Abstract
Geminiviruses constitute the largest group of known plant viruses and cause devastating losses to a wide range of crops and woody plants globally. Mulberry mosaic dwarf‐associated virus (MMDaV), identified from Chinese mulberry trees via small RNA‐based deep sequencing, is a divergent monopartite geminivirus belonging to the genus Mulcrilevirus of the family Geminiviridae. Previous studies have shown that plants employ multiple layers of defence to protect themselves from geminivirus infection. The interplay between plant and MMDaV is nevertheless less studied. This study presents evidence that MMDaV triggers hypersensitive response (HR)‐mediated antiviral defence in Nicotiana benthamiana plants. We show that the RepA protein of MMDaV is engaged in HR‐type cell death induction. We find that the RepA mutants with compromised nuclear localization ability impair their capabilities of cell death induction. Virus‐induced gene silencing of the key components of the R protein‐mediated signalling pathway reveals that down‐regulation of the nucleus‐targeting NbWRKY1 alleviates the cell death induction activity of RepA. We further demonstrate that RepA up‐regulates the transcript level of NbWRKY1. Furthermore, expression of RepA in N. benthamiana confers plant resistance against two begomoviruses. We propose that plant resistance against RepA can be potentially used to improve plant defence against geminiviruses in crops.
Keywords: antiviral response, cell death, geminivirus, MMDaV, RepA, WRKY
A group I WRKY transcription factor is up‐regulated by mulberry mosaic dwarf‐associated virus (MMDaV) and modulates MMDaV‐triggered cell death in Nicotinana benthamiana.
1. INTRODUCTION
Plant viruses are obligate intracellular pathogens that are constantly causing significant losses to crop production and are continuously threatening global food production (Jones & Naidu, 2019). With a limited coding capacity, plant viruses depend on the hosts for replication, intracellular and intercellular movement, and long‐distance trafficking. Exposure to plant virus infection enables plants to develop sophisticated and efficient antiviral defence systems to respond properly at various levels (Boualem et al., 2016; Nicaise, 2014; Yang et al., 2019; Zhao & Li, 2021). A better understanding of plant defences against virus infection may help to develop novel antiviral strategies for disease control.
One successful mechanism of plant immunity against viral pathogens is antiviral RNA silencing, an evolutionarily conserved sequence‐specific mechanism that has been well demonstrated to be effective against both DNA and RNA viruses (Jin et al., 2020). In plants, Dicer‐like proteins, Argonautes, double‐stranded RNA‐binding proteins, and RNA‐dependent RNA polymerases constitute the main components of the antiviral RNA silencing pathway, which involves the production of virus‐derived small interfering RNAs or the formation of RNA‐induced silencing complexes that guide the degradation of viral RNAs. As a counterdefence, plant viruses encode suppressors of RNA silencing to manipulate the antiviral RNA silencing pathway at both the transcriptional and posttranscriptional levels (Csorba et al., 2015; Jin et al., 2020). Hypersensitive response (HR) is another common early plant antiviral defence mechanism that is characterized by rapid programmed cell death at the site of pathogen infection and in the neighbouring cells. Once initiated, HR is associated with several features, including a burst of reactive oxygen species and nitric oxide, reinforcement of cell walls, accumulation of plant hormones such as salicylic acid (SA), jasmonic acid (JA), or ethylene (ET), and activation of pathogenesis‐related genes (Dangl et al., 1996). After initiation, the HR signals can be transmitted to noninfected cells and induce systemic acquired resistance that confers broad‐spectrum resistance against diverse pathogens (Balint‐Kurti, 2019). Despite HR being often beneficial to necrotrophs that acquire nutrition from dead host tissues, it is generally related to host resistance to biotrophic pathogens that require living host tissues for survival. For plant viruses, HR is closely coupled with plant resistance, which can often be induced by virus‐encoded avirulence determinants (Avr) that are recognized by plant R proteins. The majority of the cloned R‐genes possess a nucleotide‐binding site (NBS) and/or leucine‐rich (LRR) repeat domains (NB‐LRRs or NLRs) that recognize the corresponding Avr factor in a “gene‐for‐gene” manner (Balint‐Kurti, 2019). The examples include the recognition of the Magnaporthe Avr‐Pita by rice Pi‐ta, Pseudomonas AvrPto and AvrPtoB by tomato Pto, the Cladosporium fulvum Avr2 by tomato Cf‐2, and tobacco mosaic virus p50 by the N gene (Caplan et al., 2008; Kumar et al., 2021). To date, several components, such as NDR1, RAR1, NTF6, WRKY1, and MEK2, are believed to comprise part of the signalling cascade to initiate R‐gene activation and result in HR (Liu et al., 2004).
Geminiviruses are plant‐infecting DNA viruses that encapsidate their single‐stranded DNA genome(s) in twinned icosahedral particles. They can be transmitted by whiteflies, aphids, leafhoppers, or treehoppers to cause devastating diseases worldwide and are emerging as a major constraint for the productivity of a great variety of herbaceous plants and an increasing number of woody plants such as citrus, apple, grapevine, and mulberry trees (Al Rwahnih et al., 2013; Loconsole et al., 2012; Ma et al., 2015; Qiu et al., 2020). Based on genomic organization, insect vectors, genome‐wide pairwise sequence identities, and host ranges, geminiviruses have been classified into 14 genera (Becurtovirus, Begomovirus, Capularvirus, Citlodavirus, Curtovirus, Eragrovirus, Grablovirus, Maldovirus, Mastrevirus, Mulcrilevirus, Opunvirus, Topilevirus, Topocuvirus, and Turncurtovirus) (Varsani et al., 2017; Zerbini et al., 2017; https://talk.ictvonline.org/taxonomy/). Owing to their small and compact genome, geminiviruses encode few viral products. The replication‐associated protein (Rep) encoded from the complementary sense transcript is the sole protein indispensable for the replication of geminiviruses belonging to the genera Begomovirus, Curtovirus, Maldovirus, Opunvirus, Turncurtovirus, Eragrovirus, and Topocuvirus. However, the Rep protein of geminiviruses in the genera Becurtovirus, Capulavirus, Citlodavirus, Mastrevirus, Mulcrilevirus, Grablovirus, and Topilevirus is produced from a spliced intron‐containing complementary strand transcript. Unlike Rep, the replication‐associated protein A (RepA), a c.30 kDa protein unique to these geminiviruses, is expressed from the unspliced transcript. RepA shares identical primary sequences in approximately 200 N‐terminal amino acid residues with Rep where both proteins contain the three amino acid motifs (RCR‐I, ‐II, and ‐III) required for initiation of rolling‐circle replication (Fondong, 2013). Although the RepA protein of mastreviruses is reported to be involved in the creation of a favourable cellular environment for virus replication, and the RepA protein of two mastreviruses, oat dwarf virus (ODV) and bean yellow dwarf virus, was shown to activate HR‐like cell death in Nicotiana benthamiana plants (Diamos & Mason, 2018; Qian et al., 2015), the function of the RepA protein of other geminiviruses has not been investigated.
