Since 1994, the JAMA Guides have been advising clinicians how to use articles about diagnostic tests (5, 6, 16). Consistent with the principles of evidence-based medicine (EBM), clinicians are first expected to assess the validity of a test and then to decide whether it is of importance in the care of their patients. By making use of likelihood ratios (LRs), they should be able to revise pretest probabilities (PreTPs) of an illness and convert them into significant posttest probabilities (PostTPs) simply by using a nomogram or by doing rather simple calculations. As a consequence, it seems reasonable to expect clinical microbiologists, as well as other laboratory specialists, to know the LRs of their tests, to communicate these to physicians, and to implement their exploitation.
A popular booklet dedicated to EBM (15) is supplied with several cards, allowing caregivers to make rapid calculations for numbers needed to treat, PostTPs, and numbers needed to harm, for therapy, diagnosis, and prognosis problems, respectively. In the future, more and more physicians will probably be keeping nomograms or calculators in their white-coat pockets, either to obtain near-bed probabilities or to work up to them when appraising papers in the library. We and other laboratory people should offer our partnership to them. We should try to summarize the properties of our tests in only one number, to be provided in addition to other statistics.
Actually, the LR seems to be the best candidate for this number because it can meet the principal goal of a diagnostic test, i.e., the revision of disease probability. Even properties of qualitative tests in clinical microbiology are commonly expressed as sensitivity and specificity. According to Gallagher (4), what these conventional terms actually tell us is how likely a test result is to be positive or negative, given that a patient does or does not have the target disorder for which one is testing. There is a paradoxical inversion of customary clinical logic intrinsic to this definition, since knowledge of whether the patient had the illness would clearly obviate the need for a diagnostic test in the first place. On the other hand, predictive values (PVs) tell us what we really want to know clinically, which is the probability that a patient does or does not have the illness, given that a test result is positive or negative. Unfortunately, as disease prevalence decreases in a population, the positive PV (PPV) falls and reciprocally the negative PV (NPV) rises (and vice versa). LRs give us the same information as PVs without being subject to shifts in disease prevalence.
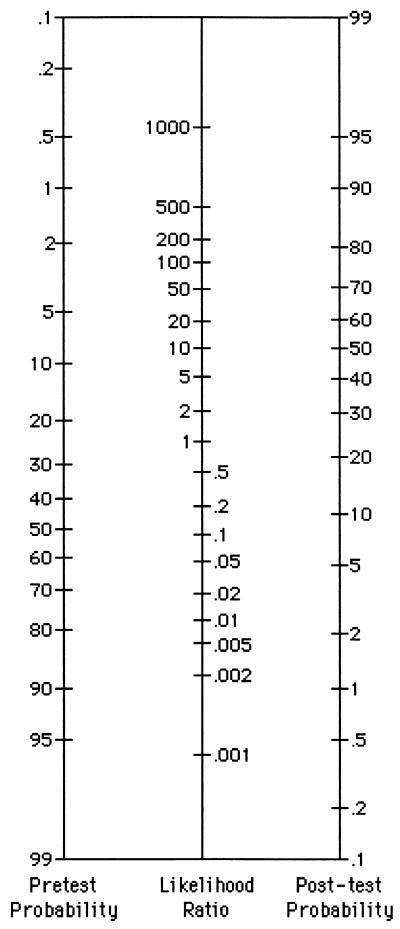
Let's imagine that an obstetrician is interested in a microbiology report on a DNA-probe method for Neisseria gonorrhoeae; it could replace the standard N. gonorrhoeae culture in the hospital laboratory. The sensitivity of the new test is 0.97 (97%), and the specificity is 0.99 (99%). The PPV and NPV are indicated as 0.99 (99%) and 0.97 (97%), respectively; they are based on a prevalence of 50%, because of the way the validation study was designed: 100 patients with positive N. gonorrhoeae cultures and 100 patients with negative cultures (see 2×2 tables and related calculations in reference 3). The physician feels confident about a positive result of this new test for a patient with a cervical purulent discharge. Fifty-fifty is a fairly close estimate of PreTPs of having N. gonorrhoeae in the local setting. But what about a positive result for a women who has undertaken a prenatal screening, if the prevalence of gonorrhoea is about 1%? To answer this question, a new PPV should be considered. LRs can provide just the same information in a quicker way. LRs were not reported by Sackett et al. (15), but it should be easy to obtain them with the aid of a pocket calculator by using the formulas LR(+) = sensitivity/(1 − specificity) and LR(−) = (1 − sensitivity)/specificity. In the examples, LR(+) = 0.97/(1 − 0.99) = 97 and LR(−) = (1 − 0.97)/0.99 = 0.03. The physician picks up the Fagan nomogram (Fig. 1) from his/her pocket (15); by extending a straightedge from the PreTPs (1%) through the LR(+) of 97, the point of intersection of the line with the PostTP axis defines a posterior probability of 49%. He/she could flip a coin and get just the same chance of knowing the truth. Afterwards, doing the same maneuver though the LR(−) of 0.03, the physian realizes that a negative result looks quite credible: the intersection with the PostTP is below 0.1%, which means that the chance of the result being truly negative is more than 99.9%. Nevertheless, the new method does not prove to be really effective. The obstetrician concludes that is is probably better to trust cultures for prenatal screenings.
