Abstract
Understanding how vectors alter the interactions between viruses and their hosts is a fundamental question in virology and disease ecology. In honey bees, transmission of deformed wing virus (DWV) by parasitic Varroa mites has been associated with elevated disease and host mortality, and Varroa transmission has been hypothesized to lead to increased viral titres or select for more virulent variants. Here, we mimicked Varroa transmission by serially passaging a mixed population of two DWV variants, A and B, by injection through in vitro reared honey bee pupae and tracking these viral populations through five passages. The DWV-A and DWV-B variant proportions shifted dynamically through passaging, with DWV-B outcompeting DWV-A after one passage, but levels of both variants becoming equivalent by Passage 5. Sequencing analysis revealed a dominant, recombinant DWV-B strain (DWV-A derived 5′ IRES region with the rest of the genome DWV-B), with low nucleotide diversity that decreased through passaging. DWV-A populations had higher nucleotide diversity compared to DWV-B, but this also decreased through passaging. Selection signatures were found across functional regions of the DWV-A and DWV-B genomes, including amino acid mutations in the putative capsid protein region. Simulated vector transmission differentially impacted two closely related viral variants which could influence viral interactions with the host, demonstrating surprising plasticity in vector-host-viral dynamics.
Keywords: Apis mellifera, bee health, deformed wing virus, evolution, transmission, virulence, RNA viruses, Varroa destructor
Impact Statement
Antagonistic relationships between organisms are fascinating, complicated, and critical to understand, particularly when these relationships result in diseases that endanger populations of critical species like insect pollinators. We investigated how the introduced and highly successful ectoparasite Varroa destructor alters the virulence (disease severity) and evolution of a pathogen that it transmits, deformed wing virus (DWV) in honey bees (Apis mellifera). Through simulated vector transmission, we found that mixed DWV populations increased in titre and displayed individual differences in proportions of variants, demonstrating that simulated vector transmission does impose selection on these viral populations. Furthermore, two main variants demonstrated differences in selection signatures and abundance of missense mutation, which can potentially affect disease outcomes and evolution. Contrary to other studies, these passaged populations were all highly virulent to developing pupae. Further studies are needed to understand the mechanisms by which simulated transmission and Varroa transmission selects for altered viral variants, and how these different master variants interact with each other and with the host.
Data Summary
All supporting qPCR and processed sequenced data can be found in the Supplementary Tables (available in the online version of this article). Raw sequencing reads have been deposited in the NCBI Sequence Read Archive (Bioproject number PRJNA731530). Inoculum consensus genome sequences for DWV-A and the recombinant DWV-B/A were deposited on NCBI (MT940255-MT940256).
Introduction
Antagonistic interactions are mediated by complex interactions between the genomes of the interacting organisms. Over time, these long-term relationships drive co-evolution through continuous ‘arms races’, and may lead to a stable system, providing that all other abiotic and biotic factors remain static [1]. In reality, these variables very rarely remain constant, and as the disease ecology context changes, the stabilized dynamics are disrupted. One such change in host-pathogen dynamics is through the introduction of a disease vector [2]. Vectors can increase pathogen horizontal transmission rates and increase virulence, or disease severity, by potentially bypassing immune defenses of the host or lessening dependency of the virus on its host for transmission. Indeed, arthropod-transmitted pathogens represent a majority of disease pressures on human health and agriculture (Institute of Medicine (US) [3]. Investigating how host, pathogen, and vector systems interact in static environments and subsequently react to altered ecological contexts can provide key insights into host-pathogen evolution.
The honey bee-virus-vector system represents a critical emerging infectious disease system in this key pollinator species [4, 5]. Deformed wing virus (DWV), a (+)ssRNA Picornavirus-like virus, is one of the major pathogens infecting the western honey bee (Apis mellifera) across the globe [6–8]. DWV infection phenotype ranges from covert, asymptomatic infection of individual bees, severe symptoms in individual bees including physical deformities [6], behavioural changes [9–11], reduced longevity [12], and, if infection is sufficiently severe at the group level, can result in colony mortality [13]. DWV is transmitted through multiple routes, including vertical transmission from parent to offspring, and horizontal transmission through contaminated food and glandular secretions [14]. In the past decades, DWV has acquired a new method of introduction, vector transmission by the Varroa destructor mite, an ectoparasitic mite that feeds on developing and adult honey bees [15–17]. Transmission typically occurs when a DWV-infected Varroa mite slices through the cuticle to feed on the hemolymph and fat body of the developing pupae [18, 19], which results in high levels of DWV infection across bee tissues [8]. If levels are high enough, pupae emerge as adults with deformed wings among other symptoms. At the colony level, in the absence of Varroa, DWV is associated with a mild infection phenotype [20], but when these two stressors are coupled together at uncontrolled levels, they are associated with significant honey bee colony losses in temperate regions [21–23] and are the most critical stressors of honey bee health globally [24, 25].
At the molecular level, DWV transmission by Varroa is associated with increased viral titres, which is correlated with increased symptoms [18, 26–28]. In geographic regions where Varroa has been more recently introduced, honey bees and other species exhibit higher DWV loads [24, 29–32]. Specific DWV ‘master variants’, DWV-A [8] and DWV-B (also known as Varroa destructor virus-1) [33], are known to be transmitted by Varroa [34], have continuously changing incidence and distribution across the globe [35, 36], and their recombinants are associated with Varroa transmission and high virulence [28, 37, 38]. Another less common variant, DWV-C [39], has also been identified within Varroa mites [40]. Attempts to definitively assign any DWV variants with increased virulence have been challenging. McMahon et al. [35] suggested that DWV-B was more virulent than DWV-A in honey bee adults in laboratory assays [35], while Mordecai et al. (2016) [38] investigated DWV populations at the colony level, and proposed that DWV-B, having an avirulent phenotype, protects colonies from the more lethal DWV-A [39]. More recently, studies suggest that these variants are equally virulent [41], or that both are lethal [42, 43].
The phenotype of increased virulence and observed viral variant shifts associated with the introduction of Varroa may be indicative of virus evolution [5, 44]. Thus, the introduction of Varroa as a vector and subsequent observation of altered DWV variant dynamics has provided an excellent and ecologically relevant model to investigate how vectors can influence disease evolution and host-pathogen interactions in real time. However, based on recent studies in which DWV was passaged across generations of pupae in controlled laboratory settings, DWV virulence was found to either decrease [45, 46], which is contrary to previously suggested virulence theory [2], or DWV virulence was found to be unaffected by passaging, perhaps suggesting that DWV has already adapted to vector transmission [47]. However, these studies either evaluated viral populations after only a single passage [45], examined only one strain of DWV [47], or included co-infecting viruses with weaker associations with Varroa [46]. As time, strain variation, and co-infecting pathogens are all critical determinants of disease evolution, it is, therefore, critical to investigate the mechanism of vector influence on viral virulence evolution in a DWV-specific context through time through stricter models to then incorporate back into full disease frameworks. Understanding how viral transmission by Varroa generates more virulent DWV is critical for developing methods for improving bee health while providing broader insights into how viruses, vectors, and hosts fundamentally interact and evolve.
Through molecular approaches, we sought to investigate how the Varroa transmission route of direct injection into the hemolymph of pupal honey bees can alter DWV-A and DWV-B variant populations. We simulated Varroa transmission by serially passaging multiple lineages of DWV populations via injection through five generations of in vitro reared honey bee pupae, assessing whether these DWV populations were associated with survival rate differences, then measuring variant levels from each passage via qPCR and next generation sequencing. We recreated the selective pressure of a Varroa transmission route, but with more stringent parameters than natural conditions, to understand if and how an introduced vector transmission route influences viral populations, from single nucleotide variation to full master variant levels, and host-virus interactions.
Methods
Honey bee samples
Bees were collected in Summer 2018 from a Penn State University research colony with a single drone inseminated (SDI) queen – this allowed for approximately 75 % relatedness between sister bees due to the honey bee’s haplodiploid sex-determination system [48], and therefore, minimized the effect of differing honey bee host genetics influencing DWV infection. This SDI colony was inspected weekly to ensure health status (i.e. low virus infection and low parasite load) and confirm the presence of the original queen. Honey bees were in vitro reared [49]. This allowed for controlled development across samples, and also limited other unknown environmental variables that may affect DWV infection (including exposure to DWV from the colony during early development).
Virus inoculum
The original starting inoculum was isolated from a single, symptomatic (i.e. deformed wings) adult bee collected in Baton Rouge, Louisiana, USA, from a colony with a high Varroa mite infestation (Simone-Finstrom, personal communication). Crude isolations were conducted to create inocula from Passages 1 through 5. Virus inocula were prepared from individual bees using the following protocol : 2 ml screw-cap microcentrifuge tubes with 2 mm zirconia beads were sterilized with a UV-cross linker for 2 min. Individual bees (flash-frozen and kept at −80 °C until processing) were placed into 2 ml screw-cap microcentrifuge tubes, and a fixed volume of 500 µl of molecular grade water was added. Bees were homogenized using a Bead Ruptor Elite (Omni International, Kennesaw, GA) at 6.5 ms for 45 s. Tubes were then placed on ice then centrifuged for 3 min at 6.5 m s−1. Supernatant was removed and passed through a sterile 0.2 µm syringe filter to separate viral particles from honey bee cells. This crude isolate was kept at 4 °C for no longer than a week (for injections and/or RNA extractions) and stored long-term at −80 °C.
