Abstract
Myosins are among the most fascinating enzymes in biology. As extremely allosteric chemomechanical molecular machines, myosins are involved in myriad pivotal cellular functions and are frequently sites of mutations leading to disease phenotypes. Human β-cardiac myosin has proved to be an excellent target for small-molecule therapeutics for heart muscle diseases, and, as we describe here, other myosin family members are likely to be potentially unique targets for treating other diseases as well. The first part of this review focuses on how myosins convert the chemical energy of ATP hydrolysis into mechanical movement, followed by a description of existing therapeutic approaches to target human β-cardiac myosin. The next section focuses on the possibility of targeting nonmuscle members of the human myosin family for several diseases. We end the review by describing the roles of myosin in parasites and the therapeutic potential of targeting them to block parasitic invasion of their hosts.
Keywords: myosin, interacting heads motif, cancer, parasites, therapeutics
INTRODUCTION
Every step we take, every heartbeat, every movement of different muscular organs in our body, and many brilliant innovations of nature such as cell division, cell migration, and vesicular transport are only possible because of force generated by the motor protein myosin as it travels over cytoskeletal actin filaments. The discovery of conventional muscle myosin in the nineteenth century (1) laid the foundation for what is now a vast field of its own: molecular motors, which include a large family of myosins, a similarly large family of kinesins, and the dyneins (2, 3).
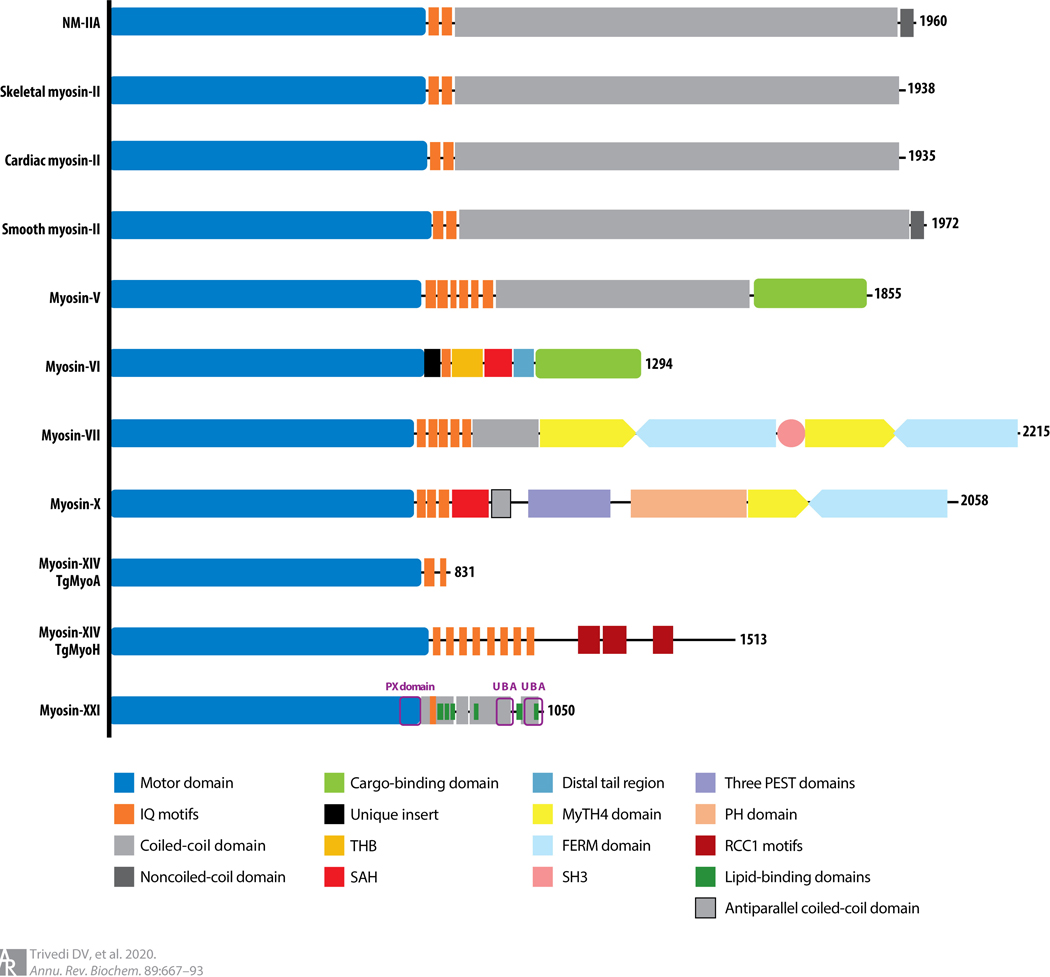
The myosin family of molecular motors is found in most eukaryotic organisms, from parasites to humans (2). In humans, there are more than 20 distinct classes and as many as 40 different myosin genes (4). Examples of some myosins across organisms are shown in Figure 1. Notable are the variety of tail domains that allow for many of the discrete functions that a particular myosin carries out, including anchoring of myosin molecules to specific sites for movement relative to actin filaments and specifying their binding to different cargoes (4). The tail also determines whether a myosin is single-headed or two-headed, usually by forming a coiled coil of two myosin heavy-chain α-helices in the tail region.
Figure 1.
Linear structure depictions of representative members of the myosin family of molecular motors. The N-terminal domain is a globular motor domain (blue), followed generally by one or more IQ motifs (dark orange) that bind either calmodulin molecules or specialized light chains similar in size to calmodulin. Myosin-VI has a unique insert (black) between the motor domain and the IQ domain that redirects the motor movement toward the minus end of the actin filament. Many myosins are dimeric owing to the assembly of two α-helices into a coiled coil (gray). Other specialized domains are highlighted by different colors: THB (light orange), SAH (red), and others as listed in the key. Abbreviations: FERM, 4.1 protein, ezrin, radixin, moesin; IQ, isoleucine-glutamine; MyTH4, myosin tail homology 4; NM-IIa, nonmuscle myosin-IIa; PEST, sequence rich in proline, glutamic acid, serine, and threonine residues; PH, pleckstrin homology; PX, phox homology; RCC1, related regulator of chromosome condensation 1; SAH, single α-helix; SH3, Src homology region type 3; TgMyo, Toxoplasma gondii myosin; THB, three-helix bundle; UBA, ubiquitin-associated.
All myosins travel along actin filaments to carry out their functions. The actin cytoskeleton, with dozens of associated accessory proteins, provides the cell with structure and organization. Actin tracks are polarized (Figure 2), with barbed or plus ends and pointed or minus ends, and the plus ends are directed toward the plasma membrane or, in the case of striated muscle, the Z-lines of the sarcomere. All known myosins except myosin-VI (5) move along actin toward the plus end.
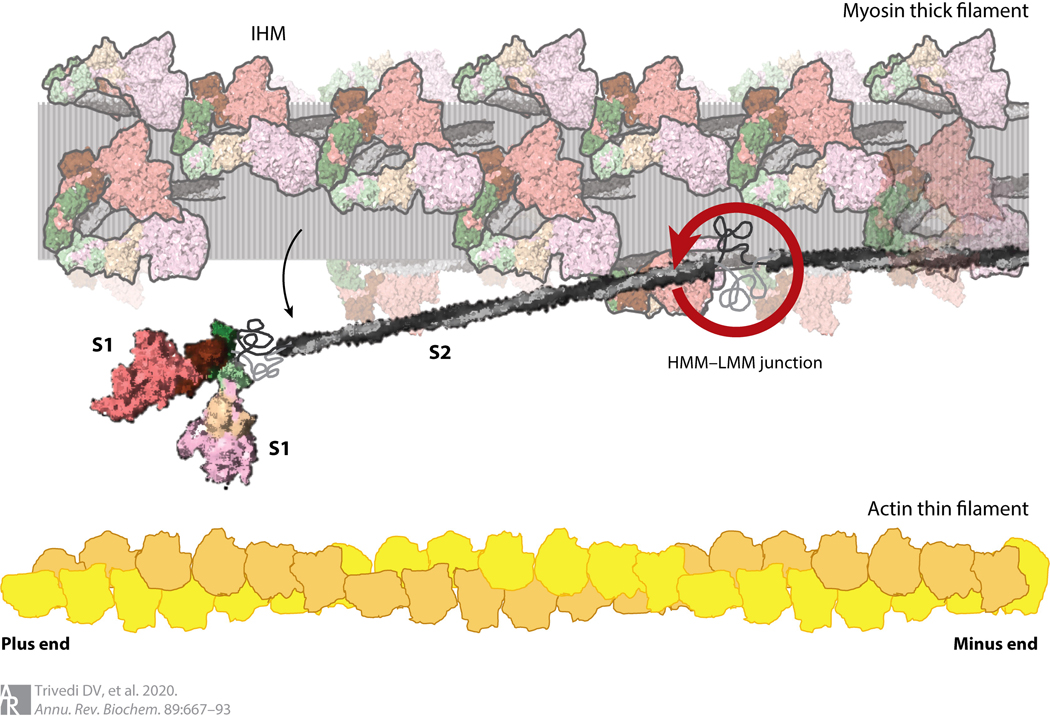
Figure 2.
Structure of cardiac myosin and arrangements in the muscle sarcomere drawn to scale. The HMM domain of the myosin consists of the two S1 domains and the S2 domain. LMM is the C-terminal portion of the coiled-coil tail to the right of the HMM–LMM junction. Interactions between the LMM portions of myosin molecules form the shaft of the thick filament. The relative sizes and spacings of the myosin-containing thick filament and the actin-containing thin filament within the muscle sarcomere are shown. The heads folded back onto the shaft of the thick filament are illustrated in their IHM state, with the S1 heads folded back onto their S2 tails (translucent myosins). A single myosin molecule released from its IHM state is shown swinging away from the thick filament, swiveling about its HMM–LMM junction. Abbreviations: HMM, heavy meromyosin; IHM, interacting heads motif; LMM, light meromyosin; S1, subfragment 1; S2, subfragment 2.
The motor domain of all myosins is the approximately 80-kDa globular myosin head (6) (approximately 780 amino acids), which has an actin-binding domain that is about 4 nm from the nucleotide-binding site, as revealed by the first crystal structure of a myosin head domain (7) (Figure 3a). This basic structure is common to all classes of myosins that have been crystallized (8). At the opposite end of the globular head domain from the actin-binding site is the converter, which is immediately distal to two cysteine-containing helices called the SH1 and SH2 helices (Figure 3b). These two helices change their configurations dramatically, associated with the movement of the converter through an extensive angle while bound to actin (Figure 3c,d). This movement is accompanied by the release of one of the ATP hydrolysis products, inorganic phosphate (Pi) (Figure 4a). Thus, myosin is a Pi sensor, and the converter rotation associated with Pi release is the basis of all movements of myosins along actin. The step size the myosin takes (its movement along actin per ATP hydrolyzed) is amplified by a long α-helical extension of the globular heavy chain, which is stabilized by association with calmodulin molecules or, in the case of muscle and some nonmuscle myosins, calmodulin-like light chains (approximately 20 kDa each). The example shown in Figure 2 is the muscle myosin globular head domain known as subfragment 1 (S1), where the lever arm length is about 10 nm, with two light chains bound: the essential light chain (ELC) and the regulatory light chain (RLC) (Figure 4b). Other myosins have longer or shorter lever arms and rotate about different angles to accommodate different functions they serve. However, the essential elements of all myosins are depicted in Figure 3.
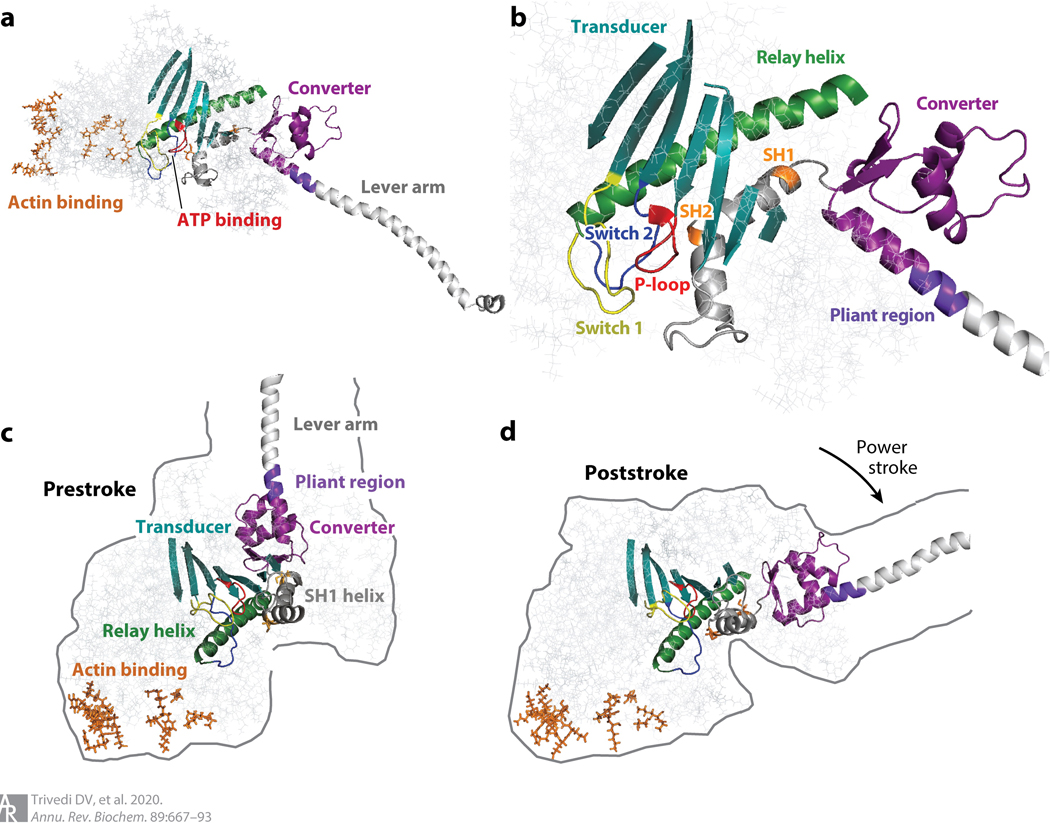
Figure 3.
