Abstract
Several existing drugs have gained initial consideration due to their therapeutic characteristics against COVID-19 (Corona Virus Disease 2019). Hydroxychloroquine (HCQ) was proposed as possible therapy for shortening the duration of COVID-19, but soon after this, it was discarded. Similarly, known antiviral compounds were also proposed and investigated to treat COVID-19. We report a pharmacophore screening using essential chemical groups derived from HCQ and known antivirals to search a natural compound chemical space. Molecular docking of HCQ under physiological condition with spike protein, 3C-like protease (3CLpro), and RNA-dependent RNA polymerase (RdRp) of SARS-CoV2 showed − 8.52 kcal/mole binding score with RdRp, while the other two proteins showed relatively weaker binding affinity. Docked complex of RdRp-HCQ is further examined using 100 ns molecular dynamic simulation. Docking and simulation study confirmed active chemical moieties of HCQ, treated as 6-point pharmacophore to screen ZINC natural compound database. Pharmacophore screening resulted in the identification of potent hit molecule [(3S,3aR,6R,6aS)-3-(5-phenylsulfanyltetrazol-1-yl)-2,3,3a,5,6,6a-hexahydrofuro[3,2-b]furan-6-yl]N-naphthalen-ylcarbamate from natural compound library. Additionally, a set of antiviral compounds with similar chemical scaffolds are also used to design a separate ligand-based pharmacophore screening. Antiviral pharmacophore screening produced a potent hit 4-[(1,5-dimethyl-3-oxo-2-phenylpyrazol-4-yl)-(2-hydroxyphenyl)methyl]-1,5-dimethyl-2-phenylpyrazol-3-one containing essential moieties that showed affinity towards RdRp. Further, both these screened compounds are docked (− 8.69 and − 8.86 kcal/mol) and simulated with RdRp protein for 100 ns in explicit solvent medium. They bind at the active site of RdRp and form direct/indirect interaction with ASP618, ASP760, and ASP761 catalytic residues of the protein. Successively, their molecular mechanics Poisson Boltzmann surface area (MMPBSA) binding energies are calculated over the simulation trajectory to determine the dynamic atomistic interaction details. Overall, this study proposes two key natural chemical moieties: (a) tetrazol and (b) phenylpyrazol that can be investigated as a potential chemical group to design inhibitors against SARS-CoV2 RdRp.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s11030-021-10358-5.
Keywords: COVID19, SARS-CoV2, Hydroxychloroquine, Docking, Pharmacophore screening, SARS-CoV2 inhibitor, RdRp
Introduction
Coronavirus has caused two epidemics in recent years. Severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003 caused more than 8000 infections and 700 deaths globally [1, 2]. This was followed by the Middle East respiratory syndrome coronavirus (MERS-CoV) [3, 4], which infected more than 2500 people and 800 deaths across 27 countries in 2012. Recently, we have witnessed the widespread pandemic caused by the novel coronavirus (SARS-CoV2) [5], which has already infected 71 million people and caused more than 1.59 million deaths (approximately a ~ 6% mortality rate). While the natural host for SARS-CoV2 is the bat, in all the cases virus managed to transmit into humans via an intermediate host. In SARS-CoV, the intermediate host is civet, while in MERS it was the camel. However, in SARS-CoV2 there is a lack of information on the intermediate host, but the pig has been hypothesized as the potential carrier. COVID19, the disease condition caused by SARS-CoV2 virus, has been declared as a pandemic by the WHO [6]. Therefore, the current situation demands an urgent need to contain and design therapeutic molecule to treat COVID19.
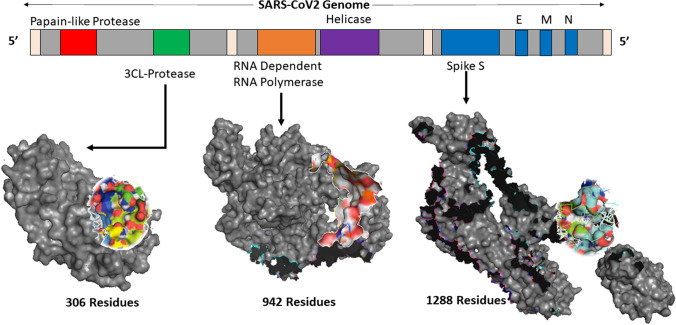
SARS-CoV2 is a RNA virus with enveloped architecture with its genetic material enclosed in an outer coat made from a small piece of the plasma membrane of the cell. The envelope may play a vital role in helping the virus to survive and infecting other cells. SARS-CoV2 has the largest genome (~ 30 Kb) compared to any other RNA virus but has similar structure protein content. These structural proteins are: (i) spike glycol protein (S), (ii) envelop small membrane protein (E), and (iii) membrane protein (M), with some viruses also having (iv) hemagglutinin-esterase (HE) [7], and (v) nucleoprotein (N) as shown in Fig. 1. These proteins are responsible for the entry of the virus into the host system. SARS-CoV2 has 6 ORFs in its genome, where ORF1a and ORF1v produce 2 polypeptides. These polypeptides are chomped by viral proteins: chymotrypsin-like protease or main protease (3CLpro) and papain protease [8]. Once the virus enters the host system, it transcribes and translates its genome. This process starts with the replicase complex [9] having RNA-dependent RNA polymerase (RdRp) playing an important role along with helicase and protease.
Fig. 1.
Three-dimensional structure rendering of 3CLpro, RdRp and spike protein, with their corresponding binding site targeted in this study. PDB ID for spike protein, 3CLpro, and RdRp is 6VSB 6M03 and 6M71, respectively
Most antiviral molecules are designed to either target the entry of the virus into the host organism or damage their replication/growth mechanism. Earlier, it has been observed that SARS-CoV binds with angiotensin-converting enzyme (ACE2) receptor [10–12], while MERS-CoV binds with DPP-4 [13] for entering the cells. SARS-CoV2 has high homology with the SARS-CoV, and therefore, it is anticipated that the spike protein of SARS-CoV2 also binds with the ACE2 receptor. This makes the spike protein one of the major drug targets for treating COVID19. Moreover, 3CLpro and RdRp are also considered as potential drug targets to inhibit the growth of the virus. Chloroquine has been established as an effective anti-malarial drug with antiparasitic activity [14]. An analogue of this compound, hydroxychloroquine (HCQ), has also been demonstrated to be an effective antibacterial compound [15, 16]. Recently, it has been found that these compounds have antiviral activity including inhibitory effect against SARS-CoV2. Initial in vitro data for chloroquine against SARS-CoV2 [17] made it a promising chemical scaffold, while later in vivo testing has been conducted to estimate its efficacy [18].
Recently, many existing drugs are tested against SARS-CoV2 to identify the potential therapeutic molecule [19–23]. In this study, we applied a computational drug design approach to find the active pharmacophore of hydroxychloroquine (HCQ) against proteins of SARS-CoV2, spike protein (including receptor-binding domain—RBD), 3CLpro, and RdRp. HCQ showed the maximum affinity with the RdRp while relatively lower binding with the spike and the 3CLpro protein. A similar computational affinity study was performed earlier where HCQ showed effective binding with the RdRp protein [24]. The exact mechanism of HCQ is still not clear, but it has been demonstrated experimentally that creates a basic environment in cytoplasm by elevating the pH that hinders virus internalization [25]. Its indirect effect on RdRp is also demonstrated in vitro where HCQ helps zinc uptake to the cells that eventually inhibit the activity of RdRp [26]. We searched active pharmacophore of the HCQ skeleton in a natural compound library to pull out potential natural hit compounds that can show inhibitory activity against SARS-CoV2 RdRp. In conjugation to HCQ active pharmacophore screening, we also performed multi-ligand pharmacophore screening using known antiviral molecules. Here, nine known compounds, lopinavir, ritonavir, ribavirin, galidesivir, favipiravir, remdesivir, tenofovir, saquinavir, and elvitegravir are used to design active pharmacophore. Resultant 3-point, 4-point, 5-point, and 6-point pharmacophores are searched in the ZINC database, and the top five hits for each pharmacophore are docked with RdRp. The best hits are further investigated in a molecular dynamic simulation where it showed a robust and stable complex with RdRp. Overall, the study showcases the potential of two natural origin compounds as a probable therapeutic agent against SARS-CoV2 RdRp.
