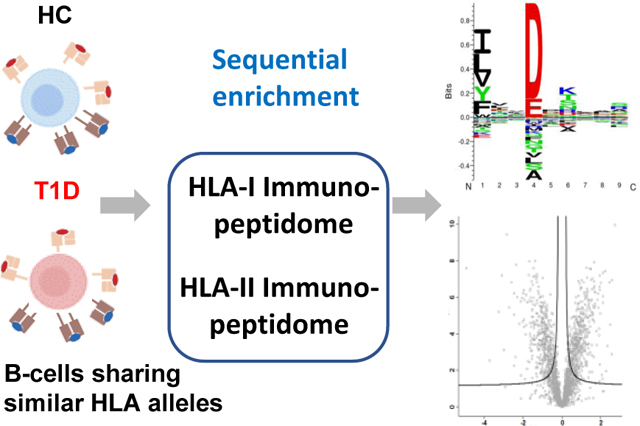
Here, we used the B lymphocytes derived from peripheral blood mononuclear cells (PBMCs) of T1D patients and healthy controls (HC) to enrich the HLA immunopeptides. Specifically, two B cell lines (2676 (T1D) and 2675 (HC)) derived from a T1D patient and a healthy person belonging to same family were selected. These two cell lines share identical four-digit HLA alleles for the genes, HLA-A, HLA-B, HLA-C, HRA-DP, HLA-DQ, and HLA-DRB1 as confirmed by the HLA allele typing analysis (supplemental Table 1). In parallel, we cultured HB-95 and HB-145 hybridoma cell lines and purified anti-HLA-I (W6/32) and anti-HLA-II (IVA12) antibodies, respectively. These HLA-I and HLA-II antibodies were cross-linked to Protein-A Sepharose 4B beads in separate columns. As depicted in Fig. 1, we followed the tandem enrichment procedure using cross-linked antibodies in which the HLA-I complexes were enriched from B cell lysates and subsequently used their flow-throughs to enrich the HLA-II complexes. Then, the HLA-I and HLA-II immunopeptides were separated from their corresponding HLA molecules using a C18 plate. The enriched immunopeptides were analyzed by LC-MS/MS for their identification, characterization, quantitation, and downstream analyses (Fig. 1) as described in next sections.
Fig. 1.
Schematic showing the basic steps of the HLA allele-specific quantitative immunopeptidomics.
*NMpans: NetMHCpan 4.1 and NetMHCIIpan 4.0; F.T.: flowthrough.
Identification and Characterization of HLA Class I Immunopeptides of T1D and Healthy B Cells
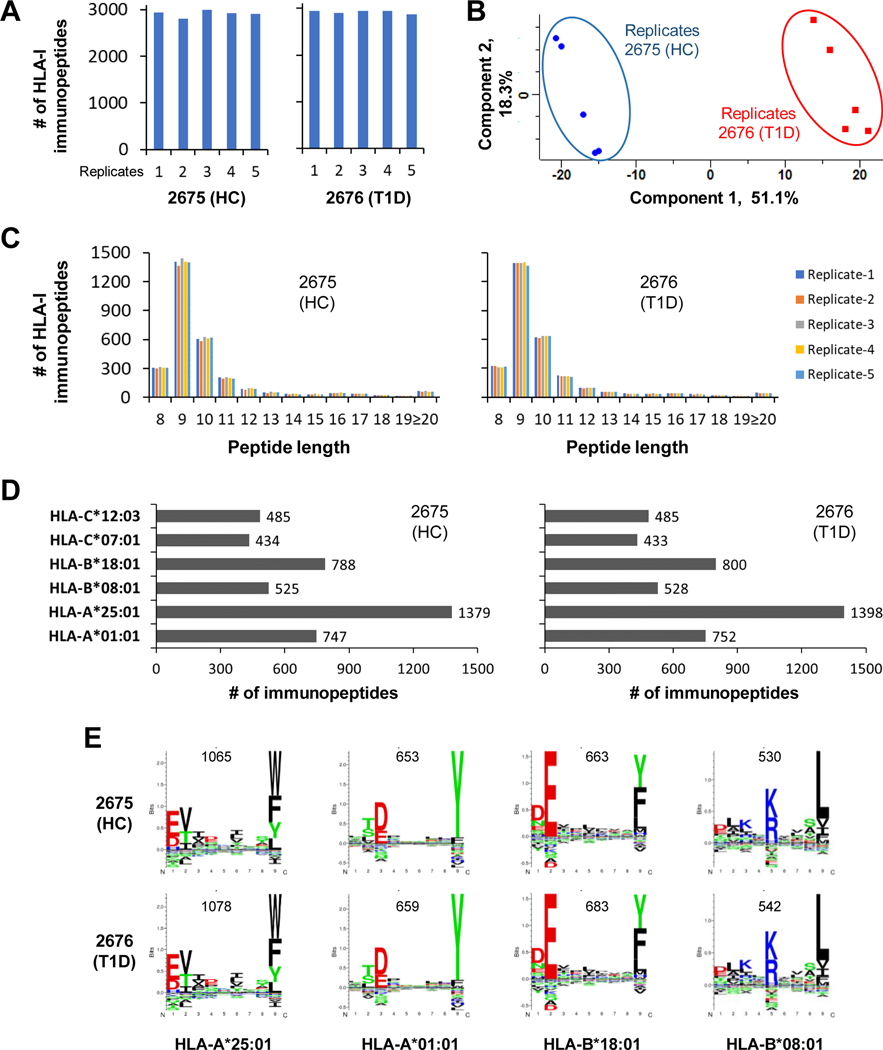
HLA-I immunopeptides were enriched using five sets of 2676 (T1D) and 2675 (HC) cells (30 ×106 cells / each set) and were analyzed in five replicates by LC-MS/MS. To assess the LC-MS/MS data quality, we examined the pairwise Pearson correlations (r) of the five replicate analyses. The immunopeptide expression data showed very good correlation within five T1D (r = 0.92 – 0.965), within five HC (r = 0.917 – 0.969) and between five T1D and five HC replicate analyses (r = 0.829 – 0.9) (supplemental Figs. S1A–S1C). The immunopeptides identified in each of the five replicate analyses are shown in Fig. 2A. A total of 3427 HLA-I immunopeptides were identified of which 3017 and 2974 were found in 2676 (T1D) and 2675 (HC), respectively (supplemental Table 2). The T1D and HC immunopeptides identified in replicates were clearly distinguished by principal component analysis (PCA) with component-1 greater than 50% (Fig. 2B). As expected, most of the HLA-I immunopeptides were found with peptide length of 8–13 amino acids, 9 being the most dominant (Fig. 2C). Next, we performed HLA binding affinity prediction of the immunopeptides using NetMHCpan 4.1. More than half of the HLA-I immunopeptides exhibited binding affinity for HLA-A*25:01 and HLA-A*01:01 alleles, and the others for HLA-B and HLA-C alleles (Fig. 2D). The total number of identified HLA-I immunopeptides (Fig. 2A) is lower than the total number of immunopeptides shown in Fig. 2D, which is attributed to the immunopeptides that showed binding affinity toward more than one HLA allele. Next, we used GibbsCluster-2.0 analysis for simultaneous alignment and clustering of the HLA-I immunopeptides. The clustering analysis found motifs for four HLA alleles, HLA-A*25:01, HLA-A*01:01, HLA-B*18:01, and HLA-B*08:01 (Fig. 2E). A relatively higher number of the immunopeptides aligned and clustered for the HLA-A*25:01 allele, which is consistent with the higher number of immunopeptides shown binding affinity for the same HLA allele (Figs. 2D–2E). Further verification of these four HLA-I motifs using Motif viewer, which shows naturally presented ligand motifs, revealed similar amino acid consensus sequence patterns suggesting the quality of immunopeptide sequences identified in this study (Supplemental Fig. S2, left panel).
Fig. 2. In-depth identification and characterization of HLA-I immunopeptidomes of T1D and healthy B cells that share same HLA alleles.
A, Bar diagram representing the number of HLA-I peptides detected in five replicate analyses of 2676 (T1D) and 2675 (HC) cells. These immunopeptides were identified with 1% FDR. B, Principle component analysis of HLA-I peptide intensities from five replicate analyses per condition. C, HLA-I peptide lengths observed in replicate analyses of T1D and HC cells. D, HLA-I peptides were assigned to different allotypes based on their binding affinities to HLA alleles. E, Alignment and clustering of HLA-I peptides of T1D and HC cells revealed consensus motif sequences of HLA-I alleles.