Virus passaging
The viral passaging paradigm is summarized in Fig. 1. The starting inoculum was injected into in vitro reared honey bee pupae at the white-eyed stage (14 days post-egg-laying). For the initial inoculum, the virus concentration was approximately 1×106 genome equivalents per microlitre, and was approximately 1.3 : 1 DWV-A : DWV-B. The inoculum did not include other common bee viruses including sacbrood virus and black queen cell virus, determined by Illumina sequencing (see below, Next Generation Sequencing). Then 1.5 µl of the inoculum was injected into each bee using a mouth aspirator with an attached 10 µl capillary tube pulled into a needle. Needles were changed between sample groups to avoid cross-contamination. To measure DWV levels from the original colony and the effect of the injection itself on DWV levels, control bees (in vitro reared, but otherwise unmanipulated) and PBS-injected bees (injected with 1× PBS) were included as controls (referred to as ‘no inject’ and ‘PBS’ controls). Bees were collected and flash-frozen 4 days post-injection (DPI), 3 days prior to emergence from pupation (i.e. eclosion). Note that typically the virus would be transmitted to other bees after emergence by the adult bee or the associated Varroa mite, and thus our protocol simulates a stricter transmission paradigm than natural conditions.
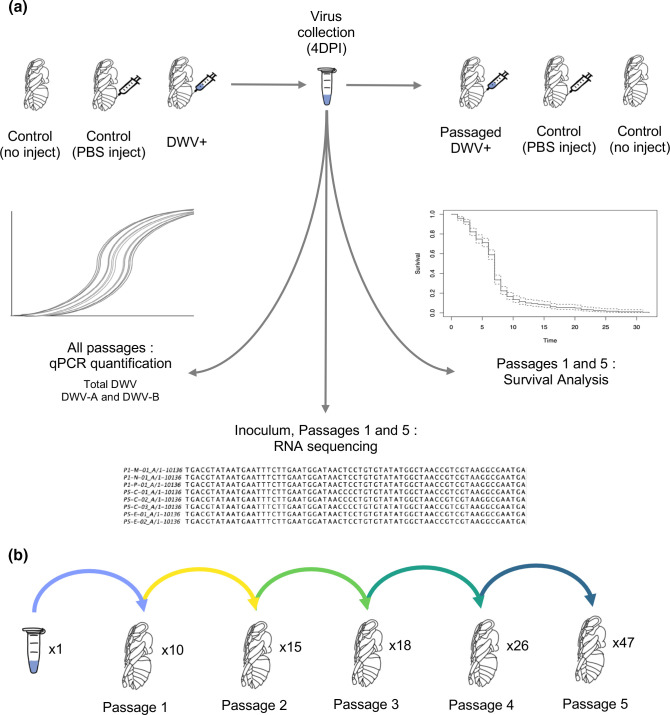
Fig. 1.
Schematic for the DWV passaging paradigm. (a) Each injection experiment included bees that were in vitro reared but unmanipulated (Control, no inject), PBS injected (Control, PBS inject), and bees injected with deformed wing virus (DWV+). At 4 days post-injection (4 DPI), pupae were collected and crude inocula were created from individual pupae. This paradigm resulted in one ‘passage’. Inocula created from single bees were subsequently injected into two new pupae, and again harvested 4 DPI, and passaged inocula were again prepared from individual pupae. This method was repeated for five passages. All passaged inocula were assessed for DWV titres via qPCR. Passage 1 inocula and a subset of inocula from Passage 5 were also assessed for virulence differences in survival assays. The starting inoculum, Passage 1 inocula, and all Passage 5 inocula were sequenced to identify nucleotide variation that shifted through passaging. (b) From one starting DWV population, multiple lineages were established in Passage 1, and a total of 47 final viral populations (Passage 5), were created through this passaging paradigm.
To passage a virus, viral inocula with confirmed DWV (see below, Virus Quantification) was injected into two new in vitro reared honey bee pupae at the white-eyed stage using the method described above and again isolated 4 DPI. Inoculums generated from passaging were not normalized by titre between passages – bees were homogenized in a fixed volume (500 ul) and directly injected to the next round of bees without titering. If bees perished prior to 4 DPI, they were removed from the study. While crude isolations were prepared from control bees (i.e. ‘no inject’ and ‘PBS’) for background DWV quantification, inocula from ‘PBS’ bees were not passaged, and ‘PBS’ bees were injected with fresh 1× PBS each experiment.
Survival assays
Survival assays were conducted in Fall 2019 with two Penn State University research SDI colonies: the original SDI colony from 2018, containing the same genotype of bees through which the virus was passaged, and an additional SDI colony naive to the passaged virus. Honey bees were in vitro reared [49] and injected at the white-eyed stage with the ‘P1’ (passaged once) and a subset of ‘P5’ (passaged five times) passaged virus, normalized to a concentration of 106 genome equivalents per microlitre. Groups also included ‘no inject’ and ‘PBS’, as before, as well as the starting inoculum (‘P0’), which appeared to have lost infectiousness after a year in storage, and can instead be considered an additional control. After injections, pupae were kept in incubators at 34.5 °C and approximately 75 % R.H. Mortality was monitored every 24 h. At eclosion (adult emergence from pupation, 7 days post-injection), surviving bees were transferred to petri dishes separated by treatment group and supplied with 30 % sucrose (ad libitum), and incubated 34.5 °C and approximately 85 % R.H. This paradigm was repeated across seven trials total.
Virus quantification
RNA was extracted from 30 µl of each virus inoculum using a Direct-zol RNA Miniprep kit (Zymo Research, Irvine, CA) following the manufacturer’s protocol, using 90 µl QIAZOL reagent (Qiagen, Hilden, Germany), and eluted in 25 ul molecular grade water. RNA concentration was quantified using a NanoDrop 1000 Spectrophotometer (ThermoFisher Scientific, Maltham, MA). cDNA was prepared from each sample using a High-Capacity cDNA Reverse Transcription Kit with RNase Inhibitor (ThermoFisher Scientific) using the following reaction mix: 1 µl of 10× Buffer, 0.4 µl of 25× dNTP mix, 1 µl of 10× random primers, 0.5 µl of Reverse Transcriptase, 0.5 µl of RNAse Inhibitor, and 6.6 µl of RNA. cDNA was diluted 1 : 20× to allow a sufficient amount of cDNA for all real-time quantitative PCR (qPCR) reactions. qPCR was conducted using PowerUp SYBR Green Master Mix (ThermoFisher) using the following reaction mix : 5 µl SYBR, 1 µl of 10× Forward Primer, 1 µl of 10× Reverse primer, 1 µl of water, and 2 µl cDNA. qPCR was conducted using a 7900HT Fast Real-Time PCR system (Applied Biosystems) under the following parameters: 50 °C for 2 min, 95 °C for 10 min, then cycle 40×95 °C for 15 s and 60 °C for 1 min, and then a dissociation stage step for melting curve analysis. qPCR primers can be found in Table S11. GAPDH (P1) and eIF3-S8 (P2-P5) are included as the internal reference to confirm successful RNA extraction and cDNA synthesis for each sample.
DWV levels were calculated from qPCR experiments as absolute quantification (genome equivalents) per nanogram RNA based off standard curves created from serial dilutions (102 - 106) of gBlocks (Integrated DNA Technologies, Coralville, IA) designed to match the PCR amplicon for each primer set. Individual levels of DWV-A and DWV-B targeting the capsid (CP) and non-structural (NS) regions were measured using strain specific primers [28]. Raw and cleaned qPCR data can be found in Supplementary Tables.
Next generation sequencing
RNA extracted from the starting inoculum, Passage 1, and Passage 5 viral populations was sent to the Pennsylvania State Genomics Core Facility (University Park, PA, USA) for library preparation and sequencing. Libraries were prepared from viral populations of individual bee samples, and named using the following nomenclature : Passage-Lineage-Sample (e.g. P1-M-01 is Sample one from the M lineage from Passage 1). The 55 samples were sequenced on the Illumina Miseq platform, resulting in 150 nucleotide single end stranded mRNA reads. Total reads per sample ranged between 307 630–599 780 (Table S15). Reads were assessed for quality with FastQC (version v0.11.9) and quality trimmed with Trimmomatic (version 0.39, option SLIDINGWINDOW:4 : 30).
Inoculum consensus DWV-A and DWV-B genomes were created by aligning reads from the inoculum sample (P0-X-01) to DWV-A and -B reference genomes from NCBI (Ref. NC_004830.2 and NC_006494.1, respectively) using Hisat2 (version 2.1.0) [50]. Variants were then called, indexed, and normalized, and consensus fastq sequence file generated with bcftools (version 1.8) [51]. The fastq file was converted to fasta with bcftools vcfutils.pl vcf2fq, and bases with qualities less than 20 were converted to Ns using seqtk (version 1.3-r106) [52]. This method successfully constructed a consensus DWV-A genome, approximately 97 % similar to the DWV-A NCBI reference (NC_004830.2, determined by blast (2.10.1+), default settings), and the protein coding sequence of our DWV-B consensus genome approximately 99 % similar to the DWV-B NCBI reference (NC_006494.1), with a 5′ end of approximately 1 kb undetermined due to low quality. De novo assembly by Trinity (version v2.1.1) [53] and blast did not identify any contigs that sufficiently covered this missing region. We therefore instead assembled contigs using Trinity from the Passage 1 samples, and created a consensus ‘P1’ genome by taking contigs >8500 bp, converting to all the same strand using seqkit seq (options -r -p), and creating a multisequence alignment using Clustal Omega (version 1.2.3) [54]. This alignment was loaded into Jalview (version 2.11.1.4) [55], and a consensus genome was calculated from this alignment. The ‘P1’ consensus genome was then used as a reference (rather than NC_006494.1) for constructing the inoculum DWV-B consensus genome. This reconstruction revealed that the missing region, the putative IRES region, of our DWV-B consensus genome aligns closer to DWV-A (97 % similarity to NC_004830.2) compared to only 83 % similar to DWV-B (NC_006494.1), indicating this appears to be a recombinant DWV-B inoculum genome (for simplicity, we refer to this recombinant strain as DWV-B, as most of the genome, including the protein coding region, is DWV-B).
Reads from all samples were then aligned to the inoculum consensus DWV-A and -B genomes using Hisat2. No reads in inoculum or passaged samples aligned to DWV variant C (CEND01000001.1), and less than 0.01 % of reads per sample aligned to other common bee viruses in the USA (chronic bee paralysis virus (NC_010711.1), Israeli acute paralysis virus (NC_009025.1), Lake Sinai virus 2 (NC_035467.1), sacbrood virus (NC_002066.1), black queen cell virus (NC_003784.1)), and thus these samples did not appear to host active infections by any of these viruses and were therefore considered free of other co-infecting viruses.