Materials and methods
The study is composed of four major modules: (i) pharmacophore generation, (ii) pharmacophore screening (iii) molecular docking, and (iv) molecular dynamic simulation.
Protein structure collection
Three proteins from SARS-CoV2 that have been used as drug targets in this study are (a) spike protein, (b) 3CLpro protease, and (c) RdRp. Protein structures of the spike protein and the 3CLpro have been recently solved and deposited in the protein data bank (PDB) [27], PDB IDs: 6VSB and 6M03, respectively [28]. Spike protein (6VSB) is a Cryo-EM structure solved at 3.46 Å resolution. The structure consists of 3 three chains (A, B, and C) consisting of 1288 amino acids. It contains the receptor-binding domain (RBD) in upward conformation responsible for binding with the human ACE2 receptor. In the case of 3CLpro, crystal structure (6M03) is available at 2.0 Å. It has three domains: d-I (residues 8–101), d-II (residues 102–184), and d-III (residues 201–303). The structure is available in apo form, but its catalytic triad SER-HIS-ASP can guide to search the binding site. RdRp structure is also solved using electron microscopy and deposited in the protein databank with PDB ID: 6M71 [29]. We considered this experimental structure of RdRp for docking and simulation. The experimental structure consists of four chains representing three units of RNA polymerase complex; these units are nsp12 (chain A), nsp7 (chain C), and nsp8 (chains B, D). Chain A is the main RNA polymerase unit, so it is extracted from the complex to employ in this study. Binding sites for spike protein and the 3CLpro proteins are predicted using the Fpocket tool [30]. An online version of this tool is available on the Mobyle web portal. However, detection of the binding region of RdRp protein is guided using Zhang’s website COACH [31] server that used the template structure (PDB ID: 3H5S) of hepatitis C virus and the protein structure deposited in the database (6M71).
Molecular docking
Molecular docking between small compounds and SARS-CoV2 proteins was performed using Autodock-4 [32]. Protein and ligand are prepared using Autodock tool (ADT) and Marvin sketch, and multiple docking is performed using raccoon application of Autodock. Hydrogen atoms are added to the system, and gasteiger charges are assigned to prepare the ligand. pKa value of ligand is determined using ChemAxon tool integrated with Marvin sketch application. Best scored binding pocket predicted by the Fpocket that is used in molecular docking. A large grid box (126 × 126 × 126 Å) is created around the detected binding pocket with spacing 0.375 Å. This implies blind docking where the ligand can hit the large surface of the protein and find the most appropriate position to interact. Docking used the genetic algorithm to explore the binding space. The number of individuals generated in each cycle of the genetic algorithm is 150, while 2,500,000 maximum number of energy evaluations are performed. Docking program used 27,000 maximum number of generations where only the top 1 individual survives to the next generation with a 0.02 rate of mutation with 0.8 rate of crossover.
Molecular dynamic simulation
MD simulation of docked complex is performed using GROMACS 4.6.2 [33] having CHARMM27 force field [34]. Simulations are carried out under physiological conditions. Topology and parameters for small molecules are generated using CHARMM all atoms force field on SwissParam online tool [35]. Hydrogen atoms are added to the RdRp protein under physiological conditions and placed in a solvated box at a 1.4-nm distance from the wall. Protein–ligand solvated complex was energetically minimized using 5000 steps of the steepest deepest method. A time step of 2 fs was used using the SHAKE algorithm. Constant temperature (NVT) and pressure ensemble (NPT) conditions are applied to the system for 100 ps and 1 ns, respectively, to attain the equilibrium state. Ligand and protein molecules are constrained during the equilibrium phase. Post-equilibrium phase, a 100-ns all-atom simulation was performed using V-rescale temperature coupling [36] for external heat bath with 0.1 ps time constant for protein and ligand while pressure coupling was performed using Parrinello-Rahman [37] with a time constant of 2 ps, and long-range electrostatic was dealt with PME [38]. Detailed simulation methodology is given in Supplementary Text S1.
Pharmacophore generation and screening
Ligand-based pharmacophores are generated using Pharmagist [39] server. It generated a 3-point pharmacophore from reference molecule HCQ and 3-point, 4-point, 5-point, and 6-point pharmacophores from the known set of antiviral molecules used in this study. Individual 3D pharmacophore is searched in the library using ZINC pharmer tool. ZINC Database [40] is used to search the active pharmacophore among the compounds available in the library. Out of the various hit compounds displayed by the ZINC database, molecules with low RMSD values from the active sites of pharmacophore are selected for docking studies. Interaction analysis for the docked complex was performed using LigPlot + v1.4.5 [41, 42], free academic Schrödinger-Maestro v12.4 suite, and PLIP online server [43]. Eventually, VMD [44] and Pymol [45] are used for graphical representations of the molecules.
Results and discussion
Experimental structures of all three SARS-CoV2 proteins: (i) spike protein, (ii) 3CLpro, and (iii) RdRp are retrieved from the protein databank, and their respective PDB Ids are 6VSB 6M03 and 6M71.
Protein structure and binding sites
Three-dimensional coordinates of these three SARS-CoV2 proteins (spike protein, 3CLpro and RdRp) have been recently solved and submitted to the PDB database. Figure 1 shows the cartoon representation of these proteins with their respective binding sites. Fpocket predicted multiple binding sites for each protein, and the best scored (rank 1) site is used for grid design under docking. Binding information for RdRp is directly imported from COACH webserver and aligned to the PDB structure 6M71. A sufficiently large grid box (126 × 126 × 126 Å) is created around the detected binding pocket with the spacing 0.375 Å. A large grid box allows the ligand to traverse a larger conformational space on the protein surface and perform the blind docking process.
In spike protein, binding site residues predicted by Fpocket are F58, F59, V289, L293, D294, P295, L296, S297, E298, T299, K300, C301, T302, L303, S305, F306, T307, V308, E309, K310, G311, I312, Y313, Q314, T315, S596, V597, I598, T599, P600, G601, T602, N603, T604, S605, N606, Q607, V608, A609. RBD of spike protein does not contain any cavity; thus, the binding site predicted here is spatially distant from RBD. Predicted binding site residues for protease are T24, T25, T26, L27, H41, C44, M49, Y54, F140, L141, N142, G143, S144, C145, H163, H164, M165, E166, L167, P168, H172, F181, D187, R188, Q189, T189, and A190. Active residues of viral protease have catalytic dyad H41 and C141. Both these residues are found within the predicted binding site. These residues in the docking grid would maximize the possibility of ligand binding at the native catalytic site to inhibit the function of protease. However, in RdRp, a binding site is directly sourced from its template homologous structure (Hepatitis C virus polymerase) which is co-crystalized with the saccharin inhibitor and aligned with the SARS-CoV2 RdRp. Saccharin position is superposed on RdRp, and the area around it is treated as a binding site. This binding site consists of K593, W598, M601, T687, A688, A691, S759, D760, D761 C813, Q815, and P830 as binding site residues. Catalytic residues of RdRp are responsible for the entry and processing of natural substrate—NTP. D618, D760, and D761 are critical catalytic residues that play a major role in the function of SARS-CoV2 RdRp protein. As mentioned above, D760 and D761 are detected within the binding site of RdRp sourced from COACH webserver. Generation of sufficiently large grid box (126 × 126 × 126 Å) around the centre of these binding sites would estimate the likelihood of ligand to interact with the core catalytic residues of RdRp.