Mulberry mosaic dwarf‐associated virus (MMDaV) is a highly divergent monopartite geminivirus belonging to the genus Mulcrilevirus. The genome of MMDaV encodes seven open reading frames (ORFs), five on the virion sense (V1, V2, V3, V4, and V5) and two on the complementary sense (C1/RepA and C2/Rep) (Ma et al., 2015). Similar to viruses derived from the genera Becurtovirus, Capulavirus, Citlodavirus, Mastrevirus, Grablovirus, and Topilevirus, RepA and Rep proteins of MMDaV are expressed from unspliced (C1 transcript) and spliced (C1:C2 transcript) viral RNAs, respectively. In a previous study, we identified that V2 of MMDaV functions as an RNA silencing suppressor to inhibit RNA silencing‐based plant antiviral immunity (Yang et al., 2018). In this study, we constructed an infectious clone of MMDaV and found that MMDaV induces HR‐like resistance response in N. benthamiana plants. We show that the RepA protein of MMDaV triggers HR‐like cell death in N. benthamiana plants and localization to the nucleus is required for RepA to elicit HR. We also provide evidence that WRKY1 is involved in the defence response against MMDaV. Furthermore, expression of RepA could confer plant resistance against two begomoviruses. We propose that plant resistance against RepA can be potentially used to improve plant defence against geminiviruses in crops.
2. RESULTS
2.1. MMDaV induces cell death in N. benthamiana
As the long lifespan of mulberry trees hinders the study of the pathogenesis of MMDaV, we took advantage of the N. benthamiana plants that have been widely used as an excellent model plant to investigate plant–virus interactions. We set out to construct an infectious clone of MMDaV harbouring two tandem repeats of MMDaV. Agroinfiltration of the infectious MMDaV clone into 4‐week‐old N. benthamiana plants induced HR‐like cell death on the infiltrated patches of N. benthamiana leaves at 3 days postinfiltration (dpi), as visualized under visible light and ultraviolet (UV) conditions (Figure 1a). Because HR accompanies reactive oxygen species generation, including H2O2, we further carried out 3,3′‐diaminobenzidine (DAB) staining to examine the accumulation of H2O2 in the infiltrated N. benthamiana leaves. Compared with the leaves infiltrated with Agrobacterium containing 35S‐GFP, leaves infiltrated with the MMDaV infectious clone displayed brown patches (Figure 1a). Measurement of the electrolyte leakage, a common definition of cell death, from the infiltrated N. benthamiana leaf patches, showed that the ion leakage of MMDaV‐induced necrotic leaves was significantly higher than that of control leaves (Figure 1b). PCR analysis of viral DNA from infiltrated leaf patches at 36 dpi by using MMDaV‐specific primers indicated the accumulation of viral DNA in infiltrated leaf patches (Figure 1c). However, analysis of infiltrated N. benthamiana plants at 30 dpi showed that systemic leaves of all 30 infiltrated plants were free of MMDaV (data not shown). These results suggest that the infectious MMDaV clone triggers a host immune response coupled with HR‐like cell death in N. benthamiana plants.
FIGURE 1.
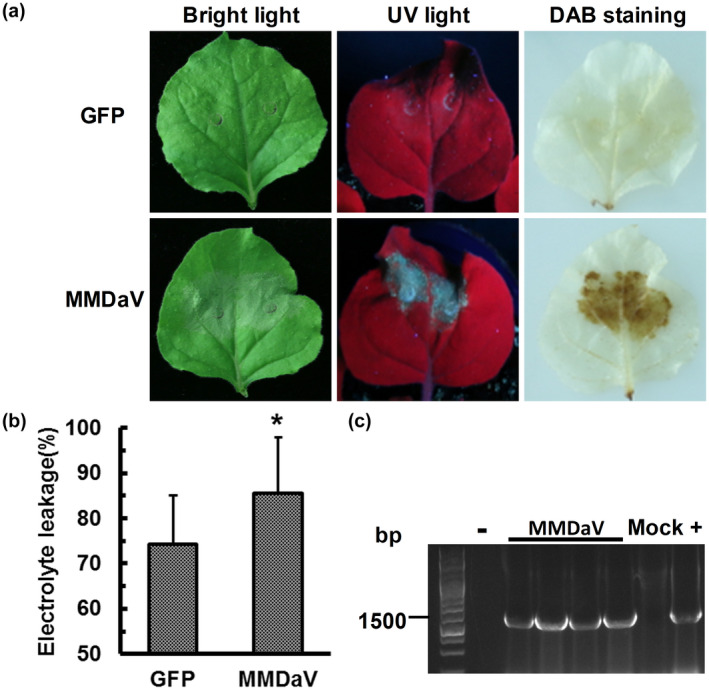
MMDaV infectious clone triggers cell death in Nicotiana benthamiana. (a) Phenotypes of N. benthamiana leaves infiltrated with Agrobacterium tumefaciens harbouring the construct expressing only the green fluorescent protein (GFP) (the upper panel) or the construct of MMDaV infectious clone (the lower panel). Photographs were captured under bright light and ultraviolet light at 3 days postinfiltration (dpi). Production of H2O2 is shown as 3,3′‐diaminobenzidine (DAB) staining. At least three biological replicates with five plants were performed for each treatment. (b) Quantification of cell death by measuring electrolyte leakage at 2 dpi. Means and standard errors are shown from four independent replicates. Asterisks denote statistically significant differences evaluated with Student's t test, *p < 0.05. Error bars represent standard deviation of each data set. (c) PCR analysis of MMDaV accumulation in infiltrated leaf patches. Specific primers were used to amplify the c.1500 bp product from the total DNA extracted from infiltrated leaf patches
2.2. The RepA and Rep proteins of MMDaV trigger cell death in N. benthamiana
To evaluate whether any of the seven proteins encoded by MMDaV accounts for MMDaV‐induced cell death, all the seven ORFs, V1, V2, V3, V4, V5, RepA, and Rep (Figure 2a), were independently inserted into the green fluorescent protein (GFP)‐containing pEarleyGate 103 vector and were infiltrated into different patches of the same N. benthamiana leaf. Notably, expression of the RepA protein induced HR‐like cell death in infiltrated N. benthamiana leaves at 3 dpi (Figure S1). Further analysis of the infiltrated leaves up to 5 dpi showed that Rep also triggered HR‐like cell death at 5 dpi (Figure 2b). However, no necrotic symptoms were observed in N. benthamiana leaves infiltrated with the empty vector expressing only the GFP or in N. benthamiana leaves infiltrated with any of the other five proteins of MMDaV, V1, V2, V3, V4, and V5 up to 5 dpi (Figure 2b). Visualization of the infiltrated N. benthamiana leaves under UV light and DAB staining further confirmed that cell death occurred only in the regions infiltrated with either RepA or Rep (Figures 2b and S1). Immunoblot analysis of plant soluble proteins extracted from infiltrated leaf patches using the anti‐GFP antibody confirmed the stable expression of all the seven MMDaV proteins (Figure 2c). To exclude the possibility that the GFP tag impairs the cell death‐inducing activity of the viral proteins, all the seven MMDaV proteins expressed from the pCHF3 vector were assessed in N. benthamiana plants. Consistent with the GFP‐fusion construct, the RepA and Rep proteins elicited HR‐like cell death in N. benthamiana leaves. None of the other five MMDaV proteins was capable of inducing HR‐like cell death (data not shown). Electrolyte leakage experiments further demonstrated that the ion permeability of N. benthamiana leaves inoculated with RepA‐GFP or Rep‐GFP was significantly higher than that of N. benthamiana leaves inoculated with 35S‐GFP (Figure 2d). These results collectively suggest that the RepA and Rep proteins trigger HR‐like cell death in N. benthamiana plants. The RepA protein is unique to geminiviruses belonging to the genera Becurtovirus, Capulavirus, Citlodavirus, Mastrevirus, Mulcrilevirus, Grablovirus, and Topilevirus. Given that RepA induced stronger necrosis symptoms than the Rep protein, we opted to select RepA for subsequent analyses.