Identification and Characterization of HLA Class II Immunopeptides of T1D and Healthy B Cells
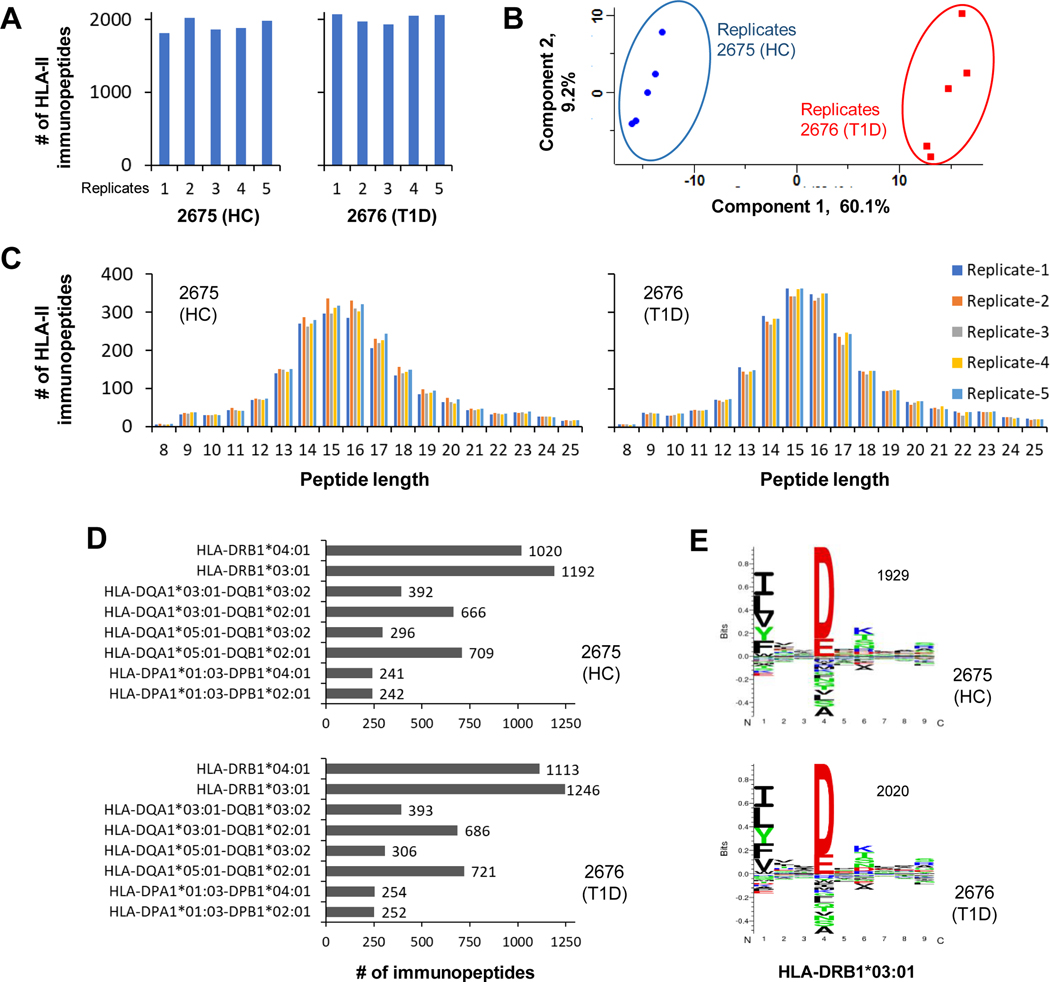
Followed by the HLA-I immunopeptides analysis, the five sets of HLA-II immunopeptides of the B cells (2676 (T1D) and 2675 (HC)) were analyzed in five replicates by LC-MS/MS (Fig. 3A). The quality of LC-MS/MS data was assessed by examining the pair-wise Pearson correlations (r) of the five replicate analyses. The immunopeptides expression data showed very good correlation within five T1D (r = 0.931 – 0.96), within five HC (r = 0.925 – 0.966) and between five T1D and five HC replicate analyses (r = 0.83 – 0.875) (supplemental Figs. S3A–S3C). This resulted in identification of 2477 HLA-II immunopeptides of which 2327 and 2222 were identified in 2676 (T1D) and 2675 (HC), respectively (supplemental Table 3). Similar to the HLA-I results, the PCA clearly distinguished the HLA-II immunopeptidomes of T1D and HC with component-1 greater than 60% (Fig. 3B). In case of HLA-II, most of the immunopeptides observed with peptide length of 12–19 amino acids as expected (Fig. 3C). Next, we performed HLA binding affinity prediction of the immunopeptides using NetMHCIIpan 4.0. More than a thousand of the identified HLA-II immunopeptides shown binding affinity for HLA-DRB1*04:01 and HLA-DRB1*03:01 alleles in both T1D and healthy B cells. In addition, more than 500 immunopeptides exhibited binding affinity for HLA-DQA1*03:01-DQB1*02:01 and HLA-DQA1*05:01-DQB1*02:01. However, a relatively lower number of immunopeptides showed binding affinity for the remaining HLA-II alleles (Fig. 3D). Then we used GibbsCluster-2.0 for simultaneous alignment and clustering of the HLA-II immunopeptides that revealed a significant motif for the allele, HLA-DRB1*03:01 in T1D and healthy B cells (Fig. 3E). The motif identification for HLA-DRB1*03:01 allele is in line with the higher number of immunopeptides shown binding affinity for that allele (Fig. 3D). Further verification of the identified motif of HLA-DRB1*03:01 using Motif viewer exhibited similar naturally presented ligand amino acid sequence pattern representing the quality of HLA-II immunopeptide sequences identified in this study (supplemental Fig. S2, right panel).
Fig. 3. In-depth identification and characterization of HLA-II immunopeptidomes of T1D and healthy B cells that share same HLA alleles.
A, Bar diagram representing the number of HLA-II peptides detected in five replicate analyses of 2676 (T1D) and 2675 (HC) cells. These immunopeptides were identified with 1% FDR. B, Principle component analysis of HLA-II peptide intensities from five replicate analyses per condition. C, HLA-II peptide lengths observed in replicate analyses of T1D and HC cells. D, HLA-II peptides were assigned to different allotypes based on their binding affinities to HLA alleles. E, Alignment and clustering of HLA-II peptides of T1D and HC cells revealed consensus motif sequences of HLA-II alleles.
HLA Allele-Specific Quantitation of the Immunopeptidome of T1D and Healthy B Cells
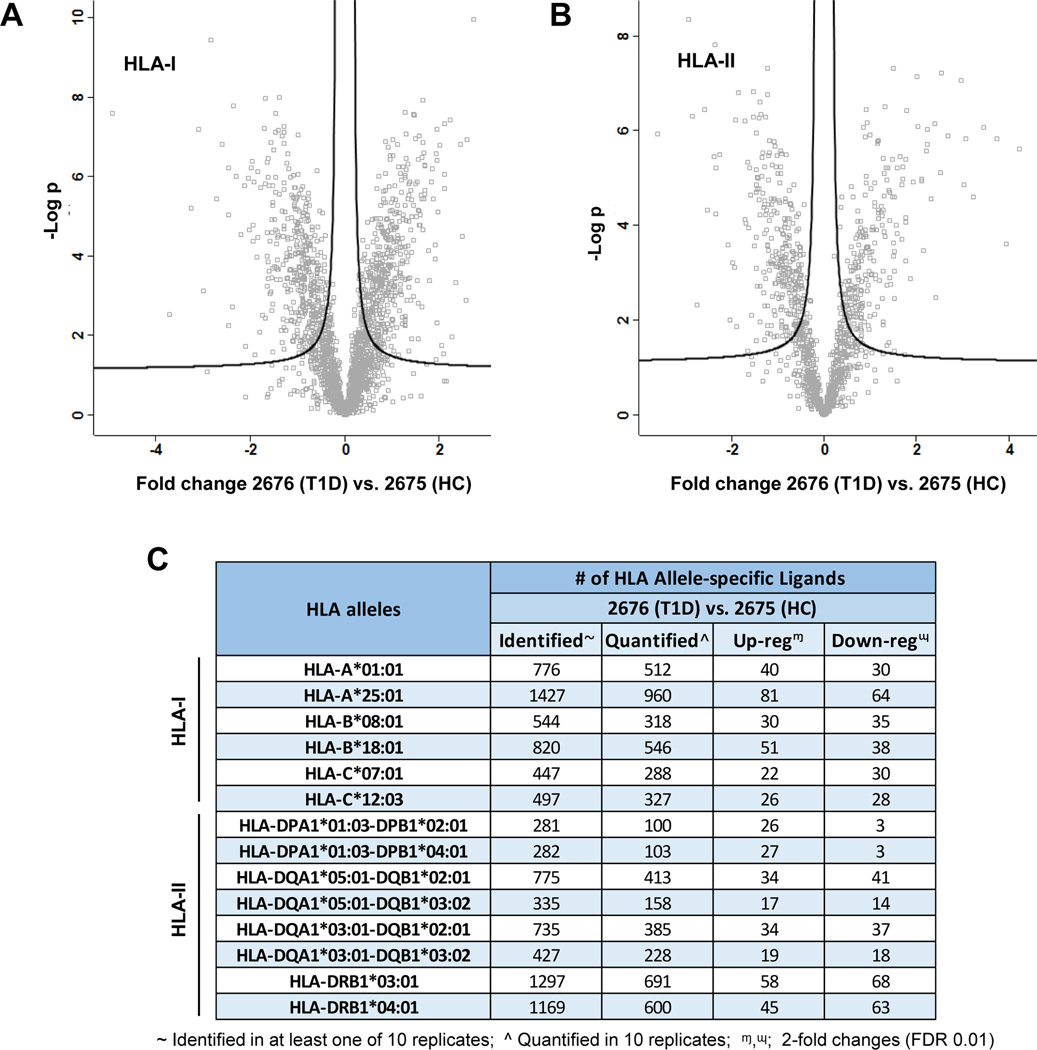
To quantify the HLA immunopeptides dysregulated in T1D B cells, we examined the 3427 HLA-I and 2477 HLA-II peptides identified in at least one of the ten replicate analyses of 2676 (T1D) and 2675 (HC) B cells (Fig. 2A and 3A). Then, for accurate quantification with statistical significance, we considered the 2131 HLA-I and 1204 HLA-II immunopeptides that were identified in all 10 replicate analyses without any missing values (Fig. 4A–4B). Label-free quantitation revealed 316 HLA-I and 216 HLA-II differentially regulated immunopeptides (DIPs) in T1D B cells with 2-fold changes (FDR, 0.01) (supplementary Tables 4 and 5). Next, the quantified immunopeptides were integrated with their HLA allele information based on their binding affinities to HLA alleles (Fig. 2D and 3D) and HLA allele typing results (supplementary Table 1). The detailed numbers of identified, quantified, and DIPs for all HLA-I and HLA-II alleles were listed in Fig. 4C and supplementary Tables 6 and 7. Among the HLA-I alleles, HLA-A*25:01 was observed with a high number of DIPs (81 up- and 64 down-regulated) (Fig. 4C). In addition, we observed a low number of DIPs for HLA-C alleles when compared with HLA-A and HLA-B alleles. In case of HLA-II alleles, HLA-DRB1*03:01 was found with most DIPs (58 up- and 68 down-regulated) followed by HLA-DRB1*04:01 allele (45 up- and 63 down-regulated). Interestingly, two alleles, HLA-DPA1*01:03-DPB1*02:01 and HLA-DPA1*01:03-DPB1*04:01 showed mostly upregulated DIPs. However, the other HLA alleles were observed with quite similar number of up and downregulated DIPs.