Binding scores and interactions
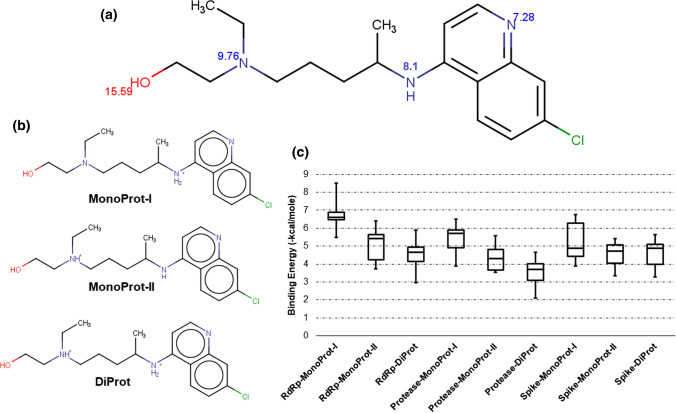
After identification of respective binding sites for all three SARS-CoV2 protein targets, HCQ molecule was prepared under physiological condition and docked within the binding grid box of each protein. Docked pose could be used to identify the interacting functional groups of HCQ and further guide the pharmacophore design and screening. HCQ has three amine functional groups; the first group is the part of the pyridine ring, while the second is immediately attached outside the pyridine ring and the third is the tertiary amine group. HCQ also has a hydroxyl group (–OH) at the terminal position. pKa values of all the ionizable groups are predicted using ChemAxon integrated into MarvinSketch tool shown in Fig. 2a. Hydroxyl (-OH) group has the highest pKa; thus, it would certainly protonate under physiological pH conditions. Pyridine ring nitrogen has a minimum pKa, so it would not protonate at pH 7.4. However, the other two nitrogen atoms have higher pKa and they will have more chance to protonate at physiological pH. An earlier study conducted by Schroeder et al. [46]. suggested that HCQ has three basic functional groups with pKa values of < 4.0, 8.3, and 9.7. Individual pKa values for all ionizable groups with the possible protonated states for HCQ molecule are shown in Fig. 2a, b. Various possible protonated states are labelled as ‘MonoProt-I’ (4-amino pyridine nitrogen is protonated), ‘MonoProt-II’ (tertiary amine nitrogen is protonated), and ‘DiProt’ (both nitrogen atoms are protonated) as shown explicitly in Fig. 2b.
Fig. 2.
HCQ molecule preparation and docking: a pKa values for all ionizable groups of HCQ predicted using ChemAxon integrated tool with MarvinSketch; b protonated states formed under physiological conditions (pH 7.4) are shown as MonoProt-I, MonoProt-II, and DiProt; c box plot for binding energy scores of RdRp, protease, and spike proteins with MonoProt-I, MonoProt-II, and DipProt states of HCQ. The negative sign of binding energy scores is ignored for better readability
All three protonated forms are prepared in MarvinSketch followed by Autodock preparation for docking with three different proteins of SARS-CoV2. Autodock4 is used for docking, a large rectangular grid box of (126 × 126 × 126) Å is prepared around the centre of each predicted binding site, and exhaustive sampling was performed to find the best pose of the ligand HCQ among all the protonation states. Binding scores for the top 10 poses are recorded using the Autodock4 scoring function. Figure 2c shows the binding energy box plots of 10 docked poses of HCQ against each protein with its three different protonated forms. Negative signs in the binding energies are ignored to provide better readability; this implies the largest binding energy value corresponds to the highest binding affinity. All three proteins showed the best docking score for Monoprot-I state of HCQ. The average binding energy for MonoProt-I complexed with RdRp is − 6.68 kcal/mol, while MonoProt-II and DiProt showed − 5.09 and − 4.57 kcal/mol average binding energies. Similarly, protease showed the best average binding energy − 5.43 kcal/mol with MonoProt-I, while the other two protonation states are − 4.31 kcal/mol and − 3.51 kcal/mol, respectively. Finally, spike protein also followed the same pattern and showed its best affinity for MonoProt-I with an average binding energy score − 4.64 kcal/mol, while − 4.50 kcal/mol and − 4.61 kcal/mol for the other two protonation states. Collectively, Fig. 2 shows the better performance of MonoProt-I for each protein and among them, RdRp showed the best binding score ( − 8.52 kcal/mol). Earlier studies on the Autodock binding scores suggested that compounds that showed a binding score < − 6.0 kcal/mole are accepted as active compounds, while any score > − 6.0 kcal/mole might be considered as an inactive compound [47]. Here, RdRp showed an average binding score better than − 6.0 kcal/mole and thus considered as a potential binding partner for HCQ in MonoProt-I state under physiological pH. Recently, we have published a docking study that showed known inhibitors of RdRp that served as a control for this study [48].
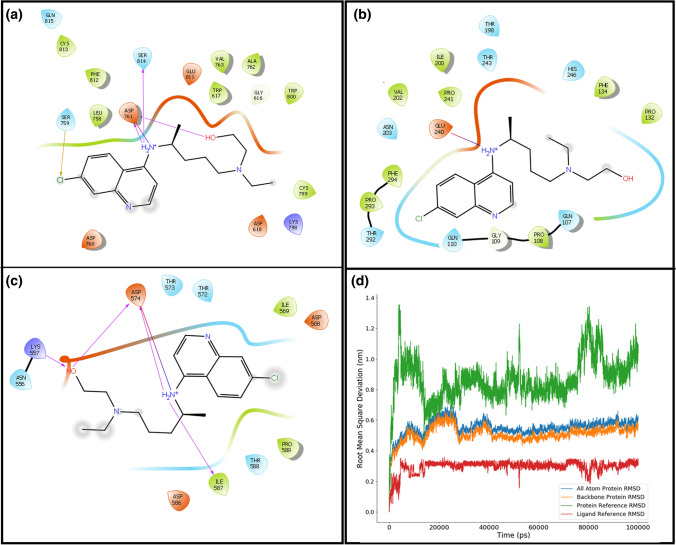
Docking exercise of HCQ with these three proteins suggested strongly that it prefers RdRp compared to the other two proteins. Although the average binding scores of HCQ with spike and 3CLpro do not fall in the accepted range (< − 6.0 kcal/mol), its best pose passed binding energy criteria; therefore, their interacting chemical moieties can still assist in generating pharmacophore. Schrödinger-Maestro v12.4 suite tool is used to plot the interactions of HCQ with respective proteins. Figure 3a–c shows the interaction plot of all three proteins with monoprotonated HCQ for their best-docked poses. The top binding score of RdRp with MonoProt-I of HCQ has − 8.52 kcal/mol binding energy, while protease and spike proteins showed − 6.49 kcal/mol and − 6.76 kcal/mol binding energy for their respective best-docked pose (shown in Fig. 2c). Figure 3a–c shows that the protonated basic amine group of HCQ forms hydrogen bonds with all three proteins in its best docked conformation. RdRp and Spike protein formed 4 and 5 hydrogen bonds with HCQ while protease could form only one hydrogen bond. Negative charged Asp761 from RdRp and Asp574 from spike protein are among the binding site lining residue, and they interact strongly with the positively charged amine group of HCQ. However, protease has negative charged residue Glu240 in the neighbouring environment of the ligand to form a hydrogen bond. RdRp has a predominantly polar binding environment compared to the other two proteins that ensure strong binding with the charged HCQ entity. In all the complexes (Fig. 3a–c), both the cyclic rings of HCQ contribute to hydrophobic interactions.
Fig. 3.
Binding interaction plot of MonoProt-I protonated state of HCQ with a RdRp, b protease, and c spike proteins of SARS-CoV2, d structural variation of RdRp:HCQ complex during 100-ns simulation. RMSD of all-atom and backbone atoms for the protein (blue and orange), RMSD of HCQ when protein is taken for alignment (green line) and when ligand (HCQ) is taken as a reference molecule for alignment (red lines)
Later, protein–ligand interaction was also confirmed using an online tool, PLIP. Results generated by PLIP agree with the interference drawn from Schrödinger-Maestro v12.4 suite. However, a few additional interactions are detected by PLIP that were not found earlier. The best pose for the spike protein HCQ complex under PLIP server showed a salt bridge between the oxygen atom of Asp586 and the nitrogen atom of HCQ. Similarly, the protease complex has additional hydrogen bonds with Thr198 and His246 and salt-bridge between the nitrogen atom of Glu240 and the nitrogen atom of HCQ. RdRp complex showed two salt-bridges involving Asp761 and Glu811 residues in the interaction table of the PLIP tool and three additional H-bonds each with Trp617, Ala762, and Trp800 are detected in PLIP but not in Schrödinger-Maestro v12.4 suite interaction analysis.