For phylogenetic analyses, consensus DWV-A and DWV-B genomes for all samples were generated using methods above, but instead utilizing the inoculum DWV-A and DWV-B consensus genomes as references. Multi-sequence alignments of generated consensus genomes and additional reference genomes (DWV-A reference (NC_004830.2), DWV-A from Pennsylvania (AY292384.1), kakugo virus (AB070959.1), DWV-B reference (NC_006494.1), DWV-B from Vespa velutina (MN565037.1), two recombinant DWV-A/DWV-B genomes (KX373900.1, MN538210.1), and DWV-C (CEND01000001.1)) were then generated with Clustal Omega using default settings. Multisequence alignment was then imported into MEGAX (version 10.1.8) for phylogenetic tree construction, calculating nucleotide diversity, and selection analysis [56]. Maximum Likelihood trees, nucleotide diversity, and codon-based Z-tests for selection were calculated with default settings and bootstrapped using 1000 replicates.
Variants within DWV-A and -B populations were called using bcftools and filtered with vcftools (Minor Allele Frequency >0.02, Minimum Quality >20, Minimum Depth >100; similar to [57]) and annotated using SNPeff (version 5.0) [58], and potential functions were predicted by identifying variable positions on the DWV-A crystal structure PDB 5MV6 [59] in Chimaera (version 1.5) [60]. Nucleotide diversity across the DWV-A and -B genomes was calculated utilizing vcftools (version 0.1.16). Consensus genomes constructed from the starting inoculum, as well as raw sequence reads can be found on NCBI (Reference: MT940255- MT940256, BioProject: PRJNA731530).
Statistical analyses
Statistical analyses were conducted in RStudio (version 1.2.5033) using the ‘stats' package (version 3.6.3). Wilcoxon rank-sum test for total DWV ~Passage was calculated after Shapiro-Wilk normality test (shapiro.test()), indicated non-normally distributed data, using the pairwise.wilcox.test() with Bonferroni correction. For DWV variants ~Passage, quantities were first transformed to normalize the data (NS quantities 1/3, CP quantities 1/4), then two-way Analysis of Variance (ANOVA) was calculated. Kaplan-Meier survival analysis and Cox proportional hazard were conducted in R using the ‘survival’ (version 2.38) and ‘survminer’ packages [61–63].
Results
An overview of the study design can be found in Fig. 1. For these experiments, a successful ‘passage’ is defined as injecting the DWV population into a white-eye stage pupa, and collecting the subsequent virus 4 days post-infection (DPI). The virus populations used for passaging will be referred to as ‘inoculum’ or ‘inocula’ (plural). Starting with one DWV inoculum, we successfully infected 10/14 individual pupae. Each of the ten resulting inocula was passaged a second time, injecting each inoculum into two bees each, and 8/10 were successfully passaged in at least 1/2 bees injected, creating 15 inocula. In Passage 3, 13/15 inocula from Passage 2 were successfully passaged, creating 18 inocula; in Passage 4, 17/18 from Passage 3 were successfully passaged into 26 inocula; in the final Passage, Passage 5, all 26 inocula from Passage 4 were successfully passaged into 47 final inocula. Note that for Passage 1, 10/14 bees were successfully infected, and 4/14 pupae had low to no DWV; for Passages 2 onward, any unsuccessful inocula were not due to lack of infection, but instead due to pupae perishing prior to collection at 4DPI.
Survival assays indicate no virulence differences between the passaged virus populations
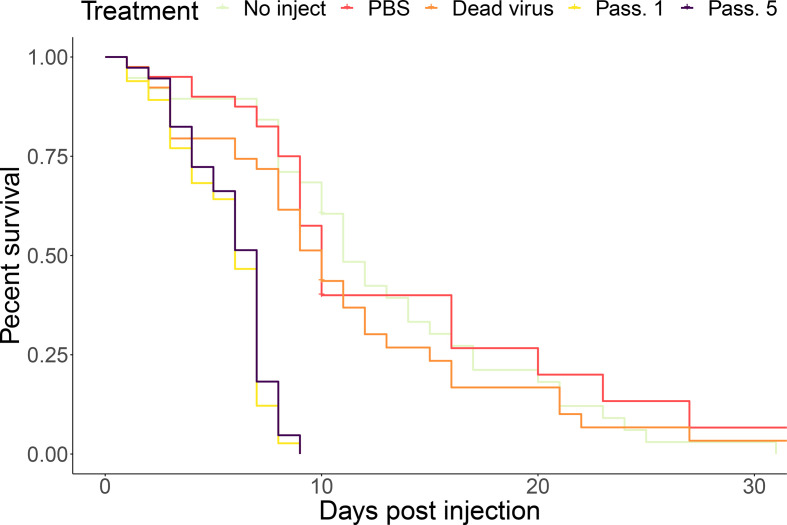
To assess the effect of passaging on the virulence of the viral populations, representative inocula from the eight maintained lineages were chosen from Passage 1 and Passage 5, and inocula were normalized to 106 genome equivalents per microlitre to assess for genetically coded differences in virulence (versus differences due to overall viral load). Across seven trials, inoculum passaged one and five times caused significantly more rapid mortality than the controls (Fig. 2, P-value <2e-12). However, the passaged populations did not significantly differ in mortality (P-value=0.24). The sharp drop in mortality at 7 days post-injection coincided with adult eclosion (i.e. emergence from the pupal stage). There was no significant difference in mortality across seven trials and across the two colonies assessed (Fig. S1a, b). We did not collect information on wing deformities, as most infected bees perished during development.
Fig. 2.
Percent survival across groups. Survival rates of virus passaged 1 and 5× were significantly different than control groups (P-value<2e-12). Nearly all mortality in the 1 and 5× passaged virus groups occurred on or before 7 days post-injection, coinciding with adult eclosion.
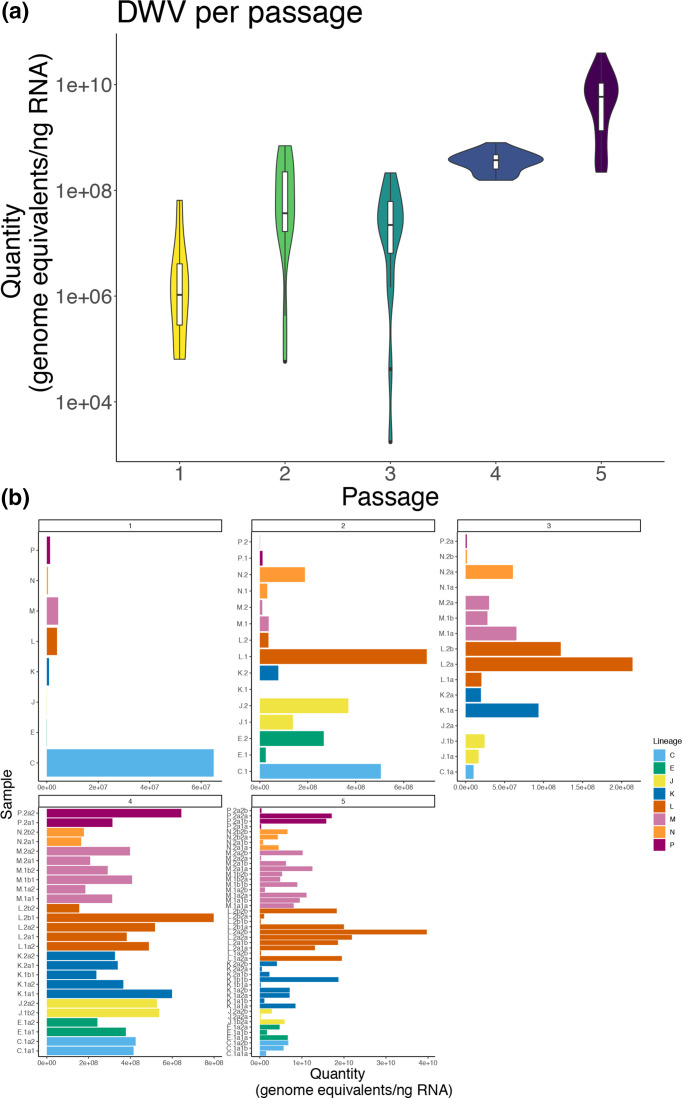
Total DWV levels increase through passaging
Each individual inoculum was tested for total DWV (i.e. DWV-A +B) via qPCR. Average DWV levels increased through passages (Fig. 3a). DWV Levels of Passages 4 and 5 were significantly higher than levels of the earlier passages (pairwise Wilcoxon Rank Sum, P<0.005), with no significant difference between the earlier passages. The distribution of DWV levels across individual inocula across passages can be found in Fig. 3(b). There was low to no DWV in control samples (see Fig. S3, Supplemental Tables).
Fig. 3.
Total DWV levels significantly increase after five passages. (a) Total DWV (targeting both DWV-A and DWV-B) levels by passage determined by qPCR. DWV levels were significantly higher in Passage 4 and 5 compared to all earlier passages (Wilcox Rank Sum test, P<0.005) (b) Distribution of total DWV levels in individual samples across passages (coloured by lineage). (a, b) DWV levels in controls (no-inject, PBS) were low to no detection, and therefore are not shown here, but can be found in Fig. S1 and Supplemental Tables.