Fig. 4. HLA allele-specific quantitative immunopeptidomics of T1D and healthy B cells revealed the HLA-I and HLA-II immunopeptides dysregulated in T1D B cells.
A, Volcano plots representing the HLA-I immunopeptidomics analysis of 2676 (T1D) and 2675 (HC) cells in five replicates per condition (t-test, FDR 0.01). B, Volcano plots representing the HLA-II immunopeptidomics analysis of five replicates per condition (t-test, FDR 0.01). C, Detailed list of identified, quantified, up or downregulated allele-specific HLA-I and HLA-II immunopeptides (t-test, 2-fold regulated with FDR 0.01).
Modeling of the Cellular Functions of Candidate HLA-I and -II Antigens
Next, we used Proteomaps to investigate the cellular functions associated with the antigens of the dysregulated HLA immunopeptides identified in T1D. These dysregulated HLA-I and HLA-II immunopeptides were identified in 2676 (T1D) when compared with the healthy (2675 (HC)) B cells (2-fold changes, t-test FDR 0.01). The Proteomaps represent the quantitative composition of the cellular functions, which are based on KEGG pathways associated with HLA-I and HLA-II antigens (Supplemental Fig. S4). The analysis clearly distinguished the cellular functions of the HLA-I and HLA-II antigens. The HLA-I antigens were mainly associated with RNA transport, chromosome-related, DNA replication, spliceosome, ubiquitin-mediated proteolysis, MAPK signaling pathway, chemokine signaling pathway, and lipid and steroid metabolism (Supplemental Fig. S4A). The HLA-II antigens were linked with ubiquitin labeling, chaperones and folding catalysts, CD molecules, HIF-1 signaling pathway, NF-Kappa B signaling pathway, antigen processing and presentation, glycan metabolism, phagosome, and lysosome (Supplemental Fig. S4B). This helped us to distinguish the B cell HLA-I and HLA-II antigens by quantitatively mapping their cellular functions. These results might serve as a resource to understand the B cell immune regulation functions in autoimmune diseases.
Verification of HLA Class I and II Immunopeptide Sequences by in-Silico and PRM Analyses
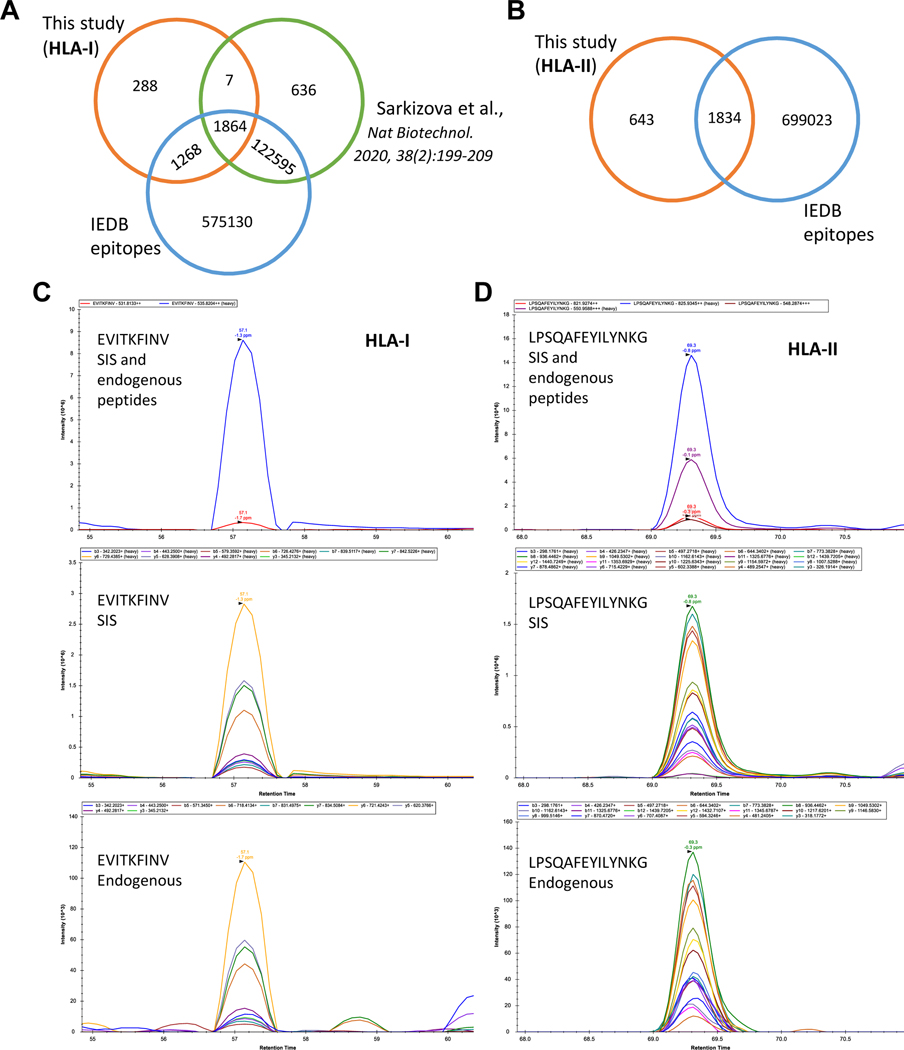
To verify the HLA-I and HLA-II immunopeptides dysregulated in T1D, we followed two different approaches (Fig. 5). First, the in-silico verification was carried out by comparing the HLA-I immunopeptides identified in this study with the HLA epitopes reported in IEDB and the HLA-I immunopeptides published in a recent study by Sarkizova et al.12 (Fig. 5A). Most of the HLA-I peptides identified in this study (91.5%) were also reported by either IEDB or Sarkizova et al. study. However, 288 HLA-I peptides were not reported and were considered as potential novel HLA-I peptides. In case of HLA-II, 74% of the peptides identified in this study were reported in IEDB (Fig. 5B). We would consider the remaining 643 of the HLA-II immunopeptides as a resource for their sequence verification by the future studies.
Fig. 5. Verification of HLA-I and HLA-II immunopeptides dysregulated in T1D B cells by in silico and PRM analyses.
A, Venn diagram illustrates the overlap of HLA-I immunopeptides investigated in this study, IEDB and Sarkizova et al. study. B, Venn diagram shows the overlap of HLA-II immunopeptides identified in this study and IEDB. C, A representative example for the PRM verification of HLA-I peptides is shown. The stable isotope labeled synthetic (SIS) peptides spiked as internal standards. While the top panel illustrates the precursor ions of endogenous and SIS peptides, the middle and bottom panels depict the fragment ions of SIS and endogenous peptides, respectively. D, A representative example for the PRM verification of HLA-II peptides is shown. The high intensity precursors were chosen (top panel) to exhibit their fragment ions in middle and bottom panels. The other details are as described in Fig. 5C.