Information collected from Schrödinger-Maestro v12.4 suite interaction plot and PLIP suggests that minimum 1–2 H-bond donors/acceptors, 1–2 charged groups for salt bridge formation, and 2–3 hydrophobic groups are key interacting pharmacophores that are required for binding with SARS-CoV2 proteins.
Conformational variation of HCQ
Further, a molecular dynamic simulation was performed on the docked complex to overcome the non-flexibility limitation of rigid docking. RdRp complex is selected for the molecular dynamic simulation based on Autodock binding energy score. A 100-ns all-atom explicit solvent MD simulation is performed for the best-docked pose of HCQ with RdRp. In this process, protein–ligand complex is minimized using steepest descent algorithm to achieve the thermodynamically stable complex. Supplementary Figure S1 (blue line) shows the energy minimization curve for the protein–ligand HCQ complex. Later, the system is equilibrated for 100 ps under NVT and 1000 ps under NPT ensembles before the production phase. RdRp-HCQ complex is sufficiently equilibrated under the solvent medium. Supplementary Figures S2 and S3 show the temperature and pressure plot for NPT and NVT equilibrium process. These thermodynamic properties showed satisfactorily behaviour in the equilibrium phase. Once the system is minimized and equilibrated, a 100-ns production phase is performed.
All-atoms and backbone-atoms RMSD for protein is shown in Fig. 3d, the initial conformation of protein deviated to 0.5–0.6 nm at the outset of the simulation. Once it crossed 20 ns time, it showed a steady behaviour that continued till the end of simulation. This deviation characteristic confirmed the conformational stability of RdRp protein during the 100 ns simulation time. Figure 3d also shows the ligand HCQ deviation within the binding site of RdRp. It showed the RMSD pattern when a protein (green) and ligand (red) are considered separately as a reference molecule for alignment. Initially, protein is taken for alignment fit where HCQ conformation deviated to ~ 1.2 nm compared to starting pose within 10 ns of simulation, but it drops down to 0.6 nm in the later phase. After 20 ns simulation time, HCQ shows minor variation till the end of the simulation. Here, the standard deviation (std) of the RMSD for the last 80 ns of simulation is 0.11 nm that confirms the steady-state behaviour for the protein–ligand complex. When protein is considered as a reference alignment molecule, the RMSD value consists of both the rotational and translational movement of the ligand. Here, a high value of RMSD ~ 1.0–1.2 nm in the last 80 ns of simulation articulates the translocation of HCQ to another region of the binding pocket. Once it relocates to a new position within the binding site, it attains stability in this conformation for the rest 80 ns of simulation. In contrast, when the ligand is used as a reference molecule for alignment, it only accounts for the rotational movement in RMSD calculation. In this case, RMSD showed a very stable pattern (red line) around 0.3 nm for the entire simulation. Figure 3d shows that in RdRp-HCQ complex, protein attains a steady state in short time, while ligand HCQ changes its binding location in the initial phase, but then it also acquires higher conformational stability in the final phase of simulation.
Supplementary Figure S4a shows the average root mean square fluctuation (RMSF) for the individual residue for RdRp protein. This plot explains that 87% of the residues have fluctuation less than 0.3 nm, while 67% of them are even lower than 0.2 nm, only 1.5% residues showed RMSF above 0.5 nm. Overall, 14 residues have relatively higher fluctuation; 12 of these are terminal residues. In addition to these terminal residues, Arg74 and Lys50 have RMSF greater than 0.4 nm. None of these residues belong to the catalytic/active site of RdRp that upholds the protein–ligand complex stability. Among all the atomic interaction, hydrogen bonding is considered as a major factor for protein–ligand complex stability. Figure S4b shows the number of hydrogen bonds in a different frame of the simulation; these numbers fluctuated between 1 and 3. H-bond number calculation showed that 86% of frames have one or more protein–ligand H-bonding. Hydrogen bond analysis was performed additionally for the binding site residues in RdRp-HCQ complex. It showed that there are several frames during the simulation where binding site residues can form hydrogen bond with HCQ molecules. Distance distribution plot for donors and acceptors atoms between HCQ molecule and binding site residues confirms the presence of 20–30 pairs that fall under the hydrogen bond distance criteria. However, their angular orientation may not allow all of them to successfully form hydrogen bonds. This also advocates that there is always a higher chance of forming more hydrogen bonds as shown in Supplementary Fig. S5.
Most probable binding pose
Investigating the conformational behaviour of HCQ within the binding site of RdRp, a clustering method is applied. The complete trajectory is clustered using 0.3-nm RMSD criteria deploying `gromos` method of clustering. This resulted in seven distinct clusters. However, top four cluster constitutes 98% of the total population. These four clusters consist of 7952, 963, 640, and 297 structures, respectively. The middle structure of each cluster is considered for analysis. The smallest cluster has eight structures. We compared the starting coordinate with the middle structures of four most populated cluster to examine the movement of HCQ. Supplementary Figure S6 shows that in each cluster HCQ position is shifted from the starting position. These four poses shown in Supplementary Fig. S6 could be considered as the most probable pose of the HCQ. Hence, it can be concluded that during simulation, HCQ showed translational and rotational movement but always reside within the binding region of RdRp.
Complete trajectory when clustered using 3 Å RMSD criteria and middle structure of each cluster is compared with the starting complex, and then, it is observed that middle structures from two clusters have shown highest RMSD of greater than 6 Å with the starting complex. These two complexes are studied to find the position of HCQ with respect to the active site residues LEU758, SER759, ASP760 and ASP761 of RdRp protein. Both the complexes showed the HCQ in proximity of active site residue that suggest that even at higher deviation HCQ does not loose contact with the active site residues as shown in Supplementary Fig. S7.
MMPBSA binding energy
The Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA) binding energy between protein and ligand is calculated using the g_mmpbsa tool. This tool gives three binding energy components: (1) molecular mechanics, composed of van der Waal component and electrostatic energy, (2) Poisson Boltzmann-based solvation polar energy, and (3) SASA-based non-polar energy. RdRp is a big protein with 931 amino acids with 72 missing residues in its solved structure. However, the binding site is relatively smaller, and thus, it can accommodate nucleotide as a natural substrate. Here, MMPSA method is applied to estimate the binding free energy of HCQ and protein complex. Earlier, the complete molecular dynamic simulation trajectory of best-docked pose of HCQ is clustered that formed seven distinct clusters. The middle structures of these most populated clusters are considered as most promising poses for HCQ in the binding site of RdRp. MMPBSA energy is calculated for these most promising poses using its binding site residues. The average MMPBSA binding energy over all the seven representative structure is -23.48 kcal/mol consisting of four binding energy components (vdW, electrostatic, polar solvation energy, and non-polar solvation energy). A high value of binding energy is due to the absence of entropy component in the final energy, but the negative value indicates the stability of the complex. Polar solvation energy is positive during the simulation, but it is neutralized by the molecular mechanics (vdW + elec) component. Non-polar SASA binding energy majorly has small negative values which make the total binding energy stable for these poses. The mean of MMPBSA binding energies across all the poses with its various components is shown in Supplementary Fig. S8a. The binding energy for the individual middle structure for each cluster and the size of the cluster are shown in Supplementary Fig. S8b. Most populated clusters with 7952 structures have the highest binding free energy that confirms the stable formation of HCQ-RdRp complex.
