Abstract
The ribosome is one of the main antibiotic targets. Many different types of inhibitors can stop cells from growing by binding at functional centers of the ribosome and interfering with its ability to synthesize proteins. These antibiotics were usually viewed as general protein synthesis inhibitors, which indiscriminately stop translation at every codon of every mRNA preventing the ribosome from making any protein. However, at each step of the translation cycle, the ribosome interacts with multiple ligands (mRNAs, tRNA substrates, translation factors, etc.), and as the result, the properties of the translation complex vary from codon to codon and from gene to gene. Therefore, rather than being indiscriminate inhibitors, many of the ribosomal antibiotics impact protein synthesis in a context-specific manner. This review presents a snapshot of the growing body of evidence that some, and possibly most, of the ribosome-targeting antibiotics manifest site-specificity of action, which is modulated by the nature of the nascent protein, the mRNA or the tRNAs.
Keywords: ribosome, translation, protein synthesis, antibiotic, resistance, chloramphenicol, linezolid, macrolides, ketolides, erythromycin, kasugamycin, pactamycin, peptidyl transferase center, nascent peptide exit tunnel
INTRODUCTION
Antibiotics, drugs that inhibit the growth of bacteria, including pathogenic ones, are the most successful medicines found by humans and have saved millions of lives. Unfortunately, the broad use of antibiotics, both legitimate and frivolous, has resulted in the spread of resistant strains, against which many antibiotics no longer work. Therefore, finding new natural and synthetic antibiotics, or modifying the strategies to use the known compounds, is one of the most urgent tasks of the modern health sciences.
The absolute majority of currently known antibiotics have been found serendipitously, by testing secondary metabolites produced by soil or marine microorganisms or by screening immense libraries of synthetic compounds. Optimization of prototype antibiotics has been also largely driven not by knowledge of the mechanism of action, but by synthesizing a large number of derivatives and testing them in various assays. Although these approaches have been occasionally successful in yielding some excellent antibiotics currently used in the clinic, the paucity of new drugs in the pipeline calls for unorthodox and innovative approaches that would revitalize the discovery process. A clear understanding of how antibiotics work at the molecular level and actively exploiting this knowledge to make better medicines should become the guiding principle of drug discovery endeavors.
Our current knowledge of the mechanisms of antibiotic action is in fact very limited. In many instances the commonly accepted views on how the antibiotic achieves its inhibitory effect on the target and cell growth are based on research carried out decades ago, using the fairly limited arsenal of methods available at the time. Furthermore, the findings of those studies, many of which date back to the 60s and 70s of the 20th century, were inevitably interpreted within the framework of the then contemporary understanding of the biology and chemistry of the cell and its components. Breakthrough discoveries of the subsequent years in the areas of gene expression, cell physiology, and structural biology since then have not necessarily been incorporated into the current models of the mechanisms of action of many antibiotic families.
The majority of antibiotics inhibit growth and proliferation of bacteria by targeting one of the essential enzymes. Most of such enzymes catalyze fairly simple biochemical reactions, where either two substrates are united into a single molecule, an individual compound is split into two products, or a certain molecule is transformed into its isoform. In these cases, enzyme inhibition generally means that the catalyzed reaction becomes too slow to sustain cell growth. However, the straightforward rules of enzyme inhibition do not necessarily apply to one of the main antibiotic targets in the bacterial cell, the ribosome, because it handles a multiplicity of substrates and functional partners during the complex process of protein synthesis.
A great variety of clinically used and investigational antibiotics achieve their therapeutic effects by interfering with the functions of the ribosome, the central component of the gene expression apparatus indispensable for sustaining life. In a fast growing cell, the majority of the ribosomes are involved in translation and therefore, antibiotics encounter ribosomes executing varying steps of protein synthesis (Figure 1). By analogy with the conventional enzymes, it has been largely assumed that ribosomal antibiotics arrest translation equally efficiently at every codon of a gene. However, one should keep in mind that at every codon, the translation complex is different and antibiotics encounter ribosomes engaged with different mRNAs and translation factors, associated with diverse combination of tRNA substrates, and carrying distinct nascent peptides. Thus, during progression through the mRNAs coding sequences, the ribosome catalyzes peptide bond formation between 400 possible combinations of donor and acceptor substrates (only counting the major 20 amino acids) and is transiently occupied with an even greater number of tRNA combinations. Thus, even without considering the contribution of the ribosome-associated nascent proteins and mRNA structures, which change as the ribosome moves from codon to codon, the variety of ribosomal complexes potentially available in the cell to interact with an antibiotic is enormous.
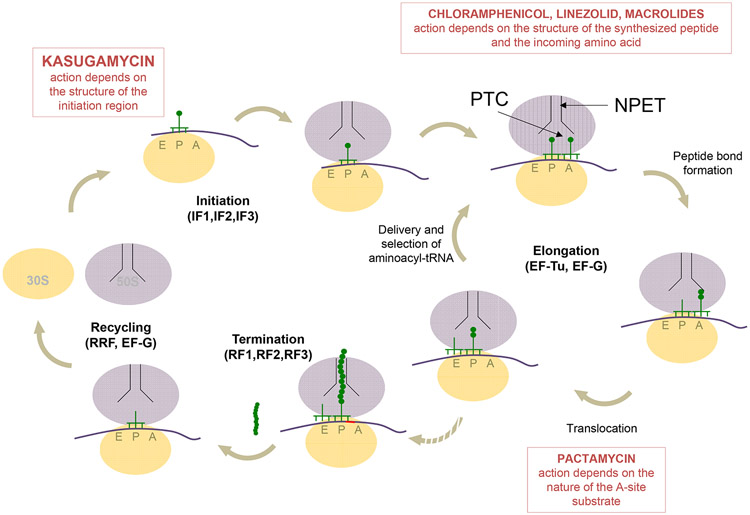
Figure 1.
An overview of protein synthesis and the steps inhibited by antibiotics with context-specific mode of action. At the initiation step, the ribosome, assisted by initiation factors (IF1-3), locates the start codon in mRNA and binds the initiator tRNA in the P site. Kasugamycin selectively interferes with initiation of translation of some mRNAs depending on the structure of their ribosome binding site. During translation elongation, EF-Tu-delivered aminoacyl-tRNAs are selected and then accommodated in the A site. As the peptide bond is formed in the peptidyl transferase center (PTC), the growing nascent protein is transferred to the A site tRNA. The antibiotic-mediated direct (chloramphenicol and linezolid) or allosteric (macrolides) inhibition of peptide bond formation depends on the structure of the nascent protein. Subsequent translocation of the ribosome is promoted by EF-G. Inhibition of translocation by pactamycin may depend on the nature of the A site substrate. During translation elongation, the growing nascent protein is threaded through the nascent peptide exit tunnel (NPET). When the ribosome encounters a stop codon, it enters the termination phase, during which the completed protein is released with the help of termination factors (RF1 or RF2, and RF3). Finally, at the recycling phase, the combined action of ribosome recycling factor (RRF) and EF-G splits the ribosome into its subunits.
It is well established that the nature of the ligands can significantly affect the properties of the ribosome. The different tRNAs can bind with distinct kinetics (18; 27) and accuracies (16; 78) and can be translocated with different efficiencies and rates (23; 28; 44). In addition, the efficiency of peptide bond formation is critically influenced by the nature of the donor and acceptor amino acids (39; 106). Because these factors modify the properties of the ribosome as a decoder and catalyst, they should also have a profound impact on the action of the antibiotics that interfere with ribosomal functions. With all these considerations in mind, the concept of a universal inhibitor of protein synthesis, whose action would be independent of the nature of the substrates and conformations of the ribosome, appears highly improbable.
In this review, we will discuss the accumulating evidence that several classes of ribosomal antibiotics inhibit translation in a ligand- or context-specific way. We will present several well-documented examples of major classes of ribosome-targeting antibiotics whose activities are context specific. We will also discuss the current experimental tools that are suitable for investigating the codon- and gene-specific action of antibiotics.
NEW GENOMIC AND BIOCHEMICAL TECHNIQUES PROVIDE TOOLS FOR REVEALING CONTEXT-SPECIFICITY OF ANTIBIOTIC ACTION
Why has the concept that the action of ribosomal antibiotics could be ligand- or context-specific not been explored in early studies? A simple explanation is that the techniques available at the dawn of the antibiotics era were inadequate for either detecting or examining the gene-specific and moreover codon-specific action of ribosomal inhibitors. Studying inhibition of bulk protein synthesis in vivo or of translation of a single reporter gene in vitro does not tell whether the effect of the drug is the same on all of the cellular proteins. Furthermore, biochemical studies, commonly involving surrogates of tRNA substrates, could tell little about the possible influence of various ribosomal ligands on drug action.
The major advances in our understanding of how ribosomal antibiotics work have been brought about by several recently developed powerful in vivo and in vitro techniques. Arguably, the most notable among them are ribosome profiling (Ribo-Seq) and primer-extension inhibition (toeprinting) analysis.
Ribo-Seq shows the distribution of ribosomes along translated mRNAs in the cell (36; 59) (Figure 2a). The technique is based on isolation and deep-sequencing of the ribosome-protected mRNA fragments and mapping such ribosome footprints to the genome. The Ribo-Seq data reveal the average ribosome occupancy of every single codon of the gene. Comparison of individual codon occupancy between the antibiotic-treated cells and the untreated controls reveals the mRNA sites where the drugs exert the most pronounced inhibition of translation: the occupancy of such codons will be increased in antibiotic-treated cells. Conversely, the codons where the ribosome could successfully evade the inhibitory action of the drug will show decreased occupancy.