In the second approach, we selected several HLA-I and HLA-II immunopeptides for their sequence verification by PRM. These immunopeptide candidates were selected based on the significant fold changes observed in T1D when compared with HC cells (2676 (T1D) vs. 2675 (HC)) as well as the biological significance of their corresponding antigens (supplemental Fig. S5–S6). The stable isotope labeled synthetic (SIS) peptides of the candidates were spiked as internal standards in real samples ahead of PRM analysis. Of the selected 27 HLA-I and 25 HLA-II candidates, 27 and 19, respectively were verified with SIS peptides. The representative examples of PRM-verified HLA-I and HLA-II immunopeptide sequences were illustrated in Fig. 5C and 5D, respectively. The remaining PRM-verified HLA-I and HLA-II candidates were organized in supplemental Figs. S7 and S8. In addition, MixMHC2pred was used to assess the confidence of the HLA-II peptides, except four peptides with their HLA alleles not included in the MixMHC2pred database, %Rank_best score ranged from 0.00412 – 0.757, with eight of the peptides having scores less than 0.1, which showed the confidence of these HLA-II peptides and the strong binding affinity to their respective HLA alleles (supplemental Fig. S6).
Selection of Additional B Cells Derived from T1D Patients and Healthy Controls
To validate HLA-I and HLA-II immunopeptides dysregulated in T1D, we searched for additional B cell lines that possess one or more HLA-I and HLA-II alleles similar to both 2675 (HC) and 2676 (T1D) cells. We selected a set of six B cell lines, of which 2679 (HC) and 2678 (T1D), 2812 (HC) and 2765 (T1D) belong to same family, respectively. Here, same family means the B cells derived from a T1D patient and a healthy person who were from the same biological family. However, the remaining two cell lines (2741 (HC) and 3027 (T1D)) were selected from two different families. HLA genotyping was performed using the DNA extracted from these cell lines and HLA allele information has been presented in supplemental Table 1. The genotyping results confirmed that four of the HLA-I alleles were similar between the six additional cell lines and the two cell lines (2675 (HC) and 2676 (T1D)) used in the proof-of-concept experiment. They were HLA-A*01:01, HLA-B*08:01, HLA-C*07:01, and HLA-C*12:03. The additional B cell lines also showed seven similar HLA-II alleles as observed in the 2676 (T1D) and 2675 (HC) lines, HLA-DPA1*01:03-DPB1*04:01, HLA-DQA1*05:01-DQB1*02:01, HLA-DQA1*05:01-DQB1*03:02, HLA-DQA1*03:01-DQB1*02:01, HLA-DQA1*03:01-DQB1*03:02, HLA-DRB1*03:01, and HLA-DRB1*04:01 (supplemental Table 1).
Enrichment and Characterization of HLA Class I and II Immunopeptides of Additional B Cells
Following the selection of additional B cell lines, their HLA-I and HLA-II immunopeptides were enriched and investigated in two replicates by LC-MS/MS to validate the immunopeptides dysregulated in T1D. We identified a total of 6928 HLA-I and 1044 HLA-II immunopeptides in six cell lines (supplemental Tables 8 and 9) and the numbers of peptides identified in each cell line were represented in supplemental Fig. S9A. In agreement with the observed results (Figs. 2C and 3C), most of the HLA-I and HLA-II immunopeptides showed peptide length of 8–13 and 12–19 amino acids, respectively (supplemental Fig. S9B). Next, the HLA binding affinity prediction revealed the percentages of immunopeptides associated with different HLA-I and HLA-II alleles (supplemental Fig. S10A–S10B). These percentages were ranging from 9–28% and 3–29% for different alleles in cases of HLA-I and HLA-II, respectively. Among the HLA-I alleles, HLA-A*01:01 and HLA-A*03:01 were found with the highest and lowest number of binding affinity percentages for immunopeptides (supplemental Fig. S10A). In case of HLA-II, a relatively higher percentage of immunopeptides showed binding affinity for HLA-DRB1*03:01 or HLA-DRB1*04:01 alleles (supplemental Fig. S10B). Next, the simultaneous alignment and clustering of the immunopeptides revealed the corresponding motifs of the HLA-I and HLA-II alleles (supplemental Fig. S11–S12). We further verified the identified motifs using Motif viewer which revealed similar pattern of naturally presented ligand sequences for most of the HLA alleles (supplemental Fig. S11–S12). The above results demonstrate the quality of immunopeptidome data of the six B cell lines selected for validation of HLA immunopeptides dysregulated in T1D B cells.
Validation of HLA Class I and II Immunopeptides Dysregulated in T1D Using Additional B Cells
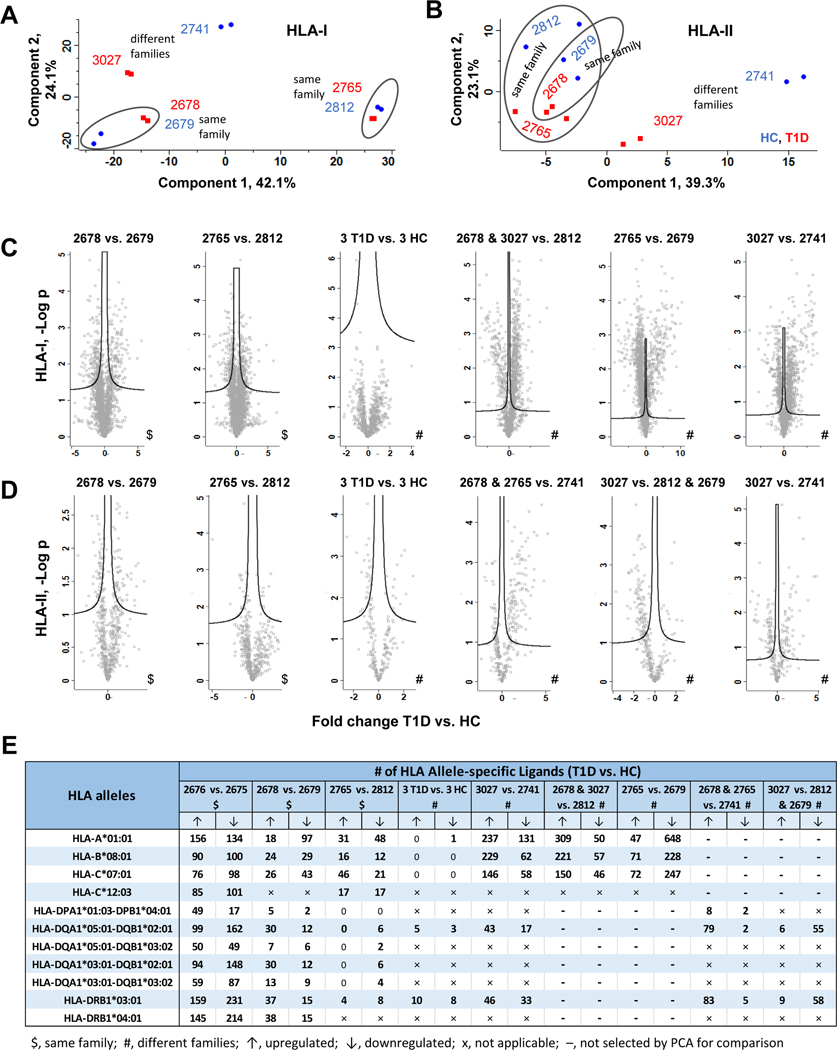
The HLA-I and HLA-II immunopeptides differentially regulated in proof-of-concept experiment (2676 (T1D) vs. 2675 (HC)) were validated using additional six B cell lines. To this end, we considered comparing the two cell line pairs that belong to the same families (family 1, 2679 (HC) vs. 2678 (T1D); and family 2, 2812 (HC) vs. 2765 (T1D)), and a pair of cell lines that belong to the different families (family 3, 2741 (HC) vs. family 4, 3027 (T1D)). In addition, we considered comparing all T1D and HC cell lines as this is the common approach in large scale omics studies (2678, 2675, 3027 (3 T1D) vs. 2679, 2812, 2741 (3 HC)). Furthermore, based on PCA results, the T1D and HC cell lines that were clearly distinguished by component 1 were selected for the quantitative comparison (Figs. 6A–B). This includes 2678, 3027 (2 T1D) vs. 2812 (HC) and 2765 (T1D) vs. 2679 (HC) in case of HLA-I, and 2678, 2765 (2 T1D) vs. 2741 (HC) and 3027 (T1D) vs. 2812, 2679 (2 HC) in case of HLA-II (Figs. 6A–B). The immunopeptides identified without missing values were included in all these selected quantitative comparisons to detect the DIPs (Figs. 6C–D).
Fig. 6. Validation of HLA immunopeptides dysregulated in T1D B cells using additional B cell lines.