Structures of pivotal sites and structural elements relevant to all myosin motors. (a) PyMOL visualization of the myosin head (S1 domain) without the light chains bound to the heavy-chain α-helix (lever arm). Actin-binding residues (orange) are in stick mode. The ATP-binding pocket (red) is about 4 nm away from the actin-binding surface. The converter (purple) links the lever arm (light gray, without light chains shown) to the motor domain. (b) Enlarged view of the structure shown in panel a. The switch 1 (yellow), switch 2 (blue), and P-loop (red) form the nucleotide-binding pocket. The relay helix (green), transducer (dark cyan), and SH1 and SH2 (orange residues) helices (gray) are between the nucleotide-binding pocket and the converter (purple). A short pliant region (purple-blue) links the converter to the lever arm (light gray). (c) Arrangement of structural elements in the prestroke state of the S1. (d) Arrangement of structural elements in the poststroke state of the S1. Abbreviation: S1, subfragment 1.
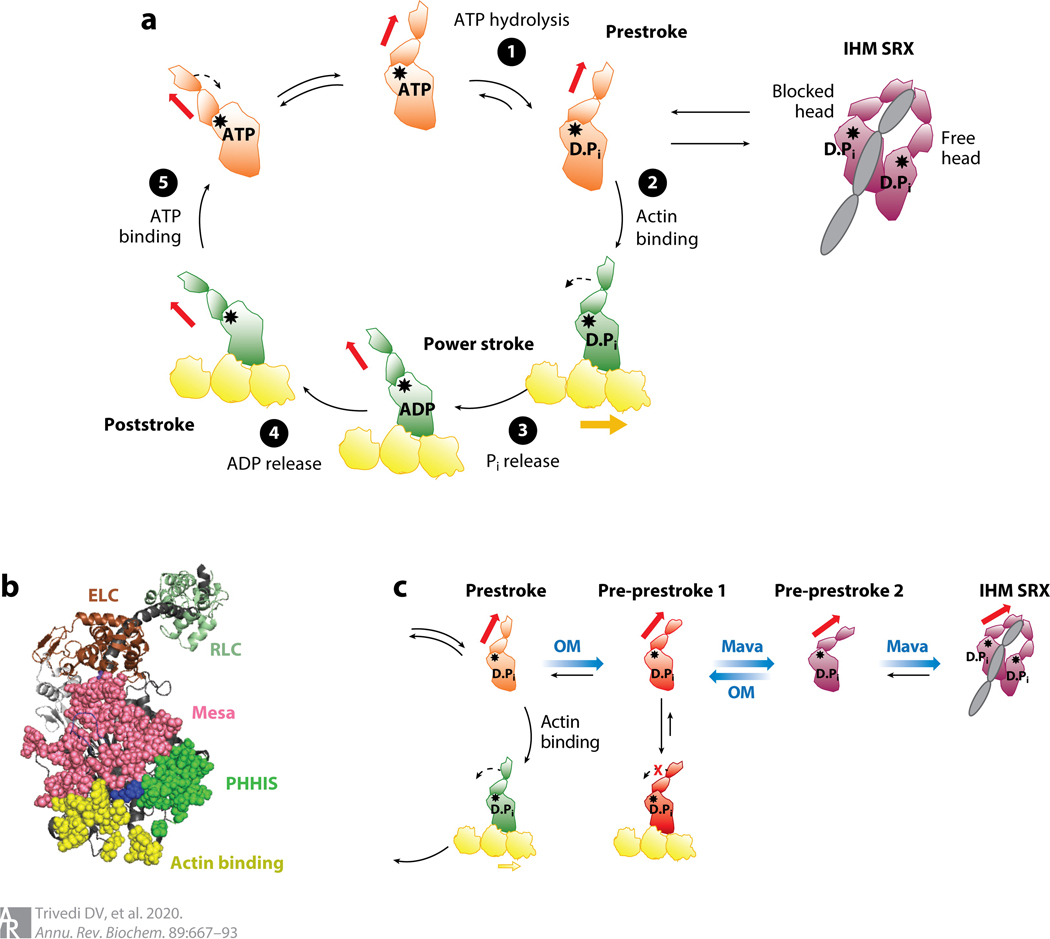
Figure 4.
S1 heads of myosin-II in various states. (a) Chemomechanical cycle of the interaction of myosin heads with actin. ❶ The S1, free of actin, hydrolyzes ATP, and the products remain bound to the active site. ❷ The prestroke S1 with bound ADP and Pi binds to actin (yellow). ❸ While bound to actin, associated with Pi release, the lever arm swings to the left (black dashed arrow) about a fulcrum point (black star), moving the actin filament to the right (bold yellow arrow) with respect to the myosin thick filament.❹ ADP release allows a further small stroke to the poststroke position and frees the active site for binding of ATP. ❺ ATP binding weakens the interaction of the S1 with actin. The prestroke state can be pulled into the folded IHM SRX state (purple). Actin-bound and actin-unbound myosin heads are shown in green and orange, respectively. Red arrow denotes direction of the lever arm. (b) PyMOL structure showing three critical surfaces of the S1: the actin-binding face (yellow), the mesa (pink), and the PHHIS (green). At the apex of the pyramid formed by these three surface domains is residue Arg403 (blue), which, when mutated to a glutamine, causes hypertrophic cardiomyopathy. The ELC (brown) and RLC (pale green) are shown attached to the lever arm heavy-chain α-helix (dark gray). This homology model of the prestroke state of human β-cardiac S1 can be downloaded from our website (http://spudlab.stanford.edu/homology-models). (c) Predicted different conformational states for OM-bound (red) and Mava-bound (purple) S1 heads. Abbreviations: D.Pi, ADP.Pi; ELC, essential light chain; IHM, interacting heads motif; Mava, mavacamten; OM, omecamtiv mecarbil; PHHIS, primary head-head interaction site; Pi, inorganic phosphate; RLC, regulatory light chain; S1, subfragment 1; SRX, super relaxed.
The transition of the head from a prestroke to a poststroke configuration results from an ensemble of structural changes that add up to a new, lower-energy state of the S1, associated with Pi and ADP release. The S1 returns to its higher-energy prestroke state upon ATP binding and hydrolysis, with the ADP and Pi products remaining bound at the active site. There are many noteworthy changes in the structure of the S1 associated with prestroke to poststroke transition. An important structural element in the motor domain is a long α-helix known as the relay helix (Figure 3b). A primary structural difference between the prestroke and poststroke states of myosin is the presence of a kink in the relay helix at the beginning of the power stroke (Figure 3c), and upon transitioning to the poststroke state, subsequent straightening of the relay helix ac-commodates a 60–70° rotation of the converter domain and the attached lever arm for muscle myosin-II (Figure 3d). Associated with the prestroke to poststroke transition is a change in the nucleotide pocket that allows Pi to be released from the active site. The changes in the affinities of ATP, ADP, and Pi for the active site are associated with movements of a P-loop, a switch 1 loop, and a switch 2 loop, which are critical elements of the nucleotide-binding site (Figure 3b). These same three elements are found in the G proteins, which are evolutionarily related to myosin (9, 10).
Muscle myosin and nonmuscle myosin-IIs (NM-IIs) self-assemble into bipolar thick filaments with heads arranged in a helical pattern (Figure 2). This self-assembly involves interactions between the light meromyosin (LMM) portion of the coiled-coil tail. The subfragment 2 (S2) portion of the tail is free to swing away from the shaft of the thick filament to allow the S1 heads to interact with actin. Figure 2 shows the structure of the arrangement of myosin with respect to actin in human cardiac muscle drawn to scale. The myosin heads will not reach the actin filament without the S2 swinging away from the thick filament. The ability to swing out derives from a hinge region at the heavy meromyosin (HMM)–LMM junction, presumably a somewhat melted-out region of the coiled-coil tail (11) (Figure 2).
While the HMM–LMM hinge is specific to the class II type of myosins, a critical feature for all myosins is a similar disordered hinge at the S1–tail junction (Figure 2). This hinge region, which, like the HMM–LMM junction, is protease sensitive in muscle myosins (12, 13), provides the S1 heads the needed rotational freedom to more easily make productive interactions with actin.
During muscle relaxation, many of the myosin heads are believed to be flush against the shaft of the thick filament. These heads are thought to be mainly in a state that involves the two S1 heads asymmetrically folded back onto their S2 tail and interacting with one another in the interacting heads motif (IHM) (14–22) (Figures 2 and 4a). The blocked head (its actin-binding site is buried in the structure) has a mesa surface (15, 23, 24) (Figure 4b) that interacts with the proximal S2 tail and a primary head-head interaction site, which is distinct from the actin-binding interface and binds to the converter domain on the free head (actin-binding site exposed). Both heads in the IHM are believed to be in an OFF state. Heads in this state are effectively pulled out of the cycle of interaction between myosin and actin (Figure 4a). This level of control of myosin function is believed to operate across all myosin-IIs (25, 26) and probably applies to other members of the myosin family as well. While they do not explicitly fold into an IHM configuration, myosin-Va (27), a type I myosin (28), and myosin-VIIa (29, 30) appear to involve the folding of the tail domain to interact with the head and inhibit its ATPase activity.
Many molecules of the single-headed myosins, such as myosin-I (31), have to act in concert to ensure continuous association with actin during movement. Myosin molecules need to let go of the actin to hydrolyze another ATP for another cycle of interaction with actin, and other molecules need to be actin-associated during this time to keep the actin and myosin ensemble engaged. This is also true of some two-headed myosins, such as muscle myosin-II. To achieve the high velocities needed during muscle contraction, each myosin head must spend only a very short time (1–10 ms) in a strongly bound force-producing state with actin. Thus, muscle contraction involves many myosin heads assembled into a thick filament so that during sliding of the myosin-containing thick filaments past the actin-containing thin filaments, there is at least one head, and in reality more, bound to actin at any moment during contraction to keep the two filaments engaged. Such myosins are called nonprocessive motors, meaning that a single molecule cannot take multiple steps along actin before dissociating. They generally spend <10% of their ATPase cycle time in a state strongly bound to actin (that is, their duty ratio is <0.1).
Some two-headed myosin molecules are processive, meaning that one molecule can carry cargo along an actin filament over considerable distances. Thus, myosin-V, which carries cargo from one place to another in nonmuscle cells, can take more than 20 steps of about 36 nm each (a total distance of >0.7 μm) toward the plus end of actin filaments, and myosin-VI can carry cargo similar distances toward the minus end. Myosin-VI reverses the normal directionality of myosins by inserting a new mechanical element between the converter and the light chain–binding domain (32–34) (Figure 1). Processive myosin motors generally spend >50% of their ATPase cycle time in a state strongly bound to actin (that is, their duty ratio is >0.5).
ALL MYOSINS ARE HIGHLY ALLOSTERIC SENSORS
The binding of ATP is associated with three significant changes in the myosin molecule that point to the highly developed allostery used by this magnificent enzyme. First, the switch elements close and interact with the γ-phosphate of ATP. This conformational change positions the ATP for hydrolysis to ADP and Pi (35, 36), which remain bound at the active site (Figure 4a). Second, the changes at the active site upon ATP binding are allosterically associated with changes in the actin-binding domain approximately 4 nm away, which dramatically weakens the affinity of myosin for actin (37) (Figure 4a). Third, the ATP and ADP.Pi configurations at the active site are associated with changes in the converter and lever arm, again several nanometers away from the active site. The binding of ATP and ADP.Pi results in a prestroke configuration of the converter and associated lever arm, accommodated by the kink in the relay helix. Thus, the formation of the prestroke state of myosin occurs after the myosin is dissociated from actin.
Upon strong binding to actin, changes in the conformation of myosin are again allosterically transmitted to the nucleotide site and the converter–lever arm (38–40). An important structural analysis of the nucleotide-free myosin-V head by Coureux et al. (41) shows that the relay helix can straighten with the converter–lever arm now positioned in the poststroke state while the switch 2 element remains closed. In this structure, the straightening of the relay helix is associated with a bending of a seven-stranded β-sheet known as the transducer (42) (Figure 2c,d), and the active site is structurally altered to enhance the release first of Pi then of ADP. Thus, the power stroke occurs, appropriately, only after actin is bound (Figure 4a).