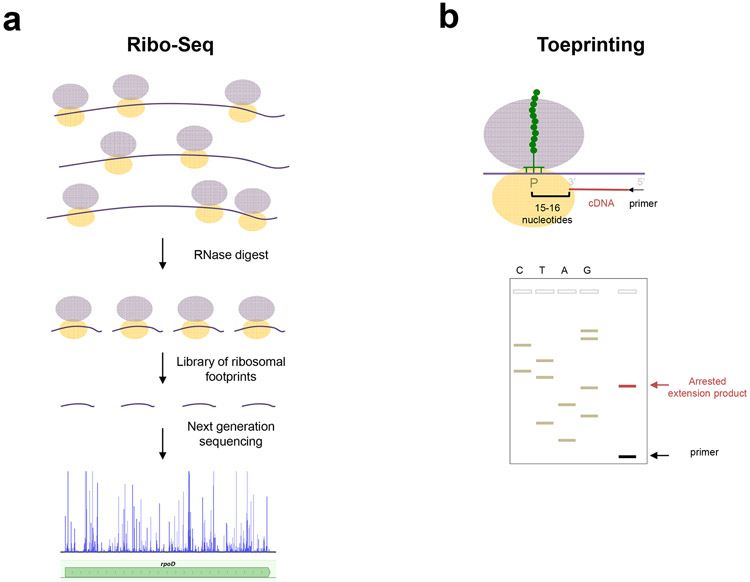
Figure 2.
Methodologies for studying the context specific action of ribosomal antibiotics. (a) Ribo-Seq, also known as ribosome profiling, involves the isolation, deep-sequencing and mapping to the genome of ribosomal footprints, the mRNA fragments associated with the translating ribosomes in the living cell. Analysis of antibiotic-induced changes in the ribosomal footprints pattern at the codon or gene level reveals that the action of several antibiotic families is context specific. (b) In vitro toeprinting analysis allows for mapping the position of an arrested ribosome in mRNA with codon precision. Top: extension of a DNA primer (black arrow) by reverse transcriptase (not shown) is interrupted by the presence of a stalled ribosome. The 1st nucleotide of the codon occupying the P site and the 3’-terminal end of the truncated-cDNA are separated by a distance of 15-16 nucleotides. Bottom: the cDNA products, along with sequencing reactions, are resolved in a sequencing gel. The position of the stalled ribosome is inferred from the migration of the toeprint band. Antibiotic-dependent appearance of toeprint bands reveals the specific codons where translation is arrested.
In vitro toeprinting analysis allows locating the ribosome within an mRNA with codon precision (33). The assay is based on annealing a DNA primer to the in vitro-translated mRNA template and extending it with reverse transcriptase. The reverse transcriptase stops the primer extension when it encounters a ribosome paused on the mRNA and mapping the 3’ end of the synthesized complementary DNA allows the precise determination of the codon at which the ribosome has stalled (Figure 2b). The application of toeprinting for studying ribosome progression along mRNA has been made possible by the development of the so-called PURE system, where each of the proteins required for translation, tRNAs and ribosomes are individually purified and then combined together (84). The toeprinting technique has been successfully applied to studies of intrinsic and drug-induced translation arrest (56; 100).
Several other technologies can also advance the understanding of antibiotic specificity. The single-molecule fluorescence resonance energy transfer (smFRET) technique monitors the progression of a single ribosome through a number of mRNA codons by monitoring the intersubunit rotation or substrate binding (14; 79). Analysis of smFRET traces makes it possible to extract antibiotic-induced changes in the kinetics of the ribosome traversing mRNA, as well as the effect of the drugs on the life-time of the individual functional states of the ribosome at individual codons. Finally, the recent progress in increasing the resolution of cryo electron microscopic (Cryo-EM) reconstructions of ribosomal complexes, as well as continuing improvement of the crystallographic structures of the ribosome, makes it possible to analyze the interactions of antibiotics with the ribosome associated with functional substrates (4; 5; 72; 73; 75). The application of innovative technics has made it possible to gain insights into the specificity of action of several classes of ribosomal inhibitors that we discuss in the following sections.
ACTION OF PEPTIDYL TRANSFERASE-TARGETING ANTIBIOTICS DEPENDS ON THE NATURE OF THE DONOR AND ACCEPTOR SUBSTRATES
Formation of peptide bonds, that takes place in the peptidyl transferase center (PTC), is the only reaction catalyzed by the ribosome that involves rearrangement of covalent bonds. Although ribosomal RNA is involved in the proton relay required for peptidyl transfer (26; 74), the ribosome acts primarily as an entropic catalyst and the major acceleration of the reaction comes from the proper placement of the donor (peptidyl-tRNA) and acceptor (aa-tRNA) substrates (69; 86; 108).
The PTC is a prevalent target for protein synthesis inhibitors (70; 104). Most of the PTC drugs sterically hinder accommodation of the substrates in the active site. One of the oldest known PTC-targeting antibiotics is chloramphenicol (CHL), a natural compound produced by several Streptomyces species (Figure 3a) (96). CHL has always been considered a classic PTC inhibitor (55; 67; 103), a view compatible with structural studies that show its binding in the A site, occupying the space required for the placement of the acceptor amino acid esterifying tRNA (12; 24; 81) (Figure 3b). Because the proper placement of the acceptor substrate is critical for peptide bond formation, CHL was thought to efficiently block formation of any and every peptide bond.
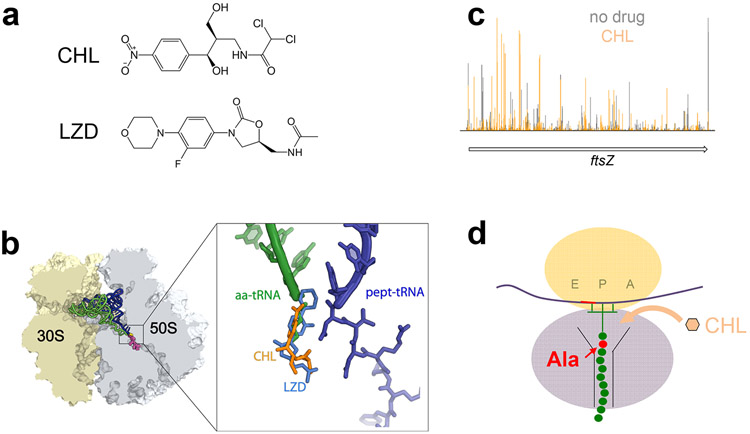
Figure 3.
The context specific action of PTC inhibitors chloramphenicol (CHL) and linezolid (LZD). (a) The chemical structures of CHL and LZD. (b) The binding site of CHL and LZD overlaps with that of the aminoacyl moiety of the A site aa-tRNA. (c) RiboSeq analysis shows that CHL (and LZD) causes redistribution of ribosomes on mRNA (51). In the presence of either one of the inhibitors, ribosomes are preferentially arrested at the codons that immediately follow Ala codons. (d) Cartoon representation of the preferred target of CHL (or LZD), a ribosome carrying a nascent peptide whose penultimate residue is an alanine.
Although the model of CHL action as a universal inhibitor of peptide bond formation has been broadly accepted and was supported by the ability of the drug to inhibit peptidyl transfer in some model assays, it could not easily account for a number of experimental results. For example, in vivo synthesis of specific polypeptides was seemingly less sensitive to the presence of CHL than translation of the majority of the proteins (reviewed in (67)). Furthermore, appreciable in vitro inhibition of peptide bond formation between some substrates required strikingly high (≥ 1 mM) concentrations of CHL (13; 31; 66).
A generally similar controversy has been accumulating about the mode of action of another PTC-targeting drug, linezolid (LZD), a synthetic oxazolidinone antibiotic introduced in the clinic 50 years after CHL (52) (Figure 3a). LZD binds to a ribosomal site that closely matches that of CHL (9; 37; 46; 105) (Figure 3b). Therefore, similar to CHL, LZD was thought to act as a universal inhibitor of peptidyl transfer because it distorts the placement of the acceptor amino acid in the PTC. However, in model assays utilizing fMet-tRNAiMet as a donor substrate, LZD either completely failed to inhibit peptide bond formation (43) or produced only modest inhibition even at high concentrations (65). Yet, it could readily inhibit in vitro synthesis of reporter proteins or the overall protein synthesis in bacterial cells (85).
Many of the discrepancies regarding the action of CHL and LZD have been resolved by understanding that neither CHL nor LZD are universal inhibitors of peptide bond formation. Instead, these PTC-binding drugs interfere with translation elongation in a context-specific manner, that is by preferentially arresting ribosomes at specific mRNA sites. The key experimental data that revealed context-specificity of action of these drugs came from Ribo-Seq experiments, designed to understand the action of CHL and LZD in vivo (51; 57). In these experiments, E. coli cells were exposed to high concentrations of CHL or LZD for a short period of time that was sufficient to completely inhibit protein synthesis and the distribution of ribosomes along mRNAs was analyzed. If, as the classical model would have predicted, CHL and LZD were to inhibit equally well formation of any peptide bond, the drugs would have simply ‘frozen’ the ribosomes on the mRNA codons where the encounter with the antibiotic had occurred. In this case, the distribution of the ribosome footprints along mRNA would not have changed relative to untreated control cells. However, analysis of the Ribo-Seq data revealed something very different: exposure to CHL or LZD caused reallocation of the ribosomes along mRNAs (Figure 3c). It became evident that upon encountering the antibiotic, the majority of the ribosomes still polymerize several peptide bonds and progress through several codons before being arrested at specific sites.
