Abstract
Dengue virus (DENV), like other viruses, closely interacts with the host cell machinery to complete its life cycle. Over the course of infection, DENV interacts with several host factors with pro-viral activities to support its infection. Meanwhile, it has to evade or counteract host factors with anti-viral activities which inhibit its infection. These molecular virus-host interactions play a crucial role in determining the success of DENV infection. Deciphering such interactions is thus paramount to understanding viral fitness in its natural hosts. While DENV-mammalian host interactions have been extensively studied, not much has been done to characterize DENV-mosquito host interactions despite its importance in controlling DENV transmission. Here, to provide a snapshot of our current understanding of DENV-mosquito interactions, we review the literature that identified host factors and cellular processes related to DENV infection in its mosquito vectors, Aedes aegypti and Aedes albopictus, with a particular focus on DENV-mosquito omics studies. This knowledge provides fundamental insights into the DENV life cycle, and could contribute to the development of novel antiviral strategies.
Keywords: dengue virus, mosquitoes, virus-host interactions, viral fitness
Dengue is arguably the most important mosquito-borne viral infectious disease in humans. This acute disease is caused by four genetically related but antigenically distinct dengue viruses (DENV-1 to -4). All four DENVs are transmitted by Aedes (Ae.) mosquitoes, principally Ae. aegypti. As the global footprint of Ae. aegypti continues to expand from the tropics to the subtropics [1], well over half of the world’s population lives in areas at risk of DENV infection [2]. An estimated 100 million people develop dengue and thousands die from this disease each year [3]. Presently, the only licensed dengue vaccine, Dengvaxia, comes with some limitations prior to usage and there are no specific therapeutics to treat dengue [4]. Dengue prevention has and will continue to rely on vector population suppression. To strengthen conventional mosquito control strategies, the development of novel insecticide-free strategies for vector control has been a major focus of research in recent years. The use of natural bacterium Wolbachia infection to reduce the vectorial capacity of Ae. aegypti has shown promising results in a recently completed randomized controlled trial [5]. Other promising approaches include the use of genetic engineering technology to develop vector populations which are refractory to DENV infection [6]. Advances in our understanding of Ae. aegypti at the molecular level will allow us to improve these strategies and use them in combination to effectively reduce the global disease burden of dengue.
The DENV genome is composed of a positive-sense, single-stranded RNA (+ssRNA). The genome is approximately 10.7 kb in length and constitutes a single open reading frame (ORF) flanked with 5′ and 3′ untranslated regions (UTRs). The ORF of the genome encodes three structural proteins (capsid (C), pre-membrane/membrane (prM/M) and envelope (E)) and seven non-structural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5). Given that the DENV genome encodes only 10 proteins, the virus is inevitably reliant on its host dependency (sometimes also referred to as pro-viral) factors (HDFs) to complete its life cycle. At the same time, it has to evade or suppress host restriction (or anti-viral) factors (HRFs) that would otherwise suppress or inhibit infection. The availability of genomic and proteomic tools has enabled the identification of HDFs and HRFs of DENV in mammalian cells [7–12]. These studies have collectively revealed a complex network of DENV-mammalian host interactions as well, summarized in previous reviews [13–17]. While the understanding between DENV and its mammalian host has made remarkable progress, the interaction of DENV with the mosquito host is less well defined.
The life cycle of DENV in a mosquito begins when it ingests a blood meal from a viremic person, whereupon DENV must successfully infect the mosquito midgut before it can disseminate systematically in the mosquito, including to the salivary glands. Once the salivary glands are infected, the mosquito is then capable of transmitting DENV to other humans via its saliva during subsequent blood meals. The virus-mosquito interactions that underpin the DENV life cycle in the mosquito vectors, however, are not well understood even though such critical interactions could shape viral fitness in epidemiological and clinical settings [18, 19]. This knowledge could pave the way for future mechanistic studies, and would be crucial to identify novel targets that may disrupt DENV transmission from the mosquito vectors to humans. Here, we review the literature to establish a snapshot of our current understanding of DENV-mosquito interactions, with a focus on DENV-mosquito omics studies.
Due to the lack of resources to perform functional genomics in both Ae. aegypti and Ae. albopictus, the secondary vector of DENVs, the first high-throughput screen to identify host factors that are required for DENV infection in insects was conducted in Drosophila melanogaster cells [20]. A genome-wide loss of function screen based on RNA interference (RNAi) targeting 22632 genes identified 116 HDFs. These include genes involved in endocytosis, vesicular transport, the unfolded protein response, and RNA-binding proteins potentially involved in viral genome replication, transcription, translation or/and packaging. This study provided the first comprehensive list of DENV HDFs in insects, and identified remarkable conservation of host factors required for DENV infection between human and insect hosts.
DENV-mosquito transcriptomic studies
Innate immune response
With the advent of publicly available complete genome sequences of Ae. aegypti [21] and more recently Ae. albopictus [22], omics studies for DENV infection in its natural vectors became possible. Using microarray and RNA sequencing (RNAseq), various groups have identified differentially expressed genes (DEGs) in DENV-infected mosquito cells and mosquitoes related to innate immunity, metabolism, and stress response. Among the immunity-related genes, the more prominently represented ones include those associated with the Toll pathway and to a lesser extent the JAK-STAT pathway.
The midgut, salivary glands and carcass of Ae. aegypti infected with DENV consistently showed upregulation of genes in the Toll pathway, a major component of the mosquito innate immune system [23–25]. In the follow-up studies, targeted RNAi-based gene knockdown identified the signal transduction adaptor protein MyD88 of the Toll pathway as a HRF whose knockdown increased viral load in DENV-infected Ae. aegypti midgut [26, 27]. Meanwhile, knockdown of its negative regulator Cactus reduced viral load [25]. The importance of the Toll pathway in controlling DENV infection has also been observed across different DENV serotypes and Ae. aegypti strains [26, 27].