BINDING OF THERAPEUTIC SMALL-MOLECULE EFFECTORS OF CARDIAC MYOSIN ACTIVITY REFLECTS MYOSIN’S ALLOSTERY
Small molecules binding to the catalytic head domain of cardiac myosin favor different conformational states of myosin and alter the number of myosin molecules functionally accessible for interaction with actin. These effects are also highly allosteric, with none of the small-molecule effectors binding at the active nucleotide site. Different small molecules have different effects on the functional parameters of myosin and trap the molecule in different conformational states. Besides the conventional prestroke and poststroke states, several myosin head crystal structures show the lever arm in positions that are different from these conventional configurations (for a review, see 43), revealing considerable flexibility of the lever arm position. At least one intermediate position between the conventional prestroke and poststroke configurations was also defined from dynamic studies on myosin using time-resolved fluorescence energy transfer (44).
Omecamtiv mecarbil (OM), developed by Cytokinetics and now in phase 3 clinical trials for heart failure, is an activator of cardiac muscle contraction (45–48) that binds near the SH1 helix and close to the surface of the S1 near the converter domain (49). OM accelerates Pi release in the presence of actin, and this is proposed to result in more heads entering the strongly bound force-producing state, as discussed by Planelles-Herrero et al. (49). Consistent with this is the finding that OM positions the lever arm in a conformation close to the prestroke state (49), thus pulling the equilibrium from the poststroke configuration to the ADP.Pi-bound prestroke configuration (Figure 3c,d). Increasing the number of heads in the ADP.Pi prestroke state should provide more heads ready to interact with actin (49). In addition, the binding of OM may move heads out of their sequestered OFF state, causing an additional increase in the number of heads in their ADP.Pi state, ready to interact with actin (43, 50). Furthermore, cardiac contraction is regulated by the Ca2+ - sensitizing tropomyosin–troponin complex bound to the actin filaments. An additional activating effect of the OM-bound heads could be that their more rapid Pi release and hence more rapid than normal binding to actin help push tropomyosin out of its position that blocks myosin binding. This may result in the recruitment of more normal, OM-free heads.
Interestingly, OM-bound heads have a reduced rate of the prestroke to poststroke transition (approximately 2 s−1 rather than the normal approximately 15 s−1) (51) and a reduced stroke size (52). Since stroke completion is associated with ADP release followed by ATP rebinding and release from actin, the OM-bound heads remain bound to actin for a much longer time than normal heads (approximately 80 ms rather than approximately 10 ms) (52, 53). This longer binding time and the reduced stroke size reduces the velocity of actin filament movement in vitro (53–56). If the OM-bound heads did not stroke at all while bound, they would produce zero work. Since the OM-bound heads do still undergo a small stroke, however, work is performed, adding to the force production by the normal, OM-free heads. Systole is observed to be extended (45), probably due to the increased number of myosin heads bound as well as the longer binding time of the OM-bound heads, giving an overall longer force-producing period.
During cardiac muscle contraction, any given myosin head is likely used only once during a particular systolic/diastolic contraction cycle (57). This means that a primed head with ADP.Pi bound is ready to go onto actin in a force-producing state, bleeding off that force as the lever arm swings, providing an amount of work that is the average force produced times the distance moved. Then that head releases, not to be used again in that heartbeat. This means it has plenty of time to hydrolyze its newly bound ATP and get ready for binding again to actin before the next heartbeat occurs.
Another allosteric effector, this time an inhibitor of contractility, is mavacamten, developed by MyoKardia for treatment of hypertrophic cardiomyopathy (HCM). Mavacamten, also in phase 3 clinical trials, appears to position the S1 heads in a structural state that easily fits into a folded-back sequestered state (58), possibly the IHM configuration. Either the mavacamten-bound S1 takes up a head configuration that is very near the head configurations in the folded-back sequestered state, or it has the compliance to easily adopt the conformation(s) of the heads in the folded-back sequestered state (43, 59). Mavacamten binding to bovine cardiac HMM (60) and human β-cardiac 25-hep HMM (58) causes inhibition of the basal ATPase of myosin as well as its actin-activated ATPase activity (61).
MAMMALIAN NONMUSCLE MYOSIN-IIs HAVE BEEN PROPOSED AS A POSSIBLE THERAPEUTIC TARGET
NM-IIs have very similar structures to muscle myosin (2). In mammalian cells, there are three different isoforms—NM-IIA, NM-IIB, and NM-IIC—with similar head domains and shared ELCs and RLCs. The head domains of these three isoforms show 90–92% similarity (62). As in muscle myosin, the tail regions of the NM-IIs are responsible for both dimerization of the heavy chains and formation of functional filaments (63). The C-terminal, short, nonhelical tailpiece of these three isoforms exhibits the most sequence divergence (75–77% similarity) and is thought to be responsible for their differential subcellular localizations.
Two different domains on NM-II can be phosphorylated: the RLC and the tail. Phosphorylation of the RLC by myosin light-chain kinase and Rho-associated protein kinase (ROCK) transforms the molecule from a folded, assembly-incompetent, inactive conformation into an asymmetric, unfolded active form that can form bipolar thick filaments, hydrolyze MgATP, and slide actin filaments (64, 65). However, phosphorylation of the coiled-coil region of the tail and the nonhelical tailpiece by kinases such as protein kinase C, casein kinase II, and transient receptor potential melastatin 7 dissociate the bipolar filaments and prevent further filament formation (66). In NM-IIA, phosphorylation of the tail domain is associated with the epithelial to mesenchymal transition in mouse epithelial cells (67).
In general, NM-II acts as a principal controller of cell morphology, with roles in several vital processes, including cellular migration and postsynaptic dendritic plasticity in neurons. NM-II also generates forces that remodel biochemical signaling by inducing changes in interactions between actin-associated proteins that can eventually regulate gene transcription. Additionally, NM-II has recently surfaced as a critical regulator of diverse, genetically complex diseases, including neuronal disorders, cancers, and vascular disease. Many of these functions have been discussed in detail in a review by Newell-Litwa et al. (68). Here, we discuss the emerging role of NM-II activity in cancer and briefly touch on its role in neurological and vascular disorders.
Nonmuscle Myosin-II in Cancer
NM-II plays an essential role in cytokinesis by forming a contractile ring at the furrow region of the dividing cell and applying forces that drive cell division. Over the past decade, many different types of cancers have been associated with altered NM-II regulation and localization. In these cancer models, NM-II is thought to be a crucial driver for processes that are necessary for tumor invasion and metastasis (reviewed in 69). Among the different types of cancer, breast cancer, glioblastoma, and squamous cell carcinoma have been the most studied. These cancer cells are thought to have differential expression and activation of NM-II isoforms, which lead to cell division and migration patterns that underlie tumorigenesis and invasion (68).
The current hypothesis is that many upstream regulatory signaling pathways converge down to the NM-II family of molecular motors, and hence NM-II has been proposed as a potential therapeutic target. For example, downregulation of the tumor suppressor gene BRCA2 leads to mislocalization of NM-II in mitosis, leading to chromosome instability and aneuploidy (70, 71). Similarly, the oncogene Ras and its downstream target BRAF, which regulate cell survival and propagation, also increase NM-II activation, leading to tumor invasion in vitro and in a mouse melanoma model in vivo (72–75). In both in vitro and ex vivo models, it has been shown that gliomas use NM-II–dependent contractility to migrate within the brain, and NM-II inhibition by blebbistatin prevents glioma invasion (76, 77). In a rodent glioblastoma model, genetic deletion of both NM-IIA and NM-IIB genes decreases tumorigenesis and weakens tumor proliferation (78). Targeting NM-IIA alone impairs glioblastoma invasion but, in fact, enhances glioblastoma growth and lethality (78). In conjunction with a previous report (79), the Picariello et al. (78) study suggests that effective treatment of glioblastoma will require inhibiting targets that drive both invasion and proliferation separately. However, two reports have shown that targeting NM-IIA induces squamous cell carcinoma in mice (80, 81); while one report (80) found that NM-IIA stabilizes the tumor suppressor p53, a second did not observe this (81).
Another study (82) showed that stress-sensitive proteins such as NM-IIA and NM-IIC are highly upregulated in pancreatic ductal adenocarcinoma cancer and patient-derived pancreatic cancer cell lines. NM-IIB, by contrast, virtually disappears with pancreatic ductal adenocarcinoma cancer. In such disease models, NM-IIC is involved in the formation of actin arcs in single cells and cortical actin belts in tissue spheroids. The authors used this to their advantage by targeting NM-IIC with a small-molecule activator, 4-hydroxyacetophenone, which increases NM-IIC assembly and stiffens cells. This decreases propagation, induces cortical actin belts, and slows retro-grade actin flow in spheroids. Similar treatment in a pancreatic cancer mouse model shows a reduction in liver metastases. The authors argue that increasing the activity of these mechanoresponsive proteins by increasing NM-IIC assembly to overcome the capacity of cells to polarize and penetrate may be an effective strategy to improve the survival rate of patients with pancreatic cancer.
Initial stages of tumor cell metastasis require an epithelial to mesenchymal transition that involves activation of amoeboid migration and loss of cell–cell adhesion. NM-II has roles in cell body translocation and retraction of the posterior of the cell during migration. In a study involving MDA-MB-231 breast cancer cells, both NM-IIA and NM-IIB were recruited to the lamellar margin during active spreading on fibronectin (83). There was also a transient increase in RLC phosphorylation associated with the recruitment. Furthermore, pharmacological or small interfering RNA–mediated inhibition of NM-II and myosin light-chain kinase indicates that both NM-IIA and NM-IIB contribute to overall cell migration but that the NM-IIB isoform contributes explicitly to the mechanics of lamellar spreading on extracellular matrix.
Given these observations and others, as reviewed in Reference 68, NM-II has been suggested as a possible target to arrest tumor metastasis. However, based on the roles of NM-IIs in other aspects of cell biology (e.g., see the sections titled Nonmuscle Myosin-II in Neurological Disorders and Nonmuscle Myosin-II in Other Disorders) and the widespread localization of NM-IIs in most cells in the body, toxicity from small-molecule effectors may be an important issue to consider.
Nonmuscle Myosin-II in Neurological Disorders
NM-II plays significant roles in neuronal cell biology by mediating growth cones, which are actin-enriched structures in neurites, axons, or dendrites that drive their motility toward desired targets in response to chemical cues. Among the three isoforms, NM-IIB is highly enriched in the brain, where it is located at both the presynaptic terminals and postsynaptic dendritic spines (84). The findings that NM-IIB mediates synaptic vesicle recycling and regulates the maturation of spines and the clustering of glutamate receptors in the postsynaptic terminals led researchers to label it as an essential regulator of stimuli-induced changes in spine morphology that underlie learning and memory formation (85–87). In addition to a direct role in neuronal cell biology, NM-II also plays a vital role in glial cell function (76, 88), the integrity of the blood-brain barrier (89, 90), and microglia activation in neuroinflammation (91).
Unsurprisingly, NM-II has been implicated in diverse neurological disorders (reviewed in 92). These include neurodevelopmental disorders such as autism, neurodegenerative disorders such as Alzheimer’s disease, and neuronal migration disorders such as lissencephaly and disorders of axon regeneration following central nervous system injuries (93). De novo mutations in MYH9, which codes for NM-IIA, are associated with autism, schizophrenia, and intellectual disability, and de novo mutations in MYH10, which codes for NM-IIB, are related to both schizophrenia and autism (94). Direct targeting of NM-II using blebbistatin and inhibiting the NM-II ROCK restored axon growth on inhibitory substrates, in chick dorsal root and retinal ganglion cells, in rat cerebellar granule neurons (93, 95–99), and in an in vivo rat spinal cord injury model. These observations have led to the suggestion that NM-II may be a potential therapeutic target for axon regeneration in devastating spinal cord injuries and central nervous system lesions.
In glial biology, NM-II plays a significant role in preserving the function of astrocytes, oligodendrocytes, and microglia (reviewed in 100). In microglia, NM-II contributes to neuroinflammation resulting from microglia activation and release of inflammatory cytokines (reviewed in 101). In a rodent model of Parkinson’s disease, ROCK inhibition prevents microglia activation and the phagocytosis of disintegrating dopaminergic neurons (102). Similarly, amyotrophic lateral sclerosis results in increased proinflammatory cytokine production, which is reduced by ROCK inhibition, leading to increased mouse motoneuron survival (103–105), also suggesting a role of NM-II in such disease progression.
The above complex roles of NM-IIs in neuronal functions suggest that toxicity from small-molecule effectors may be a hurdle when considering NM-IIs as a target for disease.
Nonmuscle Myosin-II in Other Disorders
NM-II activity in endothelial and smooth muscle cells is essential for blood vessel integrity of arteries, veins, and capillaries and therefore may contribute to many vascular disorders, such as atherosclerosis and edema. Autosomal dominant NM-IIA mutations have also been reported in many macrothrombocytopenia disorders such as May-Hegglin anomaly, Epstein syndrome, Fechtner syndrome, and Sebastian syndrome. Other than those, NM-IIA-related disorders also often present with deafness, cataracts, and nephropathy in affected individuals (106). Given the widespread localization of NM-IIs in most cells of the body, more work will be required to assess the toxicity profile of targeting NM-IIs.