FIGURE 2.
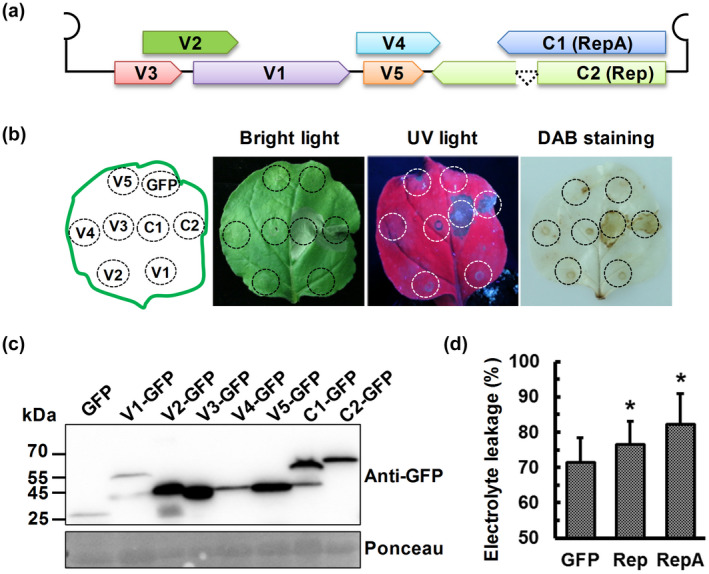
Identification of the MMDaV‐encoded protein(s) that could induce cell death in Nicotiana benthamiana. (a) Schematic diagram of the genome organization of MMDaV in a linearized form. The seven proteins encoded by MMDaV were shown in different colours. (b) Transient expression of the seven MMDaV‐encoded proteins in N. benthamiana by agroinfiltration. Green fluorescent protein (GFP) and GFP fusion constructs corresponding to the agroinfiltrated leaves were indicated in the schematic diagram in the left panel. Photographs were captured under bright light and ultraviolet (UV) light at 5 days postinfiltration (dpi). 3,3′‐diaminobenzidine (DAB) staining was performed to evaluate the production of H2O2. Each experiment was repeated three times and at least five plants were used per biological replicate. (c) Western blot analysis of GFP and GFP fusion protein accumulation in N. benthamiana leaves using an anti‐GFP monoclonal antibody. Ponceau staining of RuBisCO serves as a loading control. (d) Quantification of cell death by measuring electrolyte leakage at 2 dpi for RepA and 4 dpi for Rep. Means and standard errors were shown from four independent replicates. Error bars represent standard deviation of each data set. Asterisks indicate statistically significant differences determined with Student's t test, *p < 0.05
2.3. Mapping of domains indispensable for RepA‐induced cell death
To map domain(s) responsible for RepA‐induced cell death, the amino acid sequences of MMDaV RepA were aligned to that of other geminiviruses, and the functional domains of MMDaV RepA were predicted using Interpro (http://www.ebi.ac.uk/interpro/). Sequence analysis showed that MMDaV RepA has seven conserved domains, namely (a) the catalytic domain required for DNA/RNA binding, (b) the rolling‐circle replication (RCR) motif I (FLTFP) involved in specific dsDNA binding, (c) the RCR motif II (HFH), a metal‐binding site for protein folding and DNA cleavage, (d) the RCR motif III (YIQKE), a potential catalytic site for DNA cleavage, (e) the central domain that is crucial for oligomerization of the RepA protein, (f) the Walker A (GPTRSGKT), and (g) the Walker B (LYNVIDDI) domains that are related to the DNA helicase activity (Figure 3a). To delineate the functional domains of RepA, eight deletion mutants were constructed (Figure 3a), and their ability to induce cell death was evaluated via agroinfiltration in N. benthamiana leaves. Concomitant analysis of the cell death symptoms and the H2O2 level using DAB staining showed that the deletion mutants RepADM1 (amino acids 1–7 deleted), RepADM7 (Walker A domain deleted), and RepADM8 (amino acids 218–264 deleted) could still induce cell death in N. benthamiana leaves comparable to wild‐type RepA‐induced cell death (Figure 3b). Conversely, the deletion of the catalytic domain (RepADM2, amino acids 7–120 deleted) or the central domain (RepADM3, amino acids 122–218 deleted) completely abolished the ability of RepA to trigger cell death (Figure 3b). In addition, the three RCR motif‐deletion mutants, RepADM4 (amino acids 15–19 deleted), RepADM5 (amino acids 57–63 deleted), and RepADM6 (amino acids 103–108 deleted), were found to elicit cell death in N. benthamiana leaves, albeit to a substantially lesser degree than the wild‐type RepA (Figure 3b). Western blot analysis of the proteins extracted from infiltrated N. benthamiana leaves using the anti‐GFP antibody showed that all the eight deletion mutants were well expressed (Figure S2). These results demonstrated that the N‐terminal seven amino acids and the C‐terminal region encompassing the amino acids 218 to 264 are not required by RepA to induce cell death, whereas the RCR motifs of RepA are involved in the induction of cell death.
FIGURE 3.