What distinguishes the sites of preferential action of CHL and LZD? Is it the nature of the amino acids participating in peptide bond formation, the structure of tRNAs or a unique folding of the mRNA? Unexpectedly, bioinformatics analysis of the Ribo-Seq data showed that the primary factor that defined the sites of the most pronounced inhibition of translation by CHL or LZD was none of those factors, but the nature of the nascent peptide. Specifically, CHL most efficiently arrested ribosomes when the penultimate amino acid of the growing protein was an alanine (Figure 3d) and, somewhat less frequently, a threonine or a serine (51). Because of that, the ribosomes were most readily arrested by CHL at the codons that followed Ala, Thr or Ser codons. The trend was even more pronounced with LZD, where the majority of the arrest sites occurred at the codons that were preceded specifically by an Ala codon. While the penultimate residue of the nascent peptide played the major role in defining the sites of CHL and LZD action, the identities of the amino acids at the P and A sites also influenced CHL and LZD site-specificity. Analysis of the codons at which ribosome density was reduced in antibiotic-treated cells showed that the presence of certain amino acids, in particular glycine, in the P or A sites of the PTC prevented LZD and CHL from arresting translation (51).
For a number of sites, the trend revealed by Ribo-Seq could be reproduced in cell-free translation assays. Toeprinting analysis confirmed that CHL and LZD most efficiently arrest translation when the ribosome carries a nascent chain with an alanine in the penultimate position and mutagenizing this Ala to other amino acids decreased antibiotic-mediated arrest (51). In agreement with the Ribo-Seq data, glycine residues in the P or A sites of the PTC counteract the in vitro action of either of the inhibitors.
Noteworthy, toeprinting experiments also showed that neither CHL nor LZD could efficiently inhibit formation of the first peptide bond. In view of the aforementioned findings this is not really surprising: the initiating ribosome does not carry a nascent peptide with a penultimate alanine. Corroborating this observation, metagene analysis of the Ribo-Seq data shows that in cells treated with CHL or LZD, the ribosomes redistribute from the initiator codons of the genes and are eventually arrested at downstream codons (51). In retrospect, because most of the classic in vitro assays of peptide bond formation employed fMet-tRNAiMet as a donor substrate, it is not surprising that very high concentrations of CHL or LZD were required to achieve even marginal inhibition of the transpeptidation reaction involving this substrate (43; 55).
The mechanistic underpinnings of site-specificity of action of the PTC-targeting CHL and LZD still remain to be elucidated. One plausible scenario is that the presence of Ala, Thr or Ser in the penultimate position of the nascent protein favors drug binding due to a direct interaction of the A site bound antibiotic with the short side chain of these amino acids. Other amino acids may directly or allosterically hinder antibiotic binding or, in the case of glycine, not provide the contacts that would increase the drug’s affinity for the PTC. The inability of CHL or LZD to efficiently inhibit peptide bond formation involving glycine as an acceptor could be explained by co-accommodation of the antibiotic and an amino acid with the simplest possible side chain (a hydrogen atom) in the PTC A site.
MACROLIDE ANTIBIOTICS ARREST THE RIBOSOME AT SITES WITH SPECIFIC SEQUENCE MOTIFS
Macrolides are among the most clinically successful ribosome-targeting drugs. The prototype macrolide, erythromycin (ERY) (Figure 4a), as well as drugs of the second generation (such as azithromycin (AZI) or clarithromycin) have been broadly used for decades for the treatment of infections caused by Gram-positive pathogens (2; 22). The medical use of macrolides of the third generation, the ketolides, (e.g., telithromycin (TEL)) (Figure 4a), is thus far limited due to toxicity issues, but their significantly increased efficacy compared to that of previous macrolides attracts important efforts for developing them into clinically-useful antibacterials (reviewed in (11; 29)). A recent, highly innovative chemical platform, which allows combinatorial synthesis of a great variety of new macrolide compounds from their basic building blocks (83), increases the plausibility of finding new clinically efficient compounds of this class.
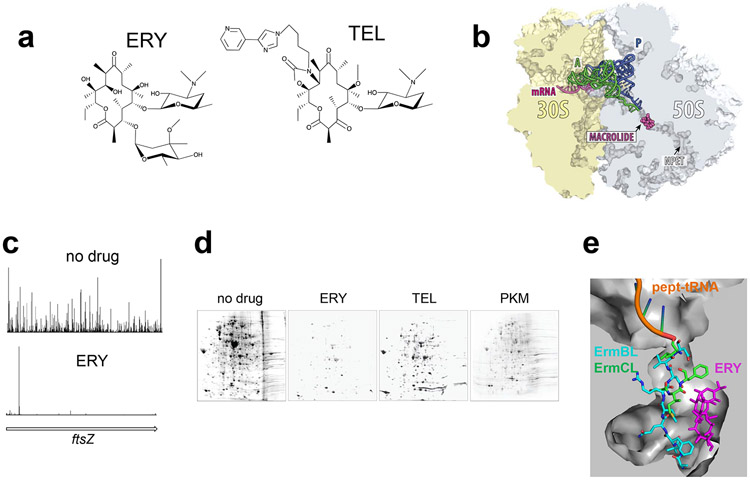
Figure 4.
Macrolide antibiotics halt translation of proteins carrying specific sequence motifs. (a) The chemical structures of the prototype macrolide erythromycin (ERY) and the ketolide telithromycin (TEL). (b) Macrolides and ketolides bind at the NPET and partially obstruct the passage of the newly made protein. A and P site tRNAs are also shown. (c) Ribo-Seq data demonstrate that TEL (as well as other macrolides) causes translation arrest at discrete and specific sites along mRNAs (41). (d) Two-dimensional electrophoretic analysis of the proteins synthesized in E. coli cells exposed to saturating concentrations of ERY, TEL, or the natural ketolide pikromycin (PKM). (e) The different paths of the ErmBL and ErmCL nascent peptides in the NPET partially obstructed by the presence of an ERY molecule (4).
Macrolides bind at a short distance from the PTC in the nascent peptide exit tunnel (NPET) (12; 24; 81; 94), the conduit through which the newly made protein leaves the ribosome (Figure 4b). Because macrolides are bulky and the NPET is rather narrow, it has been thought that these drugs obstruct the NPET so significantly that translation of every protein should be aborted when the nascent chain becomes 4-10 amino acids long and its N-terminus reaches the macrolide binding site (91). Strangely, however, macrolides are unable to completely inhibit bulk protein synthesis, as estimated by incorporation of radiolabeled [35S]-Met into polypeptides. Even when E. coli cells are exposed to ERY concentration exceeding the minimal inhibitory concentration, MIC, by 100-fold translation continues for several hours at around 7% of the level of the untreated control (42). The incomplete inhibition effect is even more striking with ketolides. Residual translation remains at around 20% in cells treated with 100-fold MIC of TEL (42) and at equivalently high concentrations, the natural ketolides pikromycin or methymycin inhibit proteins synthesis by only 60% (1). Some in vitro experiments indicated that, while synthesis of some reporters (e.g. poly-Lys) is highly sensitive to macrolide inhibition, production of other polypeptides (poly-Phe or GFP) is nearly indifferent to the presence of specific macrolides (58; 63; 89). It was equally hard to explain the ability of short peptides with defined sequences to displace macrolide molecules from their binding site in the NPET (49; 92; 93).
Ribo-Seq experiments carried out in Gram-positive Staphylococcus aureus and Gram-negative E. coli cells treated with macrolides offered an explanation for many of these paradoxes (20; 41). It turned out that macrolides arrest the ribosomes only at a limited number of discrete mRNA sites defined by the presence of specific sequence motifs (Figure 4c).
One of the most prevalent macrolide-arrest motifs revealed by Ribo-Seq is Arg/Lys–x –Arg/Lys (where x represents any amino acid). The macrolide-bound ribosome halts progression at the second codon of the motif, when it needs to catalyze the transfer of the nascent protein carrying Arg or Lys in its penultimate position to the acceptor residue of the incoming Lys- or Arg-tRNA. Polymerizing this sequence, which was called the +x+ motif, because of the positive charge of Arg and Lys, represents a problem for the ribosome bound to any of the tested macrolides (20; 41; 88). Other sequences could also become troublesome, but only when specific macrolides are bound to the ribosome. For example, ERY or AZI, but not so much TEL, inhibit peptide bond formation between Pro and either positively- or negatively charged amino acids, and also impede the transfer of a nascent chain ending with Asp to Lys-tRNA (20; 41). In addition, some stretches of hydrophobic amino acids, such as the IFVI sequence of the ErmCL peptide encoded in the regulatory ORF of the inducible ermC macrolide resistance gene, causes translation arrest of the ribosome complexed with cladinose-containing macrolides, but not with ketolides (35; 97; 100). Importantly, not every occurrence of the main stalling motif +x+ is necessarily associated with macrolide-induced translation arrest: in some instances, a more extended context, likely involving more distal segments of the nascent chain, could modulate the inhibitory action of the macrolide antibiotic (Sothiselvam, Mankin, Vázquez-Laslop, unpublished).