THE UNCONVENTIONAL MOLECULAR MOTOR MYOSIN-X HAS ALSO BEEN PROPOSED AS A POSSIBLE THERAPEUTIC TARGET
Myosin-X, an unconventional molecular motor located at the tips of filopodia, is essential for wound healing (107), filopodia formation (108), angiogenesis (109), the regulation of growth cone formation (110), and invadopodia formation (111). Myosin-X is widely expressed in low abundance in vertebrate tissues. It is prominent in the developing brain, endothelial tissues, and many epithelial tissues (112). Like other myosins, myosin-X has a head domain that binds actin, hydrolyzes ATP, and generates force. Its lever arm consists of three isoleucine-glutamine (IQ) binding motifs, and it has a tail that endows class-specific properties such as dimerization and cargo binding. Myosin-X is unusual among actin-based motor proteins in that its tail can provide a direct link to microtubules. A unique feature of the myosin-X tail is its three pleckstrin homology domains, one of which binds to the vital signaling lipid phosphatidylinositol (3,4,5)-trisphosphate (PIP3) (113, 114). In the absence of PIP3 binding, myosin-X can exist as a monomer in which the tail folds back onto the head to form a compact and auto-inhibited OFF state (115). Once activated to dimerize, myosin-X can undertake its biological roles, including transporting cargo and forming filopodia.
Myosin-X in Cancer
One of the first pieces of evidence of filopodia involvement in cancer biology was the discovery of both upregulated fascin, a filopodial crosslinking protein, and filopodial structures themselves in aggressive metastatic cancer cases (116). Both fascin and myosin-X have been identified as critical components in structures of a different type called invadopodia, which are actin-rich protrusions with proteolytic activity (111) capable of digesting surrounding extracellular matrix, an escape mechanism for primary tumor cells. Myosin-X knockdown and silencing results in the blockage of invadopodia function by inhibiting matrix digestion ability (111). Similarly, suppression of myosin-X in filopodium-like protrusions significantly decreases proliferation rates and invasiveness of cancer cells in breast cancer cell models (117).
Myosin-X in Infectious Disease
Recent evidence from filoviruses such as Marburg virus suggests that myosin-X may also be involved in filopodia-dependent viral budding. Expression of a dominant-negative myosin-X construct caused a significant reduction in virus-like particle production (118). The spread of the diarrhea-causing bacterium Shigella flexneriis also myosin-X driven (119).
In thinking about myosin-X as a potential therapeutic target, one runs into the same problem as for the NM-II family. Myosin-X is widely distributed and plays multiple roles in cell biology. Small-molecule effectors against myosin-X are therefore likely to lead to significant toxicity.
MAMMALIAN MYOSIN-V, MYOSIN-VI, AND MYOSIN-VII ARE ALSO DISEASE ASSOCIATED
Many other unconventional mammalian myosins such as myosin-V, myosin-VI, and myosin-VII localize to several intracellular compartments and participate in transporting and anchoring events (2). While the catalytic heads of these myosins share significant similarities with conventional myosins, their tail regions are highly divergent (120) (Figure 1). The diversity in the tail domain of these myosins is in the specific domains that bind to different adaptor binding proteins, which in turn can bind to different cargoes, such as an organelle, vesicle, or protein complex (121).
Mutations in such unconventional myosins have been linked to many human diseases. For example, different classes of myosins are associated with hearing through their contribution to the structure of the stereocilia of the inner ear (122). Mutations in class Ia, IIc, IIIa, VI, VIIa, and XV myosins can lead to Usher syndrome, the leading cause of genetic deaf-blindness (2). Similarly, myosin-Vb might transport apical endosomes in brush border cells (123), which could explain the association of mutations in this gene with microvillus inclusion disease (124). Mutations in myosin-Va are linked to Griscelli syndrome, which is characterized by defects in pigmentation and neuronal malfunction (125). Many of these myosins—other than myosin-XV, which is specifically known to be present only in the stereocilia of the inner ear—are distributed throughout different organs of the body, and therefore targeting these myosins therapeutically could lead to toxic effects.
THE SPECIALIZED MYOSINS FOUND IN PARASITES MAY REPRESENT EFFECTIVE THERAPEUTIC TARGETS
Myosin-XIV Is a Class of Specialized Myosin Found in Many Mammalian Parasites
Class XIV myosins are present in the protist group Alveolata, and specifically the phylum Apicomplexa, which includes a broad variety of obligate intracellular parasites that infect many different animal species. Some of these parasites cause devastating human diseases such as malaria (Plasmodium spp.), toxoplasmosis (Toxoplasma gondii), or enteric infections (Cryptosporidium), notably in the developing world. Malaria is responsible for half a million deaths each year worldwide, and toxoplasmosis affects around 33% of the human population. A 2011 study (126) put toxoplasmosis as the second leading cause of foodborne-related deaths and the fourth leading cause of foodborne-related hospitalizations in the United States. Apart from infecting humans, these parasites infect livestock and cause significant economic losses.
Host cell penetration, an essential step in the infection process, is considered to be an active process governed by a unique actomyosin-based structure in the parasite called the glideosome (127). The glideosome, located between the plasma membrane and the inner membrane complex of the parasite, is critical for the gliding motility in these parasites that propels them across biological barriers and helps them invade and egress from host cells. Several apicomplexans fail to glide or invade when treated with drugs that interfere with actin dynamics (e.g., latrunculin B, cytochalasins, jasplakinolide) or with myosin activity (e.g., 2,3-butanedione monoxime). Knockdown of some critical actomyosin components of the glideosome, using a tetracycline-inducible transactivation system, led to a reduction in host-cell invasion (128). However, knockout of some parasite invasion machinery genes also led to the discovery of other redundant pathways the parasite can employ for host cell invasion (129).
Like every other myosin, myosin-XIVs have a conserved head domain that houses the actin-and ATP-binding sites and a lever arm that binds accessory light chains (Figure 1). Myosin-XIVs lack a tail region that typically allows for dimerization. Hence, these are monomeric myosins. The highly studied MyoA of class XIV myosin is essential for maintaining the gliding motility and invasion of the Plasmodium and Toxoplasma parasites (127, 128, 130, 131). MyoA is a low-duty-ratio, fast, monomeric motor and is highly conserved in all apicomplexans (4, 132–134). Both T. gondii and Plasmodium falciparum appear to express MyoA through all life cycles of the parasite. The T. gondii MyoA binds a regulatory light chain called MLC1 and an essential light chain called ELC1. A functionally redundant ELC2 was also discovered, and both ELC1 and ELC2 were found to be important in invasion, egress, and motility of T. gondii (135). Recent work from Trybus and colleagues (136, 137) demonstrates cochaperone-mediated expression and purification of T. gondii and P. falciparum MyoA motors. The T. gondii MyoA moved actin at fast velocities in the in vitro motility assay when both the RLCs and ELCs were bound to the lever arm (136). The P. falciparum MyoA also moved actin at higher velocities only when the light chain called myosin A tail domain interacting protein and the P. falciparum specific ELC were bound (137).
Recently, a significant breakthrough was achieved by Powell et al. (138) when they crystalized the structure of the T. gondii MyoA motor domain at 2.6 Å. The structure demonstrates interesting strategies for chemomechanical coupling and efficient force transduction. The unique sequence elements discovered in this structure are located in known binding sites for inhibitors of myosin function and suggest that specific inhibitors may be able to be developed to target myosin-XIV motors of apicomplexan parasites. Following up, another breakthrough from Houdusse and colleagues (139) came when they determined the crystal structure of MyoA from P. falciparum. Combining structural, kinetic, and parasitological analyses, the authors proved that MyoA is essential for erythrocyte invasion by the parasite. They demonstrate that phosphorylation of the N-terminal extension of this MyoA tunes the motor either for high force (required for optimal invasion) or fast motility speed (required for parasite dissemination). This is a clear example of how the myosin family of motors shares the same global architecture but the evolutionary addition and deletion of structural elements makes them serve very distinct purposes. These evolutionary strategies can be exploited to design novel classes of therapeutics targeted to specific myosins in these parasites. The MyoA motor, along with its light chains and other proteins, forms a central part of the glideosome, which helps to propel the parasites and aids in infecting the host cells. The light chain and the associated proteins target the MyoA to the inner membrane complex of the T. gondii parasites. The P. falciparum species contain an analogous glideosome assembly with its different components (140, 141). Ca2+ -dependent phosphorylation of T. gondii MyoA has also been reported that aids in parasite motility and egress from the host cell (142, 143). Phosphorylation sites on the T. gondii myosin light chain 1 are near a site on this light chain that covalently binds a small molecule and inhibits parasite motility (144).
MyoB, another member of the myosin-XIV family, is not associated with the glideosome but rather is at the apical region of the P. falciparum cell. It has a very distinct light chain with a calmodulin-like domain at its C terminus and an extended N-terminal region, which is thought to help localize the motor to its appropriate cellular location (145). MyoC, a third member of the family, is a product of alternate splicing of a single gene that encodes both MyoB and MyoC in T. gondii. They are mostly similar except for their tail domains, which give them different solubilities and different subcellular localizations. MyoC in T. gondii assembles into distinct glideosomes from MyoA and can compensate for the MyoA glideosomes, and vice versa (146). This redundancy helps parasite survival.
From a potential therapeutic point of view, MyoH is particularly interesting. MyoH is indispensable for the survival of T. gondii, and its conditional depletion completely blocks gliding motility, invasion, and egress. This occurs despite a functional MyoA glideosome assembly. MyoH appears to be the principal initiator of the gliding motility, which is then communicated to the MyoA-associated glideosome (147). It is found in all apicomplexan parasites except P. falciparum and Gregarina polymorpha. Its extended neck region has eight IQ domains bound to accessory light chains. The MyoH features tail domains with similarity to alpha-tubulin suppressor 1 (ATS1) and related regulator of chromosome condensation 1 (RCC1) proteins. RCC1 binds chromatin, and ATS1 may be involved in coordinating the cell cycle (148, 149) (Figure 1). The MyoH tail region establishes contact between the tubulin-based and actin-based cytoskeletons. This may hint at the role of this myosin during mitosis, cytokinesis, or other cell-cycle related events. The RLCs of MyoH regulate the activity of myosin in a Ca2+ -dependent way (150).
MyoD and MyoE are associated with T. gondii, but they are not essential for survival. MyoD is the smallest motor, structurally similar to MyoA, and functions as a monomeric, fast, low-duty-ratio myosin. Both MyoD and MyoE lack tail domains. An unclassified T. gondii motor, myosin-L, is also thought to belong to class XIV, and a recent CRISPR gene disruption study pointed out that the myosin-L gene was essential for the parasite viability (151).
Myosin-XIVs are also found in ciliates such as Tetrahymena thermophila. These myosins rely on phosphorylation-dependent changes in myosin activity. Myosin-XIVs diverge from other myosins in that they have a specific His-Tyr-Ala-Gly sequence near the SH1 and SH2 helices. While this sequence has a tyrosine or phenylalanine at the second position in other myosins, myosin-XIVs, as a hallmark, possess either serine or threonine at this location. The smaller serine or threonine residues here facilitate a tight coupling between the active site and the lever arm of myosin-XIVs, which imparts desirable functionalities for gliding and invasion (138).
Other Classes of Parasite Myosins
The apicomplexan MyoF, together with myosins from a particular diatom, forms a separate class of myosins called myosin-XXII (4). This broadly conserved apicomplexan class of myosins is predicted to feature multiple IQ motifs similar to those in class V, VIII, XI, and XIII. The apicomplexan myosin-XXIIs have a long tail predicted to contain four to six WD40 repeats at the C-terminal end. The WD40 repeats form β-propeller structures known to be involved in transcriptional regulation and signal transduction. These repeats are known to coordinate multiprotein assemblies and may target to centrosomes or the nucleolus (152, 153). The N-terminal regions of these tails are predicted to form coiled-coil structures. MyoF of T. gondii is important in centrosome positioning, apicoplast inheritance, and organelle trafficking, thus making it particularly crucial for parasite survival (154). T. gondii depends on intracellular protein secretion via secretory vesicles called dense granules, and the trafficking of these dense granules is undertaken by the MyoF motor (155).
Two myosins of T. gondii and some other apicomplexans are part of the myosin XXIII and XXIV classes. MyoG and MyoJ of T. gondii and Eimeria tenella form class XXIII, and MyoI from T. gondii and a homolog from a Cryptosporidium species belong to class XXIV (4). These sequences are predicted to feature one or two IQ motifs, and the MyoG tail contains a single myosin tail homology 4 (MyTH4) domain. MyTH4 domains, found in several myosin classes, bind microtubules. However, the exact role of this domain in the apicomplexan myosins is unknown. T. gondii MyoG is found in the parasite periphery, and its gene disruption led to no change in the parasite activity (156). MyoJ is found in the basal region of the parasite, and parasites lacking this motor displayed an overall reduced fitness. A mouse model of infection showed a loss of virulence when the parasites lacked MyoJ (156). E. tenella is an apicomplexan capable of causing coccidiosis in poultry, sheep, and goats. Eimeria infections are severely damaging to the poultry industry and cost the United States approximately $1.5 billion in annual losses (157). Several Eimeria species infect various fishes, reptiles, birds, and a wide variety of mammals.