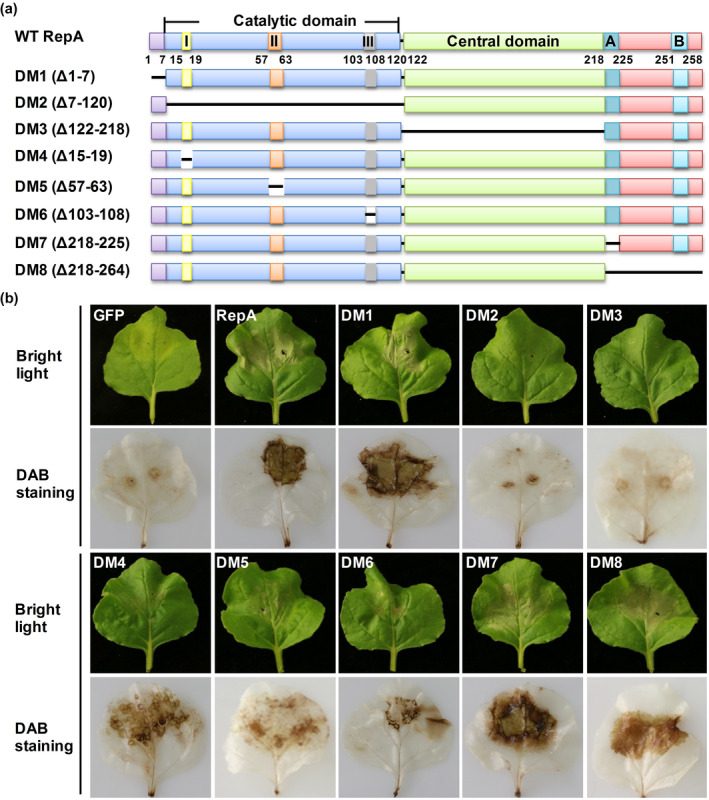
Effects of MMDaV RepA deletion mutations on RepA cell death‐inducing activity. (a) Schematic representation of wild‐type (WT) RepA and the eight RepA deletion mutants. The predicted conserved domains were shown as indicated. The catalytic domain contains the conserved motifs I (DNA binding), II (metal binding), and III (DNA cleavage and ligation). A and B in the C‐terminus of RepA represent the Walker A and Walker B motif, respectively. (b) Analysis of the cell death‐inducing activity of RepA deletion mutants. Top panels, visualization of the cell death response in agroinfiltrated Nicotiana benthamiana leaves under bright light at 3 days postinfiltration (dpi). Bottom panels, the representative images of agroinfiltrated N. benthamiana leaves stained with 3,3′‐diaminobenzidine (DAB) at 3 dpi
2.4. Nuclear accumulation is critical for RepA to induce cell death
Geminiviruses replicate in the nuclei of infected plant cells. To determine the subcellular localization of MMDaV RepA, Agrobacterium tumefaciens containing construct expressing GFP‐fused RepA (RepA‐GFP) protein or only the GFP protein was infiltrated into leaves of transgenic N. benthamiana plants overexpressing the nuclear marker red fluorescent protein (RFP)‐histone 2B (H2B) (hereafter referred to RFP‐H2B plants) (Martin et al., 2009), respectively. Confocal microscopy analysis showed that the GFP signal was observed only in the nucleus of the epidermal cells expressing RepA‐GFP, while GFP was localized to the nucleus and the cytoplasm of the epidermal cells expressing only the GFP protein (Figure 4), suggesting that RepA localizes to the nucleus on transient expression in N. benthamiana plants.
FIGURE 4.
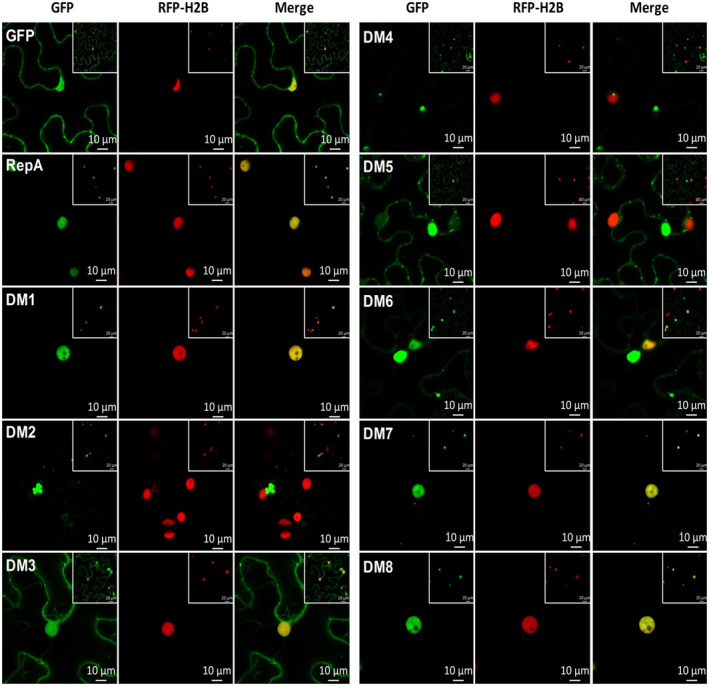
Subcellular localization of MMDaV RepA and RepA deletion mutants. Red fluorescent protein‐histone 2B (RFP‐H2B) transgenic plant leaves were infiltrated with Agrobacterium tumefaciens harbouring constructs to express green fluorescent protein (GFP) or GFP fusion proteins. The fluorescence of the epidermal cells of RFP‐H2B plant leaves was observed using a confocal laser scanning microscope at 36–48 hours postinfiltration. RFP‐H2B serves as a marker for the nucleus. The experiment was done three times, at least 30 cells were observed per sample and replicate. Scale bar: 10 μm
To determine whether deletion of the functional domains of RepA affects the subcellular localization of RepA, eight GFP‐fused RepA deletion mutants were individually infiltrated into the leaves of RFP‐H2B plants and the subcellular localization of each mutant was visualized by confocal microscopy at 36 hpi. Three expression patterns were observed for the eight GFP‐fused RepA mutants: nucleus‐only, nucleus and cytoplasm, and cytoplasm only. Similar to the GFP‐fused wild‐type RepA, the three GFP‐fused RepA mutants, RepADM1‐GFP, RepADM7‐GFP, and RepADM8‐GFP, were observed to target the nucleus on transient expression in RFP‐H2B plants. In contrast, the majority of RepADM2‐GFP, RepADM4‐GFP, and RepADM5‐GFP exhibited GFP signal mainly in the cytoplasm. For RepADM3‐GFP and RepADM6‐GFP, the detected GFP signal was distributed both to the nucleus and the cytoplasm (Figure 4).
Interestingly, the nuclear accumulation of RepA seems to be essential for its cell death induction activity; the RepA mutants RepADM2‐GFP, RepADM3‐GFP, RepADM4‐GFP, RepADM5‐GFP, and RepADM6‐GFP with localization to the cytoplasm displayed impaired cell death induction activity. On the other hand, the deletion mutants RepADM1‐GFP, RepADM7‐GFP, and RepADM8‐GFP, which localized mostly to the nucleus, exhibited high ability to induce cell death. These results imply that there might be a correlation between nuclear accumulation and cell death induction of RepA. Furthermore, to determine whether RepA‐induced cell death requires its nuclear localization, an artificial nuclear export sequence (NES; PKILALKLAGLDIN) and a nonfunctional NES mutant (nes; PKILAAKAAGADIN) were tagged with RepA‐GFP fusion at its N‐terminus and their subcellular localization and abilities to induce cell death were evaluated. Even though the fusion protein of NES‐RepA‐GFP was distributed both in the nucleus and cytoplasm at 36 hpi (Figure S3a), cell death induced by NES‐RepA‐GFP was attenuated when compared to that induced by RepA‐GFP. However, the nuclear localization and the cell death induction ability of nes‐RepA‐GFP were not altered when compared to RepA‐GFP (Figure S3a,b). Western blot analysis showed that the introduction of NES or nes did not affect the expression of RepA (Figure S3c). These results indicate that the nuclear localization of RepA is likely to be crucial for its activity in inducing cell death.