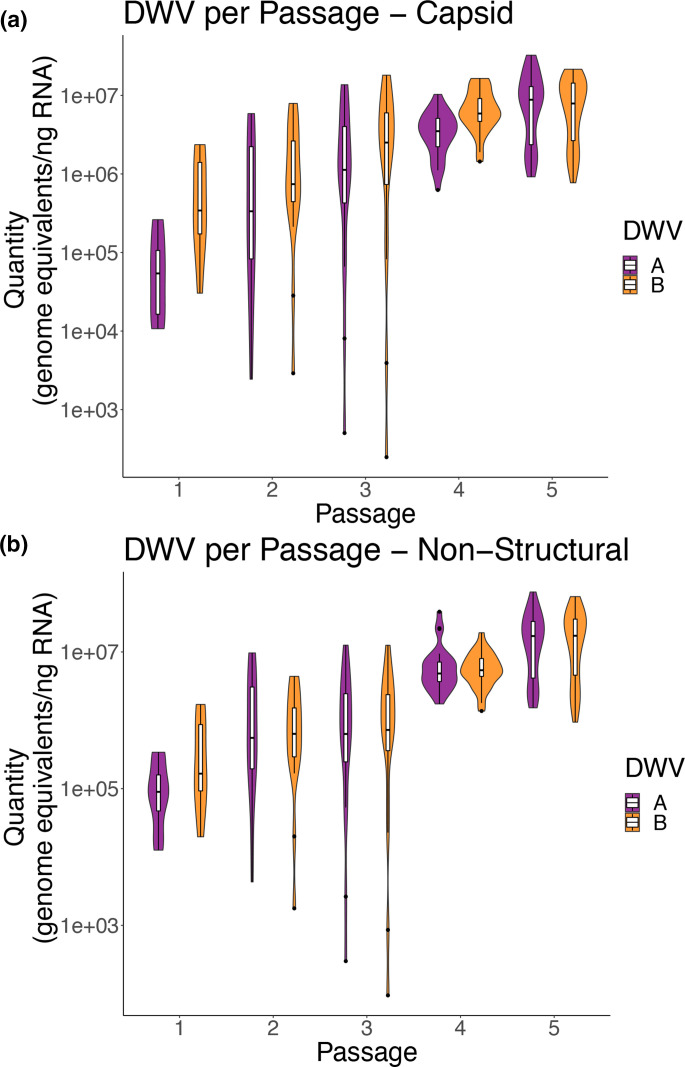
Total DWV variant levels also increase through passaging
In addition to total DWV (DWV-A +B), inocula were tested for two variants of DWV at two regions of the genome, using previously established variant-specific primers to target the DWV-A or DWV-B genome at approximately 4800–5000 and 8600–8880, henceforth referred to as the ‘Capsid’ (‘CP’) or ‘Non-structural’ (‘NS’) regions (as in [28], respectively). Levels of DWV variants at both regions mirrored a similar rise through passaging as levels of total DWV (Figs 4a, b and S4a, b). In general, levels of DWV-A and DWV-B in Passage 5 at both regions were higher than the DWV variants in Passage 1 and 2 (Fig. 4a, b, two-way ANOVA, P<0.0011).
Fig. 4.
DWV variant levels significantly increase after five passages (a, b) DWV-A and DWV-B levels by passage targeting two regions of the DWV genome, (a) non-structural region and (b) capsid region, determined by variant-specific qPCR. DWV levels were significantly higher in Passage 5 (P5) compared to all other passages (two-way ANOVA, P<0.01) (c, d) Scatter plots of DWV-A and DWV-B levels by passage targeting two regions of the DWV genome, (c) Non-structural region (Pearson correlation=0.884 (0.836–0.919, 95 % confidence, P-value<2.2e-16) and (d) Capsid region (Pearson correlation=0.837 (0.771–0.885, 95 % confidence, P-value<2.2e-16), determined by variant-specific qPCR. (a–d) DWV levels in controls (no-inject, PBS) were low to no detection, and therefore are not shown here, but can be found in Supplemental Tables.
Correlation assessment demonstrate associations between passages, variants, and regions
Correlations between levels of DWV (DWV-A, DWV-B, and total DWV) within passages, with the previous passage, and with their starting levels in Passage 1 can be found in Fig. S4. All DWV levels were positively associated with Passage; thus, as Passage increased, so too did total levels of total DWV or levels of DWV–A and DWV–B as measured at both regions, confirming our previous analysis (Figs 3a and 4). Levels of DWV-A, DWV-B, and total DWV were significantly positively correlated to one another within their passage. When comparing DWV-A and DWV-B in a given passage to the variant levels of their previous passage, all but DWV-B at the CP region were significantly positively associated with DWV populations in the previous passage; thus, higher DWV levels in a given passage were associated with higher DWV levels in the subsequent passage (Fig. S4).
Overview of sequencing results of the inoculum, Passage 1, and Passage 5
Analysis of the starting inoculum revealed a DWV-A consensus genome approximately 97 % similar to the NCBI reference genome (NC_004830.2, Fig. 5). Our DWV-B consensus genome was revealed to be a recombinant strain (see ‘Methods’ for more information), with the protein coding region most similar to DWV-B, while the first 1 kb (the putative internal ribosome entry site (IRES)) was more similar to DWV-A (Fig. 5). When aligning reads from the inoculum sample to these inoculum consensus genomes, we find approximately 1.3 : 1 DWV-A to DWV-B reads. Patterns of DWV-A : DWV-B ratios in the passaged samples similarly tracked what was observed in the qPCR experiments - higher counts for DWV-B than DWV-A in Passage 1 samples, with Passage 5 samples at an average ratio of 0.8 : 1 reads aligning to consensus DWV-A to DWV-B. Read depth was fairly even across the genome, with notable spikes in read depth across all samples (Fig. S5a, Table S15). Depth at the 5′ end is high in all samples, likely due to the similarity between DWV-A and the recombinant end of the DWV-B genome.
Fig. 5.
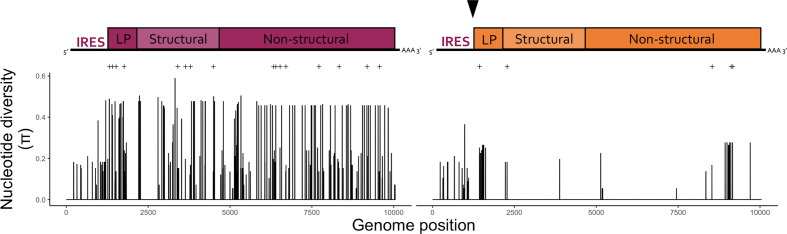
Variation across DWV-A and DWV-B genome populations. Reference maps for DWV-A (purple) and recombinant DWV-B (orange), labelled with genomic region used in nucleotide diversity analysis. The recombinant DWV-B genome consisted of a DWV-A derived 5' IRES region (97 % similarity to the DWV-A reference NC_004830.2, compared to only 83 % similar to the DWV-B reference NC_006494.1) and the rest of genome consisting of DWV-B. The caret in the DWV-B figure indicates the recombination point. Nucleotide diversity across DWV-A and DWV-B consensus genomes across Passage 1 and Passage 5 is shown below. Variable sites with missense mutations resulting in amino acid changes are denoted with (+).
DWV-A has a higher nucleotide diversity than DWV-B
When examining the sub-consensus variation within our DWV populations, we found that the DWV-A populations have higher nucleotide diversity across the genome compared to the DWV-B populations (Fig. 5). DWV-B, conversely, only exhibited increased nucleotide diversity near the RNA dependent RNA polymerase coding region, and at the 5′ end (Fig. 5). As the 5′ end of the recombinant DWV-B represents a region more similar to DWV-A, it is not clear whether this is representative of increased diversity in this region of the recombinant DWV-B genome, or increased variation within DWV-A populations that is being misclassified due to sequence similarity. Percent variation was not significantly associated with increased levels of DWV-A or DWV-B (Supplemental Figure 5b), nor associated with average read depth for either variant (Fig. S5c). Lineages differ in their DWV-A percent variation through passaging, with some samples increasing through passaging (e.g. Lineage M), some decreasing (e.g. Lineage J), while others remain similar through passaging (e.g. Lineage C).
Relationships between consensus DWV populations across passages
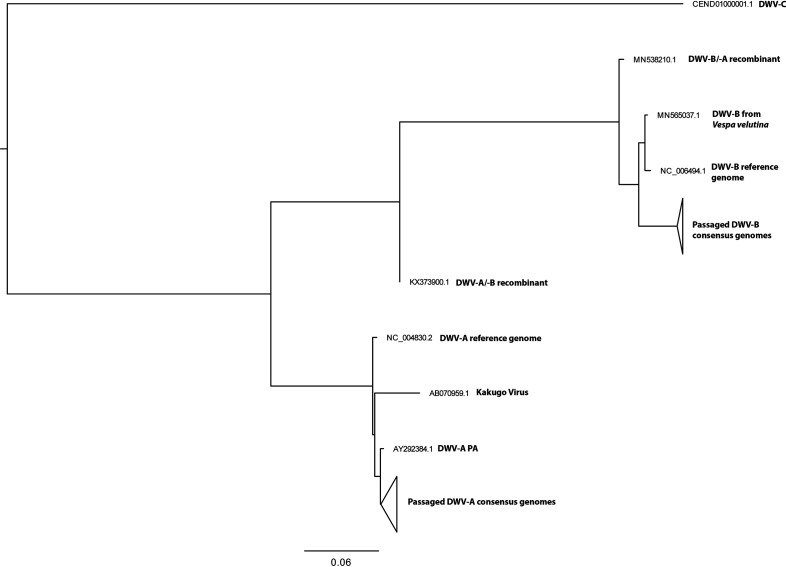
Phylogenetic analysis of whole genome DWV-A and DWV-B consensus genomes across samples can be found in Fig. 6. The two main clusters represent DWV-A and DWV-B clades. The DWV-A clade further resolves by lineage, but not by Passage. Due to the high similarity across the consensus DWV-B genomes, we observe much less resolution within this clade (Fig. S6).
Fig. 6.
Phylogeny of DWV-A and DWV-B passaged populations. Maximum likelihood trees with 1000 bootstrap replicates generated from full genome nucleic acid sequences for all consensus DWV-A and DWV-B populations. Clades representing Passaged populations were collapsed for clarity; non-collapsed trees can be found in Fig. S6.