A, Principle component analysis of HLA-I peptide intensities from six B cell lines (two replicate analyses per T1D or healthy cell line). B, Principle component analysis of HLA-II peptide intensities from six B cell lines (two replicate analyses per T1D or healthy cell line). C, Volcano plot comparisons of HLA-I immunopeptidomes from different sets of T1D and healthy B cells belong to same or different families. The differentially regulated HLA-I immunopeptides were identified by t-test with FDR 0.05. D, Volcano plot comparisons of HLA-II immunopeptidomes from different sets of T1D and healthy B cells belong to same or different families. The differentially regulated HLA-II immunopeptides were identified by t-test with FDR 0.05. E, Detailed list of up or downregulated allele-specific HLA-I and HLA-II immunopeptides detected in T1D when compared to healthy B cells (t-test with FDR 0.05, S0 0.1).
The DIPs were investigated from the above defined comparisons of additional B cell lines using S0, 0.1 and FDR, 0.05 in both HLA-I and HLA-II cases (Figs. 6C–D), which are the same parameters and cutoff as for comparison between the two cell lines (2676 (T1D) vs. 2675 (HC)) used in the proof-of-concept experiment (supplementary Tables 10 and 11). The immunopeptides observed in these comparisons were then integrated with HLA allele information based on their binding affinities to specific HLA alleles and HLA allele typing results. The detailed numbers of up and down regulated HLA-I and HLA-II immunopeptides identified in all comparisons were listed in Fig. 6E (supplementary Tables 12 and 13).
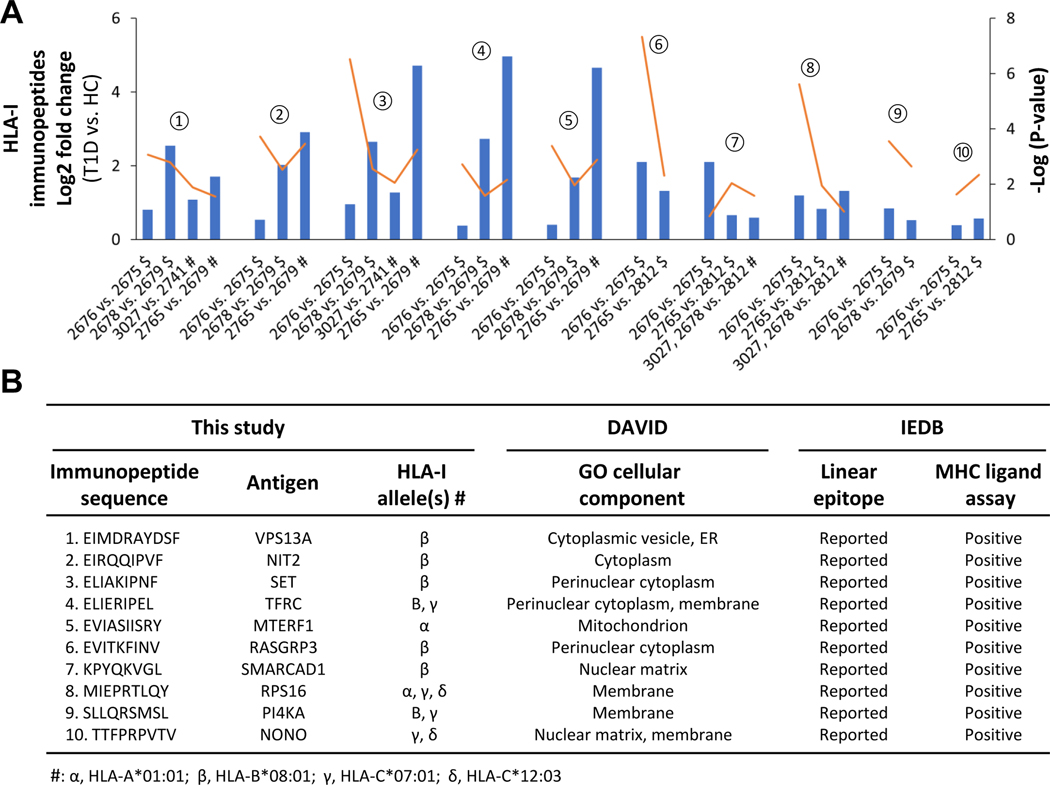
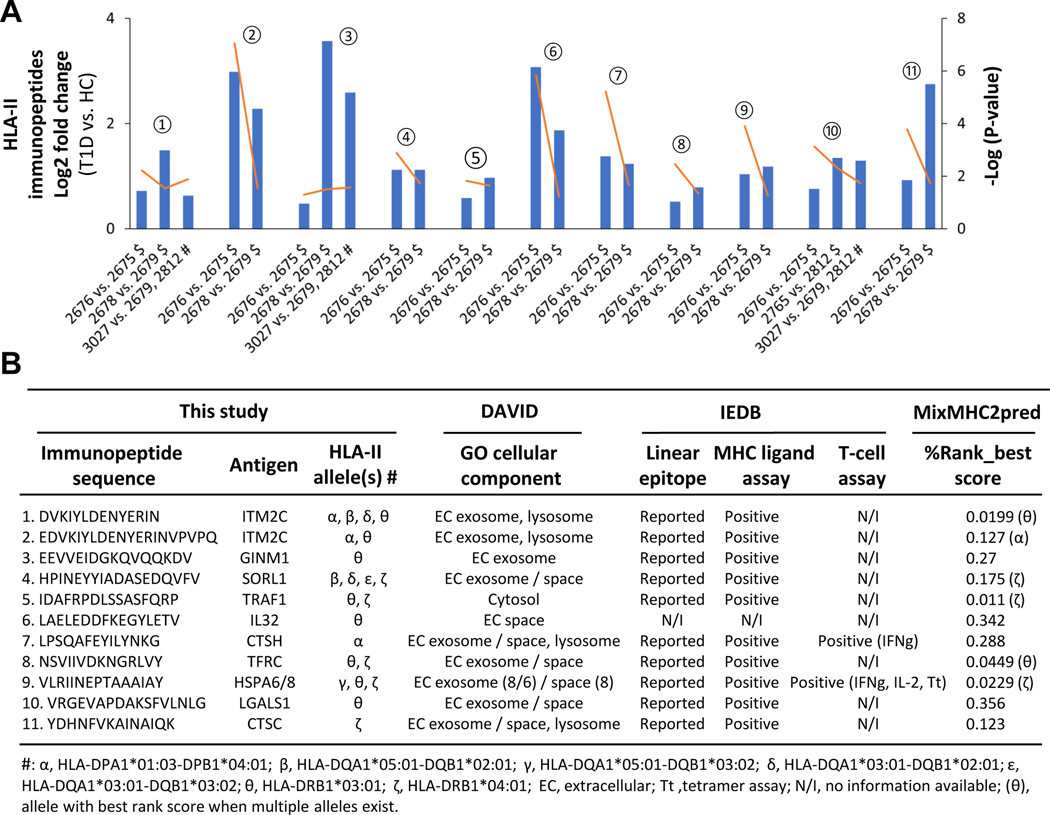
Next, we validated the HLA-I and II immunopeptides dysregulated in T1D ((2676 (T1D) vs. 2675 (HC)) based on the required and optional criteria as described below. If the HLA immunopeptides upregulated in proof-of-concept experiment ((2676 (T1D) vs. 2675 (HC)) were also found upregulated in at least one of the two comparisons made between the additional T1D and healthy cell lines belonging to the same families (2679 (HC) vs. 2678 (T1D) or 2812 (HC) vs. 2765 (T1D)) then they were considered as potential candidates – this is the required criterion to validate the regulation status of HLA candidates. In addition, the HLA immunopeptides upregulated in proof-of-concept experiment ((2676 (T1D) vs. 2675 (HC)) could also be found upregulated in any comparisons made between the additional T1D and healthy cell lines belonging to different families then those results were considered as supporting validation data – this is the optional criterion to support the regulation status of the HLA candidates. Following the required and optional criteria, we identified ten HLA-I and eleven HLA-II immunopeptides as dysregulated candidates in T1D B cells (Fig. 7 and 8). The fold changes and significance p-values of the ten HLA-I candidates are depicted in column-line graph (Fig. 7A), and their additional details such as sequences and allele information were summarized in Fig. 7B. Similarly, the details of eleven HLA-II candidates dysregulated in T1D are shown in Fig. 8. We also used MixMHC2pred to assess the confidence of these HLA-II peptides, %Rank_best score ranged from 0.011 – 0.356, with four of the peptides having scores less than 0.05 (Fig. 8B), which showed the confidence of these HLA-II peptides and the strong binding affinity to their respective HLA alleles. In addition, peptides belong to the HLA molecules, which may be degradation products of HLA molecules, identified with above mentioned required and optional criteria were listed in Supplemental Fig. S13.
Fig. 7. Evaluation of candidate HLA-I immunopeptides dysregulated in T1D B cells.
A, Column-line graph depicting the significant fold changes of candidate HLA-I immunopeptides in T1D compared to HC (t-test: FDR, 0.05 and S0, 0.1). The symbols, $ and # indicate the comparisons of T1D and healthy B cells belonging to same and different families, respectively. The orange lines indicate the p-values. The numbers from 1 to 10 help connecting the candidate HLA-I immunopeptides between Figs. 7A and 7B. B, Details of candidate HLA-I immunopeptide sequence, allele information, cellular localization and IEDB information are shown.