2.5. RepA and MMDaV elicit plant immune responses
Plant cell death is often associated with plant immune responses and involvement of hormone signalling molecules, such as SA, JA, and ET, or by combinations of these signalling compounds (Choi & Hwang, 2015). To understand whether RepA or MMDaV‐induced cell death is related to plant immune responses, reverse transcription quantitative PCR (RT‐qPCR) was performed to examine the expression of immune response‐related marker genes. These include NbPR‐1a and NbPR2, well‐known marker genes for SA‐mediated plant immunity (Dean et al., 2005; Loake & Grant, 2007), NbNPR1, the master regulator of SA‐dependent signalling, NbPR4 and NbLOX, marker genes for JA‐mediated plant immunity (Asai & Yoshioka, 2009; Rodriguez et al., 2014), and Nb ERF1, one of the marker genes for the ET‐dependent plant immunity (Pieterse et al., 2012). Agrobacteria expressing RepA‐GFP and MMDaV infectious clone were individually infiltrated into N. benthamiana leaves using agroinfiltration‐mediated expression of GFP and RepADM2‐GFP as controls. Initially, four housekeeping genes, glyceraldehyde 3‐phosphate dehydrogenase (GAPDH), Actin, elongation factor 1‐α (EF1α), and protein phosphatase 2A (PP2A), were selected to evaluate whether they are suitable for use as reference genes. By using three algorithms, geNorm, NormFinder, and BestKeeper, we found that GAPDH, Actin, and EF1α were suitable as reference genes for gene expression analysis in N. benthamiana infiltrated with RepA‐GFP or MMDaV infectious clone (data not shown). Therefore, GAPDH was chosen for the normalization of quantitative expression of the immune response‐related marker genes. Further RT‐qPCR analysis of the transcript levels of the selected immune response‐related marker genes showed that the expression of NbPR‐1a, NbPR2, NbPR4, and NbLOX transcripts was up‐regulated in N. benthamiana leaves inoculated with RepA and MMDaV infectious clones at 36 hpi when compared to the control plants that were infiltrated with GFP or RepADM2‐GFP. In contrast, no significant changes were observed for the NbNPR1 and NbERF 1 transcripts in response to RepA or MMDaV infectious clone (Figure 5). The response of PTI‐related marker genes NbCYP71D20 (Heese et al., 2007), NbACRE31, NbWRKY7, and NbWRKY8 (McLellan et al., 2013) to the expression of RepA and MMDaV was also detected using RT‐qPCR. As shown in Figure 5, the expression levels of NbACRE31, NbWRKY7, and NbWRKY8 were significantly increased in N. benthamiana plants infiltrated with RepA‐GFP and MMDaV when compared to those infiltrated with GFP and RepADM2‐GFP (Figure 5). These results suggest that RepA and MMDaV triggered HR‐associated plant immune responses.
FIGURE 5.
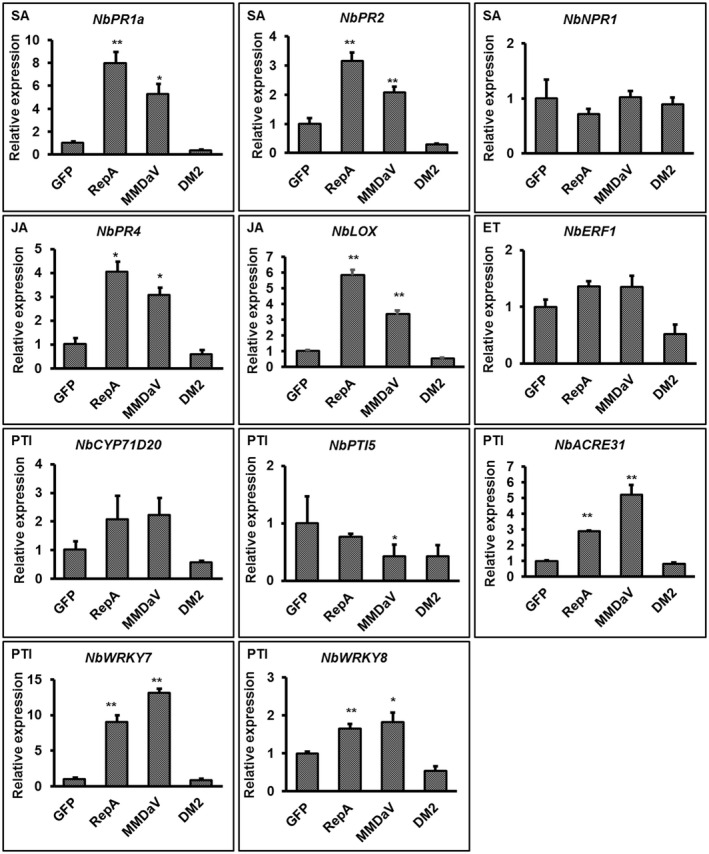
Relative expression of immunity‐associated genes by reverse transcription quantitative PCR (RT‐qPCR) in Nicotiana benthamiana leaves infiltrated with the control constructs, MMDaV or RepA‐GFP. The expression of NbGAPDH was used as a normalizer. The expression of each gene is represented relative to NbGAPDH using the comparative C t method (2−ΔΔ C t). Error bars represent standard deviation of each data set. Asterisks denote statistically significant differences as evaluated with Student's t test, *p < 0.05, **p < 0.01. A similar trend was obtained from three biological replicates. SA, salicylic acid; JA, jasmonic acid; ET, ethylene; PTI, pathogen‐associated molecular pattern‐triggered immunity
2.6. WRKY1 positively regulates RepA and MMDaV‐induced cell death in N. benthamiana
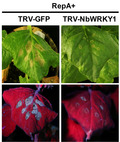
Signalling cascade components such as NDR1, RAR1, COI, CTR, NTF6, WRKY1, NPR1, and MEK2 play a critical role in cell death induction during plant interactions with bacteria and oomycetes (Meng & Zhang, 2013; Menke et al., 2005; del Pozo et al., 2004; Zhang et al., 2012). Because most pathogens might trigger a common/interconnected plant signalling network (Eulgem & Somssich, 2007), we explored tobacco rattle virus (TRV)‐induced gene silencing to assess whether any of these signalling components is involved in RepA‐elicited cell death. TRV2‐derived gene silencing constructs were generated and infiltrated together with TRV1 to individually suppress the expression of NDR1, RAR1, COI, CTR, NTF6, WRKY1, NPR1, and MEK2 in N. benthamiana leaves. After 10 days of infiltration when silencing is initiated, RepA was transiently expressed in the newly emerging three leaves of the silenced plants by infiltration of the RepA‐GFP construct. Silencing of NDR1, RAR1, COI, CTR, NTF6, NPR1, and MEK2 showed no apparent influence on RepA‐induced cell death (Figure S4). Nevertheless, RepA‐induced cell death was markedly compromised in WRKY1‐silenced plants compared with the control plants that were inoculated with TRV‐GFP (Figure 6a). RT‐qPCR analysis revealed that the transcript abundance of WRKY1 was reduced by about 82% in N. benthamiana plants inoculated with TRV‐WRKY1 when compared with the control plants, suggesting a successful silencing of WRKY1 (Figure 6c). To obviate the possibility that WRKY1 silencing affects agroinfiltration‐mediated transient expression of RepA‐GFP, immunoblot was performed to detect the expression of RepA using an anti‐GFP antibody. As shown in Figure 6e, RepA accumulated in WRKY1‐silenced N. benthamiana plants, suggesting that WRKY1 is required for RepA‐induced cell death.