Cryptosporidium is a genus of the apicomplexan family that causes cryptosporidiosis, which leads to severe gastrointestinal problems in both immunocompetent and immunocompromised individuals. It is one of the most common causes of waterborne disease in the United States. Between 2009 and 2017, cryptosporidium outbreaks in the United States increased by 13% annually (158). As of 2015, there is only one drug approved for its treatment. Cryptosporidium parvum is known to have two class XIV, one class XII, one class XXIV, and two class VI myosins. MyoI in C. parvum is predicted to dimerize based on coiled-coil–forming regions. MyoI, belonging to class XXIV in T. gondii, maintains cell–cell communication of the parasites within a vacuole and ensures proper cell division. It contains two IQ motifs, a coiled-coil domain (ensuring dimerization), and a long tail without any known protein domains (159). Deletion of MyoI in T. gondii showed no defects in the parasite lytic cycle, but it led to asynchronous division in the host cell vacuole (160). It is not known if MyoI plays an active role in the transport of material between parasites within the vacuole. Apicomplexans also contain myosins whose exact functions are unknown. These myosins are similar to the minus end–directed myosin-VI motors found in humans. A recent systematic gene deletion study demonstrated that MyoF, MyoK (class XXIII), and the MLC-B light chain of MyoB are likely essential in the asexual blood stages of Plasmodium berghei, a parasite used in rodent models of malaria (161).
The fungus-like oomycete family of plant pathogens also has myosin genes. For example, the pathogen Phytophthora ramorum, known to cause the disease sudden oak death, has 25 myosin genes that encode 13 different kinds of myosins (162). Some of these myosins appear to belong to class XXII, XXIII, or XXIV (4). P. ramorum leads to mortality in several oak tree species and also causes diseases in several native and nonnative ornamental plants, shrubs, and trees within the United States. Yeast species such as Candida also have a repertoire of myosins, with a recent study (163) demonstrating the efficacy of an actin-binding drug in murine vulvovaginal candidiasis. This study revealed the importance of the yeast cytoskeleton in maintaining its life cycle. The presence of myosins in the Candida species opens avenues for designing novel therapeutics for multidrug-resistant species of Candida, such as Candida auris. This newly discovered species is tagged as an emerging global health threat by the US Centers for Disease Control and Prevention, as it is nonresponsive to any of the well-known antifungals.
Additionally, an insect-specific myosin-XX is present in Drosophila and the malaria parasite vector mosquito Anopheles gambiae. This myosin contains a well-conserved IQ motif in the neck domain. Myosins are also critical in the functioning of parasitic nematodes that infect humans with intestinal and other diseases. A recent study (164) pointed at the presence of 13–18 myosin genes in the nematode family, which includes parasitic nematodes such as Strongyloides ratti (a member of the Strongyloides family of parasites that infect humans) and Brugia malayi (which causes elephantiasis in humans). Thus, developing novel therapeutics targeted to these myosins is an open arena of exploration.
In summary, several classes of myosins—specifically myosin-XIV—that are likely to be targetable in particular ways without affecting mammalian host myosins represent an attractive target for small-molecule therapeutics for parasitic invasions. Indeed, in a recent screen, several egress and invasion inhibitors of the blood stage of P. falciparum were discovered (165).
Myosin-XXI
A similarly attractive potential target for small-molecule therapeutics is myosin-XXI, found in parasitic kinetoplastids such as Trypanosoma and Leishmania. Leishmania is transmitted by sandflies, and the human infection is caused by 21 of the 30 species of Leishmania protozoa that infect mammals. It occurs in the cutaneous and visceral forms in humans, with the visceral form being fatal. Tropical and subtropical regions are affected by leishmaniasis, including Mexico, Texas, and countries in Central and South America, southern Europe, Asia, the Middle East, and Africa. Myosin-XXI is present in both the motile and nonmotile stages of the Leishmania parasite life cycle. In the motile stage, these myosins are predominantly localized to the proximal part of the flagellum but are also found in other cell compartments. Two pools of myosin-XXI have been identified within the parasite: a membrane-bound fraction found at the base of the flagellum and a cytosolic fraction that is presumably involved in local transport (166).
Reducing the levels of Leishmania myosin-XXI expression in cultured motile stages of the parasite led to a loss of intracellular trafficking in the parasite and a loss of flagellar assembly. The heterozygotes appeared short and stumpy and had significantly shorter flagella, which were not enough to propel the parasite forward (167). Homozygous knockout of myosin-XXI in these parasites was lethal. Deletion of the myosin gene led to ploidy in these parasites, indicating that the presence of myosin-XXI is critical for the survival of Leishmania.
Recent studies have reported expression and purification of full-length constructs and a truncated minimal motor domain construct of Leishmania myosin-XXI (168, 169). The motor is activated by actin and binds calmodulin that is required for motility but not for ATPase. Myosin-XXI is structurally similar to all other myosins except that the neck region does not contain perfect IQ motifs. Rather, the converter is followed by an extended dimerization region until the C-terminal end of the molecule. However, close to the converter are binding sites of Leishmania-specific calmodulin-like proteins that bind the myosin in a Ca2+ -dependent manner (169) (Figure 1). This is very different from other myosins that bind calmodulin (such as myosin-V or myosin-VI), in which the dimerization region starts downstream of the calmodulin-binding sites. This ensures that dimerization does not disrupt calmodulin binding in these myosins, allowing the calmodulin-binding lever arm to transduce force and movement. However, in the case of myosin-XXI, binding of Ca2+ -calmodulin prevents dimerization and enables motility by forming a functional lever arm. In the absence of Ca2+ -calmodulin binding, the lever arm structure is lost, thus allowing dimerization, a loss in movement, and cross-linking of actin filaments. Negative-staining electron microscopy studies showed monomeric, full-length myosin-XXI bound to actin in the rigor state, while the dimeric myosin (lacking calmodulin) was shown to cross-link actin filaments (169). This motor binds lipids via its extended dimerization region; lipid binding and dimerization are mutually exclusive. The calmodulin-unbound motor is dimeric, nonmotile, and expected to be cytosolic, as it was unable to bind lipids. However, Ca2+ -calmodulin binding makes the motor monomeric and targets it to different lipid compartments. This monomeric motor can generate force and movement. Importantly, membrane binding of myosin-XXI is driven by a phospholipid-binding phox homology domain that overlaps with the converter region and is a unique specialty of this myosin. Thus, with lipid-binding regions scattered along the converter and tail region of this myosin, the myosin is well suited for specific targeting to the plasma membrane and membranes of specific organelles. Unlike myosin-XIV, myosin-XXI can adopt a monomeric or a dimeric state according to the dynamic needs of the parasite. The very C-terminal region of this myosin is also predicted to contain two ubiquitin-associated domains (Figure 1), whose role is not clear.
Trypanosomes are also reported to contain myosin-XXI along with myosin-I. Trypanosoma cruzi, the causative agent of Chagas disease, has expanded the kinetoplastid myosin family to several members with more diverse sequences (170, 171). It contains one myosin-I, six myosin-XXIs, and one unclassified myosin. Chagas disease is a major economic burden, with global costs of $7.1 billion per annum. More than 10% of these losses occur in the United States and Canada, where this disease is not even endemic. The losses are attributed to the loss in productivity by cardiovascular disease–induced early death (172). The chronic stage of this disease affects the nervous system, the digestive system, and the heart, where it causes dilated cardiomyopathy. Another family member, Trypanosoma brucei, the causative agent of sleeping sickness and nagana (animal trypanosomiasis), also possesses myosins belonging to classes I and XXI. The myosin-I in T. brucei localizes to the polarized endocytic pathway in the bloodstream form of the parasite. Knockdown studies of this myosin in T. brucei led to a significant reduction in endocytic activity, an inhibition of cell division, and eventual cell death (173). The knockdown effects are consistent with an imbalance of cellular trafficking in these parasites. T. brucei myosin-I is an unusual member of the short-tailed myosin-I family. The motor domain is followed by a single, degenerate IQ motif followed by a WW domain, which is involved in protein–protein interactions. This is further followed by a tail homology 1 domain, which is disrupted by a putative zinc-binding FYVE domain (173). TH1 domains are rich in basic residues and are thought to be involved in membrane interactions.
CONCLUSIONS AND PERSPECTIVES
As emphasized here, the members of the large family of myosin motors all work primarily the same and are all highly allosteric, and cardiac myosin has proved to be a useful target for small-molecule therapy. There are three reasons the cardiac modulators OM and mavacamten are proving so successful. First, as described above, cardiac myosin can be either activated or inhibited with small-molecule effectors. Second, cardiac myosin is the most downstream component of the activation of contraction of the heart. Many current therapies rely on upstream modulations that change Ca2+ transients in cells, but these have many pleiotropic effects. Both OM and mavacamten have little or no effect on Ca2+ transients, but rather, they modulate the contractile machinery directly. In the case of HCM, nearly 40% of all HCM mutations occur in the β-cardiac myosin itself, and mavacamten targets the very protein that carries the mutation that is the source of the disease. Third, work from both Cytokinetics and MyoKardia has shown that one can target cardiac myosin very specifically without significantly affecting skeletal or smooth muscle cells. This specificity, combined with the downstream nature of the target, means very little toxicity is seen with these agents. However, owing to the ubiquitous presence of NM-IIs in the body, pleiotropic side effects may arise when targeting them for diseases such as cancer. However, while this is not ideal, many current cancer drugs target proteins that are critical for tumor survival but are also present in other cells of the body, leading to significant toxicity and side effects, but these are considered acceptable if the cancer progression is halted.
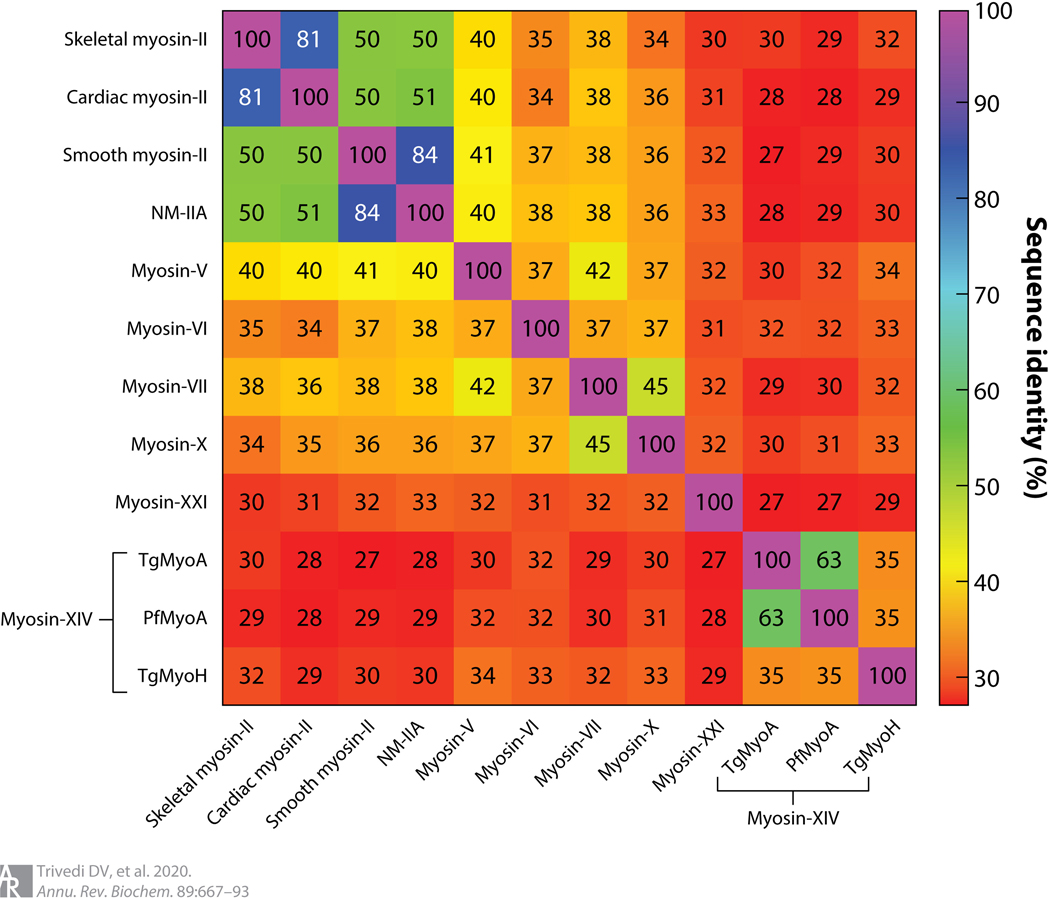
Parasite myosins, in contrast, could be promising targets. Parasite myosins are considerably more different from mammalian myosins than are the various mammalian myosins from one another (Figure 5), and as in the cases of malaria and leishmaniasis, some classes of these myosins are critical for the survival of these parasites, making them an attractive target for therapeutic intervention. Thus, it is very likely that small-molecule effectors can be designed that show high levels of specificity for parasite myosins, thereby causing little or no toxicity to the mammalian host cells. Furthermore, since many different parasites carry closely related myosin types, one could likely develop a single small-molecule effector that acts across several parasitic diseases. As the genomic revolution continues, and as we delve deeper into the genomes of these enigmatic parasites, we are bound to gain more insights into the presence of novel myosins in them that can be specifically targeted by small molecules. A recent work (174) reported on the rise of a multidrug-resistant colineage of P. falciparum that evolutionarily diversified into multiple subgroups and acquired new genetic features. From Cambodia, this genetically enhanced subgroup has rapidly spread into neighboring southeast Asian countries. This raises the terrifying prospect of such genetically fit parasites spreading into Africa, where most malaria cases occur. There is an urgent need for novel classes of antimalarials as these parasites develop resistance against current therapies (175, 176). As climate change warms the earth, typically tropical and subtropical parasitic diseases are expected to spread into now temperate areas.
Figure 5.