Screened compounds
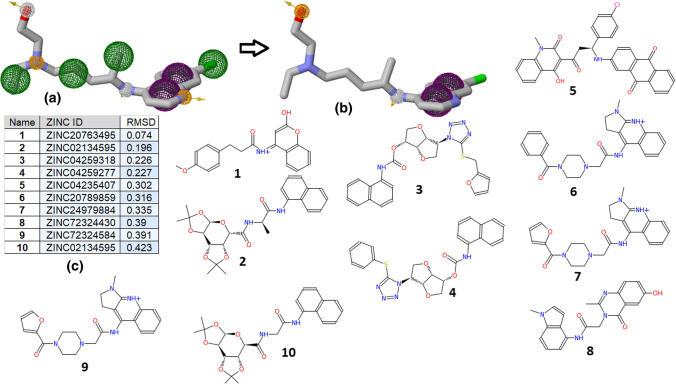
Once the binding and interacting groups are confirmed for the HCQ complexes with the SARS-CoV2 proteins, a pharmacophore generation and search are performed using the active functional group responsible for the interaction. Pharmagist and ZincPharmer online tool are used for this purpose, where HCQ is taken as a template molecule and ZINC natural compound library [39] as a screening database. HCQ has 13 pharmacophore points in total, 2 aromatic rings (ARO), 2 hydrogen bond donor (HBD), 3 hydrogen bond acceptor (HBA), and 6 hydrophobic groups (HYD). Based on the interaction and simulation study, pharmacophores are selected for screening against the ZINC natural compound database. Terminal hydroxyl group was considered an essential HBA (hydrogen bond acceptor) pharmacophore, while the nitrogen was chosen as HBD (hydrogen bond donor) pharmacophore point. Aromatic rings of HCQ are considered as HYD (hydrophobic) as well as ARO (aromatic stacking) pharmacophore. This reduced the number from a 13-point to a 6-point pharmacophore search. Total available pharmacophores of HCQ and its corresponding points used in searching are shown in Fig. 4a, b. Pharmacophore screening resulted in top 50 hits arranged as per their RMSD with reference molecule HCQ. The top 10 compounds based on their RMSD are shown in Fig. 4c), where the minimum RMSD reported is 0.07 Å (ZINC20763495). Structures of the top 10 pharmacophore screening hits are shown in Fig. 4.
Fig. 4.
Screened compounds based on HCQ active pharmacophore: a the maximum number of pharmacophores present in HCQ are 13, b 6-points pharmacophores are used for screening, c top 10 hits based on RMSD criteria, and d chemical structure of all top 10 hit compounds
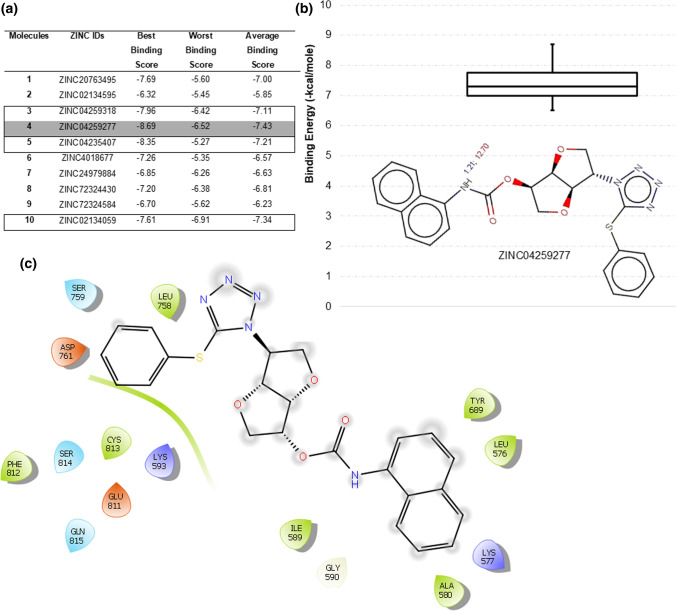
These top 10 ZINC compounds from the ZINC natural molecule database showed high pharmacophore similarity with the HCQ molecule exhibiting required ARO, HDB, HBA, and HYD functional groups. Docking study of HCQ has shown that RdRp is the best binding target for HCQ. List of these top 10 screened compounds is shown in Table S1a with their corresponding ZINC IDs and IUPAC names. As per this dogma, all top 10 compounds are docked using the Autodock4 tool with RdRp to find the binding affinity. The same docking grid o is used that employed in HCQ docking. Figure 5a shows the best, worst, and average Autodock docking binding scores of these 10 natural molecules for all the 10 poses generated in docking. Compound 4 yielded the best binding score of − 8.69 kcal/mole in its first pose, while its average binding score is − 7.43 kcal/mol. Further, compounds 3, 5, and 10 also have strong binding scores with average binding energy better than HCQ. Figure 5b shows the ionizable functional group of 4, ChemAxon predicted only one ionizable amine group with pKa 1.21 (blue) and 12.70 (red). Blue colour pKa represents the acid dissociation constant between the neutral base and its conjugated acid, while red colour pKa represents the acid dissociation constant between the neutral acid and its conjugated base as shown in Fig. 5b. This indicates that none of the nitrogen would be in a protonated or deprotonated state under the physiological condition of pH 7.4. Figure 5b also shows the box plot of the binding energies for the 10 docked poses of 4. The protein–ligand interaction plot for the best docked pose of 4 is shown in Fig. 5c; it does not form any hydrogen bond with RdRp but the core catalytic residues Ser759 and Asp761 positioned in proximity. Figure 5 summarizes that 4 might be used as a potential lead for inhibitor design against SARS-CoV2 RdRp.
Fig. 5.
Binding scores of screened natural compounds generated from HCQ pharmacophore-based search, a top poses are generated in docking. The best compound is highlighted in grey and other potential hits as outlined. b pKa value for best-identified compound 4 with the boxplot of binding energy scores for all its 10 docked poses; c interaction plot for the best identified compound 4
Antiviral compounds pharmacophore
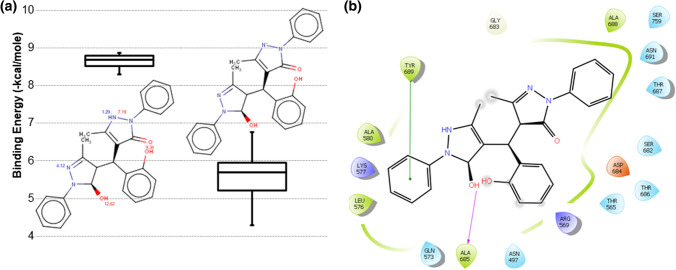
This study also used known inhibitors of RNA viruses to generate active pharmacophore; these known inhibitors are lopinavir, ritonavir, ribavirin, galidesivir, favipiravir, remdesivir, tenofovir, saquinavir, and elvitegravir as shown in Supplementary Table S1b. These molecules are selected based on their chemical scaffolds and their respective protein targets in RNA viruses. Remdesivir, galidesivir, and favipiravir are direct targeting RdRp of RNA viruses [49–51], while lopinavir targets ribosomal RNA, and its combination with Ritonavir is used for the treatment of HIV-infection [52, 53]. Ribavirin is used against RNA viruses and consists of nucleoside scaffold like remdesivir [47], galidesivir, and favipiravir. Tenofovir also belongs to the nucleoside compound family with antiviral activity [54]. Saquinavir and Elvitegravir are added to the list as they showed promising inhibitory activity against RNA viruses and share a similar scaffold [55, 56]. Many of these compounds are also proposed for inhibiting SARS-CoV2. These molecules are aligned to design the 3-point, 4-point, 5-point, and 6-point pharmacophores. These pharmacophores have HBD, HBA, HYD, and ARO as active groups. All these generated pharmacophores (3-points, 4-points, 5-points, and 6-points) are screened against the 2,17,77,093 compounds of the ZINC database. The top five hits based on the RMSD from each (3-points, 4-points, 5-points, and 6-points) screening are selected for docking, and thus, 20 compounds are docked with RdRp protein to evaluate their binding affinity. Docked scores of these 20 potential hits are shown in Supplementary Table S2. Compound 15 showed the best docking energy score − 8.86 kcal/mol, while its average docking score − 8.63 kcal/mol also surpassed others. RMSD of the best hit compound 15 with each antiviral compound is listed in Supplementary Table S1. Ionizable group of these compounds are identified, and respective pKa values are predicted. Figure 6a shows the pKa values of ionizable groups of 15, predicted pKa for one nitrogen group is 4.12, while for other nitrogen it has two values: 1.29 (blue) and 7.19 (red). Blue colour pKa represents the acid dissociation constant between the neutral base and its conjugated acid, while red colour represents the acid dissociation constant between the neutral acid and its conjugated base. Based on these two pKa values, this nitrogen has chances to deprotonate and formed negative ion species. Figure 6a shows the chemical structure and binding energy boxplot for neutral and negative variants of 15. The neutral variant showed a relatively better binding energy, hence considered for further operations. Figure 6b shows the interaction of the natural variant of 15 in its best docked pose. It showed a single hydrogen bond with Ala685 and one stacking interaction with Tyr689. Resembling 4, this compound also docked at the catalytic site of RdRp. Best docked poses of HCQ, 4 and 15 are shown simultaneously in Supplementary Fig. S7. It shows that HCQ and 4 share partial common sites/residues, while 15 is placed at a distant position, similar conclusion is drawn from their interaction plots.