FIG. 1.
Nomogram for interpreting diagnostic test results (http://cebm.jr2.ox.ac.uk).
Just the same kind of questions, though in a much more dramatic environment, could be asked by physicians in the emergency department. I do not know if anyone has done so, but I can imaging that a sequential application of LRs could be useful in critical conditions. Jaeschke and colleagues state the following about sequential testing (6): “… each item of history, or each finding on physical examination, represents a diagnostic test. We generate pretest probabilities that modify with each new finding … .” In other words, in sequential testing, each PostTP acts as the PreTP for the following step. Laboratory tests, i.e., a rapid antigen detection test, CRP, and peripheral leukocyte count, could be exploited in just that way for a patient suspected of having acute meningitis.
Where do PreTPs come from? Obviously, they can be difficult to estimate, just as a physician can encounter difficulties in making diagnoses. A PreTP has been defined as an early assessment of diagnostic possibilities before the test is performed, based on individual clinical expertise (6, 15). Physicians can derive PreTPs from their own accumulating clinical experience, specific for the setting in which they work. Early in their careers, they must learn to select both leading diagnostic hypothesis and likely alternatives (16). This implies quantifying chances that a condition exists or not, as the obstetrician has done above, in order to emphasize either specificity or sensitivity. Besides, the authors of studies of disease probability are expected to display tables listing the diagnoses made and the numbers and percentages of patients with those diagnoses, along with confidence intervals (13, 16). Moore states that clinicians and laboratory people should work together in order to obtain prevalence data of diseases, as well as test sensitivities and specificities needed for probability calculations (10).
Today, the EBM language is spoken by the most important medical journals, not to mention the Cochrane Library. The use of LRs has been described for diagnosis of venous-catheter infections (9), osteomyelitis (18), parasite infections (7), meningitis (1), sexually transmitted diseases (11), Lyme disease (17), Helicobacter pylori infection (12, 14), and prosthetic joint infections (2). In addition to their mention in cited articles and books (5, 6, 15), LRs and other epidemiological tools are fully explained in several websites (http://cebm.jr2.ox.ac.uk [Center of EBM Oxford University, Oxford, United Kingdom] and http://www.cdc.gov/global [a teaching module developed by the Centers for Disease Control and Prevention]). Nevertheless, an eager clinician is generally faced with a still scanty and sparse availability of the parameters he/she needs to critically appraise laboratory literature (8), also in the clinical microbiology field.
In my opinion, a way of implementing EBM as part of normal clinical microbiological testing could be by using LRs whenever possible. Clinicians should be aware that the real value of LRs lies in their ability to optimize the power of diagnostic tests to revise disease probability. I agree that, in order to do this, a clinician-microbiologist partnership must exist, and I recognize that this will be quite difficult. This is what our group is trying to accomplish, but we have not had much success up to now. It would be easier if major clinical microbiology journals implemented their use of LRs and if the test properties that make them suited for a critical appraisal (5, 6) were made more evident (validity, reproducibility, reduction of spectrum bias, verification bias, etc.).