Heat map showing the percent sequence identity of the motor domain across different isoforms of myosin from a variety of organisms. Multiple sequence alignment was performed using the EMBL-EBI Clustal Omega tool. Myosin-XXI is the Leishmania infantum sequence, and the mammalian myosins are all human sequences. Abbreviations: Pf, Plasmodium falciparum; Tg, Toxoplasma gondii.
Work over the past several years has led to extensive structural and mechanochemical understanding of the myosin motor. Given the promise that myosin has shown as a therapeutic target for the treatment of cardiac disease, it is likely that this understanding will be leveraged to design therapeutics for diseases that involve myosin at the core of their functioning.
ACKNOWLEDGMENTS
We thank Steven Rosenfeld, Robert Adelstein, Miguel Vicente, Claudia Veigel, Douglas Robinson, Ron Rock, and Ellen Yeh for discussions. J.A.S. is supported by National Institutes of Health grants R01 GM033289 and R01 HL1171138. D.V.T. is supported by an American Heart Association postdoctoral fellowship (17POST33411070).
Footnotes
DISCLOSURE STATEMENT
J.A.S. is a cofounder and member of the scientific advisory boards of Cytokinetics and MyoKardia, biotechnology companies developing small molecules that target the sarcomere for the treatment of various muscle diseases. K.M.R. is on the scientific advisory board at MyoKardia. S.N. is a scientist at MyoKardia.
LITERATURE CITED
- 1.Kühne W. 1864. Untersuchungen über das Protoplasma und die Contractilität. Leipzig, Ger.: Engelmann [Google Scholar]
- 2.Hartman MA, Spudich JA. 2012. The myosin superfamily at a glance. J. Cell Sci 125:1627–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vale RD. 2003. The molecular motor toolbox for intracellular transport. Cell 112:467–80 [DOI] [PubMed] [Google Scholar]
- 4.Foth BJ, Goedecke MC, Soldati D. 2006. New insights into myosin evolution and classification. PNAS 103:3681–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wells AL, Lin AW, Chen LQ, Safer D, Cain SM, et al. 1999. Myosin VI is an actin-based motor that moves backwards. Nature 401:505–8 [DOI] [PubMed] [Google Scholar]
- 6.Toyoshima YY, Kron SJ, McNally EM, Niebling KR, Toyoshima C, Spudich JA. 1987. Myosin subfragment-1 is sufficient to move actin filaments in vitro. Nature 328:536–39 [DOI] [PubMed] [Google Scholar]
- 7.Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, et al. 1993. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science 261:50–58 [DOI] [PubMed] [Google Scholar]
- 8.Sweeney HL, Houdusse A. 2010. Structural and functional insights into the myosin motor mechanism. Annu. Rev. Biophys 39:539–57 [DOI] [PubMed] [Google Scholar]
- 9.Kull FJ, Vale RD, Fletterick RJ. 1998. The case for a common ancestor: kinesin and myosin motor proteins and G proteins. J. Muscle Res. Cell Motil 19:877–86 [DOI] [PubMed] [Google Scholar]
- 10.Vale RD, Milligan RA. 2000. The way things move: looking under the hood of molecular motor proteins. Science 288:88–95 [DOI] [PubMed] [Google Scholar]
- 11.Harrington WF, Ueno H, Davis JS. 1988. Helix-coil melting in rigor and activated cross-bridges of skeletal muscle. Adv. Exp. Med. Biol 226:307–18 [PubMed] [Google Scholar]
- 12.Kominz DR ME, Nihei T, Kay CM. 1965. The papain digestion of skeletal myosin A. Biochemistry 4:2373–82 [Google Scholar]
- 13.Lowey S, Slayter HS, Weeds AG, Baker H. 1969. Substructure of the myosin molecule. I. Subfragments of myosin by enzymic degradation. J. Mol. Biol 42:1–29 [DOI] [PubMed] [Google Scholar]
- 14.Alamo L, Wriggers W, Pinto A, Bartoli F, Salazar L, et al. 2008. Three-dimensional reconstruction of tarantula myosin filaments suggests how phosphorylation may regulate myosin activity. J. Mol. Biol 384:780–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trivedi DV, Adhikari AS, Sarkar SS, Ruppel KM, Spudich JA. 2018. Hypertrophic cardiomyopathy and the myosin mesa: viewing an old disease in a new light. Biophys. Rev 10:27–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alamo L, Pinto A, Sulbaran G, Mavarez J, Padron R. 2018. Lessons from a tarantula: new insights into myosin interacting-heads motif evolution and its implications on disease. Biophys. Rev 10:1465–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wendt T, Taylor D, Messier T, Trybus KM, Taylor KA. 1999. Visualization of head-head interactions in the inhibited state of smooth muscle myosin. J. Cell Biol 147:1385–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wendt T, Taylor D, Trybus KM, Taylor K. 2001. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. PNAS 98:4361–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zoghbi ME, Woodhead JL, Moss RL, Craig R. 2008. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. PNAS 105:2386–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung HS, Komatsu S, Ikebe M, Craig R. 2008. Head-head and head-tail interaction: a general mechanism for switching off myosin II activity in cells. Mol. Biol. Cell 19:3234–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodhead JL, Zhao FQ, Craig R, Egelman EH, Alamo L, Padron R. 2005. Atomic model of a myosin filament in the relaxed state. Nature 436:1195–99 [DOI] [PubMed] [Google Scholar]
- 22.Al-Khayat HA, Kensler RW, Squire JM, Marston SB, Morris EP. 2013. Atomic model of the human cardiac muscle myosin filament. PNAS 110:318–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spudich JA. 2015. The myosin mesa and a possible unifying hypothesis for the molecular basis of human hypertrophic cardiomyopathy. Biochem. Soc. Trans 43:64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nag S, Trivedi DV, Sarkar SS, Adhikari AS, Sunitha MS, et al. 2017. The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat. Struct. Mol. Biol 24:525–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee KH, Sulbaran G, Yang S, Mun JY, Alamo L, et al. 2018. Interacting-heads motif has been conserved as a mechanism of myosin II inhibition since before the origin of animals. PNAS 115:E1991–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung HS, Burgess SA, Billington N, Colegrave M, Patel H, et al. 2008. Conservation of the regulated structure of folded myosin 2 in species separated by at least 600 million years of independent evolution. PNAS 105:6022–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trybus KM. 2008. Myosin V from head to tail. Cell Mol. Life Sci 65:1378–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoffler HE, Bahler M. 1998. The ATPase activity of Myr3, a rat myosin I, is allosterically inhibited by its own tail domain and by Ca2+ binding to its light chain calmodulin. J. Biol. Chem 273:14605–11 [DOI] [PubMed] [Google Scholar]
- 29.Umeki N, Jung HS, Watanabe S, Sakai T, Li XD, et al. 2009. The tail binds to the head-neck domain, inhibiting ATPase activity of myosin VIIA. PNAS 106:8483–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, Baboolal TG, Siththanandan V, Chen M, Walker ML, et al. 2009. A FERM domain autoregulates Drosophila myosin 7a activity. PNAS 106:4189–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McIntosh BB, Ostap EM. 2016. Myosin-I molecular motors at a glance. J. Cell Sci 129:2689–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bryant Z, Altman D, Spudich JA. 2007. The power stroke of myosin VI and the basis of reverse directionality. PNAS 104:772–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menetrey J, Bahloul A, Wells AL, Yengo CM, Morris CA, et al. 2005. The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature 435:779–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross JL, Ali MY, Warshaw DM. 2008. Cargo transport: molecular motors navigate a complex cytoskeleton. Curr. Opin. Cell Biol 20:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geeves MA, Holmes KC. 1999. Structural mechanism of muscle contraction. Annu. Rev. Biochem 68:687–728 [DOI] [PubMed] [Google Scholar]
- 36.Málnási-Csizmadia A, Pearson DS, Kovács M, Woolley RJ, Geeves MA, Bagshaw CR. 2001. Kinetic resolution of a conformational transition and the ATP hydrolysis step using relaxation methods with a Dictyostelium myosin II mutant containing a single tryptophan residue. Biochemistry 40:12727–37 [DOI] [PubMed] [Google Scholar]
- 37.Geeves MA, Conibear PB. 1995. The role of three-state docking of myosin S1 with actin in force generation. Biophys. J 68:194S–99S [PMC free article] [PubMed] [Google Scholar]
- 38.Yengo CM, De La Cruz EM, Chrin LR, Gaffney DP II, Berger CL. 2002. Actin-induced closure of the actin-binding cleft of smooth muscle myosin. J. Biol. Chem 277:24114–19 [DOI] [PubMed] [Google Scholar]
- 39.Yengo CM, De La Cruz EM, Safer D, Ostap EM, Sweeney HL. 2002. Kinetic characterization of the weak binding states of myosin V. Biochemistry 41:8508–17 [DOI] [PubMed] [Google Scholar]
- 40.Holmes KC, Angert I, Kull FJ, Jahn W, Schroder RR. 2003. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature 425:423–27 [DOI] [PubMed] [Google Scholar]
- 41.Coureux PD, Wells AL, Menetrey J, Yengo CM, Morris CA, et al. 2003. A structural state of the myosin V motor without bound nucleotide. Nature 425:419–23 [DOI] [PubMed] [Google Scholar]
- 42.Sweeney HL, Houdusse A. 2004. The motor mechanism of myosin V: insights for muscle contraction. Philos. Trans. R. Soc. B 359:1829–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spudich JA. 2019. Three perspectives on the molecular basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Pflüg. Arch. Eur. J. Physiol 471:701–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shih WM, Gryczynski Z, Lakowicz JR, Spudich JA. 2000. A FRET-based sensor reveals large ATP hydrolysis-induced conformational changes and three distinct states of the molecular motor myosin. Cell 102:683–94 [DOI] [PubMed] [Google Scholar]
- 45.Malik FI, Hartman JJ, Elias KA, Morgan BP, Rodriguez H, et al. 2011. Cardiac myosin activation: a potential therapeutic approach for systolic heart failure. Science 331:1439–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cleland JG, Teerlink JR, Senior R, Nifontov EM, Mc Murray JJ, et al. 2011. The effects of the cardiac myosin activator, omecamtiv mecarbil, on cardiac function in systolic heart failure: a double-blind, placebo-controlled, crossover, dose-ranging phase 2 trial. Lancet 378:676–83 [DOI] [PubMed] [Google Scholar]
- 47.Teerlink JR, Clarke CP, Saikali KG, Lee JH, Chen MM, et al. 2011. Dose-dependent augmentation of cardiac systolic function with the selective cardiac myosin activator, omecamtiv mecarbil: a first-in-man study. Lancet 378:667–75 [DOI] [PubMed] [Google Scholar]
- 48.Teerlink JR, Felker GM, McMurray JJ, Solomon SD, Adams KF Jr., et al. 2016. Chronic Oral Study of Myosin Activation to Increase Contractility in Heart Failure (COSMIC-HF): a phase 2, pharmacokinetic, randomised, placebo-controlled trial. Lancet 388:2895–903 [DOI] [PubMed] [Google Scholar]
- 49.Planelles-Herrero VJ, Hartman JJ, Robert-Paganin J, Malik FI, Houdusse A. 2017. Mechanistic and structural basis for activation of cardiac myosin force production by omecamtiv mecarbil. Nat. Commun 8:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kampourakis T, Zhang X, Sun YB, Irving M. 2018. Omecamtiv mecarbil and blebbistatin modulate cardiac contractility by perturbing the regulatory state of the myosin filament. J. Physiol 596:31–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rohde JA, Thomas DD, Muretta JM. 2017. Heart failure drug changes the mechanoenzymology of the cardiac myosin powerstroke. PNAS 114:E1796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woody MS, Greenberg MJ, Barua B, Winkelmann DA, Goldman YE, Ostap EM. 2018. Positive cardiac inotrope omecamtiv mecarbil activates muscle despite suppressing the myosin working stroke. Nat. Commun 9:3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu C, Kawana M, Song D, Ruppel KM, Spudich JA. 2018. Controlling load-dependent kinetics of β-cardiac myosin at the single-molecule level. Nat. Struct. Mol. Biol 25:505–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aksel T, Choe Yu E, Sutton S, Ruppel KM, Spudich JA. 2015. Ensemble force changes that result from human cardiac myosin mutations and a small-molecule effector. Cell Rep. 11:910–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y, White HD, Belknap B, Winkelmann DA, Forgacs E. 2015. Omecamtiv mecarbil modulates the kinetic and motile properties of porcine β-cardiac myosin. Biochemistry 54:1963–75 [DOI] [PubMed] [Google Scholar]