DWV diversity and selection indices across DWV variant, passaging, and regions
Nucleotide diversity was lower in Passage 5 samples than Passage 1 samples, for both DWV-A and DWV-B. Generally, there was no difference when comparing selection indices between Passage 1 and Passage 5 samples within the DWV quasispecies (i.e. DWV-A and -B consensus genomes across samples) (Table 1). When comparing DWV-A and DWV-B, both showed signatures of neutral and positive selection across the coding region (Table 1). When comparing across different regions of the genome, DWV-A maintained a consistent pattern of neutral and negative selection signatures, while DWV-B had significant neutral and negative signatures at the non-structural region, and only negative signatures in the structural region (Table S16).
Table 1.
Estimates of the nucleotide diversity and selection indices for the DWV-A and DWV-B consensus genomes, calculated in megaX
|
Diversity and selection indices across the coding sequence | |||||
|---|---|---|---|---|---|
|
DWV-A | |||||
|
Sample Passage |
Nucleotide diversity (π) |
Tajima’s D |
Codon-Based Z Test of Selection (Nei-Gojobori) |
||
|
neutral |
positive |
negative |
|||
|
Passage 1 |
0.00339 (SE 0.00043) |
−2.373 |
P=6.549E-17* |
P=1 |
P=7.131E-24* |
|
Passage 5 |
0.0009 (SE 0.0002) |
−2.368 |
P=3.4195E-12* |
P=1 |
P=1.7456E-13* |
|
DWV-B | |||||
|
Sample Passage |
Nucleotide diversity (π) |
Tajima’s D |
Codon-Based Z Test of Selection (Nei-Gojobori) |
||
|
neutral |
positive |
negative |
|||
|
Passage 1 |
0.00103 (SE 0.00024) |
−0.182 |
P=0.0007* |
P=1 |
P=0.0002* |
|
Passage 5 |
0.00051 (SE 0.00013) |
−0.298 |
P=0.0008 * |
P=1 |
P=0.0002 * |
Variants across populations were found in functional regions of the viral genomes
Variants were identified across both the DWV-A and DWV-B genome. A total of 192 variants were identified for DWV-A across all samples; these variants were not unique to individual samples, but often shared within lineages across passaging (Table S16). DWV-B contained fewer variants, only 41 identified across all samples (Table S17). A proportion of these variants identified across all DWV-A and DWV-B were classified as missense, 8.3 % (16/192) and 12.2 % (5/44), respectively (Tables 2, S16 and S17), and were identified across the genomes. Across all variants (i.e. coding and non-coding), only 20 % (8/41) of variants increased from Passage 1 to Passage 5 in DWV-B populations, whereas 58.3 % (112/192) variants increased from Passage 1 to Passage 5 in DWV-A populations. As variants are often shared across samples within lineages, this may be an artefact of some lineages are being more highly represented in Passage 5. Several variants, though, increased across multiple lineages, such as the missense variant at DWV-A position 9181 (Met2679Ile) that was detected in only the C lineage in Passage 1, but in Lineages C, K, L, M in Passage 5. Additionally, some DWV-A missense variants were identified in regions of interest, such as within the DWV-A VP1 ‘spike’ (Ala754Val, Asn832Asp, Ala884Ser, Fig. S7) [59, 64] and within conserved functional motifs of the 3C protease (His2190Leu) and the RNA-dependent RNA polymerase (RDRP) (Lys2806Arg) [8].
Table 2.
Missense variants annotated by SNPeff within DWV-A and DWV-B quasispecies populations across all samples
|
Annotated variants | |||||||
|---|---|---|---|---|---|---|---|
|
DWV-A | |||||||
|
Putative region |
Position |
Nucleotide change |
AA change |
P0 Allele frequency |
P1 Allele frequency |
P5 Allele frequency |
Additional characteristics |
|
LP |
1319 |
G>A |
Ala59Thr |
0 |
0.125 |
0.1087 |
L specific, minor variant by Passage 5 (P5) |
|
LP |
1412 |
A>G |
Thr90Ala |
0 |
0.25 |
0.2935 |
Sole variant in C lineage, minor variant in other lines |
|
LP |
1521 |
A>G |
Lys126Arg |
0 |
0 |
0.0870 |
P5 specific; Minor variant throughout |
|
LP / structural |
1765 |
G>T |
Gln207His |
0 |
0.125 |
0.0978 |
In VP2, possibly interacting with RNA genome; M lineage specific, minor variant by P5 |
|
Structural |
3405 |
C>T |
Ala754Val |
0 |
0.125 |
0.0761 |
In capsid protein spike; M lineage specific, minor variant by P5 |
|
Structural |
3638 |
A>G |
Asn832Asp |
0 |
0.125 |
0.1087 |
In capsid protein spike; L lineage specific, minor variant by P5 |
|
Structural |
3794 |
G>T |
Ala884Ser |
0 |
0.1875 |
0.0761 |
In capsid protein spike; in J lineage in Passage 1 (P1) but not P5, minor variant by P5 for M lineage |
|
Structural |
4490 |
T>G |
Ser1116Ala |
0 |
0.0625 |
0.0761 |
In VP3; Minor variant in P1 and P5 |
|
Non-structural |
6323 |
G>T |
Gly1727Cys |
0 |
0.125 |
0.1087 |
L lineage specific, minor variant by P5 |
|
Non-structural |
6398 |
G>A |
Val1752Ile |
0 |
0.1875 |
0.3804 |
Sole variant in C lineage, minor variant in other lines |
|
Non-structural |
6527 |
G>A |
Ala1795Thr |
0.5 |
0.1875 |
0.1304 |
Sole variant in N lineage, minor variant in others |
|
Non-structural |
6702 |
C>T |
Thr1853Ile |
0 |
0.0625 |
0.0761 |
Minor variant in M lineage |
|
Non-structural |
7713 |
A>T |
His2190Leu |
0 |
0 |
0.0870 |
Variant in a highly conserved residue in 3C protease - P5 specific in K lineage |
|
Non-structural |
8330 |
T>A |
Ser2396Thr |
0 |
0.125 |
0.0761 |
M specific; sole variant in P1, minor variant in Passage 5 |
|
Non-structural |
9181 |
G>A |
Met2679Ile |
0 |
0.1875 |
0.3804 |
In P1, only present in C lineage, then present in C,K,L, and M lineages by P5 |
|
Non-structural |
9561 |
A>G |
Lys2806Arg |
0 |
0.0625 |
0.0761 |
Variant in a highly conserved aa in motif VII of RDRP; M specific minor variant throughout |
|
DWV-B | |||||||
|
Putative region |
Location |
Nucleotide change |
AA change |
P0 Allele frequency |
P1 Allele frequency |
P5 Allele frequency |
Lineage or passage specific? |
|
LP |
1433 |
T>C |
Tyr106His |
0 |
0.3125 |
0.1196 |
Specific to C and E lineages (except for one P1 N) |
|
Structural |
2275 |
T>C |
Leu386Phe |
0 |
0.3125 |
0.0652 |
Only maintained in C lineage |
|
Non-structural |
8530 |
C>T |
Lys2471Asn |
0 |
0.125 |
0.0870 |
P lineage specific |
|
Non-structural |
9109 |
C>T |
Leu2664Phe |
0 |
0.25 |
0.1522 |
E and N lineage specific |
|
Non-structural |
9163 |
C>T |
Met2682Ile |
0 |
0.25 |
0.1522 |
E and N lineage specific |
Discussion
Here, we investigated how different DWV variant populations responded to vector transmission of DWV in developing honey bee pupae. Varroa transmission of DWV was simulated by directly injecting DWV into white-eyed pupae, with successfully replicating virus collected 4 days post-infection, and subsequently injected into a new white-eyed pupa. This procedure was repeated five times, creating eight lineages of virus and 47 final DWV populations. Levels of total DWV and two major DWV variants increased significantly through passages, demonstrating that the vector transmission route can indeed influence DWV populations. Next generation sequencing of viral populations collected at Passage 1 and Passage 5 demonstrated that the DWV-B population was a DWV-A/DWV-B recombinant strain, with a DWV-B protein coding region and putative internal ribosome entry site (IRES) region more similar to DWV-A. Interestingly, while there was higher genetic variation in DWV-A versus DWV-B, both variants exhibited signatures of neutral and negative selection across their genomes. While the main impacts of passaging appeared to be on the ratios of A and B in the viral populations, several lineages accumulated missense mutations across functional regions of the DWV-A which may contribute to differences in viral characteristics and host-virus interactions.
The overall increase in DWV loads is contrary to what has been observed in previous studies. Remnant et al. [46] found that levels of DWV decreased with passaging, while the levels of two other co-infecting bee viruses (sacbrood virus and black queen cell virus) increased substantially with passaging. Thus, the authors suggested that the more virulent DWV was eliminated by vector transmission [46]. Similarly, Gisder et al. [45] found decreased DWV virulence, but only measured after one passage [45]. In the case of Yañez et al. [47], DWV-A remained consistent across passages, and virulence, measured by manifested disease symptoms (i.e. presence or absence of deformed wings) was not found to be associated with either levels or genetics [47]. Our observation of consistently increasing DWV levels through passaging may be the result of selection for DWV that replicates within the 4 DPI selection window, and through this bias towards increasing amounts of faster replicating viruses, the subsequent viral passages were able to amplify to higher and higher levels. These bees were also free of any other non-DWV viral species that could have competed with and eliminated the DWV variants, allowing these variants to persist and accumulate. Moreover, we also conducted multiple passages, which intensified any consequences of our passaging paradigm. Clearly, the timing of viral collection during artificial selection (or the timing of vector transmission under natural conditions) and presence of alternative viruses or stressors can generate different selection pressures on viral populations [2, 65].