Fig. 8. Evaluation of candidate HLA-II immunopeptides dysregulated in T1D B cells.
A, Column-line graph depicting the significant fold changes of candidate HLA-II immunopeptides in T1D compared to HC (t-test: FDR, 0.05 and S0, 0.1). The symbols, $ and # indicate the comparisons of T1D and healthy B cells belong to same and different families, respectively. The orange lines indicate the p-values. The numbers from 1 to 10 help connecting the candidate HLA-II immunopeptides between Figs. 8A and 8B. B, Details of candidate HLA-II immunopeptide sequence, allele information, cellular localization, IEDB information and MixMHC2pred verification are shown.
Biological Evaluation of the Candidate HLA-I and II Immunopeptides
Next, we evaluated the biology of candidate HLA immunopeptides in terms of cellular localization, ligand binding ability, and immune-regulatory functions. The Gene Ontology cellular component analysis using DAVID revealed that HLA-I and HLA-II candidate antigens were mostly localized in cytoplasm and extracellular region, respectively as expected (Figs. 7B–8B). Further, we found that the linear epitopes of 20 of the 21 candidate HLA-I and HLA-II immunopeptides have been reported previously in IEDB, except the peptide from IL-32 (Figs. 7B–8B). Most importantly, 20 of these 21 candidates immunopeptides are positive for HLA ligand binding assay according to IEDB (Figs. 7B–8B). Interestingly, we found that two (LPSQAFEYILYNKG and VLRIINEPTAAAIAY) of the eleven HLA-II immunopeptides were positive for T-cell assay or tetramer assay and were associated with an increased release of IFN-γ or IL-2 (Fig. 8B). These two HLA-II immunopeptides belong to cathepsin H (CTSH) and heat shock proteins (HSPA6/8) antigens, respectively. The list of identified HLA-I and HLA-II candidates (Figs. 7B–8B) is a basis for future investigations to explore their immunoregulatory functions.
DISCUSSION
Mass spectrometry-based immunopeptidomic studies enabled the investigation of antigens and HLA immunopeptides from different types of tissues and cell lines.3, 6, 7, 9, 10, 12, 39 Quantitative immunopeptidomics typically is performed by treating the cells with pro-inflammatory cytokines or other immune system-modulating agents to see how antigen presentation changes for the exact same cells under different treatment conditions.3, 6, 7 However, this does not apply to patient-related specimens (e.g., T1D patients and healthy controls), which generally do not share identical alleles but is fundamental to understand the natural immunoregulatory functions in patients. Here, we studied the immunopeptides of eight B-lymphocytes of T1D patients and healthy controls, which are matching well with the confounding factors such as age, gender, and race (supplemental Table 1).
We looked for what may contribute to the quantitative changes observed in the immunopeptides between T1D and HC B cells. Because quantitation was performed on the immunopeptides commonly identified in all samples, the differences are unlikely due to single nucleotide polymorphisms (SNPs). We next looked at whether the quantitative differences are a result of differences in antigen expression level between T1D and HC. In this respect, we did not see similar trend in differential regulation status of the multiple immunopeptides identified from the antigens presented in Figs. 7 and 8. For example, of the 207 immunopeptides identified for the 10 HLA-II antigens (Fig. 8, supplemental Table S3-HLA-II), 82 were differentially regulated (57 up- and 25 down-regulated) and the remaining 125 were unchanged (supplemental Table S11_2676vs2675-HLA-II). Similar results were also observed for the HLA-I antigens. This points out that the antigens expression level may not be the major cause of their differential regulation. This brings an interesting question of what causes the change. Since a few of the antigens presented in Figs. 7 and 8, such as cathepsins (CTSC, CTSH) are involved in proteasome/immunoproteasome and antigen presentation machinery, differences in antigen processing and antigen presentation between T1D and HC may explain some of these findings. But a more detailed study is needed for a better understanding. Although not likely the major contributor to the findings in this study, other factors such as single nucleotide polymorphisms (SNPs) and expression levels of antigens could be different between T1D and healthy controls, particularly in a large cohort of samples. In this regard, leveraging SNP genotyping, exome and RNA sequencing can further improve the comprehensiveness of HLA immunopeptidome profiling and provide a better understanding of the origin of DIPs and further elucidate the roles of gene regulation in the pathogenesis of T1D.
In T1D, CD4+ T-cell immune response, which is mediated by HLA-II antigen presentation, plays important role in autoimmune destruction of pancreatic beta cells. However, exploring the antigen cross-presentation to CD8+ cells via HLA-I antigen is also important to understand the mechanisms of T1D pathogenesis. While we used T1D and healthy B cells with identical HLA alleles in the proof-of-concept experiment (Figs. 2–4), six additional B cell lines with similar HLA alleles were exploited to validate the HLA-I and II immunopeptides dysregulated in T1D (Fig. 6), which confirmed several candidate HLA-I and HLA-II immunopeptides dysregulated in T1D B cells (Figs. 7–8). Two HLA-II immunopeptides belonging to cathepsin H (CTSH) and heat shock proteins (HSPA6/8) antigens, respectively, were reported in the IEDB to have validated T-cell assays. Indeed CTSH was reported to be a T1D risk gene and an important regulator of beta cell function during T1D progression,40, 41 and heat shock proteins are intracellular chaperones and involved in autoimmune diseases,42 in particular the 70 kDa HSPA6 was reported to stimulate macrophage inflammation synergistically with the B11–23 peptide segment of proinsulin.43 Functional annotation of the dysregulated antigens indicates that CTSH and CTSC are associated with T cell mediated toxicity, and seven proteins (CTSC, CTSH, HSPA8/6, IL-32, LGALS1, NIT2, and NONO) are involved in immune response. Furthermore, transferrin receptor (TFRC) is known to be linked with T cell/B cell activation,44, 45 and soluble transferrin receptor levels was reported to have an association with gastric autoimmunity in T1D.46 A couple of studies also revealed the potential involvement of CTSH, HSPA8, IL-32 in T1D development.40–42, 47 The epitopes of all these antigens were identified as HLA-II immunopeptides and were dysregulated in T1D (Fig. 8). In the future, it would be interesting to investigate the roles of these HLA-II immunopeptides and their antigens to understand the mechanisms associated with beta cell destruction during the development and progression of T1D.
In conclusion, we demonstrated the feasibility to use the HLA allele-specific quantitative immunopeptidomics to profile the changes of immunopeptides between T1D and healthy B cell lines that belong to different families with partially shared HLA alleles. Although this study has limitations in that the cell lines are not the same as B lymphocytes freshly isolated from the PBMCs of patients, and therefore the roles of the dysregulated B cell-immunopeptides in T1D pathogenesis remain to be defined, we believe that our method, by exploring the expression of HLA immunopeptides naturally derived from the diseased and healthy controls at the allele level, would help to accelerate the clinical applications of this approach to immune-related diseases.
Supplementary Material
Fig S1. Scatter plots of HLA-I immunopeptidome of 2676 (T1D) and 2675 (HC) B cells.
Fig S2. Binding motifs for the HLA-I and -II immunopeptides of two B cell lines (2676 (T1D) and 2675 (HC)).
Fig S3. Scatter plots of HLA-II immunopeptidome of 2676 (T1D) and 2675 (HC) B cells.
Fig. S4. Cellular functions of the HLA-I and -II antigens dysregulated in T1D.
Fig S5. List of HLA-I immunopeptides selected for verification by PRM.
Fig S6. List of HLA-II immunopeptides selected for verification by PRM.
Fig S7. Representative chromatograms of verified HLA-I immunopeptides.
Fig S8. Representative chromatograms of verified HLA-II immunopeptides.
Fig S9. Number of immunopeptides enriched from six additional B cell lines.
Fig S10. Composition of immunopeptides identified in six B cell lines according to HLA alleles.
Fig S11. Binding motifs for the HLA-I immunopeptides of six additional B cell lines.
Fig S12. Binding motifs for the HLA-II immunopeptides of six additional B cell lines.
Fig S13. Peptides identified from HLA molecules.
Table S1.Clinical data and HLA-I and -II allele information of B cells.
Table S2. HLA-I immunopeptidome of 2676 (T1D) and 2675 (HC) B cells.
Table S3. HLA-II immunopeptidome of 2676 (T1D) and 2675 (HC) B cells.
Table S4. Differentially regulated HLA-I immunopeptides in 2676 (T1D) B cells relative to 2675 (HC) B cells.
Table S5. Differentially regulated HLA-II immunopeptides in 2676 (T1D) B cells relative to 2675 (HC) B cells.
Table S6. Differentially regulated allele-specific HLA-I immunopeptides in 2676 (T1D) B cells relative to 2675 (HC) B cells.
Table S7. Differentially regulated allele-specific HLA-II immunopeptides in 2676 (T1D) B cells relative to 2675 (HC) B cells.