- 56.Swenson AM, Tang W, Blair CA, Fetrow CM, Unrath WC, et al. 2017. Omecamtiv mecarbil enhances the duty ratio of human β-cardiac myosin resulting in increased calcium sensitivity and slowed force development in cardiac muscle. J. Biol. Chem 292:3768–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spudich JA. 2014. Hypertrophic and dilated cardiomyopathy: four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophys. J 106:1236–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anderson RL, Trivedi DV, Sarkar SS, Henze M, Ma W, et al. 2018. Deciphering the super relaxed state of human β-cardiac myosin and the mode of action of mavacamten from myosin molecules to muscle fibers. PNAS 115:E8143–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robert-Paganin J, Auguin D, Houdusse A. 2018. Hypertrophic cardiomyopathy disease results from disparate impairments of cardiac myosin function and auto-inhibition. Nat. Commun 9:4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rohde JA, Roopnarine O, Thomas DD, Muretta JM. 2018. Mavacamten stabilizes an autoinhibited state of two-headed cardiac myosin. PNAS 115:E7486–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kawas RF, Anderson RL, Ingle SRB, Song Y, Sran AS, Rodriguez HM. 2017. A small-molecule modulator of cardiac myosin acts on multiple stages of the myosin chemomechanical cycle. J. Biol. Chem 292:16571–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marigo V, Nigro A, Pecci A, Montanaro D, Di Stazio M, et al. 2004. Correlation between the clinical phenotype of MYH9-related disease and tissue distribution of class II nonmuscle myosin heavy chains. Genomics 83:1125–33 [DOI] [PubMed] [Google Scholar]
- 63.Côté GP, Robinson EA, Appella E, Korn ED. 1984. Amino acid sequence of a segment of the Acanthamoeba myosin II heavy chain containing all three regulatory phosphorylation sites. J. Biol. Chem 259:12781–87 [PubMed] [Google Scholar]
- 64.Craig R, Smith R, Kendrick-Jones J. 1983. Light-chain phosphorylation controls the conformation of vertebrate non-muscle and smooth muscle myosin molecules. Nature 302:436–39 [DOI] [PubMed] [Google Scholar]
- 65.Pecci A, Ma X, Savoia A, Adelstein RS. 2018. MYH9: structure, functions and role of non-muscle myosin IIA in human disease. Gene 664:152–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dulyaninova NG, Bresnick AR. 2013. The heavy chain has its day: regulation of myosin-II assembly. Bioarchitecture 3:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beach JR, Hussey GS, Miller TE, Chaudhury A, Patel P, et al. 2011. Myosin II isoform switching mediates invasiveness after TGF-β-induced epithelial–mesenchymal transition. PNAS 108:17991–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Newell-Litwa KA, Horwitz R, Lamers ML. 2015. Non-muscle myosin II in disease: mechanisms and therapeutic opportunities. Dis. Model. Mech 8:1495–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Friedl P, Alexander S. 2011. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell 147:992–1009 [DOI] [PubMed] [Google Scholar]
- 70.Daniels MJ, Wang Y, Lee M, Venkitaraman AR. 2004. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science 306:876–79 [DOI] [PubMed] [Google Scholar]
- 71.Takaoka M, Saito H, Takenaka K, Miki Y, Nakanishi A. 2014. BRCA2 phosphorylated by PLK1 moves to the midbody to regulate cytokinesis mediated by nonmuscle myosin IIC. Cancer Res. 74:1518–28 [DOI] [PubMed] [Google Scholar]
- 72.Arozarena I, Calvo F, Crespo P. 2011. Ras, an actor on many stages: posttranslational modifications, localization, and site-specified events. Genes Cancer 2:182–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Helfman DM, Pawlak G. 2005. Myosin light chain kinase and acto-myosin contractility modulate activation of the ERK cascade downstream of oncogenic Ras. J. Cell Biochem 95:1069–80 [DOI] [PubMed] [Google Scholar]
- 74.Zhong C, Kinch MS, Burridge K. 1997. Rho-stimulated contractility contributes to the fibroblastic phenotype of Ras-transformed epithelial cells. Mol. Biol. Cell 8:2329–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen JC, Zhuang S, Nguyen TH, Boss GR, Pilz RB. 2003. Oncogenic Ras leads to Rho activation by activating the mitogen-activated protein kinase pathway and decreasing Rho-GTPase-activating protein activity. J. Biol. Chem 278:2807–18 [DOI] [PubMed] [Google Scholar]
- 76.Beadle C, Assanah MC, Monzo P, Vallee R, Rosenfeld SS, Canoll P. 2008. The role of myosin II in glioma invasion of the brain. Mol. Biol. Cell 19:3357–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Salhia B, Hwang JH, Smith CA, Nakada M, Rutka F, et al. 2008. Role of myosin II activity and the regulation of myosin light chain phosphorylation in astrocytomas. Cell Motil. Cytoskelet 65:12–24 [DOI] [PubMed] [Google Scholar]
- 78.Picariello HS, Kenchappa RS, Rai V, Crish JF, Dovas A, et al. 2019. Myosin IIA suppresses glioblastoma development in a mechanically sensitive manner. PNAS 116:15550–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dhruv HD, McDonough Winslow WS, Armstrong B, Tuncali S, Eschbacher J, et al. 2013. Reciprocal activation of transcription factors underlies the dichotomy between proliferation and invasion of glioma cells. PLOS ONE 8:e72134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schramek D, Sendoel A, Segal JP, Beronja S, Heller E, et al. 2014. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science 343:309–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Conti MA, Saleh AD, Brinster LR, Cheng H, Chen Z, et al. 2015. Conditional deletion of nonmuscle myosin II-A in mouse tongue epithelium results in squamous cell carcinoma. Sci. Rep 5:14068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Surcel A, Schiffhauer E, Thomas D, Zhu Q, DiNapoli K, et al. 2017. Harnessing the adaptive potential of mechanoresponsive proteins to overwhelm pancreatic cancer dissemination and invasion. bioRxiv 190553. 10.1101/190553 [DOI] [Google Scholar]
- 83.Rahman KW, Li Y, Wang Z, Sarkar SH, Sarkar FH. 2006. Gene expression profiling revealed survivin as a target of 3,3′-diindolylmethane-induced cell growth inhibition and apoptosis in breast cancer cells. Cancer Res. 66:4952–60 [DOI] [PubMed] [Google Scholar]
- 84.Chandrasekar I, Huettner JE, Turney SG, Bridgman PC. 2013. Myosin II regulates activity dependent compensatory endocytosis at central synapses. J. Neurosci 33:16131–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hodges JL, Newell-Litwa K, Asmussen H, Vicente-Manzanares M, Horwitz AR. 2011. Myosin IIb activity and phosphorylation status determines dendritic spine and post-synaptic density morphology. PLOS ONE 6:e24149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ryu J, Liu L, Wong TP, Wu DC, Burette A, et al. 2006. A critical role for myosin IIb in dendritic spine morphology and synaptic function. Neuron 49:175–82 [DOI] [PubMed] [Google Scholar]
- 87.Rex CS, Gavin CF, Rubio MD, Kramar EA, Chen LY, et al. 2010. Myosin IIb regulates actin dynamics during synaptic plasticity and memory formation. Neuron 67:603–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rusielewicz T, Nam J, Damanakis E, John GR, Raine CS, Melendez-Vasquez CV. 2014. Accelerated repair of demyelinated CNS lesions in the absence of non-muscle myosin IIB. Glia 62:580–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beard RS Jr., Haines RJ, Wu KY, Reynolds JJ, Davis SM, et al. 2014. Non-muscle Mlck is required for β-catenin- and FoxO1-dependent downregulation of Cldn5 in IL-1β-mediated barrier dysfunction in brain endothelial cells. J. Cell Sci 127:1840–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Srivastava K, Shao B, Bayraktutan U. 2013. PKC-β exacerbates in vitro brain barrier damage in hyperglycemic settings via regulation of RhoA/Rho-kinase/MLC2 pathway. J. Cereb. Blood Flow Metab 33:1928–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Janssen S, Gudi V, Prajeeth CK, Singh V, Stahl K, et al. 2014. A pivotal role of nonmuscle myosin II during microglial activation. Exp. Neurol 261:666–76 [DOI] [PubMed] [Google Scholar]
- 92.Nadif Kasri N, Van Aelst L. 2008. Rho-linked genes and neurological disorders. Pflüg. Arch. Eur. J. Physiol 455:787–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hur EM, Yang IH, Kim DH, Byun J, Saijilafu, et al. 2011. Engineering neuronal growth cones to promote axon regeneration over inhibitory molecules. PNAS 108:5057–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li J, Cai T, Jiang Y, Chen H, He X, et al. 2016. Genes with de novo mutations are shared by four neuropsychiatric disorders discovered from NPdenovo database. Mol. Psychiatry 21:298. [DOI] [PubMed] [Google Scholar]
- 95.Borisoff JF, Chan CCM, Hiebert GW, Oschipok L, Robertson GS, et al. 2003. Suppression of Rho-kinase activity promotes axonal growth on inhibitory CNS substrates. Mol. Cell. Neurosci 22:405–16 [DOI] [PubMed] [Google Scholar]
- 96.Kubo T, Yamaguchi A, Iwata N, Yamashita T. 2008. The therapeutic effects of Rho-ROCK inhibitors on CNS disorders. Ther. Clin. Risk. Manag 4:605–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Monnier PP, Sierra A, Schwab JM, Henke-Fahle S, Mueller BK. 2003. The Rho/ROCK pathway mediates neurite growth-inhibitory activity associated with the chondroitin sulfate proteoglycans of the CNS glial scar. Mol. Cell. Neurosci 22:319–30 [DOI] [PubMed] [Google Scholar]
- 98.Yu Z, Liu M, Fu P, Xie M, Wang W, Luo X. 2012. ROCK inhibition with Y27632 promotes the proliferation and cell cycle progression of cultured astrocyte from spinal cord. Neurochem. Int 61:1114–20 [DOI] [PubMed] [Google Scholar]
- 99.Kilinc D, Blasiak A, O’Mahony JJ, Lee GU. 2014. Low piconewton towing of CNS axons against diffusing and surface-bound repellents requires the inhibition of motor protein-associated pathways. Sci. Rep 4:7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rossi D. 2015. Astrocyte physiopathology: at the crossroads of intercellular networking, inflammation and cell death. Prog. Neurobiol 130:86–120 [DOI] [PubMed] [Google Scholar]
- 101.Schwartz M, Kipnis J, Rivest S, Prat A. 2013. How do immune cells support and shape the brain in health, disease, and aging? J. Neurosci 33:17587–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barcia C, Ros CM, Annese V, Carrillo-de Sauvage MA, Ros-Bernal F, et al. 2012. ROCK/Cdc42-mediated microglial motility and gliapse formation lead to phagocytosis of degenerating dopaminergic neurons in vivo. Sci. Rep 2:809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ding J, Li QY, Wang X, Sun CH, Lu CZ, Xiao BG. 2010. Fasudil protects hippocampal neurons against hypoxia-reoxygenation injury by suppressing microglial inflammatory responses in mice. J. Neurochem 114:1619–29 [DOI] [PubMed] [Google Scholar]
- 104.Parisi C, Arisi I, D’Ambrosi N, Storti AE, Brandi R, et al. 2013. Dysregulated microRNAs in amyotrophic lateral sclerosis microglia modulate genes linked to neuroinflammation. Cell Death Dis. 4:e959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tonges L, Gunther R, Suhr M, Jansen J, Balck A, et al. 2014. Rho kinase inhibition modulates microglia activation and improves survival in a model of amyotrophic lateral sclerosis. Glia 62:217–32 [DOI] [PubMed] [Google Scholar]
- 106.Balduini CL, Pecci A, Savoia A. 2011. Recent advances in the understanding and management of MYH9-related inherited thrombocytopenias. Br. J. Haematol 154:161–74 [DOI] [PubMed] [Google Scholar]
- 107.Wood W, Martin P. 2002. Structures in focus—filopodia. Int. J. Biochem. Cell Biol 34:726–30 [DOI] [PubMed] [Google Scholar]
- 108.Berg JS, Cheney RE. 2002. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat. Cell Biol 4:246–50 [DOI] [PubMed] [Google Scholar]
- 109.Gerhardt H, Golding M, Fruttiger M, Ruhrberg C, Lundkvist A, et al. 2003. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J. Cell Biol 161:1163–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yu H, Wang N, Ju X, Yang Y, Sun D, et al. 2012. PtdIns (3,4,5) P3 recruitment of Myo10 is essential for axon development. PLOS ONE 7:e36988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schoumacher M, Goldman RD, Louvard D, Vignjevic DM. 2010. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol 189:541–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Berg JS, Derfler BH, Pennisi CM, Corey DP, Cheney RE. 2000. Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J. Cell Sci 113:3439–51 [DOI] [PubMed] [Google Scholar]
- 113.Mashanov GI, Tacon D, Peckham M, Molloy JE. 2004. The spatial and temporal dynamics of pleckstrin homology domain binding at the plasma membrane measured by imaging single molecules in live mouse myoblasts. J. Biol. Chem 279:15274–80 [DOI] [PubMed] [Google Scholar]
- 114.Lu Q, Yu J, Yan J, Wei Z, Zhang M. 2011. Structural basis of the myosin X PH1N-PH2-PH1C tandem as a specific and acute cellular PI(3,4,5)P3 sensor. Mol. Biol. Cell 22:4268–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Umeki N, Jung HS, Sakai T, Sato O, Ikebe R, Ikebe M. 2011. Phospholipid-dependent regulation of the motor activity of myosin X. Nat. Struct. Mol. Biol 18:783–88 [DOI] [PubMed] [Google Scholar]
- 116.Vignjevic D, Schoumacher M, Gavert N, Janssen KP, Jih G, et al. 2007. Fascin, a novel target of β-catenin-TCF signaling, is expressed at the invasive front of human colon cancer. Cancer Res. 67:6844–53 [DOI] [PubMed] [Google Scholar]