Ratios of DWV-A and DWV-B changed dynamically through the course of the experiment. After the first passage, a higher proportion of the viral population corresponded to DWV-B, despite the original DWV inoculum being comprised of 1.3 : 1 DWV-A : DWV-B. This may be evidence of strain variation in replication rates, with DWV-B replicating faster than DWV-A. However, a previous demonstrated slightly higher levels of DWV-A over DWV-B during the exponential replication phase (<48 h post-injection) [66]. Thus testing this hypothesis would require more detailed time course studies, ideally in cell culture. Through the additional passages, however, our proportions of DWV-A and DWV-B across the lineages become again balanced. Norton et al. [66] similarly found that co-infecting pupae also resulted in higher levels of DWV-B than DWV-A after one passage [66], but Gisder et al. [45] found an overall shift in the population from DWV-B to DWV-A after one passage [45]. These differing results between studies may potentially be due to the differing methodology (some with DWV-A only, some with other co-infecting viruses and unequal starting variant levels; different collection and passaging paradigms; etc.), but serves to demonstrate the fundamental complexity of RNA virus dynamics and evolution regarding specific strains, timing, presence of other co-infecting viruses, and likely other factors associated with host physiological condition.
In these populations, there was much lower nucleotide diversity of DWV-B compared to DWV-A. This may be the result of initiating the study with samples collected from a single, symptomatic bee, and not be reflective of the full viral population across the globe. However, the DWV-B variant was only recently introduced to US bee populations [36], and thus may have not diversified as much as the DWV-A variant in the honey bee population from which the original samples were collected. Furthermore, since our virus population was collected in the US where Varroa is endemic, this particular viral population may already have been adapted to vector transmission, and serial passaging through vector mimicking methods may not result in extreme changes in variant population or virulence. To ensure infection in our bees, we also injected with a high titre of virus (106 genome equivalents), likely higher than the number of particles introduced through natural Varroa transmission. Our passaging paradigm was therefore designed to be stricter than normal ecology – DWV was injected at the white-eyed stage with a high titre of virus, and was collected 4 days post-injection, 3 days prior to what would be the ‘natural’ timeline of honey bee adult eclosion, Varroa emergence, and potential virus transmission. This may be why it was possible to observe the progressively increasing viral levels, shifting variants, signatures of selection, and overall high virulence.
When examining the whole genome between passages, both DWV-A and DWV-B showed signatures of neutral and negative selection. This suggests that the passaging regime used in our study did impose indeed a selective pressure on DWV, similar to Yañez et al., who also found signatures of negative selection in their adult bee DWV populations [47]. While nucleotide variation was identified across both DWV-A and DWV-B genomes, many functional regions, such as within the helicase protein (previously found to be highly conserved [67]), did not obtain any measurable amino acid changes. A subset of missense variants was identified within functional regions of the coding region with the potential to affect viral infection. Particularly for DWV-A, non-conserved amino acid changes across multiple lineages were identified within the VP1 ‘spike’ region, which is hypothesized to be involved in interaction with the host cell and/or RNA virus genome release [64], and could therefore be influencing infection dynamics at the cellular level. Other minor variants were identified within conserved regions of non-structural protein motifs, such as the 3C protease and the within the RDRP. While the DWV-A variant did compete more effectively with DWV-B after Passage 1, further studies would be needed to determine if these sequence changes influenced DWV-A’s ability to compete with and/or the ability of the dual viral population to reach higher titres in later passages. Coupling newly developed infectious clones [68, 69], honey bee cell lines [70] and improved long read sequencing accuracy to assess viral recombination will allow for more comprehensive investigations of DWV and other RNA virus molecular dynamics.
Previous studies have found that viruses with small genomes have higher mutation rates [71] [72], and virus adaptation can occur rapidly [73]. Furthermore, DWV in particular has been shown to readily form and accumulate recombinants amongst variants [69]. Viral populations can be considered to be ‘viral quasispecies’, or collections of related viral genomes reproducing with high mutation and recombination rates to high population sizes [74]. While there were cases of accumulations over new variants in the lineages in our study, the more dramatic changes appear to be the result of shifts in DWV-A versus DWV-B abundance. The shifting variants as well as negative selection signatures may have been a result of our strict selection paradigm: by removing other forms of DWV transmission (e.g. oral transmission), there may have been selection against variants that support other aspects of DWV disease ecology [75]. Additionally, both DWV-A and/or DWV-B could be mutating, and any beneficial de novo mutations may not have been present at high enough levels by Passage 5, or be recombining [37] at undetected levels. Indeed, as our DWV-B consensus strain was already recombinant, and other studies have also identified DWV recombinants [66, 67]. Additionally, both qPCR and deep sequencing methods demonstrated increasing viral levels and accumulation of both DWV variants through passaging, again demonstrating that these viral populations show classic characteristics of RNA virus quasispecies.
DWV-A and DWV–B share approximately 85 % nucleotide similarity. Co-infections with similar strains may lead to competition between strains [76], or cooperation among strains, if they act as one, highly diverse, viral quasispecies [77]. Initially, there was competition, with one master variant (DWV-B) reaching higher levels than the other after one passage, but levels quickly re-balanced in subsequent passages. With both variants reaching higher titres in later passages, this may provide evidence for cooperation. Both variants experienced negative selection, though despite this selection, DWV-A resulted in greater protein-sequence variation overall. It remains to be determined how genetic variation between the strains and within populations of these strains potentially influence viral-viral interactions. While our study did not aim to select for virulence, our viral populations were highly virulent, with infected bees dying significantly earlier than uninfected controls. However, between once and five-time passaged virus, there were no observed differences in survival. All viral inocula were normalized to the same titre, but had different proportions of DWV-A and DWV-B (Fig. 3) and differing nucleotide variation (Tables 1 and 2) across the populations. These differing DWV populations did not result in observable differences in virulence, similar to other studies where no virulence differences were detected between DWV-A and DWV-B [41–43]. This may be further evidence that DWV virulence is not specific to any certain DWV master variant, but acts through the quasispecies population as a whole, resulting in higher titres associated with worsened disease [78]; alternatively, one or both DWV master variants could simply be present in these passaged populations at sufficiently high levels to result in high virulence. Through normalizing the amount of DWV between the once and five times passaged virus populations, we were able to investigate whether there were any genetically encoded differences in virulence across these passaged samples (either master variants or single nucleotide polymorphisms), but we may have also removed an essential characteristic of the viral population (viral levels during exposure) that may underlie variation in virulence. As our original starting inoculum was no longer infectious during survival assays, we cannot conclude how initial passaging affected virulence, only that continued passaging does not result in observable altered virulence. Interestingly, we did not observe a difference in virulence when measuring survival across different colonies – the survival of the colony genotype naïve to these experimentally-derived DWV populations was not observed to be better nor worse than the colony through which the virus was passaged. Heritable differences against stressors like Varroa have been observed in bees [79, 80], and other genotype by genotype interactions have been observed in other insects and their pathogens [81–83]; thus, additional studies investigating genotype by genotype interactions between DWV and honey bees should be conducted.
The passaging paradigm used in this study provides insights into how viral variants can respond to different selection pressures. However, the results of this lab-based protocol cannot necessarily be used to predict DWV dynamics under complex ecological conditions. First, there may be other DWV-A and B variants circulating in the USA or around the globe that behave differently than the variants we used in this study. Second, with both qPCR and sequencing, we cannot directly say whether all genome copies were indeed infectious particles without cell culture based tests such as plaque forming assays. Third, as we used injections as an approximation for vector transmission, which allowed for standardized injections across samples and passages, but did not incorporate all features of Varroa vector transmission and parasitization, such as its disruption of the honey bee immune response while Varroa feeds [18]. Fourth, there are other routes of transmission that occur in honey bee colonies which may favour the maintenance of variants or combinations of variants. Our model simulated a system in which Varroa acts as a vector in a non-propagative manner [84], and instead focused on the effect of the shorter incubation time (4 days) and the effect of the viral population directly into the open circulatory system of the developing bee, and how this direct transmission route and shortened incubation time affected the viral population and resulting virulence within and across master variants. Thus, while we demonstrated that direct transmission alone can drive viral population differences, further studies incorporating additional complexities, such as Varroa’s capacity to immunocompromise, or by incorporating Varroa itself, are needed to fully understand how the introduction of Varroa has affected DWV, and how vector transmission can alter disease evolution.
Varroa, historically a parasite of the eastern honey bee Apis cerana, was introduced to A. mellifera populations in the mid-20th century, and with no co-evolved defenses in A. mellifera against this ectoparasite, it rapidly spread unchecked to vulnerable honey bee populations across the globe, transmitting DWV along the way [17, 25]. Due to these two unchecked stressors, in addition to other factors including pesticide usage and landscape alteration, US beekeepers lose an average of 30–40 % of their colonies each year [85], causing economic strain on beekeepers and potentially threatening agricultural production. With such a high rate of evolution and interdependence with its host and community, and additional factors like Varroa recently and dramatically altering disease ecology, bee viruses are an obvious and critical avenue for continued investigation in pollinator health [7]. Furthermore, the DWV/Varroa epidemic is just one example of widespread, unsustainable emerging infectious disease affecting agricultural and ecological systems. There is an abundance of vector-borne viruses that influence human, animal, and plant health across the globe, and detailed laboratory studies can provide valuable information and perhaps insights into the fundamental principles through which vectors selected for altered viral populations and influence host-pathogen interactions and evolution.
Supplementary Data
Funding information
This work was supported by a grant from the US Department of Agriculture Animal and Plant Health Inspection Service (#16-8130-0501) and USDA Hatch funds (#4716, ‘Sustainable Solutions to Problems Affecting Bee Health.’) awarded to CMG, USDA Hatch funds (Accession #1010032; Project #PEN04608) to JLR, a Pennsylvania State Center for Pollinator Research Apes Valentes Graduate Research Award awarded to AMR, and support from the Penn State Huck Institutes Molecular Cellular and Integrative Biosciences Graduate Program and College of Agricultural Sciences for AMR.
Acknowledgements
We would like to thank Michael Simone-Finstrom, Phil Tokarz, and Chauncy Hinshaw for providing the starting DWV inoculum, and Kate Anton for expert beekeeping assistance. We would additionally like to thank Sarthok Rahman and Eugene Ryabov for suggestions on bioinformatic analyses, and to Lindsey Organtini and Nicole Hackenbrack for their assistance with DWV structural biology.