Table S8. HLA-I immunopeptidome of six additional B cells.
Table S9. HLA-II immunopeptidome of six additional B cells.
Table S10. Differentially regulated HLA-I immunopeptides in six additional B cells.
Table S11. Differentially regulated HLA-II immunopeptides in six additional B cells.
Table S12. Validation of HLA-I immunopeptides using additional six B cell lines.
Table S13. Validation of HLA-II immunopeptides using additional six B cell lines.
Acknowledgements
The authors thank Dr. Janelle A. Noble of the Children’s Hospital Oakland Research Institute for HLA genotyping of the initial B cell lines. This work was supported by the National Institute Of Diabetes And Digestive And Kidney Diseases of the National Institutes of Health under Award Number R01DK114345.
Footnotes
SUPPORTING INFORMATION:
The following supporting information is available free of charge at ACS website http://pubs.acs.org.
References
- 1.Vizcaíno JA; Kubiniok P; Kovalchik KA; Ma Q; Duquette JD; Mongrain I; Deutsch EW; Peters B; Sette A; Sirois I; Caron E, The Human Immunopeptidome Project: A Roadmap to Predict and Treat Immune Diseases. Mol Cell Proteomics 2020, 19, (1), 31–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caron E; Kowalewski DJ; Chiek Koh C; Sturm T; Schuster H; Aebersold R, Analysis of Major Histocompatibility Complex (MHC) Immunopeptidomes Using Mass Spectrometry. Mol Cell Proteomics 2015, 14, (12), 3105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chong C; Marino F; Pak H; Racle J; Daniel RT; Muller M; Gfeller D; Coukos G; Bassani-Sternberg M, High-throughput and Sensitive Immunopeptidomics Platform Reveals Profound Interferongamma-Mediated Remodeling of the Human Leukocyte Antigen (HLA) Ligandome. Mol Cell Proteomics 2018, 17, (3), 533–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reynisson B; Alvarez B; Paul S; Peters B; Nielsen M, NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res 2020, 48, (W1), W449–w454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Racle J; Michaux J; Rockinger GA; Arnaud M; Bobisse S; Chong C; Guillaume P; Coukos G; Harari A; Jandus C; Bassani-Sternberg M; Gfeller D, Robust prediction of HLA class II epitopes by deep motif deconvolution of immunopeptidomes. Nature Biotechnology 2019, 37, (11), 1283–1286. [DOI] [PubMed] [Google Scholar]
- 6.Stopfer LE; Mesfin JM; Joughin BA; Lauffenburger DA; White FM, Multiplexed relative and absolute quantitative immunopeptidomics reveals MHC I repertoire alterations induced by CDK4/6 inhibition. Nat Commun 2020, 11, (1), 2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newey A; Griffiths B; Michaux J; Pak HS; Stevenson BJ; Woolston A; Semiannikova M; Spain G; Barber LJ; Matthews N; Rao S; Watkins D; Chau I; Coukos G; Racle J; Gfeller D; Starling N; Cunningham D; Bassani-Sternberg M; Gerlinger M, Immunopeptidomics of colorectal cancer organoids reveals a sparse HLA class I neoantigen landscape and no increase in neoantigens with interferon or MEK-inhibitor treatment. J Immunother Cancer 2019, 7, (1), 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanoix J; Durette C; Courcelles M; Cossette E; Comtois-Marotte S; Hardy MP; Cote C; Perreault C; Thibault P, Comparison of the MHC I Immunopeptidome Repertoire of B-Cell Lymphoblasts Using Two Isolation Methods. Proteomics 2018, 18, (12), e1700251. [DOI] [PubMed] [Google Scholar]
- 9.Wang J; Jelcic I; Muhlenbruch L; Haunerdinger V; Toussaint NC; Zhao Y; Cruciani C; Faigle W; Naghavian R; Foege M; Binder TMC; Eiermann T; Opitz L; Fuentes-Font L; Reynolds R; Kwok WW; Nguyen JT; Lee JH; Lutterotti A; Munz C; Rammensee HG; Hauri-Hohl M; Sospedra M; Stevanovic S; Martin R, HLA-DR15 Molecules Jointly Shape an Autoreactive T Cell Repertoire in Multiple Sclerosis. Cell 2020, 183, (5), 1264–1281 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forlani G; Michaux J; Pak H; Huber F; Marie Joseph EL; Ramia E; Stevenson BJ; Linnebacher M; Accolla RS; Bassani-Sternberg M, CIITA-Transduced Glioblastoma Cells Uncover a Rich Repertoire of Clinically Relevant Tumor-Associated HLA-II Antigens. Mol Cell Proteomics 2021, 20, 100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bassani-Sternberg M; Braunlein E; Klar R; Engleitner T; Sinitcyn P; Audehm S; Straub M; Weber J; Slotta-Huspenina J; Specht K; Martignoni ME; Werner A; Hein R; D HB; Peschel C; Rad R; Cox J; Mann M; Krackhardt AM, Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun 2016, 7, 13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkizova S; Klaeger S; Le PM; Li LW; Oliveira G; Keshishian H; Hartigan CR; Zhang W; Braun DA; Ligon KL; Bachireddy P; Zervantonakis IK; Rosenbluth JM; Ouspenskaia T; Law T; Justesen S; Stevens J; Lane WJ; Eisenhaure T; Lan Zhang G; Clauser KR; Hacohen N; Carr SA; Wu CJ; Keskin DB, A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat Biotechnol 2020, 38, (2), 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atkinson MA, The pathogenesis and natural history of type 1 diabetes. Cold Spring Harb Perspect Med 2012, 2, (11), a007641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pugliese A, Autoreactive T cells in type 1 diabetes. J Clin Invest 2017, 127, (8), 2881–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noble JA, Immunogenetics of type 1 diabetes: A comprehensive review. Journal of Autoimmunity 2015, 64, 101–112. [DOI] [PubMed] [Google Scholar]
- 16.Steck AK; Rewers MJ, Genetics of type 1 diabetes. Clin Chem 2011, 57, (2), 176–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rewers M; Hyöty H; Lernmark Å; Hagopian W; She JX; Schatz D; Ziegler AG; Toppari J; Akolkar B; Krischer J, The Environmental Determinants of Diabetes in the Young (TEDDY) Study: 2018 Update. Curr Diab Rep 2018, 18, (12), 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delong T; Wiles TA; Baker RL; Bradley B; Barbour G; Reisdorph R; Armstrong M; Powell RL; Reisdorph N; Kumar N; Elso CM; DeNicola M; Bottino R; Powers AC; Harlan DM; Kent SC; Mannering SI; Haskins K, Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science 2016, 351, (6274), 711–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Babon JA; DeNicola ME; Blodgett DM; Crevecoeur I; Buttrick TS; Maehr R; Bottino R; Naji A; Kaddis J; Elyaman W; James EA; Haliyur R; Brissova M; Overbergh L; Mathieu C; Delong T; Haskins K; Pugliese A; Campbell-Thompson M; Mathews C; Atkinson MA; Powers AC; Harlan DM; Kent SC, Analysis of self-antigen specificity of islet-infiltrating T cells from human donors with type 1 diabetes. Nat Med 2016, 22, (12), 1482–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kracht MJ; van Lummel M; Nikolic T; Joosten AM; Laban S; van der Slik AR; van Veelen PA; Carlotti F; de Koning EJ; Hoeben RC; Zaldumbide A; Roep BO, Autoimmunity against a defective ribosomal insulin gene product in type 1 diabetes. Nat Med 2017, 23, (4), 501–507. [DOI] [PubMed] [Google Scholar]
- 21.Smith MJ; Simmons KM; Cambier JC, B cells in type 1 diabetes mellitus and diabetic kidney disease. Nat Rev Nephrol 2017, 13, (11), 712–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith MJ; Cambier JC; Gottlieb PA, Endotypes in T1D: B lymphocytes and early onset. Curr Opin Endocrinol Diabetes Obes 2020, 27, (4), 225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serreze DV; Fleming SA; Chapman HD; Richard SD; Leiter EH; Tisch RM, B lymphocytes are critical antigen-presenting cells for the initiation of T cell-mediated autoimmune diabetes in nonobese diabetic mice. J Immunol 1998, 161, (8), 3912–8. [PubMed] [Google Scholar]
- 24.Wong FS; Wen L; Tang M; Ramanathan M; Visintin I; Daugherty J; Hannum LG; Janeway CA Jr.; Shlomchik MJ, Investigation of the role of B-cells in type 1 diabetes in the NOD mouse. Diabetes 2004, 53, (10), 2581–7. [DOI] [PubMed] [Google Scholar]
- 25.Felton JL; Maseda D; Bonami RH; Hulbert C; Thomas JW, Anti-Insulin B Cells Are Poised for Antigen Presentation in Type 1 Diabetes. J Immunol 2018, 201, (3), 861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leete P; Willcox A; Krogvold L; Dahl-Jørgensen K; Foulis AK; Richardson SJ; Morgan NG, Differential Insulitic Profiles Determine the Extent of β-Cell Destruction and the Age at Onset of Type 1 Diabetes. Diabetes 2016, 65, (5), 1362–9. [DOI] [PubMed] [Google Scholar]