FIGURE 6.
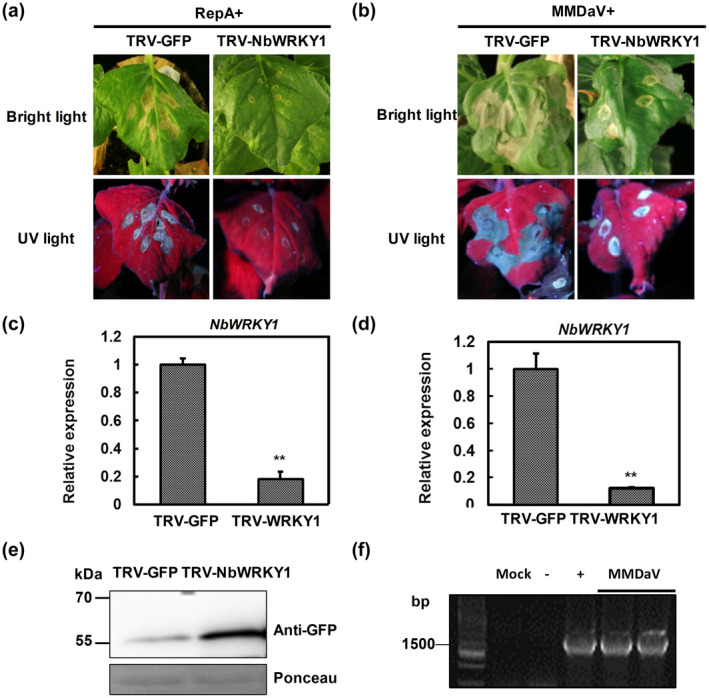
Silencing of WRKY1 impairs RepA and MMDaV‐induced cell death in Nicotiana benthamiana. (a) Effect of NbWRKY1 silencing on RepA‐induced cell death. N. benthamiana plants were first infiltrated with TRV1 and TRV2‐NbWRKY1 or TRV1 and TRV2‐GFP as a control. After 10 days, the upper leaves were infiltrated with RepA‐GFP to monitor the cell death response. Photographs were taken at 3 days postinfiltration (dpi) of RepA‐GFP. The experiment was performed three times. Six plants were used for each TRV construct per experiment. (b) Effect of NbWRKY1 silencing on MMDaV‐induced cell death in N. benthamiana. Plants were first infiltrated with TRV1 and TRV2‐NbWRKY1 or TRV1 and TRV2‐GFP as a control. After 10 days, the upper leaves were infiltrated with MMDaV to monitor the cell death response. Photographs were taken at 3 dpi of MMDaV. (c, d) Reverse transcription quantitative real‐time PCR analysis of the silencing efficiency of NbWRKY1 in plants used in (a) and (b), respectively. The expression of NbWRKY1 is represented relative to the expression of the normalizer NbGAPDH using the comparative C t method (2−ΔΔ C t). Error bars represent standard deviation of each data set. (e) Western blot analysis of RepA‐GFP fusion protein accumulation in N. benthamiana leaves used in (a) using an anti‐GFP monoclonal antibody. Ponceau staining of RuBisCO serves as a loading control. (f) PCR analysis of MMDaV accumulation in infiltrated leaf patches of plants used in (b). Specific primers were used to amplify the c.1500 bp product from the total DNA extracted from infiltrated leaf patches
To address the role of WRKY1 in MMDaV‐induced cell death, the infectious clone of MMDaV was agroinfiltrated into WRKY1‐silenced N. benthamiana plants. Interestingly, suppression of WRKY1 also impaired MMDaV‐induced cell death (Figure 6b,d,f). These results indicated that WRKY1 is involved in RepA and MMDaV‐triggered cell death.
2.7. NbWRKY1 is up‐regulated in response to RepA and MMDaV
The WRKY transcription factor (TF) superfamily consists of regulatory proteins that contain at least one conserved WRKY domain comprising the highly conserved WRKYGQK peptide sequence and a zinc finger motif (Jiang et al., 2017; Rushton et al., 2010). Although several WRKY TFs are critical players in plant resistance, less is known about their role in plant antiviral defence than their involvement in defence against fungal and bacterial pathogens (Chen et al., 2019; Jiang et al., 2017; Rushton et al., 2010). To understand how WRKY1 is involved in RepA‐induced cell death, the full‐length coding sequence of WRKY1 was cloned from N. benthamiana cDNA. The full‐length NbWRKY1 encodes a protein of 531 amino acids that contains two typical WRKY domains, structurally similar to WRKY TFs belonging to group I of the WRKY gene family. Phylogenetic analysis showed that it is closely related to WRKY1‐like TF in Nicotiana tabacum and WRKY20‐like TF in Nicotiana sylvestris, with an identity of 96% (Figure S5). Transient expression of GFP‐tagged WRKY1 fusion protein in RFP‐H2B expressing plant leaves showed that green fluorescence was exclusively detected in the nuclei of plant cells, whereas the epidermal cells expressing only the GFP protein emitted fluorescence both in the nucleus and the cytoplasm (Figure 7a). These results indicated that NbWRKY1 is targeted to the nucleus of plant cells.
FIGURE 7.