On the other hand, DENV is able to inhibit the mosquito innate immune response to favour its infection. Downregulation of several genes related to the Toll pathway was observed at different time-points during DENV infection in Ae. aegypti-derived Aag2 cells [28, 29], Ae. albopictus-derived C6/36 cells [30], and Ae. aegypti midguts and fat bodies [26]. The Toll pathway was also shown to be preferentially inhibited in the salivary glands of mosquitoes infected with DENVs containing the 3′UTR substitutions associated with high subgenomic flavivirus RNA (sfRNA) quantity and high epidemiological fitness [19]. Altogether, these data emphasize the ability of DENV to counteract the mosquito innate immune response against DENV infection.
Apart from the Toll pathway, transcriptional profiling of DENV-infected Ae. aegypti midgut, salivary glands and carcass also revealed upregulation of genes linked to the JAK-STAT pathway as well as the IMD pathway, another two major components in the mosquito innate immune system [23, 25, 31]. Targeted RNAi-mediated gene silencing specifically identified the receptor Dome and the signal transduction adaptor protein Hop/JAK of the JAK-STAT pathway as HRFs whose silencing increased viral load in Ae. aegypti midgut, and the negative regulator PIAS, together with two uncharacterized effectors, as HDFs whose silencing resulted in the opposite effect [32]. The involvement of the JAK-STAT pathway in anti-DENV defence was also observed when experiments were carried out using different Ae. aegypti strains [32]. Similar to the Toll pathway, DENV seems to be able to counteract the JAK-STAT pathway. The presence of DENV secreted NS1 (sNS1) (expressed abundantly in dengue patients) in the mosquito blood meal has been shown to be able to inhibit the JAK-STAT pathway in Ae. aegypti midgut, thus augmenting DENV infection in Ae. aegypti [33].
In contrast to the Toll and JAK-STAT pathways, the roles of the IMD pathway in controlling DENV infection remain elusive. The IMD pathway was shown to be inhibited upon DENV infection in some studies, suggesting its involvement in anti-DENV defence [28, 30]. In contrast, another study showed that while knockdown of the signal transduction adaptor protein IMD of the IMD pathway increased viral load in the midgut of DENV-infected Ae. aegypti, knockdown of its negative regulator Caspar did not reduce viral load in this organ as expected [27]. Further investigations are thus required to elucidate the significance of this innate immune pathway in anti-DENV defence.
There also appears to be crosstalk between the IMD pathway and the small interfering RNA (siRNA) pathway, a part of the RNAi machinery which forms another major component in the mosquito innate immune system [34]. Currently, there are three main RNAi-related pathways characterized in insects [35]: (i) siRNA pathway in which siRNAs are generated from double-stranded RNA (dsRNA) derived from either exogenous sources (such as viruses) or endogenous sources within cells, (ii) microRNA (miRNA) pathway in which miRNAs are generated from cell-encoded transcripts, and (iii) PIWI-interacting RNA (piRNA) pathway in which piRNAs are transcribed from the cell genome. Among the RNAi-related pathways, this review will focus on the exogeneous siRNA pathway which is the most well-characterized pathway for antiviral properties in insects [36]. It was first found that DENV-specific siRNAs were detected in Ae. aegypti midguts during DENV infection, suggesting its involvement in anti-DENV response in the mosquito [37, 38]. Indeed, the inhibition of the siRNA pathway by silencing either of its components, Dcr2, R2D2 or Ago2, increased viral load in Ae. aegypti, shortened viral incubation period, as well as enhanced viral transmissibility [37]. Furthermore, transgenic Ae. aegypti overexpressing DENV-specific dsRNAs showed significant reduction in DENV infection in the midgut, salivary glands as well as saliva, and the interruption of the siRNA pathway eliminated this reduction [39, 40]. Collectively, these findings support the involvement of the siRNA pathway in anti-DENV defence in mosquitoes.
Taken together, while the innate immune pathways are activated during DENV infection, the ability of the virus to evade these innate immune responses will determine whether DENV can successfully complete its life cycle in the mosquito vectors.
Other cellular processes
Besides mosquito innate immunity, a comparative transcriptomic study using DENV-susceptible and -refractory strains of Ae. aegypti revealed that the midgut of the susceptible strain specifically showed upregulation of genes involved in mRNA surveillance, protein processing in the endoplasmic reticulum (ER) and the proteasome [41]. In contrast, the midgut of the refractory strain showed upregulation of genes involved in metabolic processes including the Wnt signalling pathway, the glycolysis pathway and glycan biosynthesis. It would be interesting to further examine if these factors influence vector competence of Ae. aegypti. Meanwhile, another comparative transcriptional profiles of Ae. aegypti infected with DENV and other two flaviviruses including West Nile virus (WNV) and yellow fever virus (YFV) revealed a potentially conserved transcriptomic signature of flavivirus infection [42]. This includes alterations in the expression of genes involved in transport, metabolic processes, ion binding and peptidase activity. Similarly, in DENV-infected Aag2 and C6/36 cells, upregulation of genes involved in starch and sucrose metabolism, pyrimidine metabolism and drug metabolism, and downregulation of genes involved in RNA transport, purine metabolism, drug metabolism, folate biosynthesis, and valine, leucine and isoleucine degradation were observed [28, 30].
Taken together, physiological pathways related to innate immunity, metabolism, energy production and redox activity were major pathways modulated during DENV infection in the mosquito vectors. However, only a limited number of specific genes were concordant across different high-throughput studies. Depending on the species or strains of mosquitoes and infecting DENV, it is important to note that the mosquito transcriptome may also differ accordingly during infection.