- 117.Shibue T, Brooks MW, Inan MF, Reinhardt F, Weinberg RA. 2012. The outgrowth of micrometastases is enabled by the formation of filopodium-like protrusions. Cancer Discov. 2:706–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kolesnikova L, Bohil AB, Cheney RE, Becker S. 2007. Budding of Marburgvirus is associated with filopodia. Cell Microbiol. 9:939–51 [DOI] [PubMed] [Google Scholar]
- 119.Romero S, Grompone G, Carayol N, Mounier J, Guadagnini S, et al. 2011. ATP-mediated Erk1/2 activation stimulates bacterial capture by filopodia, which precedes Shigella invasion of epithelial cells. Cell Host Microbe 9:508–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Thompson RF, Langford GM. 2002. Myosin superfamily evolutionary history. Anat. Rec 268:276–89 [DOI] [PubMed] [Google Scholar]
- 121.Akhmanova A, Hammer JA 3rd. 2010. Linking molecular motors to membrane cargo. Curr. Opin. Cell Biol 22:479–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Libby RT, Steel KP. 2000. The roles of unconventional myosins in hearing and deafness. Essays Biochem. 35:159–74 [DOI] [PubMed] [Google Scholar]
- 123.Szperl AM, Golachowska MR, Bruinenberg M, Prekeris R, Thunnissen AM, et al. 2011. Functional characterization of mutations in the myosin Vb gene associated with microvillus inclusion disease. J. Pediatr. Gastroenterol. Nutr 52:307–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Müller T, Hess MW, Schiefermeier N, Pfaller K, Ebner HL, et al. 2008. MYO5B mutations cause microvillus inclusion disease and disrupt epithelial cell polarity. Nat. Genet 40:1163–65 [DOI] [PubMed] [Google Scholar]
- 125.Van Gele M, Dynoodt P, Lambert J. 2009. Griscelli syndrome: a model system to study vesicular trafficking. Pigment Cell Melanoma Res. 22:268–82 [DOI] [PubMed] [Google Scholar]
- 126.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, et al. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Frénal K, Dubremetz JF, Lebrun M, Soldati-Favre D. 2017. Gliding motility powers invasion and egress in Apicomplexa. Nat. Rev. Microbiol 15:645–60 [DOI] [PubMed] [Google Scholar]
- 128.Meissner M, Schluter D, Soldati D. 2002. Role of Toxoplasma gondii myosin A in powering parasite gliding and host cell invasion. Science 298:837–40 [DOI] [PubMed] [Google Scholar]
- 129.Andenmatten N, Egarter S, Jackson AJ, Jullien N, Herman JP, Meissner M. 2013. Conditional genome engineering in Toxoplasma gondii uncovers alternative invasion mechanisms. Nat. Methods 10:125–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dobrowolski JM, Carruthers VB, Sibley LD. 1997. Participation of myosin in gliding motility and host cell invasion by Toxoplasma gondii. Mol. Microbiol 26:163–73 [DOI] [PubMed] [Google Scholar]
- 131.Dobrowolski JM, Sibley LD. 1996. Toxoplasma invasion of mammalian cells is powered by the actin cytoskeleton of the parasite. Cell 84:933–39 [DOI] [PubMed] [Google Scholar]
- 132.Herm-Götz A, Weiss S, Stratmann R, Fujita-Becker S, Ruff C, et al. 2002. Toxoplasma gondii myosin A and its light chain: a fast, single-headed, plus-end-directed motor. EMBO J. 21:2149–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hettmann C, Herm A, Geiter A, Frank B, Schwarz E, et al. 2000. A dibasic motif in the tail of a class XIV apicomplexan myosin is an essential determinant of plasma membrane localization. Mol. Biol. Cell 11:1385–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mueller C, Graindorge A, Soldati-Favre D. 2017. Functions of myosin motors tailored for parasitism. Curr. Opin. Microbiol 40:113–22 [DOI] [PubMed] [Google Scholar]
- 135.Williams MJ, Alonso H, Enciso M, Egarter S, Sheiner L, et al. 2015. Two essential light chains regulate the MyoA lever arm to promote Toxoplasma gliding motility. mBio 6:e00845–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bookwalter CS, Kelsen A, Leung JM, Ward GE, Trybus KM. 2014. A Toxoplasma gondii class XIV myosin, expressed in Sf9 cells with a parasite co-chaperone, requires two light chains for fast motility. J. Biol. Chem 289:30832–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bookwalter CS, Tay CL, McCrorie R, Previs MJ, Lu H, et al. 2017. Reconstitution of the core of the malaria parasite glideosome with recombinant Plasmodium class XIV myosin A and Plasmodium actin. J. Biol. Chem 292:19290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Powell CJ, Ramaswamy R, Kelsen A, Hamelin DJ, Warshaw DM, et al. 2018. Structural and mechanistic insights into the function of the unconventional class XIV myosin MyoA from Toxoplasma gondii. PNAS 115:E10548–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Robert-Paganin J, Robblee JP, Auguin D, Blake TCA, Bookwalter CS, et al. 2019. Plasmodium myosin A drives parasite invasion by an atypical force generating mechanism. Nat. Commun 10:3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Baum J, Gilberger TW, Frischknecht F, Meissner M. 2008. Host-cell invasion by malaria parasites: insights from Plasmodium and Toxoplasma. Trends Parasitol. 24:557–63 [DOI] [PubMed] [Google Scholar]
- 141.Baum J, Richard D, Healer J, Rug M, Krnajski Z, et al. 2006. A conserved molecular motor drives cell invasion and gliding motility across malaria life cycle stages and other apicomplexan parasites. J. Biol. Chem 281:5197–208 [DOI] [PubMed] [Google Scholar]
- 142.Gaji RY, Johnson DE, Treeck M, Wang M, Hudmon A, Arrizabalaga G. 2015. Phosphorylation of a myosin motor by TgCDPK3 facilitates rapid initiation of motility during Toxoplasma gondii egress. PLOS Pathog. 11:e1005268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tang Q, Andenmatten N, Hortua Triana MA, Deng B, Meissner M, et al. 2014. Calcium-dependent phosphorylation alters class XIVa myosin function in the protozoan parasite Toxoplasma gondii. Mol. Biol. Cell 25:2579–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Leung JM, Tran F, Pathak RB, Poupart S, Heaslip AT, et al. 2014. Identification of T. gondii myosin light chain-1 as a direct target of TachypleginA-2, a small-molecule inhibitor of parasite motility and invasion. PLOS ONE 9:e98056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yusuf NA, Green JL, Wall RJ, Knuepfer E, Moon RW, et al. 2015. The Plasmodium class XIV myosin, MyoB, has a distinct subcellular location in invasive and motile stages of the malaria parasite and an unusual light chain. J. Biol. Chem 290:12147–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Frénal K, Marq JB, Jacot D, Polonais V, Soldati-Favre D. 2014. Plasticity between MyoC- and MyoA-glideosomes: an example of functional compensation in Toxoplasma gondii invasion. PLOS Pathog. 10:e1004504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Graindorge A, Frénal K, Jacot D, Salamun J, Marq JB, Soldati-Favre D. 2016. The conoid associated motor MyoH is indispensable for Toxoplasma gondii entry and exit from host cells. PLOS Pathog. 12:e1005388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hutchins JRA, Moore WJ, Hood FE, Wilson JSJ, Andrews PD, et al. 2004. Phosphorylation regulates the dynamic interaction of RCC1 with chromosomes during mitosis. Curr. Biol 14:1099–104 [DOI] [PubMed] [Google Scholar]
- 149.Shields CM, Taylor R, Nazarenus T, Cheatle J, Hou A, et al. 2003. Saccharomyces cerevisiae Ats1p interacts with Nap1p, a cytoplasmic protein that controls bud morphogenesis. Curr. Genet 44:184–94 [DOI] [PubMed] [Google Scholar]
- 150.Long S, Brown KM, Drewry LL, Anthony B, Phan IQH, Sibley LD. 2017. Calmodulin-like proteins localized to the conoid regulate motility and cell invasion by Toxoplasma gondii. PLOS Pathog. 13:e1006379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sidik SM, Huet D, Ganesan SM, Huynh MH, Wang T, et al. 2016. A genome-wide CRISPR screen in Toxoplasma identifies essential apicomplexan genes. Cell 166:1423–35.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Grasberger H, Bell GI. 2005. Subcellular recruitment by TSG118 and TSPYL implicates a role for zinc finger protein 106 in a novel developmental pathway. Int. J. Biochem. Cell Biol 37:1421–37 [DOI] [PubMed] [Google Scholar]
- 153.Smith TF, Gaitatzes C, Saxena K, Neer EJ. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci 24:181–85 [DOI] [PubMed] [Google Scholar]
- 154.Jacot D, Daher W, Soldati-Favre D. 2013. Toxoplasma gondii myosin F, an essential motor for centrosomes positioning and apicoplast inheritance. EMBO J. 32:1702–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Heaslip AT, Nelson SR, Warshaw DM. 2016. Dense granule trafficking in Toxoplasma gondii requires a unique class 27 myosin and actin filaments. Mol. Biol. Cell 27:2080–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Frénal K, Jacot D, Hammoudi PM, Graindorge A, Maco B, Soldati-Favre D. 2017. Myosin-dependent cell-cell communication controls synchronicity of division in acute and chronic stages of Toxoplasma gondii. Nat. Commun 8:15710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yun CH, Lillehoj HS, Lillehoj EP. 2000. Intestinal immune responses to coccidiosis. Dev. Comp. Immunol 24:303–24 [DOI] [PubMed] [Google Scholar]
- 158.Gharpure R, Perez A, Miller AD, Wikswo ME, Silver R, Hlavsa MC. 2019. Cryptosporidiosis outbreaks—United States, 2009–2017. MMWR Morb. Mortal. Wkly. Rep 68:568–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Frénal K, Foth BJ, Soldati D. 2008. Myosin class XIV and other myosins in protists. In Myosins: A Superfamily of Molecular Motors, ed. Coluccio LM, pp. 421–40. Dordrecht, Neth.: Springer [Google Scholar]
- 160.Periz J, Whitelaw J, Harding C, Gras S, Del Rosario Minina MI, et al. 2017. Toxoplasma gondii F-actin forms an extensive filamentous network required for material exchange and parasite maturation. eLife 6:e24119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wall RJ, Zeeshan M, Katris NJ, Limenitakis R, Rea E, et al. 2019. Systematic analysis of Plasmodium myosins reveals differential expression, localisation, and function in invasive and proliferative parasite stages. Cell. Microbiol 21:e13082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Richards TA, Cavalier-Smith T. 2005. Myosin domain evolution and the primary divergence of eukaryotes. Nature 436:1113–18 [DOI] [PubMed] [Google Scholar]
- 163.Ravichandran A, Geng M, Hull KG, Li J, Romo D, et al. 2019. A novel actin binding drug with in vivo efficacy. Antimicrob. Agents Chemother 63:e01585–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tan D, Hu H, Tong X, Han M, Zuo W, et al. 2019. Genome-wide identification and characterization of myosin genes in the silkworm, Bombyx mori. Gene 691:45–55 [DOI] [PubMed] [Google Scholar]
- 165.Dans MG, Weiss GE, Wilson DW. 2019. Screening the Medicines for Malaria Venture Pathogen Box for invasion and egress inhibitors of the blood stage of Plasmodium falciparum reveals several inhibitory compounds. bioRxiv 768648. 10.1101/768648 [DOI] [PubMed] [Google Scholar]
- 166.Katta SS, Sahasrabuddhe AA, Gupta CM. 2009. Flagellar localization of a novel isoform of myosin, myosin XXI, in Leishmania. Mol. Biochem. Parasitol 164:105–10 [DOI] [PubMed] [Google Scholar]
- 167.Katta SS, Tammana TV, Sahasrabuddhe AA, Bajpai VK, Gupta CM. 2010. Trafficking activity of myosin XXI is required in assembly of Leishmania flagellum. J. Cell Sci 123:2035–44 [DOI] [PubMed] [Google Scholar]
- 168.Batters C, Woodall KA, Toseland CP, Hundschell C, Veigel C. 2012. Cloning, expression, and characterization of a novel molecular motor, Leishmania myosin-XXI. J. Biol. Chem 287:27556–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Batters C, Ellrich H, Helbig C, Woodall KA, Hundschell C, et al. 2014. Calmodulin regulates dimerization, motility, and lipid binding of Leishmania myosin XXI. PNAS 111:E227–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, et al. 2005. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309:409–15 [DOI] [PubMed] [Google Scholar]
- 171.de Souza DAS, Pavoni DP, Krieger MA, Ludwig A. 2018. Evolutionary analyses of myosin genes in trypanosomatids show a history of expansion, secondary losses and neofunctionalization. Sci. Rep 8:1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lee BY, Bacon KM, Bottazzi ME, Hotez PJ. 2013. Global economic burden of Chagas disease: a computational simulation model. Lancet Infect. Dis 13:342–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Spitznagel D, O’Rourke JF, Leddy N, Hanrahan O, Nolan DP. 2010. Identification and characterization of an unusual class I myosin involved in vesicle traffic in Trypanosoma brucei. PLOS ONE 5:e12282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hamilton WL, Amato R, van der Pluijm RW, Jacob CG, Quang HH, et al. 2019. Evolution and expansion of multidrug-resistant malaria in southeast Asia: a genomic epidemiology study. Lancet Infect. Dis 19:P943–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ashley EA, Phyo AP. 2018. Drugs in development for malaria. Drugs 78:861–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tse EG, Korsik M, Todd MH. 2019. The past, present and future of anti-malarial medicines. Malar. J 18:93. [DOI] [PMC free article] [PubMed] [Google Scholar]