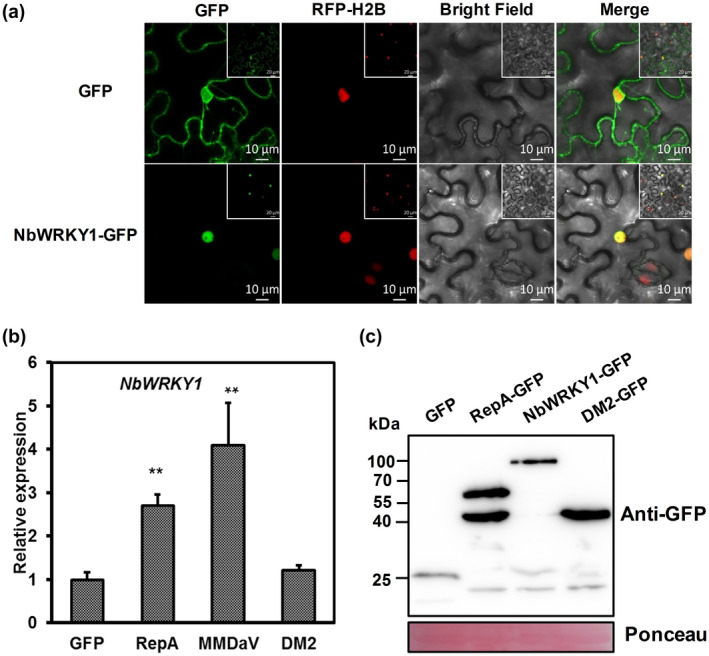
NbWRKY1 is a nucleus‐targeting protein that is up‐regulated in response to RepA and MMDaV. (a) Subcellular localization of NbWRKY1 in the epidermal cells of red fluorescent protein‐histone 2B (RFP‐H2B) transgenic plants by confocal microscopy at 36–48 hours postinfiltration. RFP‐H2B serves as a marker for the nucleus. The experiment was done three times, at least 30 cells were observed per sample and replicate. Scale bar: 10 μm. (b) Relative expression of NbWRKY1 in response to transient expression of RepA‐GFP, MMDaV, and DM2 mutant. Total RNA extracted at 3 days postinfiltration was used for reverse transcription quantitative PCR (RT‐qPCR) analysis. The relative expression of NbWRKY1 is represented to the normalizer NbGAPDH using the comparative C t method (2−ΔΔ C t). Error bars represent standard deviation of each data set. Asterisks denote statistically significant differences as evaluated with Student's t test, **p < 0.01. A similar trend was obtained from three biological replicates. (c) Western blot analysis of the expression of green fluorescent protein (GFP), RepA‐GFP, WRKY1‐GFP, and DM2 in Nicotiana benthamiana leaves used in (a) and (b). Ponceau staining of RuBisCO serves as a loading control
As RepA also localizes to the nuclei of plant cells, a yeast two‐hybrid (Y2H) assay was carried out to determine whether there is a direct interaction between RepA and WRKY1. NbWRKY1 and RepA were fused to both the GAL4 DNA‐binding domain (BD) and the GAL4 activation domain (AD) to yield BD and AD fusions. Plasmids expressing the AD and BD fusions were cotransformed to yeast strain Y2H Gold. Similar to the negative control consisting of cotransformed plasmids expressing human lamin (Lam) and simian virus 40 large T antigen (T), no growth of yeast transformants containing either AD‐RepA + BD‐NbWRKY1 or AD‐NbWRKY1 + BD‐RepA was evident on the selective medium lacking adenine, histidine, leucine, and tryptophan (data not shown), suggesting that RepA could not directly interact with NbWRKY1 in the Y2H assay.
The expression pattern of NbWRKY1 in response to RepA was then monitored in N. benthamiana leaves. RT‐qPCR analysis of the NbWRKY1 transcripts at 36 hpi showed that expression of RepA increased the NbWRKY1 transcripts by about 2.8‐fold compared with the control plants infiltrated with 35S‐GFP (Figure 7b). The level of NbWRKY1 gene expression in response to MMDaV infection was also studied. Similar to RepA, the infiltration of MMDaV infectious clone increased the NbWRKY1 transcripts by about 4.1‐fold (Figure 7b). These results suggested that both RepA and MMDaV up‐regulate the abundance of NbWRKY1 transcript.
2.8. Expression of RepA in N. benthamiana enhances plant resistance to two begomoviruses
To study whether RepA‐induced cell death can stimulate plant resistance to another pathogen, infectious clones of two begomoviruses, tomato yellow leaf curl China virus/tomato yellow leaf curl China betasatellite (TYLCCNV/TYLCCNB) (Cui et al., 2004) and tomato yellow leaf curl virus (TYLCV) (Zhang et al., 2009), were individually infiltrated into N. benthamiana plants with Agrobacterium harbouring constructs expressing GFP or RepA‐GFP, respectively. Symptoms induced by these two begomoviruses were monitored and viral DNA accumulation was detected. Disease symptoms such as downward leaf curling became visible in systemic leaves of N. benthamiana plants agroinfiltrated with GFP expression construct and TYLCCNV/TYLCCNB at 6 dpi or GFP expression construct and TYLCV at 10 dpi, whereas a significant milder symptom was observed in plants infiltrated with RepA‐GFP and TYLCCNV/TYLCCNB or RepA‐GFP and TYLCV (Figure 8a). Quantitative PCR analysis of the viral DNA level showed that the accumulation of viral DNA of both TYLCCNV and TYLCV was reduced in N. benthamiana plants expressing RepA‐GFP, which agrees with the severity of disease symptoms (Figure 8b,c). These results suggested that expression of RepA can help the plant to attenuate begomovirus infection.
FIGURE 8.
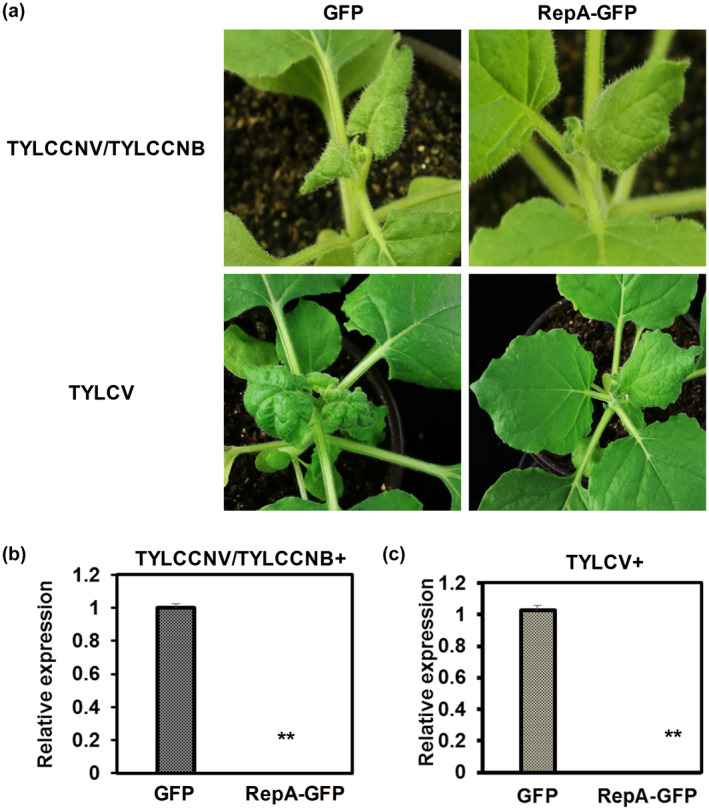
Expression of RepA enhances resistance to begomovirus infection in Nicotiana benthamiana. (a) Phenotypes of uninoculated systemic leaves of N. benthamiana plants. N. benthamiana plants were infiltrated with tomato yellow leaf curl China virus/tomato yellow leaf curl China betasatellite (TYLCCNV/TYLCCNB) with GFP or RepA‐GFP, or tomato yellow leaf curl virus (TYLCV) with GFP or RepA‐GFP as indicated. Top panels represent the uninoculated systemic leaves photographed at 6 days postinfiltration (dpi). Bottom panels represent the uninoculated systemic leaves photographed at 10 dpi. (b, c) Quantitative PCR analysis of viral DNA accumulation of TYLCCNV (b) and TYLCV (c) in systemic leaves of N. benthamiana plants as shown in (a). 25S rRNA was used as the internal reference. Means and standard errors were shown from three independent replicates. Asterisks denote statistically significant difference evaluated with Student's t test, **p < 0.01. Similar results were obtained in three independent experiments. Six plants were used per experiment
3. DISCUSSION
Recent development of metagenomics sequencing has permitted massive identification of viruses such as single‐stranded DNA viruses from both infected organisms and the environment (Zhao, Rosario, et al., 2019). However, the biological characteristics of identified viruses could not catch up with the large amount of sequence information. MMDaV, characterized from infected mulberry plants via high‐throughput sequencing, was recently classified into the genus Mulcrilevirus of the family Geminiviridae (Ma et al., 2015, https://talk.ictvonline.org/taxonomy/). Despite the V2 protein of MMDaV having been identified as a suppressor of RNA silencing and the nucleolar exclusion of V2 being promoted by RepA (Wang, Sun, et al., 2020; Yang et al., 2018), our knowledge of the biological function of MMDaV‐encoded proteins has lagged behind those of begomoviruses. Here, we found that an infectious clone of MMDaV could induce HR‐like cell death in the popular model plant N. benthamiana. We demonstrate that the RepA protein of MMDaV elicits HR‐mediated plant immune response, which has the potential to attenuate begomovirus infection.