Fig. 6.
Binding scores of 15 screened from antiviral compounds pharmacophore search, a pKa value for 15 with the boxplot of binding energy scores in its neutral and negative variants for all its 10 docked poses; b interaction plot of neutral variant for the 15 with RdRp
Molecular dynamic simulation
Best hit molecules 4 (ZINC04259277) and 15 (ZINC39444715) have molecular weights of 475.5 gm and 454.53 gm, respectively, with multiple H-bond donor and H-bond acceptor groups. The best-docked pose of compounds 4 and 15 is simulated for 100 ns like the process followed in HCQ-RdRp simulation. The other three compounds 3, 5, 10 and 17 derived from HCQ based pharmacophore search method are also proposed as a potential lead molecule, but not studied in molecular simulation. Equilibrium NVT and NPT ensembles simulation are elaborated in Figures S1, S2, and S3.
Structure fluctuation: RMSD
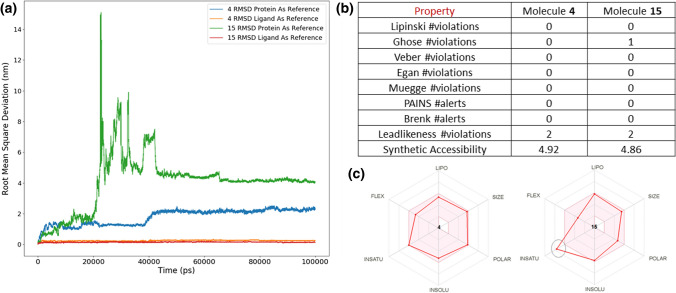
Docked complex of 4 and 15 with RdRp is simulated for 100 ns in explicit solvent medium, and RMSD of the ligand is calculated to show its translational and rotational movement. Ligand and protein both are used separately as reference aligning molecules to calculate RMSD. As explained earlier that translational variation is captured only when protein molecule is taken as reference fit. RMSD patterns of 4 and 15 are shown in Fig. 7a. Ligands RMSD under 0.2 nm for the entire simulation time when ligand molecule is taken for alignment. However, when protein is used as a reference fitting, it showed stability after 40 ns of simulation time. Molecule 4 showed relatively lesser translational motion that 15. Both the ligands showed high RMSD in the initial phase of the simulation but got stabilized during the end of the simulation. This once again shows that ligand translates inside the binding pocket, but eventually settles down and form a stable complex. It also indicates that the initial docked pose was not the most stable state and simulation helped to find the stable pose of 4 and 15.
Fig. 7.
a Root mean square 4 and 15 with RdRp. RMSD is shown when and ligand and protein are used for alignment separately b and c drug-likeness of the compounds 4 and 15
Clustering and MMPBSA energy
The clustering of the simulation trajectory of the protein–ligand complex for the best-docked pose of 4 and 15 is performed using the `gromos` algorithm with 0.3 nm RMSD cut-off criteria. This formed 15 and 32 clusters for 4 and 15, respectively, that are representative of the complete trajectory. The binding energies for the most probable complexes that are generated during the simulation are calculated using the MMPBSA method. Solute dielectric 2 and solvent dielectric 80 are used in this method for MMPBSA calculation. Clustering reduced the number of structures generated in the simulation and grouped them into different clusters, and MMPBSA energy is only calculated for the middle structure of these distinct clusters. These middle structures are the most considered as the most probable states of RdRp complexed with 4 and 15. MMPBSA energy of these middle structures of different distinct clusters is shown in Supplementary Tables S3 and S4. Polar solvation energy for the complexes is positive, but it is negated by the high negative value of molecular mechanics (vdw + elec) and non-polar. The average MMPBSA energy across all the representative structures for 4 and 15 is − 25.50 kcal/mol and − 8.27 kcal/mol, respectively, as shown in Supplementary Tables S3 and S4. The maximum populated cluster in 4 has 4205 structures and the MMPBSA binding energy for its middle structure is − 28.09 kcal/mol, while for 15 the most populated cluster has 3745 candidate structures with − 20.26 kcal/mol MMPBSA binding energy for its middle structure. These binding energies do not contain ΔS (entropy) component, so the absolute value cannot be compared with the experimental binding energy. However, negative values indicate the stability of the complex. Tables S2 and S3 show the MMPBSA binding energies of the individual cluster middle structure with the size of the cluster. Although the average MMPBSA energy for 15 is − 8.27 kcal/mol but if we closely examine the individual energies, it shows that top 3 populated cluster has high binding energies (average − 23.75 kcal/mol). These top 3 clusters occupy 60% of total conformational space. MMPBSA values indicate that there is high plausibility for 4 and 15 to interact with SARS-CoV2 RdRp. Average MMPBSA binding energy for individual residues from binding site region is calculated for the most probable poses during simulation and shown in Supplementary Fig. S9. Negative binding energy indicates their significant contribution in the binding with RdRp protein. ALA688, THR687, CYS813, and ASP761 have shown contribution in the binding of compound 15 within the binding site of RdRp protein. However, compound 4 has only one residue LYS 593 that showed negative binding energy in the protein–ligand complex.
Solvent accessible surface area (SASA) for both the compounds on the complete protein and the binding site during the simulation are calculated. SASA for both the complex is shown in Supplementary Fig. S10. Protein when complexed with 15 showed higher SASA value than 4 during initial phase of simulation, but it saturates after 40 ns of simulation both for complete protein and binding site residues. This suggest that few residues in binding site are get exposed in complex 15 when the ligand molecule translocate, but then it regained its original conformation and covered those exposed residues to lower down its SASA.
Protein conformation changes during simulation in the presence of ligand. Flexibility of protein residues allow them to form stronger interaction with the ligand atoms. An essential dynamics approach is applied to the RdRp protein bound with 4 and 15. This is achieved by dimensional reduction of the simulation trajectory by projecting first two principal components. Supplementary Figure S11 shows the collective motion for RdRp in bound state with both hit compounds. Black shows the motion of protein when it in complex with 4, while ‘red’ shows the motion 15 complex. Figure shows that collective motion of protein is more localized when it binds with 4 compared to 15. Besides, complete trajectory analysis of docked complexes, mean geometries of the last 3 ns trajectories for compounds 4 and 15 are extracted to determine the binding pose of resultant conformations. These mean structures of ligands are re-docked with RdRp protein using the Autodock tool. It generated 10 docked poses for 4 and 15, respectively, the best binding score for 4 has − 8.25 kcal/mol binding score, while − 8.42 kcal/mol for 15 as shown in Supplementary Table S5. This indicates that simulated ligands have different conformation compared to the initial state, but they still form a strong complex with the RdRp. Further, all the docked poses interactions are determined using the LigPlot tool, all poses showed 1 hydrogen bond and proximity with the residue Leu758, Ser759, Asp760, Asp761. It shows that it retains the key interaction within the binding site of the protein.