Although CHL and LZD act in a context-specific manner, they nevertheless inhibit synthesis of all cellular proteins because virtually every protein in the bacterial cell contains Ala, Thr and/or Ser which define the sites of preferential action of these drugs. When it comes to macrolide antibiotics, the situation is different, because the action of these inhibitors calls for the presence of specific amino acid sequences which occur only sparingly in the structures of the proteins. If a protein lacks the sequence that is problematic for the macrolide-bound ribosome, then it can be synthesized even in cells exposed to the macrolide. Furthermore, because the nature and prevalence of arrest motifs depends on the structure of the antibiotic, different macrolides allow for continued translation of different subsets of polypeptides. Indeed, 2D-gel electrophoresis analysis of the proteins synthesized in macrolide-treated cells shows that production of many polypeptides remains unaffected (Figure 4d). In agreement with the [35S]-Met incorporation data, cells exposed to TEL synthesize a significant number of proteins (42). As discussed above, not only the +x+ but also other motifs are problematic for the ERY-bound ribosome, therefore, fewer proteins escape inhibition by ERY (Figure 4d)(42). Conversely, many more proteins are synthesized in the cells treated with natural ketolides pikromycin or methymycin, which arrest ribosomes less efficiently even at the commonly difficult +x+ motif (Figure 4d) (1). Noteworthy, these observations demonstrate that a protein synthesis inhibitor does not need to completely abolish translation to be an active antibiotic. As long as the expression of at least one essential protein is interrupted, the bacteria would not be able to grow and proliferate.
Although we still lack a detailed understanding of the molecular mechanisms that define the context-selectivity of macrolide action, recent studies have provided several useful insights (4; 6; 7; 77; 88; 99; 100). In the case of the +x+ motif, the extensive direct interaction of the drug in the NPET with the nascent protein segment preceding the arrest motif is apparently not required for halting translation. Thus, synthesis of the MRLR peptide is arrested at the third codon of its gene, when the tripeptide MRL barely reaches the antibiotic molecule in the NPET (87). To account for this result, it was proposed that macrolides allosterically affect the properties of the PTC, interfering with its ability to catalyze peptide bond formation between certain combinations of donor and acceptor substrates (87). In agreement with this model, biochemical and structural studies show rearrangement of key PTC nucleotides in response to the binding of macrolides in the NPET (4; 6; 7; 32; 87). Direct drug-nascent chain interactions may play a more important role with other arrest motifs. Thus the stalling sequence IFVI of ErmCL failed to arrest the ERY-bound ribosome when placed at the very N-terminus of a protein, whereas it actively halts translation if preceded by five more amino acids (87; 100).
Cryo-EM studies showed that the path of the growing protein chain in the macrolide-obstructed tunnel depends on the nascent peptide sequence and the structure of the NPET-bound antibiotic (4; 6; 7) (Figure 4e). In a general scenario, macrolide-induced site-specific translation arrest requires the combined effect of the structural restrains imposed upon the nascent chain in the drug-obstructed NPET and antibiotic-triggered alterations in the PTC structure, which likely compromise the functions of the catalytic center (99,Arenz, 2016 #8528).
We are only starting to understand why certain substrates cannot properly react in the PTC when macrolides are present. Biochemical studies have shown that not only the positive charge of the key amino acids of the +x+ motif but also the length of their side chains play an important role in making this sequence particularly problematic (88). It is possible that the presence of the antibiotic bound in the vicinity of the PTC makes it energetically favorable for the extended positively-charged side chain of the penultimate Arg or Lys of the nascent peptide to protrude in the direction of the PTC A site and obstruct the accommodation of the similarly charged and equally bulky acceptor amino acid. This view aligns well with the observations of the smFRET studies where the ERY-bound ribosome, stalled after synthesizing the IFVI stalling sequence of ErmCL, was reluctant to accept the incoming aminoacyl-tRNAs (38) and with the lack of aminoacyl-tRNA or its poor A-site accommodation in the cryo-EM reconstructions of the ribosome stalled during synthesis of the ErmBL regulatory peptide (4; 7).
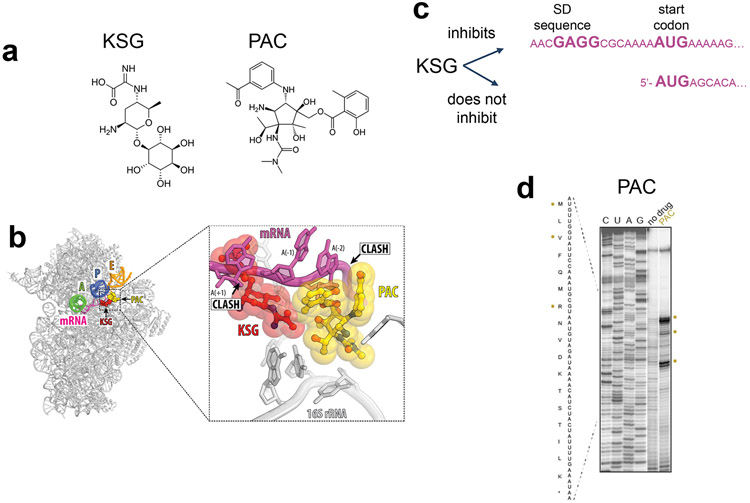
THE INITIATION INHIBITOR KASUGAMYCIN INTERFERES WITH TRANSLATION OF A SPECIFIC CLASS OF mRNAs
Kasugamycin (KSG) is an inhibitor of translation initiation (60; 71) whose binding site in the 30S subunit overlaps with the last 2 nucleotides of the E-site codon and encroaches upon the first nucleotide of the P site codon (72; 80; 82) (Figure 5a,b). The antibiotic distorts the mRNA path in the ribosome, preventing recognition of the start codon by the initiator fMet-tRNAiMet. On the basis of this proposed mechanism, one would predict that initiation of synthesis of any protein should be disrupted by KSG. However, experimental results show that KSG action strongly depends on the nature and structure of the mRNA.
Figure 5.
Kasugamycin (KSG) action depends on the structure of mRNA while the activity of pactamycin (PAC) relies on the nature of A site tRNA. (a) Chemical structures of KSG and PAC. (b) KSG and PAC bind to the small ribosomal subunit and alter the path of the mRNA in the E site. (c) KSG efficiently inhibits translation of leadered mRNAs containing SD sequence, while translation of leaderless transcripts continues in the presence of the antibiotic. (c) Toeprinting analysis gel illustrating that PAC inhibits in vitro translation of the at specific codons of ermBL mRNA (62).
Early studies showed that KSG differentially inhibit synthesis of cytoplasmic and envelope proteins in E. coli and even expression of individual envelope polypeptides was inhibited to different extent (34). Treatment of E. coli cells infected with bacteriophages MS2 or f2 with KSG inhibited the synthesis of the phage maturation proteins much stronger than that of the coat proteins (45; 61), possibly reflecting idiosyncratic structural features of the ribosome binding sites (RBS) of their mRNAs (80). In support of this notion, KSG could readily inhibit translation of the reporter lacZ gene, whose initiation region had the sequence AGGAaacacccAUG (where the Shine-Dalgarno, SD, sequence and start codon are highlighted) but not that of the gst gene whose initiation region, AGGAaacacuuAUG, differed by only two nucleotides (underlined) (82). Extrapolating these observations, one would expect that expression of genes with different RBS structures could show significantly dissimilar response to KSG treatment. Ribo-Seq analysis would be a suitable technique for illuminating at a genome-wide level the preferred RBS structures targeted by KSG.
A vivid demonstration of the mRNA-specificity of KSG action is the inability of the drug to inhibit translation of leaderless mRNAs (15; 53) (Figure 5c). Some E. coli phages and transposons carry genes that start immediately with the initiation codon and, being devoid of a 5’-untranslated region (5’-UTR), lack the SD sequence. In contrast to the canonical translation initiation, which is driven by the 30S subunit, translation of leaderless mRNAs is initiated by the 70S ribosome (8; 54; 95). In vivo and in vitro studies showed that neither ribosome binding to the start codon of the leaderless mRNAs nor initiation of translation are significantly affected by KSG (15; 53). In addition, the discrimination by KSG between leadered and leaderless mRNAs in E. coli could be further influenced by drug-induced generation of specialized ribosomes. Upon drug treatment, a fraction of the ribosomes undergoes a partial disassembly, during which the 30S subunits lose a subset of ribosomal proteins (40). The resulting 61S ribosomal particles efficiently and specifically translate leaderless mRNAs, but not mRNAs with an SD sequence. Because accumulation of the 61S particles requires simultaneous presence of KSG and leaderless mRNA, it appears that the detachment of the 30S subunit proteins occurs when the drug-bound ribosome attempts to initiate translation from the leaderless mRNA start codon.
In contrast to E. coli, whose core genome either does not contain at all, or may carry only few leaderless genes (101), the genomes of some bacterial species encode a significant number of leaderless mRNAs. It remains to be investigated whether KSG treatment of such bacteria would lead to accumulation of 61S-like ribosomal particles and differentially express leadered and leaderless mRNAs. In any event, the idea that an antibiotic can induce generation of specialized ribosomes suited for preferential expression of only a subset of cellular proteins could be developed into a strategy for drug-induced bias in proteome expression.