DENV-MOSQUITO PROTEOMIC STUDIES
In addition to the transcriptomic studies, proteomic studies have been performed. DENV-infected C6/36 cells significantly increased the expression of proteins involved in the glycolysis pathway and cellular stress response [43]. In another study, proteomic analysis of DENV-infected Ae. aegypti midguts showed an increased production of midgut proteins involved in carbohydrate and lipid metabolism, as well as the production of reactive oxygen species (ROSs) when compared to uninfected midguts [44]. These proteins potentially serve as HDFs that support energy production for viral infection and help DENV to cope with the stress induced by its infection [45]. DENV infection has also been shown to alter the expression of several salivary proteins; in particular proteins with anti-hemostatic and pain inhibitory properties [46]. The changes in the proteome of the mosquito saliva could potentially enhance successful transmission of DENV from an infected mosquito to humans during blood feeding [47].
Apart from the studies focusing on proteomic changes upon DENV infection, the virus overlay protein binding assay (VOPBA) has been used to identify cellular receptors for DENV [48–51]. This approach has identified enolase, cadherin, beta-adrenergic receptor kinase (beta-ARK) and translation elongation factor EF-1 alpha/Tu as potential DENV binding receptors in Ae. aegypti midgut homogenates and C6/36 cell lysates [50]. Other proteins such as actin, orisis, vav-1, prohibitin, ATP synthase β subunit, tubulin β chain and 70 kD heat shock cognate protein (HSC70) were also shown to bind to the membrane fractions of Ae. aegypti-derived A7 cells and C6/36 cells, and the midgut brush border membrane fraction of Ae. aegypti [51]. Independently, another group has also identified HSC70 interacting with DENV in C6/36 cells, together with 78 kDa glucose-regulated protein (GRP78 or BiP), 70 kDa heat shock protein (HSP70) and 40 kDa protein with homology to protein disulfide isomerase (PDI) [52]. However, future work is required to determine if these proteins are indeed required for DENV entry and transport into mosquito cells.
Next, the use of affinity purification assays coupled with proteomic analyses has also expanded the list of DENV-mosquito protein interactions [11, 53]. A tandem affinity purification (TAP) assay followed by proteomic analysis in DENV- or WNV-infected C6/36 cells identified 18 mosquito proteins as potential interacting partners of viral proteins [53]. Using a targeted RNAi-based knockdown approach, actin, myosin, myosin light chain kinase and PI3-kinase were identified as HDFs of DENV and WNV although their roles in the virus life cycle remain unknown. Using a similar method, a more recent study identified 28 conserved host proteins in both humans and mosquitoes as interacting partners of DENV and Zika virus (ZIKV) proteins, of which pharmacological inhibition of SEC61 inhibited both DENV and ZIKV infection in mammalian and mosquito cells [11]. Taken together, these studies showed that comparative proteomic analyses could be utilized to not only identify DENV HDFs, but also HDFs that are shared among flaviviruses in their hosts. These shared HDFs may ultimately serve as targets for broad-acting antivirals, and knowledge of these interactions may prove useful in the design of novel vector control strategies.
DENV-MOSQUITO METABOLOMIC STUDIES
Using a metabolomic analysis, DENV-infected C6/36 cells were shown to induce the production of sphingomyelins, phosphatidylcholine and ceramide which are lipids that can modify the membrane physical properties such as permeability and curvature [54]. While the changes on membrane permeability have been proposed to support the exchange of required components between the cytosol and the ER lumen during DENV replication [54], the changes on membrane curvature could potentially be exploited by DENV, to use the host membranes to form virions or/and to prevent viral RNA detection by the mosquito antiviral defence [55]. This study also found that DENV-infected C6/36 cells increased both lipid anabolism and catabolism. Although their importance on DENV infection remains unclear, lipid anabolism may be required for virus-induced membrane changes, while lipid catabolism might be essential for energy supply for efficient virus production. Recently, studies have further shown that increased production of membrane phospholipids are required to modify mosquito membrane structures to support DENV replication in mosquitoes [56, 57]. Interestingly, another recent study has shown that one of the molecular mechanisms underlying Wolbachia -mediated DENV blocking in Ae. aegypti is lipid modulation [58]. Besides, DENV infection has also been shown to induce the expression of genes encoding lipid-binding proteins – the myeloid differentiation 2-related lipid recognition protein (ML) and the Niemann Pick-type C1 (NPC1) family members – in Ae. aegypti, suggesting a role of these lipid-binding proteins as DENV host factors [24, 25]. Indeed, targeted RNAi-mediated gene silencing of a ML (AaaaegML33) and a NPC1 (AaegNPC1b) gene family member restricted DENV infection in Ae. aegypti midguts [59]. These intricate metabolic interactions between DENV and the mosquito vectors are essential in the viral life cycle and potentially represent targets to control disease transmission. Further work to study these interactions should be explored.
DENV-MOSQUITO INTERACTION NETWORKS
With the advent of computational approaches, the knowledge on DENV-mosquito interactions has been greatly expanded in terms of both prediction of novel virus-host interactions, as well as integration of known virus-host interactions into a network. This aids in the identification of significant host factors and cellular processes required during DENV infection. By using a computational method, 714 interactions between DENV and Ae. aegypti were predicted based on three high-throughput screens including genome-wide RNAi screens, transcriptomic studies and physical protein-protein interactions with the most enriched Ae. aegypti proteins involved in transport, immunity, replication/transcription/translation and metabolism [60]. Another computational study also predicted 176 DENV-Ae. aegypti interactions based on their protein structural similarities, with proteins involved in RNA processing and regulation of stress response being the most enriched [61].
PROSPECTIVE DIRECTIONS FOR RESEARCH
As reviewed above and summarized in Fig. 1, Table 1, while the knowledge on DENV-mosquito interactions is expanding, the interactions critical for DENV fitness remain poorly defined. One major challenge lies in the lack of functional validation performed on the HDFs and HRFs identified in most of these studies. Also, these studies have studied DENV-mosquito interactions using laboratory-passaged DENVs; mutations that accumulate through serial passaging could thus confound interpretation of virus-host interactions necessary for DENV replication and transmission by the mosquito vectors.