Active pharmacophores for both the screened zinc compounds from their middle structure of most populated cluster are detected; these pharmacophores play critical role in binding the ligand with protein. Here, 4 has two aromatic rings, while 15 has three. In addition, there are five donor, and seven acceptor groups present in 4, while 15 has lower number these groups with four donors and two acceptors, respectively. In case of hydrophobic pharmacophores, 15 has four such groups, while 4 has three hydrophobic groups. Overall, it shows that 4 has higher number of active pharmacophores compared to 15, but these pharmacophores are sufficient to hold the ligand molecule inside the binding site. Supplementary Figure S12 shows the active pharmacophores of these two hits.
Physicochemical properties
Moreover, the physicochemical properties and drug-likeness of 4 and 15 are calculated using SwissADME server (http://www.swissadme.ch) [57] that showed its likelihood as therapeutic molecule. Figure 7b shows the druglikeness of these compounds; four different filters are applied on these compounds to estimate their druglikeness. Lipinski filter used molecular weight, lipophilicity, donor and acceptor count, while Ghose filter used molecular weight, lipophilicity, molecular refractivity, and number of atoms to characterize the druglikeness of a given compound. Similarly, Muegge filter used molecular weight, lipophilicity, polar surface area, number of rings, number of carbon atoms, number of heteroatoms, number of rotatable bonds, donor and acceptor account in their filter criteria. Compound 4 cleared all these filters that while 15 has only one violation in Ghose filter. In the medicinal chemistry filters, both the compounds have zero alerts in PAINS (Pan-assay interference compounds). Properties listed in Fig. 7b advocate the application of 4 and 15 as therapeutic agents. SwissADME server also provides a radial graph for suitable physicochemical space for oral bioavailability. This graph covers LIPO (lipophilicity), SIZE, POLAR (polarity), INSOLU (insolubility), INSATU (insaturation) and FLEX (flexibility). Figure 7c shows the radial graph where the coloured zone represents a suitable physicochemical space. Both 4 and 15 fall completely in the favourable region expect insaturation property for 15.
In course of empirical validation of the above finding, Compound 4 is searched in the PubChem database, and it is found with ID: 11,886,000. Its IUPAC name is ‘[(3S,3aR,6R,6aS)-3-(5-phenylsulfanyltetrazol-1-yl)-2,3,3a,5,6,6a-hexahydrofuro[3,2-b]furan-6-yl] N-naphthalen-1-ylcarbamate’. Data in the PubChem show that biological testing of this molecule is performed under various cases on different samples. It has also been tested in primary in vitro assay for inhibitors of plasmodium falciparum Dd2 growth. Similarly, a PubChem ID: 2,841,659 with IUPAC name ‘4-[(1,5-dimethyl-3-oxo-2-phenylpyrazol-4-yl)-(2-hydroxyphenyl)methyl]-1,5-dimethyl-2-phenylpyrazol-3-one’ represents 15. It has also been tested in primary qHTS for delayed death inhibitors of the malarial parasite. Both the compounds have been previously proposed as anti-parasites (malarial); recently, it has also been noticed that anti-parasite compounds may have anti-viral activity. This can explain the empirical validation of the compounds found in this study.
Conclusions
Genomic constituents of SARS-CoV2 have been studied extensively to understand the impact of mutation and geographical regions on the infection rate of COVID [58]. Currently, many countries have started vaccination to control this pandemic, but recent mutation reported in SARS-CoV2 can modulate its viral transmission, replication efficiency, and virulence. Identifying peptide-based vaccine [59] and therapeutic molecule against SARS-CoV2 [60] can complement the overall control measures. HCQ has been withdrawn as a potential treatment molecule for COVID, but it showed inhibition to SARS-CoV2 in cell-based assays. EC50 0.72 µM reported in an earlier study for HCQ [45] showed its inhibition property. The application of HCQ on COVID patients is justified and discussed in many aspects [46]. Based on this principle, new compounds from the natural molecule repository are searched and validated in this study. This study combined a ligand-based and structure-based drug design approach to find a potent inhibitor against SARS-CoV2. Molecular docking and simulation of HCQ with SARS-CoV2 proteins (i) spike (with receptor-binding domain), (ii) 3CLpro protease, and (iii) RdRp guided the pharmacophore design. Screening of the active pharmacophore in the ZINC natural compound database identified ten hit compounds. Molecular docking of these hit compounds further recognized four molecules that have a binding score greater than HCQ. These compounds share similar chemical groups and showed a high affinity with the RdRp protein of SARS-CoV2. Molecule 4 showed the best result in docking and thus examined under 100-ns molecular simulation. During simulation, 4 showed its binding with RdRp protein at its catalytic site. In addition to 4, three more molecules 3, 5, and 10 also showed a significant binding score with RdRp. Supplementary figure S13 compares the best pose of HCQ with the two hits (4 and 15) identified in this study. Being natural compounds, they are likely to exhibit a significantly improved safety profile [61, 62]. HCQ pharmacophore-based drug screening used protein–ligand docked complex to determine critical chemical moiety responsible for protein–ligand interaction. We also examined the known anti-viral compounds that share common chemical scaffolds and mechanisms. In this study, nine such compounds are selected based on their activities and chemical structures. A ligand-based approach was deployed to design an active pharmacophore with multiple active features. This screening resulted in 20 hits against RdRp where docking and molecular dynamic simulation assisted to detect the most potential hit. The best docking score of 15 is − 8.86 kcal/mole, but it changed its initial conformation during MD simulation. Both 4 and 15 showed conformational stability during the last 60 ns of simulation. MMPBSA binding energies of the compounds showed thermodynamic and solvation stability. They located in the active site of the RdRp protein during the simulation with proximity to catalytic residues. Their binding score, conformation, and stability make them potential inhibitors against RdRp. The pharmacophore approach is used in this study to replace the virtual structure-based screening technique. However, the structure of RdRp is later used for docking to confirm the binding of pharmacophore hits with the protein. Hence, this study presented a combination of structure-based and ligand-based approaches to find the best hit molecule. Experimental testing of the detected compounds against RdRp activity using standard binding assay (enzyme based) or cell-based assays can confirm our computational finding.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was funded by the Department of Biotechnology, Ministry of Science and Technology (Grant Number BT/COE/34/SP15097/2015).
Author contributions
AM and AR planned the study and AM performed the computational experiments, data analysis and manuscript writing. AR supervised the project, reviewed, and edited the final paper.
Declarations
Conflict of interest
The authors declare that they do not have any conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Avinash Mishra, Email: avish2k@gmail.com.