INHIBITION OF TRANSLOCATION BY PACTAMYCIN MAY DEPEND ON THE NATURE OF THE A SITE tRNA
Pactamycin (PAC) inhibits protein synthesis in bacteria, archaea and eukaryotes by binding to the small ribosomal subunit (10; 25; 50; 107). Similar to KSG, PAC binds at the 30S subunit E site, in the mRNA channel and likely diverts the mRNA E-site codon from its regular placement (10) (Figure 5a,b). Although PAC was originally viewed as a specific inhibitor of initiation (30), subsequent studies suggested that the drug interferes with translocation (21). Strangely, however, this activity strongly depends on the nature of the pre-translocation complex, specifically, on the type of substrate in the A-site. The drug could readily inhibit translocation of the ribosome carrying tRNAiMet in the P site and the peptidyl-tRNA mimic, N-Ac-Lys-tRNALys, in the A site. However, when in a similar complex where the A site was occupied with N-Ac-Phe-tRNAPhe, no inhibition of translocation was observed even at elevated concentrations of PAC (21). None of the currently available structural data explain why the nature of the mRNA codon, the tRNA body, and/or the nascent chain esterifying the A-site tRNA would influence the action of PAC. Nevertheless, the site-specificity of PAC action has been additionally confirmed by toeprinting (62). In the presence of PAC, in vitro translation was arrested at preferential sites (Figure 5d), which varied among different mRNA templates. The effect of PAC at the genome-wide level and the molecular mechanisms responsible for site-specificity of the drug action are yet to be investigated.
AN EXPANDING NUMBER OF RIBOSOME-TARGETING ANTIBACTERIALS WITH CONTEXT-SPECIFIC ACTION
Although, as we have discussed, context specific action has been specifically investigated for only a few classes of antibiotics, this phenomenon potentially applies to several (possibly many) more classes of ribosome-targeting drugs. Studies employing in vitro toeprinting analysis suggest that several of the long-known ribosomal inhibitors (such as orthosomycins or streptogramins B) or newer antibiotics (e.g., odilorhabdins), arrest translation specifically at distinct mRNA sites (62; 64). Ongoing ribosome-profiling experiments confirm that several of these antibiotics preferentially inhibit cellular translation also at specific mRNA codons (Florin, Klepacki, Mangano, Mankin, Vázquez-Laslop, in preparation).
A SMALL MOLECULE INHIBITOR OF THE HUMAN RIBOSOME ABOLISHES PRODUCTION OF ONLY SPECIFIC PROTEINS
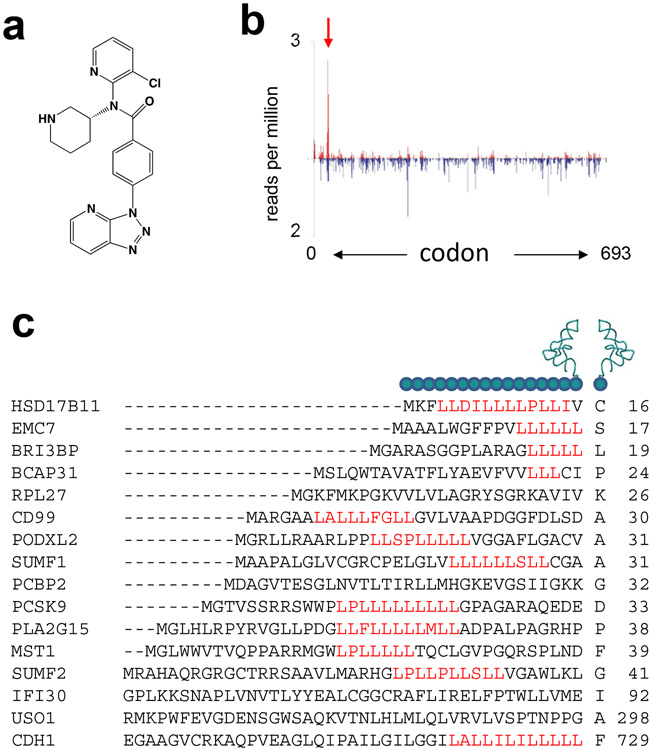
Taken to the extreme, the idea of context-specificity of antibiotic action opens the possibility of finding ribosome inhibitors that, while allowing translation of most of the cellular polypeptides, could prevent production of a handful, or potentially even only one particular protein. A spectacular validation of these seemingly far-fetched aspirations came not from the studies of antibacterials, but from an attempt to identify inhibitors of a particular human protein. The pro-protein convertase subtilisin/kexin type 9 (PCSK9) regulates the level of low density lipoprotein cholesterol (LDL-C) in the blood. Lowering PCSK9 levels result in reduction of the LDL-C concentration and decreases the incidence of cardiovascular disease (3; 17). A high-throughput screening identified a compound capable of lowering PCSK9 secretion. Subsequent cell culture studies demonstrated that reduced secretion of PSCK9 resulted from selective inhibition of its translation (68). Further optimization yielded compound PF-06446846 (Figure 6a), an even stronger inhibitor of the PCSK9 expression (47). Toeprinting and mutational analysis showed that the amino acid sequence of the nascent protein specified by the first 33 codons of the PCSK9 gene is required for PF-06446846 to be able to reduce the protein production. Treatment of cultured human cells with PF-06446846 led to specific arrest of translation at codon 34 of the PCSK9 gene as revealed by Ribo-Seq (Figure 6b). However, the Ribo-Seq data showed that the compound affected translation of another 22 proteins by arresting translation at specific codons of the corresponding genes and cell-free experiments confirmed that the expression of a number of these polypeptides was indeed sensitive to PF-06446846.
Figure 6.
A highly specific small molecule inhibitor of the human ribosome whose activity depends on the sequence of the nascent protein. (a) The chemical structure of compound PF-06446846. (b) Ribosome profiling shows that PF-06446846 specifically arrest translation at codon 34 of the human gene PCSK9 (47). (c) The amino acid sequences of the nascent peptides at the sites of translation arrest induced by PF-06446846 and the incoming A site amino acid (47). The codon number within the corresponding gene at which arrest occurs is indicated by the number to the right of the sequence.
PF-06446846 directly binds to the ribosome. Although the location of its binding site has not yet been reported, the mechanism of action is reminiscent of that of macrolide antibiotics, and the dependence of translation stalling on the nature of the nascent peptide is consistent with the binding of the inhibitor in the NPET of the human ribosome. The mechanistic details of action of PF-06446846 are unclear. Except for the prevalence of leucine-rich stretches, the sequences of the nascent peptides in the NPET of the arrested ribosome do not show any obvious similarity (Figure 6c). Furthermore, translation of many other human proteins containing comparable leucine-rich patches are unaffected by PF-06446846, indicating that either special folding of the nascent chain in the NPET or unrecognized features of the sequence context define the genes that could be inhibited by the compound.
The unwanted side effects precluded further development of PF-06446846 into a clinical drug. Nevertheless, its discovery demonstrates the conceptual feasibility of searching for (and finding!) highly context-specific ribosome inhibitors.
WHY DOES CONTEXT-SPECIFICITY OF RIBOSOMAL ANTIBIOTICS MATTER?
One may ask, how the understanding of context- or gene-specificity of antibiotic action helps to develop better drugs or expand our knowledge of cell biology? We want to offer here just three of many reasons.
1. Context specificity helps to understand and fight antibiotic resistance.
Many resistance genes are activated only when the cell is exposed to the antibiotic. Inducible expression of such genes relies on a sensing mechanism, which recognizes the presence of the inducing antibiotic, and on a response circuit that triggers gene expression when the inducer is detected. If the antibiotic is not recognized as an inducer, the resistance gene stays silent and cells remain susceptible to the drug. Therefore, understanding the mechanism of activation of resistance could lead to the development of non-inducing antibiotics that would be effective ribosome inhibitors but remain undetectable by the sensing system of the inducible resistance mechanism.
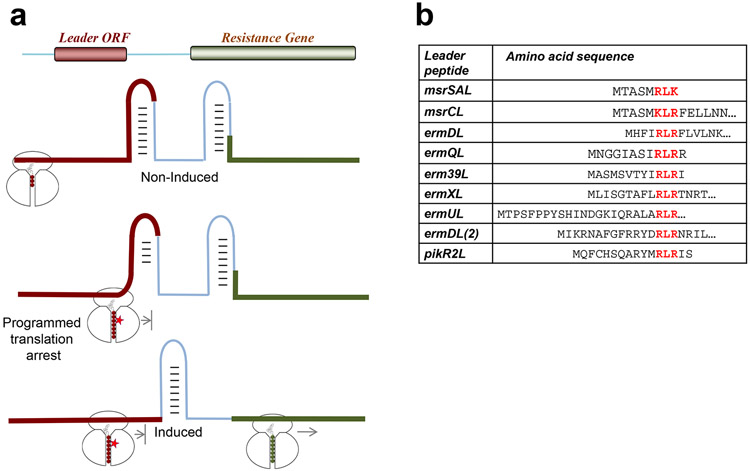
Regulation of inducible genes conferring resistance to the ribosomal inhibitors often exploits programmed translation arrest within upstream regulatory ORFs (Figure 7a) (reviewed in (48; 90; 98; 102)). In particular, this mechanism controls expression of macrolides and CHL resistance genes. In the absence of antibiotic, the leader ORF is constitutively translated while the resistance cistron remains silent because its RBS is sequestered in mRNA secondary structure (Figure 7a). When the inducing antibiotic is present, the ribosome stalls at a specific codon of the leader ORF, which triggers a conformational change in the mRNA that releases the previously occluded RBS and results in activation of the resistance gene.
Figure 7.
Activation of antibiotics resistance genes relies on the context-specific action of ribosomal-targeting antibiotics. (a) Programmed translation arrest at the leader ORF mediates the change in mRNA conformation necessary to activate expression of the resistance gene. (b) Many of the regulatory peptides responsible for activating macrolide resistance genes contain the sequence motif +x+, a prevalent arrest motif for macrolide antibiotics.