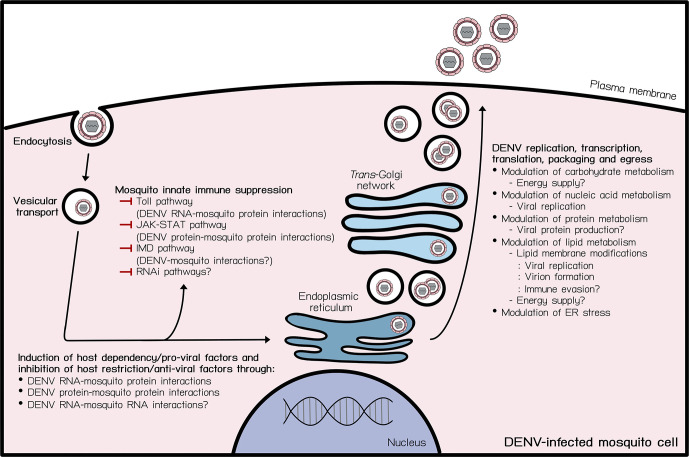
Fig. 1.
DENV-mosquito vector interactions. Schematic overview of current knowledge on DENV-mosquito interactions during the DENV life cycle in a mosquito cell. DENV depends on several interactions between its RNA and proteins with mosquito proteins to complete its life cycle in the mosquito vectors. ? indicates hypothesized relevant cellular processes. —| indicates inhibition. ER indicates endoplasmic reticulum.
Table 1.
Identification of relevant host factors and cellular processes during DENV infection in its mosquito vectors
|
Study |
Methods |
Findings |
References |
|---|---|---|---|
|
Transcriptomic studies | |||
|
Identification of the mosquito innate immune system as a part of anti-DENV defences |
Microarray Digital gene expression RNAi-based gene knockdown Microarray RNAi-based gene knockdown Microarray RNA-sequencing RT-PCR RNA sequencing RT-PCR Northern blot Immunofluorescence Transgenic Ae. aegypti RNAi-based gene knockdown |
Upregulation of genes related to the Toll, JAK-STAT and IMD pathways. The signalling components of the Toll (MyD88) and JAK-STAT (Dome and JAK/Hop) pathways are HRFs while the negative regulators of the Toll (Cactus) and JAK-STAT (PIAS) pathways are HDFs. A conserved function in anti-DENV defence of the Toll and JAK-STAT pathways across different Ae. aegypti strains. Downregulation of genes related to the Toll and IMD pathways following DENV infection. The ability of DENV sfRNA and sNS1 to inhibit the Toll and JAK-STAT pathways, respectively. The presence of the siRNA pathway and its importance in anti-DENV defence in Ae. aegypti. |
[30] [26, 28, 29], [19, 33] [37–40] |
|
Identification of other relevant cellular processes during virus infection through comparative transcriptomic studies |
Microarray Microarray RNA-sequencing |
A potential role of differentially transcriptomic changes underlying vector competence of Ae. aegypti. - DENV-susceptible Ae. aegypti strain: upregulation of genes involved in the proteasome, mRNA surveillance and protein processing in the ER. DENV-refractory Ae. aegypti strain: upregulation of genes involved in the Wnt signalling pathway, the glycolysis pathway and glycan biosynthesis. A potentially conserved transcriptomic signature of flavivirus infection. - Alteration in the expression of genes involved in metabolic processes, peptidase activity, ion binding and transport during DENV, WNV and YFV infection. A consistent upregulation of genes involved in starch and sucrose metabolism, pyrimidine metabolism and drug metabolism, and downregulation of genes involved in RNA transport, purine metabolism, drug metabolism, folate biosynthesis, and valine, leucine and isoleucine degradation in DENV-infected Aag2 and C6/36 cells. |
[41] [42] |
|
Proteomic studies | |||
|
Proteomic changes during DENV infection |
2D-DIGE MALDI-TOF MS 2D-DIGE MALDI-TOF/TOF MS 2DE LC-MS/MS |
Upregulation of proteins involved in the glycolysis pathway and the cellular stress response in C6/36 cells. Increased production of proteins involved in carbohydrate and lipid metabolism as well as the production of reactive oxygen species (ROSs) in Ae. aegypti midguts. Alteration in the expression of proteins; in particular proteins with anti-hemostatic and pain inhibitory properties in Ae. aegypti salivary glands. |
[43] [44] [46] |
|
Identification of potential cellular receptors |
VOPBA Mass spectrometry VOPBA Mass spectrometry Co-purification Mass spectrometry |
With the use of Ae. aegypti midgut homogenates and C6/36 cell lysates, cadherin, enolase, beta-adrenergic receptor kinase (beta-ARK) and translation elongation factor EF-1 alpha/Tu were identified. With the use of the membrane fractions of A7 cells, C6/36 cells and Ae. aegypti midguts, actin, orisis, vav-1, prohibitin, ATP synthase β subunit, tubulin β chain, and 70-kD heat shock cognate protein (HSC70) were identified. With the use of C6/36 cell lysates, HSC70, 78 kDa glucose-regulated protein (GRP78 or BiP), 70 kDa heat shock protein (HSP70) and 40 kDa protein with homology to protein disulfide isomerase (PDI) were identified. |
[50] [51] [52] |
|
Identification of DENV-mosquito protein interactions |
Tandem affinity purification LC-MS/MS RNAi-based gene knockdown Affinity purification LC-MS/MS Pharmacological inhibition |
18 mosquito proteins as potential interacting partners of DENV and WNV proteins. Actin, myosin, myosin light chain kinase and PI3-kinase are DENV and WNV HDFs. 28 host proteins both humans and mosquitoes as interacting partners of DENV and ZIKV proteins. SEC61 is a shared DENV and ZIKV HDF in both mammalian and mosquito cells. |
[53]. [11] |
|
Metabolomic studies | |||
|
Lipidomic changes during DENV infection |
LC-MS LC-HRMS RNAi-based gene knockdown |
Upregulation of lipid anabolism and catabolism. Enrichment of lipids that can modify the physical properties of membranes. Increased production of membrane phospholipids which are required to modify mosquito membrane structures to support DENV replication. |
[54] |
|
Potential importance of lipid modulation during DENV infection |
LC-MS RNAi-based gene knockdown Microarray RNAi-based gene knockdown |