- 27.Christie MR, Delving Into the Type 1 Diabetic Islet: Evidence That B-Cell Infiltration of Islets Is Linked to Local Hyperimmunity and Accelerated Progression to Disease. Diabetes 2016, 65, (5), 1146–8. [DOI] [PubMed] [Google Scholar]
- 28.Akashi T; Nagafuchi S; Anzai K; Kondo S; Kitamura D; Wakana S; Ono J; Kikuchi M; Niho Y; Watanabe T, Direct evidence for the contribution of B cells to the progression of insulitis and the development of diabetes in non-obese diabetic mice. Int Immunol 1997, 9, (8), 1159–64. [DOI] [PubMed] [Google Scholar]
- 29.Xiu Y; Wong CP; Bouaziz JD; Hamaguchi Y; Wang Y; Pop SM; Tisch RM; Tedder TF, B lymphocyte depletion by CD20 monoclonal antibody prevents diabetes in nonobese diabetic mice despite isotype-specific differences in Fc gamma R effector functions. J Immunol 2008, 180, (5), 2863–75. [DOI] [PubMed] [Google Scholar]
- 30.Hu CY; Rodriguez-Pinto D; Du W; Ahuja A; Henegariu O; Wong FS; Shlomchik MJ; Wen L, Treatment with CD20-specific antibody prevents and reverses autoimmune diabetes in mice. J Clin Invest 2007, 117, (12), 3857–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bassani-Sternberg M, Mass Spectrometry Based Immunopeptidomics for the Discovery of Cancer Neoantigens. Methods Mol Biol 2018, 1719, 209–221. [DOI] [PubMed] [Google Scholar]
- 32.Cox J; Mann M, MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008, 26, (12), 1367–72. [DOI] [PubMed] [Google Scholar]
- 33.MacLean B; Tomazela DM; Shulman N; Chambers M; Finney GL; Frewen B; Kern R; Tabb DL; Liebler DC; MacCoss MJ, Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, (7), 966–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tyanova S; Cox J, Perseus: A Bioinformatics Platform for Integrative Analysis of Proteomics Data in Cancer Research. Methods Mol Biol 2018, 1711, 133–148. [DOI] [PubMed] [Google Scholar]
- 35.Andreatta M; Alvarez B; Nielsen M, GibbsCluster: unsupervised clustering and alignment of peptide sequences. Nucleic Acids Res 2017, 45, (W1), W458–W463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang DW; Sherman BT; Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009, 4, (1), 44–57. [DOI] [PubMed] [Google Scholar]
- 37.Liebermeister W; Noor E; Flamholz A; Davidi D; Bernhardt J; Milo R, Visual account of protein investment in cellular functions. Proc Natl Acad Sci U S A 2014, 111, (23), 8488–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vita R; Mahajan S; Overton JA; Dhanda SK; Martini S; Cantrell JR; Wheeler DK; Sette A; Peters B, The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res 2019, 47, (D1), D339–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chong C; Muller M; Pak H; Harnett D; Huber F; Grun D; Leleu M; Auger A; Arnaud M; Stevenson BJ; Michaux J; Bilic I; Hirsekorn A; Calviello L; Simo-Riudalbas L; Planet E; Lubinski J; Bryskiewicz M; Wiznerowicz M; Xenarios I; Zhang L; Trono D; Harari A; Ohler U; Coukos G; Bassani-Sternberg M, Integrated proteogenomic deep sequencing and analytics accurately identify non-canonical peptides in tumor immunopeptidomes. Nat Commun 2020, 11, (1), 1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inshaw JRJ; Cutler AJ; Crouch DJM; Wicker LS; Todd JA, Genetic Variants Predisposing Most Strongly to Type 1 Diabetes Diagnosed Under Age 7 Years Lie Near Candidate Genes That Function in the Immune System and in Pancreatic β-Cells. Diabetes Care 2020, 43, (1), 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fløyel T; Brorsson C; Nielsen LB; Miani M; Bang-Berthelsen CH; Friedrichsen M; Overgaard AJ; Berchtold LA; Wiberg A; Poulsen P; Hansen L; Rosinger S; Boehm BO; Ram R; Nguyen Q; Mehta M; Morahan G; Concannon P; Bergholdt R; Nielsen JH; Reinheckel T; von Herrath M; Vaag A; Eizirik DL; Mortensen HB; Størling J; Pociot F, CTSH regulates β-cell function and disease progression in newly diagnosed type 1 diabetes patients. Proc Natl Acad Sci U S A 2014, 111, (28), 10305–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turturici G; Tinnirello R; Sconzo G; Asea A; Savettieri G; Ragonese P; Geraci F, Positive or Negative Involvement of Heat Shock Proteins in Multiple Sclerosis Pathogenesis: An Overview. Journal of Neuropathology & Experimental Neurology 2014, 73, (12), 1092–1106. [DOI] [PubMed] [Google Scholar]
- 43.Blasius E; Gülden E; Kolb H; Habich C; Burkart V, The Autoantigenic Proinsulin B-Chain Peptide B11–23 Synergises with the 70 kDa Heat Shock Protein DnaK in Macrophage Stimulation. Journal of Diabetes Research 2018, 2018, 4834673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bayer AL; Baliga P; Woodward JE, Transferrin receptor in T cell activation and transplantation. J Leukoc Biol 1998, 64, (1), 19–24. [DOI] [PubMed] [Google Scholar]
- 45.Neckers LM; Yenokida G; James SP, The role of the transferrin receptor in human B lymphocyte activation. J Immunol 1984, 133, (5), 2437–41. [PubMed] [Google Scholar]
- 46.De Block CE; Van Campenhout CM; De Leeuw IH; Keenoy BM; Martin M; Van Hoof V; Van Gaal LF, Soluble transferrin receptor level: a new marker of iron deficiency anemia, a common manifestation of gastric autoimmunity in type 1 diabetes. Diabetes Care 2000, 23, (9), 1384–8. [DOI] [PubMed] [Google Scholar]
- 47.Kallionpää H; Somani J; Tuomela S; Ullah U; de Albuquerque R; Lönnberg T; Komsi E; Siljander H; Honkanen J; Härkönen T; Peet A; Tillmann V; Chandra V; Anagandula MK; Frisk G; Otonkoski T; Rasool O; Lund R; Lähdesmäki H; Knip M; Lahesmaa R, Early Detection of Peripheral Blood Cell Signature in Children Developing β-Cell Autoimmunity at a Young Age. Diabetes 2019, 68, (10), 2024–2034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1. Scatter plots of HLA-I immunopeptidome of 2676 (T1D) and 2675 (HC) B cells.
Fig S2. Binding motifs for the HLA-I and -II immunopeptides of two B cell lines (2676 (T1D) and 2675 (HC)).
Fig S3. Scatter plots of HLA-II immunopeptidome of 2676 (T1D) and 2675 (HC) B cells.
Fig. S4. Cellular functions of the HLA-I and -II antigens dysregulated in T1D.
Fig S5. List of HLA-I immunopeptides selected for verification by PRM.
Fig S6. List of HLA-II immunopeptides selected for verification by PRM.
Fig S7. Representative chromatograms of verified HLA-I immunopeptides.
Fig S8. Representative chromatograms of verified HLA-II immunopeptides.
Fig S9. Number of immunopeptides enriched from six additional B cell lines.
Fig S10. Composition of immunopeptides identified in six B cell lines according to HLA alleles.
Fig S11. Binding motifs for the HLA-I immunopeptides of six additional B cell lines.
Fig S12. Binding motifs for the HLA-II immunopeptides of six additional B cell lines.
Fig S13. Peptides identified from HLA molecules.
Table S1.Clinical data and HLA-I and -II allele information of B cells.
Table S2. HLA-I immunopeptidome of 2676 (T1D) and 2675 (HC) B cells.
Table S3. HLA-II immunopeptidome of 2676 (T1D) and 2675 (HC) B cells.
Table S4. Differentially regulated HLA-I immunopeptides in 2676 (T1D) B cells relative to 2675 (HC) B cells.
Table S5. Differentially regulated HLA-II immunopeptides in 2676 (T1D) B cells relative to 2675 (HC) B cells.
Table S6. Differentially regulated allele-specific HLA-I immunopeptides in 2676 (T1D) B cells relative to 2675 (HC) B cells.
Table S7. Differentially regulated allele-specific HLA-II immunopeptides in 2676 (T1D) B cells relative to 2675 (HC) B cells.
Table S8. HLA-I immunopeptidome of six additional B cells.
Table S9. HLA-II immunopeptidome of six additional B cells.
Table S10. Differentially regulated HLA-I immunopeptides in six additional B cells.
Table S11. Differentially regulated HLA-II immunopeptides in six additional B cells.
Table S12. Validation of HLA-I immunopeptides using additional six B cell lines.
Table S13. Validation of HLA-II immunopeptides using additional six B cell lines.