Anurag S. Rathore, Email: asrathore@biotechcmz.com
References
- 1.Cheng VC, Lau SK, Woo PC, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee N, Hui D, Wu A, Chan P, Cameron P, Joynt GM, Ahuja A, Yung MY, Leung CB, To KF, Lui SF. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348:1986–1994. doi: 10.1056/NEJMoa85. [DOI] [PubMed] [Google Scholar]
- 3.Zaki AM, Van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 4.De Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, Fouchier RA, Galiano M, Gorbalenya AE, Memish ZA, Commentary PS. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the coronavirus study group. J Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO (2020) WHO Director-General's opening remarks at the media briefing on COVID-19 2020. https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---21-october-2021
- 7.De Haan CA, Kuo L, Masters PS, Vennema H, Rottier PJ. Coronavirus particle assembly: primary structure requirements of the membrane protein. J Virol. 1998;72:6838–6850. doi: 10.1128/JVI.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus AD, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RA. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012 doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sola I, Almazan F, Zuniga S, Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han DP, Penn-Nicholson A, Cho MW. Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology. 2006;350:15–25. doi: 10.1016/j.virol.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1007/s00018-004-4242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J, Li K, Wohlford-Lenane C, Agnihothram SS, Fett C, Zhao J, Gale MJ, Baric RS, Enjuanes L, Gallagher T, McCray PB. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci U S A. 2014;111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agrawal AS, Garron T, Tao X, Peng BH, Wakamiya M, Chan TS, Couch RB, Tseng CT. Generation of a transgenic mouse model of Middle East respiratory syndrome coronavirus infection and disease. J Virol. 2015;89:3659–3670. doi: 10.1128/JVI.03427-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Bari MA. Chloroquine analogues in drug discovery: new directions of uses, mechanisms of actions and toxic manifestations from malaria to multifarious diseases. J Antimicrob Chemother. 2015;70:1608–1621. doi: 10.1093/jac/dkv018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raoult D, Drancourt M, Vestris G. Bactericidal effect of doxycycline associated with lysosomotropic agents on Coxiella burnetii in P388D1 cells. Antimicrob Agents Chemother. 1990;34:1512–1514. doi: 10.1128/AAC.34.8.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raoult D, Houpikian P, Dupont HT, Riss JM, Arditi-Djiane J, Brouqui P. Treatment of Q fever endocarditis: comparison of 2 regimens containing doxycycline and ofloxacin or hydroxychloroquine. Arch Intern Med. 1999;159:167–173. doi: 10.1001/archinte.159.2.167. [DOI] [PubMed] [Google Scholar]
- 17.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao J, Tian Z, Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. BioSci Trends. 2020;14:72. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 19.Elfiky AA. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248:117477. doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Senanayake SL. Drug repurposing strategies for COVID-19. Future Sci. 2020;2:3. doi: 10.4155/fdd-2020-0010. [DOI] [Google Scholar]
- 21.Serafin MB, Bottega A, Foletto VS, da Rosa TF, Hörner A, Hörner R. Drug repositioning an alternative for the treatment of coronavirus COVID-19. Int J Antimicrob Agents. 2020;55:105969. doi: 10.1016/j.ijantimicag.2020.105969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shah B, Modi P, Sagar SR. In silico studies on therapeutic agents for COVID-19: drug repurposing approach. Life Sci. 2020;252:117652. doi: 10.1016/j.lfs.2020.117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elfiky AA. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nimgampalle M, Devanathan V, Saxena A. Screening of chloroquine, hydroxychloroquine and its derivatives for their binding affinity to multiple SARS-CoV-2 protein drug targets. J Biomol Struct Dyn. 2020;39:1–13. doi: 10.1080/07391102.2020.1782265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schrezenmeier E, Dörner T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat Rev Rheumatol. 2020;16:155–166. doi: 10.1038/s41584-020-0372-x. [DOI] [PubMed] [Google Scholar]
- 26.Lim YX, Ng YL, Tam JP, Liu DX. Human coronaviruses: a review of virus-host interactions. Diseases. 2016;4:26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Wang T, Sun Q, Ming Z, Zhang L, Ge J. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Guilloux V, Schmidtke P, Tuffery P. Fpocket: an open source platform for ligand pocket detection. BMC Bioinformatics. 2009;10:168. doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Roy A, Zhang Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics. 2013;29:2588–2595. doi: 10.1093/bioinformatics/btt447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hess B, Kutzner C, Van Der Spoel D, Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 34.MacKerell AD, Jr, Bashford D, Bellott ML, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 35.Zoete V, Cuendet MA, Grosdidier A, Michielin O. SwissParam: a fast force field generation tool for small organic molecules. J Comput Chem. 2011;32:2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- 36.Berendsen HJ, Postma JV, van Gunsteren WF, DiNola AR, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:6. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- 37.Parrinello M, Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52:8. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 38.Darden T, York D, Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:4. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 39.Schneidman-Duhovny D, Dror O, Inbar Y, Nussinov R, Wolfson HJ. Deterministic pharmacophore detection via multiple flexible alignment of drug-like molecules. J Comput Biol. 2008;15:737–754. doi: 10.1089/cmb.2007.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. ZINC: a free tool to discover chemistry for biology. J Chem Inf Model. 2012;52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 42.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 43.Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015;43:W443–W447. doi: 10.1093/protein/8.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(33–38):27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 45.Schrodinger L (2015) The PyMOL molecular graphics system, Version~1.8. 10.12691/ajidm-4-3-3
- 46.Schroeder RL, Gerber JP. Chloroquine and hydroxychloroquine binding to melanin: some possible consequences for pathologies. Toxicol Rep. 2014;1:963–968. doi: 10.1016/j.toxrep.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao X, Ye F, Zhang M, Cui C, Huang B, Niu P, Liu X, Zhao L, Dong E, Song C, Zhan S. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020;71:732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mishra A, Rathore AS. RNA dependent RNA polymerase (RdRp) as a drug target for SARS-CoV2. J Biomol Struct Dyn. 2021 doi: 10.1080/07391102.2021.1875886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsieh HP, Hsu JT. Strategies of development of antiviral agents directed against influenza virus replication. Curr Pharm Des. 2007;13:3531–3542. doi: 10.2174/138161207782794248. [DOI] [PubMed] [Google Scholar]
- 50.Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, Leist SR, Pyrc K, Feng JY, Trantcheva I, Bannister R. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017 doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tchesnokov EP, Feng JY, Porter DP, Götte M. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019 doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li F, Lu J, Ma X. CYP3A4-mediated lopinavir bioactivation and its inhibition by ritonavir. Drug Metab Dispos. 2012;40:18–24. doi: 10.1124/dmd.111.041400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hull MW, Montaner JS. Ritonavir-boosted protease inhibitors in HIV therapy. Ann Med. 2011;43:375–388. doi: 10.3109/07853890.2011.572905. [DOI] [PubMed] [Google Scholar]
- 54.Ray AS, Fordyce MW, Hitchcock MJ. Tenofovir alafenamide A novel prodrug of tenofovir for the treatment of human immunodeficiency virus. Antiviral Res. 2016;125:63–70. doi: 10.1016/j.antiviral.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 55.Kupferschmidt HH, Fattinger KE, Ha HR, Follath F, Krähenbühl S. Grapefruit juice enhances the bioavailability of the HIV protease inhibitor saquinavir in man. Br J Clin Pharmacol. 1998;45:355–359. doi: 10.1046/j.1365-2125.1998.t01-1-00687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pommier Y, Johnson AA, Marchand C. Integrase inhibitors to treat HIV/AIDS. Nat Rev Drug Discov. 2005;4:236–248. doi: 10.1038/nrd1660. [DOI] [PubMed] [Google Scholar]
- 57.Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gupta V, Haider S, Verma M, Singhvi N, Ponnusamy K, Malik MZ, Verma H, Kumar R, Sood U, Hira P, Satija S. Comparative genomics and integrated network approach unveiled undirected phylogeny patterns, co-mutational hot spots, functional cross talk, and regulatory interactions in SARS-CoV-2. Msystems. 2021;6:e00030–e000121. doi: 10.1128/mSystems.00030-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alam A, Khan A, Imam N, Siddiqui MF, Waseem M, Malik MZ, Ishrat R. Design of an epitope-based peptide vaccine against the SARS-CoV-2: a vaccine-informatics approach. Brief Bioinform. 2021;22:1309–1323. doi: 10.1093/bib/bbaa340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mishra CB, Pandey P, Sharma RD, Malik MZ, Mongre RK, Lynn AM, Prasad R, Jeon R, Prakash A. Identifying the natural polyphenol catechin as a multi-targeted agent against SARS-CoV-2 for the plausible therapy of COVID-19: an integrated computational approach. Brief Bioinform. 2021;22:1346–1360. doi: 10.1093/bib/bbaa378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Atanasov AG, Zotchev SB, Dirsch VM, Supuran CT. Natural products in drug discovery: advances and opportunities. Nat Rev Drug Discov. 2021;20:200–216. doi: 10.1038/s41573-020-00114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cragg GM, Pezzuto JM. Natural products as a vital source for the discovery of cancer chemotherapeutic and chemopreventive agents. Med Princ Pract. 2016;25:41–59. doi: 10.1159/000443404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.