Identification of lipid modulation as a molecular mechanism underlying Wolbachia -mediated DENV blocking in Ae. aegypti. Upregulation of genes encoding lipid-binding proteins – the myeloid differentiation 2-related lipid recognition protein (ML) and the Niemann Pick-type C1 (NPC1) family members in Ae. aegypti, and silencing of those gene family members restricted DENV infection in Ae. aegypti midgut. |
[58] |
|
Computational studies | |||
|
Establishment of DENV-mosquito interaction networks |
Computational approach based on available data from genome-wide RNAi screens, transcriptomic studies and physical protein-protein interactions Computational approach based on structural similarity of DENV and Ae. aegypti proteins |
714 DENV-Ae. aegypti interactions with Ae. aegypti proteins involved in transport, immunity, metabolism and replication/transcription/translation being the most enriched. 176 DENV-Ae. aegypti interactions with Ae. aegypti proteins involved in RNA processing and regulation of stress response being the most enriched. |
[60] [61] |
2D-DIGE, two-dimensional differential in-gel electrophoresis; 2DE, two-dimensional gel electrophoresis; LC-HRMS, liquid chromatography-high resolution mass spectrometry; LC-MS, liquid chromatography-mass spectrometry; LC-MS/MS, liquid chromatography-tandem mass spectrometry; MALDI-TOF MS, matrix assisted laser desorption ionisation time-of-flight mass spectrometry; MALDI-TOF/TOF MS, matrix assisted laser desorption ionisation time-of-flight/time-of-flight mass spectrometry; RNAi, RNA interference; RT-PCR, real-time polymerase chain reaction; VOPBA, virus overlay protein binding assay.
To date, only a limited number of studies have dissected DENV-mosquito interactions by comparing virulent and clinically proven attenuated DENVs. Studies have shown that genetic differences in the DENV genomes play a critical role in determining viral fitness both epidemiologically and clinically [18, 19, 62–66]. In mosquitoes, using a clinically tested vaccine strain DENV-2 PDK53, which differs from the wild-type clinical isolate DENV-2 16681 genetically by only nine nucleotides, our laboratory has recently showed that the change of glycine to aspartic acid at position 53 of the NS1 protein (NS1 G53D) attenuated 16681 infection in Ae. aegypti; NS1 G53D infection in the mosquito midgut is confined by the early induction of the mosquito immune response similar to PDK53 [18]. Besides the midgut, this early innate immune response also restricts viral dissemination to the other parts of Ae. aegypti including the salivary glands, impeding productive infection and subsequently successful DENV transmission by the mosquito.
Another study in mosquitoes compared DENV-2 strains with different epidemic potential in Puerto Rico. Nucleotide substitutions in the 3′UTR of an epidemiologically fitter strain of DENV-2 that had caused an outbreak in Puerto Rico in 1994 resulted in increased sfRNA production in Ae. aegypti salivary glands and thus increased virus transmission, at least in part, due to innate immune suppression in this organ [19], when compared to DENV-2 isolated before the outbreak. Altogether, these studies highlight that the use of DENVs with different clinical or epidemiological fitness with known genetic differences could enable a more targeted approach to efficiently identify critical DENV-mosquito interactions that govern viral fitness.
In addition, DENV exists as a collection of closely related genomes, or quasispecies due to the error prone nature of their replication machinery, the RNA dependent RNA polymerase (RdRp) enzyme. Genome diversity has previously been shown to contribute to changes in viral fitness and pathogenesis [67]. Recently, ultra-sequencing has been used to identify beneficial mutations that may facilitate host-specific DENV adaption, and enabled the assessment of the role of these mutants in viral fitness [68]. Elucidating how these mutants influence virus-mosquito interactions could also shed light into HDFs that will be the key to conceive new antiviral approaches.
To conclude, this review provides a brief summary of DENV-mosquito interactions with a focus on omics studies. It is likely that this body of information represents only the tip of the iceberg of the total DENV-mosquito interactome. Studies that are on-going and planned for the future will likely shed more light on the complexity of the DENV-mosquito interplay in the coming years. The use of DENVs with known genetic differences would significantly progress the identification of critical virus-mosquito interactions required for viral fitness and successful virus transmission, which will be useful for the identification of antiviral targets as well as the development of transgenic mosquitoes that are resistant to DENV infection.
Funding information
This work was funded by the Ministry of Education Tier 3 Grant (Singapore) and the Open-Fund-Young Individual Research Grant administered by the National Medical Research Council of Singapore.
Acknowledgement
We thank Nuchvara Chuenjit for help with illustration of the figure.
Author contributions
Conceptualization: T.S., E.E.O., M.M.C.; Writing - original draft preparation: T.S.; Writing - review and editing: T.S., E.E.O., M.M.C.; Funding: R.M.K., E.E.O, M.M.C.
Conflicts of interest
E.E.O. served as a dengue vaccine advisory board member for Takeda Vaccines, which uses DENV2 PDK53 strain as a component of their dengue vaccine candidate, TAK-003.