Author contributions
A.M.R. and C.M.G., conceived research and designed experiments. A.M.R. and S.L.D., performed experiments. A.M.R., conducted statistical analyses. A.M.R. and C.M.G., drafted the first version of the manuscript. All authors approved the final version.
Conflicts of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Abbreviations: CP, capsid; DPI, days post infection; DWV, deformed wing virus; IRES, internal ribosome entry site; LP, leader protein; NS, non-structural; RDRP, RNA-dependent RNA polymerase; SDI, single drone inseminated (honey bee queen).
Eighteen supplementary tables and seven supplementary figures are available with the online version of this article.
References
- 1.Anderson RM, May RM. Coevolution of hosts and parasites. Parasitology. 1982;85:411–426. doi: 10.1017/S0031182000055360. [DOI] [PubMed] [Google Scholar]
- 2.Schmid-Hempel P. Evolutionary Parasitology: the Integrated Study of Infections, Immunology, Ecology, and Genetics. Oxford University Press; 2011. [Google Scholar]
- 3.Institute of Medicine (US) Forum on Microbial Threats Vector-Borne Diseases: Understanding the Environmental, Human Health, and Ecological Connections, Workshop Summary. National Academies Press (US; 2008. [PubMed] [Google Scholar]
- 4.Genersch E, Aubert M. Emerging and re-emerging viruses of the honey bee (Apis mellifera L.) Vet Res. 2010;41:54. doi: 10.1051/vetres/2010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McMahon DP, Wilfert L, Paxton RJ, Brown MJF. Emerging viruses in bees: from molecules to ecologyviruses in bees: from molecules to ecology. Adv Virus Res. 2018;101:251–291. doi: 10.1016/bs.aivir.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Chen YP, Siede R. Honey Bee Viruses. Adv Virus Res. 2007;70:33–80. doi: 10.1016/S0065-3527(07)70002-7. [DOI] [PubMed] [Google Scholar]
- 7.Grozinger CM, Flenniken ML. Bee viruses: ecology, pathogenicity, and impacts. Annu Rev Entomol. 2019;64:205–226. doi: 10.1146/annurev-ento-011118-111942. [DOI] [PubMed] [Google Scholar]
- 8.Lanzi G, Miranda JRD, Boniotti MB, Cameron CE, Lavazza A, et al. Molecular and biological characterization of deformed wing virus of hobiological characterization of deformed wing virus of honeybees (Apis mellifera L.) J Virol. 2006;80:4998–5009. doi: 10.1128/JVI.80.10.4998-5009.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Annoscia D, Del Piccolo F, Covre F, Nazzi F. Mite infestation during development alters the in-hive behaviour of adult honeybees. Apidologie. 2015;46:306–314. doi: 10.1007/s13592-014-0323-0. [DOI] [Google Scholar]
- 10.Natsopoulou ME, McMahon DP, Paxton RJ. Parasites modulate within-colony activity and accelerate the temporal polyethism schedule of a social insect, the honey bee. Behav Ecol Sociobiol. 2016;70:1019–1031. doi: 10.1007/s00265-015-2019-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Traniello IM, Bukhari SA, Kevill J, Ahmed AC, Hamilton AR, et al. Meta-analysis of honey bee neurogenomic response links Deformed wing virus type A to precocious behavioral maturation. Sci Rep. 2020:1–12. doi: 10.1038/s41598-020-59808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benaets K, Geystelen AV, Cardoen D, Smet LD, Graaf DCD, et al. Covert deformed wing virus infections have long-term deleterious effects on honeybee foraging and survival. Proceedings of the Royal Society B. 2017:284. doi: 10.1098/rspb.2016.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Miranda JR, Genersch E. Deformed wing virus. J Invertebr Pathol. 2010;103:S48–S61. doi: 10.1016/j.jip.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Evans J, Feldlaufer M. Horizontal and vertical transmission of viruses in the honey bee, Apis mellifera. J Invertebr Pathol. 2006;92:152–159. doi: 10.1016/j.jip.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Bowen-Walker PL, Martin SJ, Gunn A. The transmission of deformed wing virus between honeybees (Apis mellifera L.) by the ectoparasitic mite Varroa jacobsoni oud. J Invertebr Pathol. 1999;73:101–106. doi: 10.1006/jipa.1998.4807. [DOI] [PubMed] [Google Scholar]
- 16.Nazzi F, Le Conte Y. Ecology of Varroa destructor, the major ectoparasite of the Western Honey Bee, Apis mellifera . Annu Rev Entomol. 2016;61:417–432. doi: 10.1146/annurev-ento-010715-023731. [DOI] [PubMed] [Google Scholar]
- 17.Oldroyd BP. Coevolution while you wait: Varroa jacobsoni, a new parasite of western honeybees. Trend Ecol Evol. 1999;14:312–315. doi: 10.1016/S0169-5347(99)01613-4. [DOI] [PubMed] [Google Scholar]
- 18.Annoscia D, Brown SP, Prisco GD, Paoli ED, Fabbro SD, et al. Haemolymph removal by Varroa mite destabilizes the dynamical interaction between immune effectors and virus in bees, as predicted by Volterras model. Proc Roy Soc B. 2019;286:20190331. doi: 10.1098/rspb.2019.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramsey SD, Ochoa R, Bauchan G, Gulbronson C, Mowery JD, et al. Varroa destructor feeds primarily on honey bee fat body tissue and not hemolymph. Proceed National Acad Sci. 2019 doi: 10.1073/pnas.1818371116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Locke B, Semberg E, Forsgren E, De Miranda JR. Persistence of subclinical deformed wing virus infections in honeybees following Varroa mite removal and a bee population turnover. PLoS ONE. 2017;12:1–10. doi: 10.1371/journal.pone.0180910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dainat B, Evans JD, Chen YP, Gauthier L, Neumann P. Predictive markers of honey bee colony collapse. PLoS One. 2012;7:e32151. doi: 10.1371/journal.pone.0032151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doke MA, Frazier M, Grozinger CM. Overwintering honey bees: biology and management. Curr Opin Insect Sci. 2015;10:185–193. doi: 10.1016/j.cois.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Natsopoulou ME, McMahon DP, Doublet V, Frey E, Rosenkranz P, et al. The virulent, emerging genotype B of Deformed wing virus is closely linked to overwinter honeybee worker loss. Sci Rep. 2017;7:1–9. doi: 10.1038/s41598-017-05596-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin SJ, Highfield AC, Brettell L, Villalobos EM, Budge GE, et al. Global honey bee viral landscape altered by a parasitic mite. Science. 2012;336:1304–1307. doi: 10.1126/science.1220941. [DOI] [PubMed] [Google Scholar]
- 25.Wilfert L, Long G, Leggett HC, Schmid-Hempel P, Butlin R, et al. Deformed wing virus is a recent global epidemic in honeybees driven by Varroa mites. Science. 2016;351:594–597. doi: 10.1126/science.aac9976. [DOI] [PubMed] [Google Scholar]
- 26.Di Prisco G, Annoscia D, Margiotta M, Ferrara R, Varricchio P, et al. A mutualistic symbiosis between a parasitic mite and a pathogenic virus undermines honey bee immunity and health. Proc Natl Acad Sci U S A. 2016;113:3203–3208. doi: 10.1073/pnas.1523515113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nazzi F, Brown SP, Annoscia D, Del Piccolo F, Di Prisco G, et al. Synergistic parasite-pathogen interactions mediated by host immunity can drive the collapse of honeybee colonies. PLoS Pathog. 2012;8:e1002735. doi: 10.1371/journal.ppat.1002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood GR, Fannon JM, Moore JD, Bull JC, et al. A virulent strain of deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Vvarroa destructor-mediated, or in vitro, transmission. PLoS Pathog. 2014;10:e1004230. doi: 10.1371/journal.ppat.1004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manley R, Temperton B, Doyle T, Gates D, Hedges S. Knock-on community impacts of a novel vector: spillover of emerging DWV-B from Varroa -infested honeybees to wild bumblebees. Ecol Lett. 2019;22:1306–1315. doi: 10.1111/ele.13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mondet F, de Miranda JR, Kretzschmar A, Le Conte Y, Mercer AR. On the front line: quantitative virus dynamics in Honeybee (Apis mellifera L.) colonies along a new expansion front of the parasite Varroa destructor . PLoS Pathog. 2014;10:e1004323. doi: 10.1371/journal.ppat.1004323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh R, Levitt AL, Rajotte EG, Holmes EC, Ostiguy N, et al. RNA viruses in hymenopteran pollinators: Evidence of inter-taxa virus transmission via pollen and potential impact on non-Apis hymenopteran species. PLoS ONE. 2010;5:12. doi: 10.1371/journal.pone.0014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tehel A, Brown MJF, Paxton RJ. Impact of managed honey bee viruses on wild bees. Curr Opin Virol. 2016;19:16–22. doi: 10.1016/j.coviro.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 33.Ongus JR, Peters D, Bonmatin JM, Bengsch E, Vlak JM, et al. Complete sequence of a picorna-like virus of the genus Iflavirus replicating in the mite Varroa destructor . J Gen Virol. 2004;85:3747–3755. doi: 10.1099/vir.0.80470-0. [DOI] [PubMed] [Google Scholar]
- 34.Gisder S, Aumeier P, Genersch E. Deformed wing virus: replication and viral load in mites (Varroa destructor) J Gen Virol. 2009;90:463–467. doi: 10.1099/vir.0.005579-0. [DOI] [PubMed] [Google Scholar]
- 35.McMahon DP, Natsopoulou ME, Doublet V, Fürst M, Weging S, et al. Elevated virulence of an emerging viral genotype as a driver of honeybee loss. Proceedings of the Royal Society B. 2016;283:1833. doi: 10.1098/rspb.2016.0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryabov E, Childers AK, Chen Y, Madella S, Nessa A, et al. Recent spread of Varroa destructor virus-1, a honey bee pathogen, in the United States. Sci Rep. 2017;7:1–10. doi: 10.1038/s41598-017-17802-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore J, Jironkin A, Chandler D, Burroughs N, Evans DJ, et al. Recombinants between Deformed wing virus and Varroa destructor virus-1 may prevail in Varroa destructor -infested honeybee colonies. J Gen Virol. 2011;92:156–161. doi: 10.1099/vir.0.025965-0. [DOI] [PubMed] [Google Scholar]