REFERENCES
- 1.Asawavichiiennjinda T, Sitthi-Amorn C, Tanyanont V. Serum cryptococcal antigen: diagnostic value in the diagnosis of AIDS-related cryptococcal meningitis. J Med Assoc Thail. 1999;82:65–71. [PubMed] [Google Scholar]
- 2.Atkins B L, Athanasou N, Deeks J J, Crook D W, Simpson H, Peto T E, McLardy Smith P, Berendt A R The OSIRIS Collaborative Study Group. Prospective evaluation of criteria for microbiological diagnosis of prosthetic-joint infection at revision arthroplasty. J Clin Microbiol. 1998;36:2932–2939. doi: 10.1128/jcm.36.10.2932-2939.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elder B L, Hansen S A, Kellog J A, Marsik F J, Zabransky R J. Method selection, example. In: McCurdy B W, editor. Verification and validation of procedures in the clinical microbiology laboratory (Cumitech 31) 1997. p. 14. Appendix A. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 4.Gallagher E J. Clinical utility of likelihood ratios. Ann Emerg Med. 1998;31:391–397. doi: 10.1016/s0196-0644(98)70352-x. [DOI] [PubMed] [Google Scholar]
- 5.Jaeschke R, Guyatt G, Sackett D L (EMB Working Group) User's guide to the medical literature. III. How to use an article about a diagnostic test. A. Are the results of the study valid? JAMA. 1994;271:388–91. doi: 10.1001/jama.271.5.389. [DOI] [PubMed] [Google Scholar]
- 6.Jaeschke R, Guyatt G, Sackett D L (EMB Working Group) User's guide to the medical literature. III. How to use an article about a diagnostic test. B. What are the results and will they help me in caring for my patients? JAMA. 1994;271:703–707. doi: 10.1001/jama.271.9.703. [DOI] [PubMed] [Google Scholar]
- 7.Joseph L, Gyorkos T W. Inferences for likelihood ratios in the absence of a gold standard. Med Decis Making. 1996;16:412–417. doi: 10.1177/0272989X9601600412. [DOI] [PubMed] [Google Scholar]
- 8.Ljimer J G, Mol B W, Heisterkamp S, Bonsel G J, et al. Empirical evidence of design-related bias in studies of diagnostic tests. JAMA. 1999;282:1061–1066. doi: 10.1001/jama.282.11.1061. [DOI] [PubMed] [Google Scholar]
- 9.McGeer A, Righter J. Improving our ability to diagnose infections associated with central venous catheters: value of Gram's staining and culture of entry site swabs. CMAJ. 1987;137:1009–1015. [PMC free article] [PubMed] [Google Scholar]
- 10.Moore R A. On the need for evidence-based clinical biochemistry. Evidence-Based Med. 1998;3:7–8. [Google Scholar]
- 11.Ndoye I, Mboup S, Schryver A, Van Dick E, Moran J, Samb N D, Sakho M L, Thior I, Wade A, Heymann D L, Meheus A. Diagnosis of sexually transmitted infections in female prostitutes in Dakar, Senegal. Sex Transm Infect. 1998;74(Suppl. 1):S112–S117. [PubMed] [Google Scholar]
- 12.Oderda G, Rapa A, Ronchi B, Lerro P, Pastore M, Staiano A, de Angelis G L, Strisciuglio P. Detection of Helicobacter pylori in stool specimens by non-invasive antigen immunoassay in children: multicentre Italian study. Br Med J. 2000;320:347–348. doi: 10.1136/bmj.320.7231.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orioski A, Hayes E B, Campbell G L, Dennis D T. Surveillance for Lyme disease—United States, 1992–1998. Division of Vector-Borne Infectious Diseases, National Center for Infectious Diseases. Morb Mortal Weekly Rep. 2000;49(SS03):1–11. [PubMed] [Google Scholar]
- 14.Quartero A O, Numans M E, de Melker R A, de Wit N J. In-practice evaluation of whole-blood Helicobacter pylori test: its usefulness in detecting peptic ulcer disease. Br J Gen Pract. 2000;50:13–16. [PMC free article] [PubMed] [Google Scholar]
- 15.Sackett D L, Scott Richardson W, Rosenberg W, Haynes R B. Evidence-based medicine. How to practice & teach EBM. New York, N.Y: Churchill Livingstone; 1998. p. 250. [Google Scholar]
- 16.Scott, Richardson W, Wilson M C, Guyatt G H, Cook D J, Nishikawa J (EBM Working Group) User's guide to the medical literature. XV. How to use an article about disease probability for differential diagnosis. JAMA. 1999;281:1214–1219. doi: 10.1001/jama.281.13.1214. [DOI] [PubMed] [Google Scholar]
- 17.Tugwell P, Dennis D T, Weinstein A, Wells G, Shea B, Nichol G, Hayward R, Lightfood R, Baker P, Steere A C. Labroatory evaluation in the diagnosis of Lyme disease. Ann Intern Med. 1997;127:1109–1123. doi: 10.7326/0003-4819-127-12-199712150-00011. [DOI] [PubMed] [Google Scholar]
- 18.Wrobel J S, Connolly J E. Making the diagnosis of osteomyelitis. The role of prevalence. J Am Podiatr Med Assoc. 1998;88:337–343. doi: 10.7547/87507315-88-7-337. [DOI] [PubMed] [Google Scholar]