Footnotes
Abbreviations: Ae. aegypti, Aedes aegypti; Ae. albopictus, Aedes albopictus; Beta-ARK, beta-adrenergic receptor kinase; BiP, binding immunoglobulin protein; C, capsid; DEG, differentially expressed gene; DENV, dengue virus; Dome, Domeless; dsRNA, double-stranded RNA; E, envelope; ER, endoplasmic reticulum; GRP78, 78-kDa glucose-regulated protein; HDF, host dependency factor; Hop/JAK, Hopscotch/Janus kinase; HRF, host restriction factor; HSC70, 70-kDa heat shock cognate protein; HSP70, 70-kDa heat shock protein; IMD, immune deficiency; JAK-STAT, Janus kinase signal transducers and activators of transcription; miRNA, microRNA; ML, myeloid differentiation 2-related lipid recognition protein; MyD88, myeloid differentiation primary response 88; NPC1, Niemann Pick-type C1; NS, non-structural; ORF, open reading frame; PDI, protein disulfide isomerase; PIAS, protein inhibitor of activated STAT; PI3-kinase, phosphoinositide 3-kinase; piRNA, PIWI-interacting RNA; prM/M, premembrane/membrane; RdRp, RNA-dependent RNA polymerase; RNA, ribonucleic acid; RNAi, RNA interference; ROS, reactive oxygen species; sfRNA, subgenomic flavivirus RNA; siRNA, small interfering RNA; sNS1, secreted NS1; +ssRNA, positive-sense, single-stranded RNA; TAP, tandem affinity purification; UTR, untranslated region; VOPBA, virus overlay protein binding assay; WNV, West Nile virus; YFV, yellow fever virus; ZIKV, Zika virus.
References
- 1.Kraemer MU, Sinka ME, Duda KA, Mylne AQ, Shearer FM, et al. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus . eLife. 2015;4 doi: 10.7554/eLife.08347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brady OJ, Gething PW, Bhatt S, Messina JP, Brownstein JS, et al. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl Trop Dis. 2012;6:e1760. doi: 10.1371/journal.pntd.0001760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization (WHO Dengue and severe dengue: Fact Sheet. 2019.
- 4.World Health Organization Revised SAGE recommendation on use of dengue vaccine. 2018.
- 5.Utarini A, Indriani C, Ahmad RA, Tantowijoyo W, Arguni E, et al. Efficacy of wolbachia-infected mosquito deployments for the control of dengue. N Engl J Med. 2021;384:2177–2186. doi: 10.1056/NEJMoa2030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jupatanakul N, Sim S, Angleró-Rodríguez YI, Souza-Neto J, Das S, et al. Engineered Aedes aegypti JAK/STAT pathway-mediated immunity to dengue virus. PLoS Negl Trop Dis. 2017;11:e0005187. doi: 10.1371/journal.pntd.0005187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu HC, Hannemann H, Heesom KJ, Matthews DA, Davidson AD. High-throughput quantitative proteomic analysis of dengue virus type 2 infected A549 cells. PLoS One. 2014;9:e93305. doi: 10.1371/journal.pone.0093305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hafirassou ML, Meertens L, Umaña-Diaz C, Labeau A, Dejarnac O, et al. A global interactome map of the dengue virus NS1 identifies virus restriction and dependency host factors. Cell Reports. 2017;21:3900–3913. doi: 10.1016/j.celrep.2017.11.094. [DOI] [PubMed] [Google Scholar]
- 9.Marceau CD, Puschnik AS, Majzoub K, Ooi YS, Brewer SM, et al. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature. 2016;535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ooi YS, Majzoub K, Flynn RA, Mata MA, Diep J, et al. An RNA-centric dissection of host complexes controlling flavivirus infection. Nat Microbiol. 2019;4:2369–2382. doi: 10.1038/s41564-019-0518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah PS, Link N, Jang GM, Sharp PP, Zhu T, et al. Comparative flavivirus-host protein interaction mapping reveals mechanisms of dengue and zika virus pathogenesis. Cell. 2018;175:1931–1945.:S0092-8674(18)31553-8. doi: 10.1016/j.cell.2018.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wongtrakul J, Thongtan T, Pannengpetch S, Wikan N, Kantamala D, et al. Phosphoproteomic analysis of dengue virus infected U937 cells and identification of pyruvate kinase M2 as a differentially phosphorylated phosphoprotein. Sci Rep. 2020;10:14493. doi: 10.1038/s41598-020-71407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acosta EG, Kumar A, Bartenschlager R. Advances in Virus Research. Cambridge, Massachusetts: Academic Press; 2014. Revisiting dengue virus-host cell interaction: New insights into molecular and cellular virology; pp. 1–109. [DOI] [PubMed] [Google Scholar]
- 14.Campos RK, Garcia-Blanco MA, Bradrick SS. Current Topics in Microbiology and Immunology. Springer, Cham; 2018. Roles of pro-viral host factors in mosquito-borne flavivirus infections. [DOI] [PubMed] [Google Scholar]
- 15.Krishnan MN, Garcia-Blanco MA. Targeting host factors to treat West Nile and dengue viral infections. Viruses. 2014;6:683–708. doi: 10.3390/v6020683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salazar MI, del Angel RM, Lanz-Mendoza H, Ludert JE, Pando-Robles V. The role of cell proteins in dengue virus infection. J Proteomics. 2014;111:6–15. doi: 10.1016/j.jprot.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Trammell CE, Goodman AG. Host factors that control mosquito-borne viral infections in humans and their vector. Viruses. 2021;13:748. doi: 10.3390/v13050748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choy MM, Ng DHL, Siriphanitchakorn T, Ng WC, Sundstrom KB, et al. A non-structural 1 protein G53D substitution attenuates a clinically tested live dengue vaccine. Cell Reports. 2020;31:107617. doi: 10.1016/j.celrep.2020.107617. [DOI] [PubMed] [Google Scholar]