Before the context-specificity of the action of macrolides or CHL was uncovered, it was unclear how antibiotic-induced programmed translation arrest operates: why in the presence of the inducing antibiotic can the ribosome translate through several codons of the leader ORF and then stop at one specific codon? Knowing the rules of context-specificity of antibiotics readily explains how the system works. The peptides encoded in the leader ORFs carry the sequence that is poorly translated by the ribosome bound with the corresponding drug. Thus, many leader ORFs of macrolide resistance genes carry the +x+ motif (Figure 7b) or other motifs known to be problematic for the macrolide-bound ribosome (20; 76; 87; 90). Similarly, programmed translation arrest in inducible CHL resistance genes exploits the specificity of CHL action: the site of the programmed ribosome stalling is defined by the appearance of the first Ala codon (the leader ORF of the cmlA gene) or the first Thr codon (the leader ORF of the cat86A gene) (51).
Understanding the principles of context-specificity allows to not only predict which antibiotics would remain invisible to the sensory system and fail to activate resistance, but also could direct future efforts toward developing non-inducing antibiotics by directing them to avoid arrest at the sequences that nature has embedded in the regulatory ORFs of the inducible resistance genes.
2. Protein-selective antibiotic action could lead to species-specific drugs
Context-specificity of macrolides and, perhaps several other classes of ribosomal antibiotics, is manifested as inhibition of a defined subset of cellular proteins (Figure 4d) (1; 42; 89). However, the sequences of homologous proteins could vary significantly in different bacterial phyla. Therefore, the spectra of polypeptides inhibited by the same drug in different bacteria could be dissimilar. This could be one of the reasons why antibiotics may have significantly different effects upon different bacterial species.
The context-specificity of antibiotics depends on the structure of the compound and thus is tunable by altering the drug’s chemical make-up (1; 32; 89; 97). Optimizing the antibiotic structure for inhibiting specific motifs, and thus abolishing translation of a defined subset of cellular proteins, could be a way for targeting specific pathogenic bacteria while sparing the harmless species of the human microbiome. With our knowledge of context-specificity expanding further, this concept could become a guiding principle for the rational optimization of ribosome-targeting therapies.
3. Ribosome-targeting antibiotics as translation modulators
The growing understanding of the context-specificity of ribosomal antibiotics suggests that we should view them not simply as inhibitors of protein synthesis, but rather as modulators of translation (19; 32). Therefore, antibiotics and other ribosome-targeting small molecules could be used to not simply halt translation, but to remodel the cellular proteome. The case of the specific inhibitor of PCSK9 synthesis in human cells discussed above is one manifestation of this principle. Once the context-specificity of drug action is better understood, the efforts to identify ribosome-targeting compounds capable of interfering with the synthesis of only the unwanted proteins would have the proper knowledge-based foundation. Hopefully, we are on the way to making new medicines by clearly understanding how they work rather than by expanding our ability to blindly screen millions of random compounds in search of fortuitous hits.
CONCLUDING REMARKS
The concept of context-specificity of antibiotic action is fairly new. Like many advances in science it is propelled by novel techniques that allow us to take a fresh look at drugs that have been studied for decades as well as at emerging ribosome inhibitors. So far, only a few attempts have been made to take advantage of Ribo-Seq, toeprinting, smFRET and other codon- or gene-specific techniques to expand our understanding of the mechanisms of antibiotic action. We are confident, however, that we will soon learn of new examples of ribosome-binding small molecules that act as modulators rather than general inhibitors of translation.
So far, we have only scratched the surface of the phenomenon of context-specificity of the ribosomal drugs. We still have a very limited understanding of why particular motifs are problematic for the antibiotic-bound ribosome or how changes in the structures of the drugs affect the specificity of their action. Finding answers to these questions will not only expand our arsenal of approaches for knowledge-based design of better drugs, but will likely illuminate the yet unknown important aspects of the ribosome function.
Summary Points.
1. Several ribosomal antibiotics inhibit translation in a context-specific manner
2. The action of chloramphenicol and linezolid depends on the nature of the penultimate amino acid of the nascent protein.
3. Specific amino acid sequences are difficult to synthesize for the macrolide-bound ribosome.
4. Macrolides inhibit synthesis of a subset of bacterial proteins; the spectrum, of the inhibited proteins depends on the antibiotic structure.
5. Kasugamycin differentially inhibits initiation of translation of mRNAs depending on the presence and the structure of the nucleotide sequence preceding the start codon.
6. The inhibition of translocation by pactamycin may depend on the nature of the A site substrate.
7. A small molecule inhibitor of the human ribosome selectively arrests translation of a small subset of proteins.
Future Issues.
1. Applying mRNA- and codon-specific technologies, such as Ribo-Seq and toeprinting to a broad range of ribosomal antibiotics in order to identify inhibitors whose action is context-specific.
2. Unraveling the molecular mechanisms of context-specificity.
3. Using the knowledge of context-specific action of antibiotics as a tool to learn about properties of the ribosome that depend on the nature of its associated ligands and substrates.
4. Exploiting context-specificity of antibiotic action to develop new and better medicines.
Acknowledgements
We thank the members of our laboratory for their contribution through the years to advancing the concept of context-specificity of antibiotic action. We are grateful to Dr. Yury Polikanov for help with the figures and to Elizabeth Woods for proofreading the manuscript. The work in the Mankin/Vázquez-Laslop laboratory is supported by grants R01 AI125518 from the National Institutes of Health and MCB 1615851 from the National Science Foundation.
Terms and Definitions
- PTC
peptidyl transferase center
- NPET
nascent peptide exit tunnel
- aa-tRNA
aminoacyl-tRNA
- pept-tRNA
peptidyl-tRNA
- ERY
erythromycin
- AZI
azithromycin
- TEL
telithromycin
- KSG
kasugamycin
- PAC
pactamycin
References
- 1.Almutairi MM, Svetlov MS, Hansen DA, Khabibullina NF, Klepacki D, et al. 2017. Co-produced natural ketolides methymycin and pikromycin inhibit bacterial growth by preventing synthesis of a limited number of proteins. Nucleic Acids Res 45: 9573–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez-Elcoro S, Enzler MJ. 1999. The macrolides: Erythromycin, clarithromycin, and azithromycin. Mayo Clin Proc 74: 613–34 [DOI] [PubMed] [Google Scholar]
- 3.Anderson KM, Castelli WP, Levy D. 1987. Cholesterol and mortality. 30 years of follow-up from the Framingham study. JAMA 257: 2176–80 [DOI] [PubMed] [Google Scholar]
- 4.Arenz S, Bock LV, Graf M, Innis CA, Beckmann R, et al. 2016. A combined cryo-EM and molecular dynamics approach reveals the mechanism of ErmBL-mediated translation arrest. Nat Commun 7: 12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arenz S, Juette MF, Graf M, Nguyen F, Huter P, et al. 2016. Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome. Proc Natl Acad Sci USA 113: 7527–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arenz S, Meydan S, Starosta A, Berninghausen O, Beckmann R, et al. 2014. Drug sensing by the ribosome induces translational arrest via active site perturbation. Mol Cell 56: 446–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arenz S, Ramu H, Gupta P, Berninghausen O, Beckmann R, et al. 2014. Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nat Commun 5: 3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balakin AG, Skripkin EA, Shatsky IN, Bogdanov AA. 1992. Unusual ribosome binding properties of mRNA encoding bacteriophage lambda repressor. Nucleic acids research 20:563–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belousoff MJ, Eyal Z, Radjainia M, Ahmed T, Bamert RS, et al. 2017. Structural basis for linezolid binding site rearrangement in the Staphylococcus aureus ribosome. mBio 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brodersen DE, Clemons WM Jr., Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103: 1143–54 [DOI] [PubMed] [Google Scholar]
- 11.Bryskier A, Denis A. 2002. Ketolides: novel antibacterial agents designed to overcome resistance to erythromycin A within gram-positive cocci. In Macrolide Antibiotics, ed. Schönfeld W, Kirst HA, pp. 97–140. Basel: Birkhäuser Verlag [Google Scholar]
- 12.Bulkley D, Innis CA, Blaha G, Steitz TA. 2010. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc Natl Acad Sci USA 107: 17158–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cannon M 1968. The puromycin reaction and its inhibition by chloramphenicol. Eur J Biochem 7: 137–45 [DOI] [PubMed] [Google Scholar]
- 14.Chen J, Choi J, O'Leary SE, Prabhakar A, Petrov A, et al. 2016. The molecular choreography of protein synthesis: translational control, regulation, and pathways. Q Rev Biophys 49: e11. [DOI] [PubMed] [Google Scholar]
- 15. Chin K, Shean CS, Gottesman ME. 1993. Resistance of lambda cI translation to antibiotics that inhibit translation initiation. J Bacteriol 175: 7471–3 First evidence that kasugamycin differentially inhibits translation depending of the structure of mRNA
- 16.Cochella L, Green R. 2005. An active role for tRNA in decoding beyond codon: anticodon pairing. Science 308: 1178–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dadu RT, Ballantyne CM. 2014. Lipid lowering with PCSK9 inhibitors. Nat Rev Cardiol 11: 563–75 [DOI] [PubMed] [Google Scholar]
- 18.Dale T, Uhlenbeck OC. 2005. Amino acid specificity in translation. Trends Biochem Sci 30: 659–65 [DOI] [PubMed] [Google Scholar]
- 19.Davies J, Spiegelman GB, Yim G. 2006. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol 9: 445–53 [DOI] [PubMed] [Google Scholar]
- 20. Davis AR, Gohara DW, Yap MN. 2014. Sequence selectivity of macrolide-induced translational attenuation. Proc Natl Acad Sci USA 111: 15379–84 One of the first two papers reporting that macrolides act as context-specific antibiotics in vivo
- 21. Dinos G, Wilson DN, Teraoka Y, Szaflarski W, Fucini P, et al. 2004. Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin: the universally conserved residues G693 and C795 regulate P-site RNA binding. Mol Cell 13: 113–24 First evidence that the action of pactamycin may depend on the nature of the A site substrate.