- 38.Mordecai GJ, Wilfert L, Martin SJ, Jones IM, Schroeder DC. Diversity in a honey bee pathogen: First report of a third master variant of the Deformed Wing Virus quasispecies. ISME J. 2016;10:1264–1273. doi: 10.1038/ismej.2015.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mordecai GJ, Brettell LE, Martin SJ, Dixon D, Jones IM, et al. Superinfection exclusion and the long-term survival of honey bees in Varroa-infested colonies. ISME J. 2016;10:1182–1191. doi: 10.1038/ismej.2015.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brettell LE, Schroeder DC, Martin SJ. RNAseq analysis reveals virus diversity within Hawaiian apiary insect communities. Viruses. 2019;11:397. doi: 10.3390/v11050397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tehel A, Vu Q, Bigot D, Gogol-döring A, Koch P, et al. The two prevalent genotypes of an emerging equally low pupal mortality and equally high wing deformities in host honey bees. Viruses. 2019;11:1–18. doi: 10.3390/v11020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brettell LE, Mordecai GJ, Schroeder DC, Jones IM, Da Silva JR, et al. A comparison of deformed wing virus in deformed and asymptomatic honey bees. Insects. 2017;8 doi: 10.3390/insects8010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kevill JL, Souza DFS, Sharples C, et al. DWV-A Lethal to Honey Bees (Apis mellifera): A Colony Level Survey of DWV Variants (A, B, and C) in England, Wales, and 32 States across the US. Viruses. 2019;11 doi: 10.1057/s41271-016-0043-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neumann P, Yañez O, Fries I, De Miranda JR. Varroa invasion and virus adaptation. Trends Parasitol. 2012;28:353–354. doi: 10.1016/j.pt.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 45.Gisder S, Möckel N, Eisenhardt D, Genersch E. In vivo evolution of viral virulence: switching of deformed wing virus between hosts results in virulence changes and sequence shifts. Environ Microbiol. 2018;20:4612–4628. doi: 10.1111/1462-2920.14481. [DOI] [PubMed] [Google Scholar]
- 46.Remnant EJ, Mather N, Gillard TL, Yagound B, Beekman M, et al. Direct transmission by injection affects competition among RNA viruses in honeybees. Proc Royal Soc B. 2019;286:20182452. doi: 10.1098/rspb.2018.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yañez O, Chávez-galarza J, Tellgren-roth C, Pinto MA, Neumann P, et al. The honeybee (Apis mellifera) developmental state shapes the genetic composition of the deformed wing virus-a quasispecies during serial transmission. Sci Rep. 2020;10:1–12. doi: 10.1038/s41598-020-62673-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winston ML. The Biology of the Honey Bee. Harvard University Press; 1991. [Google Scholar]
- 49.Schmehl DR, Tomé H, Mortensen AN, Martins GF, Ellis JD. Protocol for the in vitro rearing of honey bee (Apis mellifera L.) workers. J Apic Res. 2016;55:113–129. [Google Scholar]
- 50.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H. Seqtk: a fast and lightweight tool for processing FASTA or FASTQ sequences. 2013.
- 53.Grabherr MG, Hass BJ, Yassour M, Levin JZ, Thompson DA, et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 2013;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2 — a multiple sequence alignment editor and analysis workbench. Bioinform. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bowen CD, Renner DW, Johnston CM, Szpara ML. In vitro evolution of herpes simplex virus 1 (HSV-1) reveals selection for syncytia and other minor variants in cell culture. Virus Evol. 2020;6:1–15. doi: 10.1093/ve/veaa013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Škubník K, Nováček J, Füzik T, Přidal A, Paxton RJ, et al. Structure of deformed wing virus, a major honey bee pathogen. Proc Natl Acad Sci U S A. 2017;114:3210–3215. doi: 10.1073/pnas.1615695114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. UCSF Chimera — A Visualization System for Exploratory Research and Analysis. J Comput Chem. 2004 doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 61.Kassambara A, Kosinski M, Biecek P. survminer: Drawing Survival Curves using “ggplot2.”. 2020.
- 62.Core Team R. R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria: 2020. [Google Scholar]
- 63.Therneau T. A Package for Survival Analysis in S. 2015. [Google Scholar]
- 64.Organtini LJ, Shingler KL, Ashley RE, Capaldi EA, Durrani K, et al. Honey bee deformed wing virus structures reveal that conformational changes accompany genome release. J Virol. 2017;91:10–13. doi: 10.1128/JVI.01795-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cooper VS, Reiskind MH, Miller JA, Shelton KA, Walther BA, et al. Timing of transmission and the evolution of virulence of an insect virus. Proc Roy Soc London Ser B Biol Sci. 2002;269:1161–1165. doi: 10.1098/rspb.2002.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Norton AM, Remnant EJ, Buchmann G, Beekman M. Accumulation and competition amongst deformed wing virus genotypes in naïve aaustralian honeybees provides insight into the increasing global prevalence of gcompetition amongst deformed wing virus genotypes in naïve australian honeybees provides insight into the increasing global prevalence of genotype b. Front Microbiol. 2020;11:1–14. doi: 10.3389/fmicb.2020.00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dalmon A, Desbiez C, Coulon M, Thomasson M, Le Conte Y, et al. Evidence for positive selection and recombination hotspots in Deformed wing virus (DWV) Sci Rep. 2017;7:1–12. doi: 10.1038/srep41045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lamp B, Url A, Seitz K, Eichhorn J, Riedel C, et al. Construction and rescue of a molecular clone of Deformed wing virus (DWV) PLoS ONE. 2016;11:1–18. doi: 10.1371/journal.pone.0164639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ryabov E, Childers AK, Lopez D, Grubbs K, Posada-Florez F, et al. Dynamic evolution in the key honey bee pathogen deformed wing virus: Novel insights into virulence and competition using reverse genetics. PLoS Biol. 2019;17:10. doi: 10.1371/journal.pbio.3000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo Y, Goodman CL, Stanley DW, Bonning BC. Cell lines for honey bee virus research. Viruses. 2020;12:1–17. doi: 10.3390/v12020236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Retel C, Markle H, Becks L, Feulner PGD. Ecological and evolutionary processes shaping viral genetic diversity. Viruses. 2019;11:220. doi: 10.3390/v11030220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanjuán R, Domingo-Calap P. Mechanisms of viral mutation. Cell Mol Life Sci. 2016;73:4433–4448. doi: 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Duffy S, Shackelton L, Holmes E. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 74.Andino R, Domingo E. Viral quasispecies. Virology. 2015;479–480:46–51. doi: 10.1016/j.virol.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Domingo E, Escarmís C, Sevilla N, Moya A, Elena SF, et al. Basic concepts in RNA virus evolution. FASEB j. 1996;10:859–864. doi: 10.1096/fasebj.10.8.8666162. [DOI] [PubMed] [Google Scholar]
- 76.Alizon S, DeRoode JC, Michalakis Y. Multiple infections and the evolution of virulence. Ecol Lett. 2013;16:556–567. doi: 10.1111/ele.12076. [DOI] [PubMed] [Google Scholar]
- 77.Shirogane Y, Watanabe S, Yanagi Y. Cooperation between different variants: A unique potential for virus evolution. Virus R. 2019;264:68–73. doi: 10.1016/j.virusres.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 78.Chen YP, Higgins JA, Feldlaufer MF. Quantitative Real-Time Reverse Transcription-PCR analysis of deformed quantitative Real-Time Reverse Transcription-PCR analysis of deformed wing virus infection in the honeybee (Apis mellifera L.) Appl Environ Microbiol. 2005;71:436–441. doi: 10.1128/AEM.71.1.436-441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harbo JR, Harris JW. Heritability in honey bees (Hymenoptera: Apidae) of characteristics associated with resistance to Varroa jacobsoni (Mesostigmata: Varroidae) J Econ Entomol. 1999;92 doi: 10.1603/0022-0493-94.6.1319. [DOI] [PubMed] [Google Scholar]
- 80.Mondet F, Beaurepaire A, McAfee A, Locke B, Alaux C, et al. Honey bee survival mechanisms against the parasite Varroa destructor: a sys‐tematic review of phenotypic and genomic research efforts. Int J Parasitol. 2020;50:433–447. doi: 10.1016/j.ijpara.2020.03.005. [DOI] [PubMed] [Google Scholar]
- 81.Barribeau SM, Sadd BM, du Plessis L, Schmid-Hempel P. Gene expression differences underlying genotype-by-genotype specificity in a host–parasite system. Proc Natl Acad Sci USA. 2014;111:3496–3501. doi: 10.1073/pnas.1318628111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Roode JC, Altizer S. Host – parasite genetic interactions and virulence-transmission relationships in natural populations of monarch butterflies. Evolution. 2009;64:502–514. doi: 10.1111/j.1558-5646.2009.00845.x. [DOI] [PubMed] [Google Scholar]
- 83.Lambrechts L, Chevillon C, Albright RG, Thaisomboonsuk B, Richardson JH, et al. Genetic specificity and potential for local adaptation between dengue viruses and mosquito vectors. BMC Evol Biol. 2009;9:1–11. doi: 10.1186/1471-2148-9-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Posada-Florez F, Childers AK, Heerman MC, Egekwu N, Cook SC, et al. Deformed wing virus type A, a major honey bee pathogen, is vectored by the mite Varroa destructor in a non- propagative manner. Sci Rep. 2019;9:1–10. doi: 10.1038/s41598-019-47447-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bee Informed Partnership National Management Survey. 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.