- 19.Pompon J, Manuel M, Ng GK, Wong B, Shan C, et al. Dengue subgenomic flaviviral RNA disrupts immunity in mosquito salivary glands to increase virus transmission. PLoS Pathog. 2017;13:e1006535. doi: 10.1371/journal.ppat.1006535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nene V, Wortman JR, Lawson D, Haas B, Kodira C, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X-G, Jiang X, Gu J, Xu M, Wu Y, et al. Genome sequence of the Asian tiger mosquito, aedes albopictus, reveals insights into its biology, genetics, and evolution. Proc Natl Acad Sci U S A. 2015;112:E5907–15. doi: 10.1073/pnas.1516410112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luplertlop N, Surasombatpattana P, Patramool S, Dumas E, Wasinpiyamongkol L, et al. Induction of a peptide with activity against a broad spectrum of pathogens in the Aedes aegypti salivary gland, following infection with dengue virus. PLoS Pathog. 2011;7:e1001252. doi: 10.1371/journal.ppat.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sim S, Ramirez JL, Dimopoulos G, Diamond MS. Dengue virus infection of the Aedes aegypti Salivary gland and chemosensory apparatus induces genes that modulate infection and blood-feeding behavior. PLoS Pathog. 2012;8:e1002631. doi: 10.1371/journal.ppat.1002631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xi Z, Ramirez JL, Dimopoulos G, Schneider DS. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008;4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramirez JL, Dimopoulos G. The Toll immune signaling pathway control conserved anti-dengue defenses across diverse Ae. aegypti strains and against multiple dengue virus serotypes. Dev Comp Immunol. 2010;34:625–629. doi: 10.1016/j.dci.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sim S, Jupatanakul N, Ramirez JL, Kang S, Romero-Vivas CM, et al. Transcriptomic profiling of diverse Aedes aegypti strains reveals increased basal-level immune activation in dengue virus-refractory populations and identifies novel virus-vector molecular interactions. PLoS Negl Trop Dis. 2013;7:e2295. doi: 10.1371/journal.pntd.0002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li MJ, Lan CJ, Gao HT, Xing D, Gu ZY, et al. Transcriptome analysis of aedes aegypti aag2 cells in response to dengue virus-2 infection. Parasit Vectors. 2020;13:421. doi: 10.1186/s13071-020-04294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sim S, Dimopoulos G. Dengue virus inhibits immune responses in Aedes aegypti cells. PLoS One. 2010;5:e10678. doi: 10.1371/journal.pone.0010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Xing D, Su D, Wang D, Gao H, et al. Transcriptome analysis of responses to dengue virus 2 infection in aedes albopictus (Skuse) c6/36 cells. Viruses. 2021;13:343. doi: 10.3390/v13020343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Behura SK, Gomez-Machorro C, Harker BW, deBruyn B, Lovin DD, et al. Global cross-talk of genes of the mosquito Aedes aegypti in response to dengue virus infection. PLoS Negl Trop Dis. 2011;5:e1385. doi: 10.1371/journal.pntd.0001385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Souza-Neto JA, Sim S, Dimopoulos G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc Natl Acad Sci U S A. 2009;106:17841–17846. doi: 10.1073/pnas.0905006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J, Liu Y, Nie K, Du S, Qiu J, et al. Flavivirus NS1 protein in infected host sera enhances viral acquisition by mosquitoes. Nat Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paradkar PN, Duchemin JB, Voysey R, Walker PJ. Dicer-2-dependent activation of culex vago occurs via the TRAF-Rel2 signaling pathway. PLoS Negl Trop Dis. 2014;8:e2823. doi: 10.1371/journal.pntd.0002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonning BC, Saleh MC. The Interplay between viruses and RNAi pathways in insects. Annu Rev Entomol. 2021;66:61–79. doi: 10.1146/annurev-ento-033020-090410. [DOI] [PubMed] [Google Scholar]
- 36.Kingsolver MB, Huang Z, Hardy RW. Sect antiviral innate immunity: Pathways, effectors, and connections. J Mol Biol. 2013;425:4921–4936.:S0022-2836(13)00632-3. doi: 10.1016/j.jmb.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sánchez-Vargas I, Scott JC, Poole-Smith BK, Franz AWE, Barbosa-Solomieu V, et al. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito’s RNA interference pathway. PLoS Pathog. 2009;5:e1000299. doi: 10.1371/journal.ppat.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott JC, Brackney DE, Campbell CL, Bondu-Hawkins V, Hjelle B, et al. Comparison of dengue virus type 2-specific small RNAs from RNA interference-competent and –incompetent mosquito cells. PLoS Negl Trop Dis. 2010;4:e848. doi: 10.1371/journal.pntd.0000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franz AWE, Sanchez-Vargas I, Adelman ZN, Blair CD, Beaty BJ, et al. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proceedings of the National Academy of Sciences. 2006;103:4198–4203. doi: 10.1073/pnas.0600479103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathur G, Sanchez-Vargas I, Alvarez D, Olson KE, Marinotti O, et al. Transgene-mediated suppression of dengue viruses in the salivary glands of the yellow fever mosquito, Aedes aegypti . Insect Mol Biol. 2010;19:753–763. doi: 10.1111/j.1365-2583.2010.01032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chauhan C, Behura SK, deBruyn B, Lovin DD, Harker BW, et al. Comparative expression profiles of midgut genes in dengue virus refractory and susceptible Aedes aegypti across critical period for virus infection. PLoS ONE. 2012;7:e47350. doi: 10.1371/journal.pone.0047350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colpitts TM, Cox J, Vanlandingham DL, Feitosa FM, Cheng G, et al. Alterations in the Aedes aegypti transcriptome during infection with west nile, dengue and yellow fever viruses. PLoS Pathog. 2011;7:e1002189. doi: 10.1371/journal.ppat.1002189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patramool S, Surasombatpattana P, Luplertlop N, Sévéno M, Choumet V, et al. Proteomic analysis of an Aedes albopictus cell line infected with Dengue serotypes 1 and 3 viruses. Parasites Vectors. 2011;4 doi: 10.1186/1756-3305-4-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tchankouo-Nguetcheu S, Khun H, Pincet L, Roux P, Bahut M, et al. Differential protein modulation in midguts of Aedes aegypti infected with chikungunya and dengue 2 viruses. PLoS One. 2010;5:e13149. doi: 10.1371/journal.pone.0013149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mehrbod P, Ande SR, Alizadeh J, Rahimizadeh S, Shariati A, et al. The roles of apoptosis, autophagy and unfolded protein response in arbovirus, influenza virus, and HIV infections. Virulence. 