- 22.Dinos GP. 2017. The macrolide antibiotic renaissance. Br J Pharmacol 174: 2967–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorner S, Brunelle JL, Sharma D, Green R. 2006. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol 13: 234–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunkle JA, Xiong L, Mankin AS, Cate JH. 2010. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci USA 107: 17152–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egebjerg J, Garrett RA. 1991. Binding sites of the antibiotics pactamycin and celesticetin on ribosomal RNAs. Biochimie 73: 1145–49 [DOI] [PubMed] [Google Scholar]
- 26.Erlacher MD, Lang K, Shankaran N, Wotzel B, Huttenhofer A, et al. 2005. Chemical engineering of the peptidyl transferase center reveals an important role of the 2'-hydroxyl group of A2451. Nucleic Acids Res 33: 1618–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fahlman RP, Dale T, Uhlenbeck OC. 2004. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol Cell 16: 799–805 [DOI] [PubMed] [Google Scholar]
- 28.Fei J, Richard AC, Bronson JE, Gonzalez RL, Jr. 2011. Transfer RNA-mediated regulation of ribosome dynamics during protein synthesis. Nat Struct Mol Biol 18: 1043–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandes P 2015. Use of antibiotic core structures to generate new and useful macrolide antibiotics. In Antibiotics Current Innovations and Future Trends, ed. Sánchez S, Demain AL. pp. 375–93. Norfolk, UK: Caister Academic Press [Google Scholar]
- 30.Gale EF, Cundliffe E, Reynolds PE, Richmond MH, Waring MJ 1981. The Molecular Basis of Antibiotic Action. London: John Wiley & Sons [Google Scholar]
- 31.Gottesman ME. 1967. Reaction of ribosome-bound peptidyl transfer ribonucleic acid with aminoacyl transfer ribonucleic acid or puromycin. J Biol Chem 242: 5564–71 [PubMed] [Google Scholar]
- 32.Gupta P, Liu B, Klepacki D, Gupta V, Schulten K, et al. 2016. Nascent peptide assists the ribosome in recognizing chemically distinct small molecules. Nat Chem Biol 12: 153–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hartz D, McPheeters DS, Traut R, Gold L. 1988. Extension inhibition analysis of translation initiation complexes. Methods Enzymol 164: 419–25 Introduction of toeprinting analysis, one of the methodologies that can reveal context-specificity of ribosomal inhibitors
- 34.Hirashima A, Childs G, Inouye M. 1973. Differential inhibitory effects of antibiotics on the biosynthesis of envelope proteins of Escherichia coli. J Mol Biol 79: 373–89 [DOI] [PubMed] [Google Scholar]
- 35.Horinouchi S, Weisblum B. 1980. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci USA 77: 7079–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Balakin AG, Skripkin EA, Shatsky IN, Bogdanov AA. 1992. Unusual ribosome binding properties of mRNA encoding bacteriophage lambda repressor. Nucleic acids research 20:563–71 Introduction of Ribo-Seq, an approach able to reveal context-specific action of antibiotics in vivo
- 37.Ippolito JA, Kanyo ZF, Wang D, Franceschi FJ, Moore PB, et al. 2008. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J Med Chem 51: 3353–6 [DOI] [PubMed] [Google Scholar]
- 38.Johansson M, Chen J, Tsai A, Kornberg G, Puglisi JD. 2014. Sequence-dependent elongation dynamics on macrolide-bound ribosomes. Cell Rep 7: 1534–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johansson M, Ieong KW, Trobro S, Strazewski P, Aqvist J, et al. 2011. pH-sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A-site aminoacyl-tRNA. Proc Natl Acad Sci U S A 108: 79–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kaberdina AC, Szaflarski W, Nierhaus KH, Moll I. 2009. An unexpected type of ribosomes induced by kasugamycin: a look into ancestral times of protein synthesis? Mol Cell 33: 227–36 Report that kasugamycin induces formation of specialized ribosomes that preferentially translate a subset of proteins
- 41. Kannan K, Kanabar P, Schryer D, Florin T, Oh E, et al. 2014. The general mode of translation inhibition by macrolide antibiotics. Proc Natl Acad Sci USA 111: 15958–63 Along with ref. 19, first evidence of the context-specificity of macrolide antibiotics action in vivo
- 42.Kannan K, Vázquez-Laslop N, Mankin AS. 2012. Selective protein synthesis by ribosomes with a drug-obstructed exit tunnel. Cell 151: 508–20 [DOI] [PubMed] [Google Scholar]
- 43.Kloss P, Xiong L, Shinabarger DL, Mankin AS. 1999. Resistance mutations in 23S rRNA identify the site of action of protein synthesis inhibitor, linezolid, in the ribosomal peptidyl transferase center. J. Mol. Biol. 294: 93–101 [DOI] [PubMed] [Google Scholar]
- 44.Konevega AL, Fischer N, Semenkov YP, Stark H, Wintermeyer W, Rodnina MV. 2007. Spontaneous reverse movement of mRNA-bound tRNA through the ribosome. Nat Struct Mol Biol 14: 318–24 [DOI] [PubMed] [Google Scholar]
- 45.Kozak M, Nathans D. 1972. Differential inhibition of coliphage MS2 protein synthesis by ribosome-directed antibiotics. J Mol Biol 70: 41–55 [DOI] [PubMed] [Google Scholar]
- 46.Leach KL, Swaney SM, Colca JR, McDonald WG, Blinn JR, et al. 2007. The site of action of oxazolidinone antibiotics in living bacteria and in human mitochondria. Mol Cell 26: 393–402 [DOI] [PubMed] [Google Scholar]
- 47. Lintner NG, McClure KF, Petersen D, Londregan AT, Piotrowski DW, et al. 2017. Selective stalling of human translation through small-molecule engagement of the ribosome nascent chain. PLoS Biol 15: e2001882. Description of a ribosomal inhibitor which specifically arrests translation of a handful of human proteins
- 48.Lovett PS. 1996. Translation attenuation regulation of chloramphenicol resistance in bacteria - A review. Gene 179: 157–62 [DOI] [PubMed] [Google Scholar]
- 49.Lovmar M, Nilsson K, Vimberg V, Tenson T, Nervall M, Ehrenberg M. 2006. The molecular mechanism of peptide-mediated erythromycin resistance. J Biol Chem 281: 6742–50 [DOI] [PubMed] [Google Scholar]
- 50.Mankin AS. 1997. Pactamycin resistance mutations in functional sites of 16S rRNA. J Mol Biol 274: 8–15 [DOI] [PubMed] [Google Scholar]
- 51. Marks J, Kannan K, Roncase EJ, Klepacki D, Kefi A, Orelle C, Vázquez-Laslop N, Mankin AS 2016. Context-specific inhibition of translation by ribosomal antibiotics targeting the peptidyl transferase center. Proc Natl Acad Sci USA 113: 12150–55 First report demostrating that the action of PTC inhibitors chloramphenicol and linezolid is context-specific
- 52.Moellering RC. 2003. Linezolid: the first oxazolidinone antimicrobial. Ann Intern Med 138: 135–42 [DOI] [PubMed] [Google Scholar]
- 53.Moll I, Blasi U. 2002. Differential inhibition of 30S and 70S translation initiation complexes on leaderless mRNA by kasugamycin. Biochem Biophys Res Commun 297: 1021–26 [DOI] [PubMed] [Google Scholar]
- 54.Moll I, Hirokawa G, Kiel MC, Kaji A, Blasi U. 2004. Translation initiation with 70S ribosomes: an alternative pathway for leaderless mRNAs. Nucleic Acids Res 32: 3354–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monro RE, Vazquez D. 1967. Ribosome-catalysed peptidyl transfer: effects of some inhibitors of protein synthesis. J Mol Biol 28: 161–5 [DOI] [PubMed] [Google Scholar]
- 56.Muto H, Nakatogawa H, Ito K. 2006. Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Mol Cell 22: 545–52 [DOI] [PubMed] [Google Scholar]
- 57.Nakahigashi K, Takai Y, Shiwa Y, Wada M, Honma M, et al. 2014. Effect of codon adaptation on codon-level and gene-level translation efficiency in vivo. BMC Genomics 15: 1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Odom OW, Picking WD, Tsalkova T, Hardesty B. 1991. The synthesis of polyphenylalanine on ribosomes to which erythromycin is bound. Eur J Biochem 198: 713–22 [DOI] [PubMed] [Google Scholar]
- 59.Oh E, Becker AH, Sandikci A, Huber D, Chaba R, et al. 2011. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147: 1295–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okuyama A, Machiyama N, Kinoshita T, Tanaka N. 1971. Inhibition by kasugamycin of initiation complex formation on 30S ribosomes. Biochem Biophys Res Commun 43: 196–9 [DOI] [PubMed] [Google Scholar]
- 61.Okuyama A, Tanaka N. 1972. Differential effects of aminoglycosides on cistron-specific initiation of protein synthesis. Biochem Biophys Res Commun 49: 951–7 [DOI] [PubMed] [Google Scholar]
- 62.Orelle C, Carlson S, Kaushal B, Almutairi MM, Liu H, et al. 2013. Tools for characterizing bacterial protein synthesis inhibitors. Antimicrob Agents Chemother 57: 5994–6004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Otaka T, Kaji A. 1975. Release of (oligo) peptidyl-tRNA from ribosomes by erythromycin A. Proc Natl Acad Sci USA 72: 2649–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pantel L, Florin T, Dobosz-Bartoszek M, Racine E, Sarciaux M et al. 2018. Odilorhabdins, new antibacterial agents that cause miscoding by binding at a new ribosomal site. Mol Cell, in press [DOI] [PubMed] [Google Scholar]
- 65.Patel U, Yan YP, Hobbs FW Jr., Kaczmarczyk J, Slee AM, et al. 2001. Oxazolidinones mechanism of action: inhibition of the first peptide bond formation. J Biol Chem 276: 37199–205 [DOI] [PubMed] [Google Scholar]
- 66.Pestka S 1972. Studies on transfer ribonucleic acid-ribosome complexes. XIX. Effect of antibiotics on peptidyl puromycin synthesis on polyribosoms from Escherichia coli. J Biol Chem 247: 4669–78 [PubMed] [Google Scholar]
- 67.Pestka S 1975. Chloramphenicol. In Antibitoics III: Mechanism of Action of Antimicrobial and Antitumor Agents, ed. Corcoran JW, Hahn FE, pp. 370–95. Berlin, Heidelberg, New York: Springer-Verlag [Google Scholar]
- 68.Petersen DN, Hawkins J, Ruangsiriluk W, Stevens KA, Maguire BA, et al. 2016. A small-molecule anti-secretagogue of PCSK9 targets the 80S ribosome to inhibit PCSK9 protein translation. Chem Biol 23: 1362–71 [DOI] [PubMed] [Google Scholar]
- 69.Polacek N, Gaynor M, Yassin A, Mankin AS. 2001. Ribosomal peptidyl transferase can withstand mutations at the putative catalytic nucleotide. Nature 411: 498–501 [DOI] [PubMed] [Google Scholar]
- 70.Polacek N, Mankin AS. 2005. The ribosomal peptidyl transferase center: structure, function, evolution, inhibition. Crit Rev Biochem Mol Biol 40: 285–311 [DOI] [PubMed] [Google Scholar]
- 71.Poldermans B, Van Buul CP, Van Knippenberg PH. 1979. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3' end of 16 S ribosomal RNA of Escherichia coli. II. The effect of the absence of the methyl groups on initiation of protein biosynthesis. J Biol Chem 254: 9090–3 [PubMed] [Google Scholar]
- 72.Polikanov YS, Osterman IA, Szal T, Tashlitsky VN, Serebryakova MV, et al. 2014. Amicoumacin a inhibits translation by stabilizing mRNA interaction with the ribosome. Mol Cell 56: 531–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Polikanov YS, Starosta AL, Juette MF, Altman RB, Terry DS, et al. 2015. Distinct tRNA accommodation intermediates observed on the ribosome with the antibiotics hygromycin A and A201A. Mol Cell 58: 832–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Polikanov YS, Steitz TA, Innis CA. 2014. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat Struct Mol Biol 21: 787–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Polikanov YS, Szal T, Jiang F, Gupta P, Matsuda R, et al. 2014. Negamycin interferes with decoding and translocation by simultaneous interaction with rRNA and tRNA. Mol Cell 56: 541–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramu H, Mankin A, Vazquez-Laslop N. 2009. Programmed drug-dependent ribosome stalling. Mol Microbiol 71: 811–24 [DOI] [PubMed] [Google Scholar]
- 77.Ramu H, Vázquez-Laslop N, Klepacki D, Dai Q, Piccirilli J, et al. 2011. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol Cell 41: 321–30 [DOI] [PubMed] [Google Scholar]
- 78.Ranjan N, Rodnina MV. 2017. Thio-modification of tRNA at the wobble position as regulator of the kinetics of decoding and translocation on the ribosome. J Am Chem Soc 139: 5857–64 [DOI] [PubMed] [Google Scholar]
- 79.Rosenblum G, Chen C, Kaur J, Cui X, Zhang H, et al. 2013. Quantifying elongation rhythm during full-length protein synthesis. J Am Chem Soc 135: 11322–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schluenzen F, Takemoto C, Wilson DN, Kaminishi T, Harms JM, et al. 2006. The antibiotic kasugamycin mimics mRNA nucleotides to destabilize tRNA binding and inhibit canonical translation initiation. Nat Struct Mol Biol 13: 871–8 [DOI] [PubMed] [Google Scholar]
- 81.Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, et al. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413: 814–21 [DOI] [PubMed] [Google Scholar]
- 82.Schuwirth BS, Day JM, Hau CW, Janssen GR, Dahlberg AE, et al. 2006. Structural analysis of kasugamycin inhibition of translation. Nat Struct Mol Biol 13: 879–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seiple IB, Zhang Z, Jakubec P, Langlois-Mercier A, Wright PM, et al. 2016. A platform for the discovery of new macrolide antibiotics. Nature 533: 338–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, et al. 2001. Cell-free translation reconstituted with purified components. Nat Biotechnol 19: 751–5 [DOI] [PubMed] [Google Scholar]
- 85.Shinabarger DL, Marotti KR, Murray RW, Lin AH, Melchior EP, et al. 1997. Mechanism of action of oxazolidinones: effects of linezolid and eperezolid on translation reactions. Antimicrob Agents Chemother 41: 2132–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sievers A, Beringer M, Rodnina MV, Wolfenden R. 2004. The ribosome as an entropy trap. Proc Natl Acad Sci U S A 101: 7897–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sothiselvam S, Liu B, Han W, Ramu H, Klepacki D, et al. 2014. Macrolide antibiotics allosterically predispose the ribosome for translation arrest. Proc Natl Acad Sci USA 111: 9804–09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sothiselvam S, Neuner S, Rigger L, Klepacki D, Micura R, et al. 2016. Binding of macrolide antibiotics leads to ribosomal selection against specific substrates based on their charge and size. Cell Rep 16: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Starosta AL, Karpenko VV, Shishkina AV, Mikolajka A, Sumbatyan NV, et al. 2010. Interplay between the ribosomal tunnel, nascent chain, and macrolides influences drug inhibition. Chem Biol 17: 504–14 [DOI] [PubMed] [Google Scholar]
- 90.Subramanian SL, Ramu H, Mankin AS. 2011. Inducible resistance to macrolide antibiotics. In Antibiotic Drug Discovery and Development, ed. Dougherty TJ, Pucci MJ . New Yoork, NY: Springer Publishing Company [Google Scholar]
- 91.Tenson T, Lovmar M, Ehrenberg M. 2003. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J Mol Biol 330: 1005–14 [DOI] [PubMed] [Google Scholar]
- 92.Tenson T, Mankin AS. 2001. Short peptides conferring resistance to macrolide antibiotics. Peptides 22: 1661–68 [DOI] [PubMed] [Google Scholar]
- 93.Tenson T, Xiong L, Kloss P, Mankin AS. 1997. Erythromycin resistance peptides selected from random peptide libraries. J Biol Chem 272: 17425–30 [DOI] [PubMed] [Google Scholar]
- 94.Tu D, Blaha G, Moore PB, Steitz TA. 2005. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121: 257–70 [DOI] [PubMed] [Google Scholar]
- 95.Udagawa T, Shimizu Y, Ueda T. 2004. Evidence for the translation initiation of leaderless mRNAs by the intact 70 S ribosome without its dissociation into subunits in eubacteria. J Biol Chem 279: 8539–46 [DOI] [PubMed] [Google Scholar]
- 96.Vazquez D 1979. Inhibitors of protein biosynthesis. Berlin, Heidelberg, New York: Springer-Verlag [Google Scholar]
- 97.Vazquez-Laslop N, Klepacki D, Mulhearn DC, Ramu H, Krasnykh O, et al. 2011. Role of antibiotic ligand in nascent peptide-dependent ribosome stalling. Proc Natl Acad Sci USA 108: 10496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vázquez-Laslop N, Mankin AS. 2014. Triggering peptide-dependent translation arrest by small molecules: ribosome stalling modulated by antibiotics. In Regulatory Nascent Polypeptides, ed. Ito K, pp. 165–86. New York: Springer [Google Scholar]
- 99.Vazquez-Laslop N, Ramu H, Klepacki D, Kannan K, Manksin AS 2010. The key role of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J. 29: 3108–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vazquez-Laslop N, Thum C, Mankin AS. 2008. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell 30: 190–202 [DOI] [PubMed] [Google Scholar]
- 101.Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, et al. 2011. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 147: 147–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Weisblum B 1995. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob Agents Chemother 39: 797–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Weissbach H, Redfield B, Brot N. 1968. Studies on the reaction of N-acetyl-phenylalanyl-tRNA with puromycin. Arch Biochem Biophys 127: 705–10 [DOI] [PubMed] [Google Scholar]
- 104.Wilson DN. 2009. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol 44: 393–433 [DOI] [PubMed] [Google Scholar]
- 105.Wilson DN, Schluenzen F, Harms JM, Starosta AL, Connell SR, Fucini P. 2008. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc Natl Acad Sci USA 105: 13339–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wohlgemuth I, Brenner S, Beringer M, Rodnina MV. 2008. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J Biol Chem 283: 32229–35 [DOI] [PubMed] [Google Scholar]
- 107.Woodcock J, Moazed D, Cannon M, Davies J, Noller HF. 1991. Interaction of antibiotics with A- and P-site-specific bases in 16S ribosomal RNA. EMBO J 10: 3099–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Youngman EM, Brunelle JL, Kochaniak AB, Green R. 2004. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117: 589–99 [DOI] [PubMed] [Google Scholar]