2019;10:376–413. doi: 10.1080/21505594.2019.1605803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chisenhall DM, Christofferson RC, McCracken MK, Johnson AMF, Londono-Renteria B, et al. fection with dengue-2 virus alters proteins in naturally expectorated saliva of Aedes aegypti mosquitoes. Parasites Vectors. 2014;7 doi: 10.1186/1756-3305-7-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fontaine A, Diouf I, Bakkali N, Missé D, Pagès F, et al. Implication of haematophagous arthropod salivary proteins in host-vector interactions. Parasites Vectors. 2011;4 doi: 10.1186/1756-3305-4-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chee HY, AbuBakar S. Identification of a 48kDa tubulin or tubulin-like C6/36 mosquito cells protein that binds dengue virus 2 using mass spectrometry. Biochem Biophys Res Commun. 2004;320:11–17. doi: 10.1016/j.bbrc.2004.05.124. [DOI] [PubMed] [Google Scholar]
- 49.Mercado-Curiel RF, Esquinca-Avilés HA, Tovar R, Díaz-Badillo A, Camacho-Nuez M, et al. The four serotypes of dengue recognize the same putative receptors in Aedes aegypti midgut and Ae. albopictus cells. BMC Microbiol. 2006;6:85. doi: 10.1186/1471-2180-6-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muñoz M de L, Limón-Camacho G, Tovar R, Diaz-Badillo A, Mendoza-Hernández G, et al. Proteomic identification of dengue virus binding proteins in Aedes aegypti mosquitoes and Aedes albopictus cells. BioMed Research International. 2013;2013:1–11. doi: 10.1155/2013/875958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paingankar MS, Gokhale MD, Deobagkar DN. Dengue-2-virus-interacting polypeptides involved in mosquito cell infection. Arch Virol. 2010;155:1453–1461. doi: 10.1007/s00705-010-0728-7. [DOI] [PubMed] [Google Scholar]
- 52.Vega-Almeida TO, Salas-Benito M, De Nova-Ocampo MA, Del Angel RM, Salas-Benito JS. Surface proteins of C6/36 cells involved in dengue virus 4 binding and entry. Arch Virol. 2013;158:1189–1207. doi: 10.1007/s00705-012-1596-0. [DOI] [PubMed] [Google Scholar]
- 53.Colpitts TM, Cox J, Nguyen A, Feitosa F, Krishnan MN, et al. Use of a tandem affinity purification assay to detect interactions between West Nile and dengue viral proteins and proteins of the mosquito vector. Virology. 2011;417:179–187. doi: 10.1016/j.virol.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perera R, Riley C, Isaac G, Hopf-Jannasch AS, Moore RJ, et al. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog. 2012;8:e1002584. doi: 10.1371/journal.ppat.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.den Boon JA, Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol. 2010;64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 56.Vial T, Tan W-L, Wong Wei Xiang B, Missé D, Deharo E, et al. Dengue virus reduces AGPAT1 expression to alter phospholipids and enhance infection in Aedes aegypti. PLoS Pathog. 2019;15:e1008199. doi: 10.1371/journal.ppat.1008199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vial T, Tan WL, Deharo E, Missé D, Marti G, et al. Mosquito metabolomics reveal that dengue virus replication requires phospholipid reconfiguration via the remodeling cycle. Proc Natl Acad Sci U S A. 2020;117:27627–27636. doi: 10.1073/pnas.2015095117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koh C, Islam MN, Ye YH, Chotiwan N, Graham B, et al. Dengue virus dominates lipid metabolism modulations in Wolbachia-coinfected Aedes aegypti . Commun Biol. 2020;3:518. doi: 10.1038/s42003-020-01254-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jupatanakul N, Sim S, Dimopoulos G. Aedes aegypti ML and Niemann-Pick type C family members are agonists of dengue virus infection. Dev Comp Immunol. 2014;43:1–9.:S0145-305X(13)00293-0. doi: 10.1016/j.dci.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo X, Xu Y, Bian G, Pike AD, Xie Y, et al. Response of the mosquito protein interaction network to dengue infection. BMC Genomics. 2010;11:380. doi: 10.1186/1471-2164-11-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doolittle JM, Gomez SM. Mapping protein interactions between dengue virus and its human and insect hosts. PLoS Negl Trop Dis. 2011;5:e954. doi: 10.1371/journal.pntd.0000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bustos-Arriaga J, Gromowski GD, Tsetsarkin KA, Firestone C-. Y, Castro-Jiménez T, et al. Decreased accumulation of subgenomic RNA in human cells infected with vaccine candidate DEN4Δ30 increases viral susceptibility to type I interferon. Vaccine. 2018;36:3460–3467.:S0264-410X(18)30607-8. doi: 10.1016/j.vaccine.2018.04.087. [DOI] [PubMed] [Google Scholar]
- 63.Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science. 2015;350:217–221. doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodpai A, Pulmanausahakul R, Midoeng P, Ubol S, Yoksan S. Contribution of monocyte-derived dendritic cells infected with virulent- or attenuateddengue virus to tnf-α reactogenicity and association of il-1β and il-8 with attenuated phenotype. 2018. p. 49. p. [Google Scholar]
- 65.Sessions OM, Tan Y, Goh KC, Liu Y, Tan P, et al. Host cell transcriptome profile during wild-type and attenuated dengue virus infection. PLoS Negl Trop Dis. 2013;7:e2107. doi: 10.1371/journal.pntd.0002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Syenina A, Vijaykrishna D, Gan ES, Tan HC, Choy MM, et al. Positive epistasis between viral polymerase and the 3’ untranslated region of its genome reveals the epidemiologic fitness of dengue virus. Proc Natl Acad Sci U S A. 2020;117:11038–11047. doi: 10.1073/pnas.1919287117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dolan PT, Taguwa S, Rangel MA, Acevedo A, Hagai T, et al. Elife; 2021. Principles of dengue virus evolvability derived from genotype-fitness maps in human and mosquito cells. [DOI] [PMC free article] [PubMed] [Google Scholar]