Despite being relatively conserved, the sequences required for RepA to induce cell death apparently have diversified. Mutagenesis analysis of the RepA mutants of MMDaV revealed that the motifs I, II, and III, which are predicted to be involved in DNA binding and cleavage, were required for RepA to induce cell death in N. benthamiana plants. A seven amino acid deletion at the N‐terminal of MMDaV RepA did not impair its contribution to trigger cell death. However, it has been shown that either a nine or a 29 amino acid deletion at the N‐terminal of ODV RepA compromises its ability to induce cell death (Qian et al., 2015). The differentiation of the sequence context involved in triggering plant immune response has also been reported for the Rep protein encoded by begomoviruses. An R2S mutation of African cassava mosaic virus Rep fails to trigger the HR defence mechanism (Jin et al., 2008), whereas the ability to trigger a strong defence response against a highly virulent Sri Lankan cassava mosaic virus has been mapped to the seven amino acid motif at the C‐terminal of Rep (Wang, Zhang, et al., 2020). This study emphasizes again that not all positional homologs of geminivirus‐encoded proteins are made equal (Luna & Lozano‐Duran, 2020).
Notably, the RepA mutants that were not able to completely localize to the nucleus were also unable to elicit severe cell death in N. benthamiana plants (Figures 3 and 4). This implies that the ability of RepA to induce HR probably correlates with its nuclear localization. Nuclear localization has been implicated in the manifestation of plant disease resistance against bacterial and oomycete effectors. For example, the nuclear localization of the Phytophthora sojae effector Avh238 is essential for Avh238‐triggered cell death (Yang et al., 2017), and a nuclear localization signal (NLS) of the RxLR effector PITG_22798 secreted by Phytophthora infestans is required for both nuclear localization and cell death‐inducing activity (Wang et al., 2017). Analysis of RepA protein with online prediction software (PSORT) failed to identify basic amino acids with a consensus NLS sequence, suggesting the presence of an atypical NLS. Interestingly, as has been observed for TYLCV Rep (Maio et al., 2019), fusion of an NES to the N terminus of MMDaV RepA did not result in complete nuclear exclusion and caused only a partial redistribution of the protein outside the nucleus (Figure S3). Despite that, a partially relocalized NES‐RepA impaired its cell death‐inducing activity. Thus, the complete nuclear localization of MMDaV RepA is critical for its activity to induce cell death. It is not well understood how the partially impaired nuclear localization of RepA affects its ability to induce cell death. Experiments with a replication reporter system demonstrated that the conserved lysine residue located within the DNA‐binding domain of TYLCV Rep is essential for both initiation of viral rolling‐circle replication and control of nuclear localization of the TYLCV Rep protein (Maio et al., 2019). Interestingly, the lysines analysed in TYLCV Rep are also conserved in the motifs I, II, and III studied here. Because RepA and Rep share the same N‐terminus, it is possible that initiation of rolling circle replication during geminivirus infection triggers the cell death response.
An earlier study showed that JA‐dependent signalling is engaged in ODV RepA‐induced cell death as silencing of the F‐box protein responsible for JA perception COI1 using virus‐induced gene silencing (VIGS) leads to delayed RepA‐induced cell death (Qian et al., 2015). However, suppressed expression of COI1 in this study did not result in alleviated cell death induction by MMDaV RepA. Our results showed that NbWRKY1 contributed to MMDaV RepA‐induced cell death as MMDaV RepA failed to induce cell death in Nb WRKY1‐silenced N. benthamiana plants. Down‐regulation of Nb WRKY1 did not alleviate the accumulation of RepA protein, excluding the possibility that sustained Nb WRKY1 silencing leads to deficiency in Agrobacterium‐mediated transient expression as reported for SGT1 (Yu et al., 2019). This discrepancy further demonstrated the diversified interplays between plant and the RepA protein encoded by different geminiviruses.
WRKY proteins comprise a large family of TFs that participate in plant response to biotic and abiotic stresses. Three major groups have been classified based on the number of WRKY domains and the features of their zinc finger‐like motifs. The particular WRKY members involved in plant antiviral response vary depending on specific combinations of host and viruses. Silencing of WRKY1‐WRKY3 in N. benthamiana compromised N‐mediated resistance to tobacco mosaic virus (TMV) (Liu et al., 2004), while WRKY8 of Arabidopsis mediates antiviral response against TMV‐cg in the compatible Arabidopsis–TMV‐cg interaction (Chen et al., 2013). In the case of geminiviruses, six members of the WRKY group III subfamily are important for TYLCV infection, but they function as positive and negative regulators (Huang et al., 2016). In a recent study, the βC1 protein encoded by begomovirus‐associated betasatellite interacts with and hijacks AtWRKY20, one member belonging to the WRKY group I subfamily, to benefit whitefly vectors but deters two nonvector competitors (Zhao, Yao, et al., 2019). The expression of WRKY TFs themselves can be fine‐tuned at the transcript level by auto‐ and cross‐regulation among WRKYs or other members of TF families, epigenetic modification, and posttranscriptional regulation mediated by small RNAs. WRKY TFs can also be affected at the protein level by nuclear translocation, protein–protein interaction, and posttranslational modifications (Ng et al., 2018). As master regulators of diverse biological processes, WRKY proteins regulate downstream signalling by recognizing the TTGACC/W‐box sequences in the promoter regions of putative downstream target genes or by physically interacting with a substantial number of proteins involved in signalling, transcription, plant defence, and other cellular processes (Chi et al., 2013; Wani et al., 2021). The NbWRKY1 cloned in this study belongs to the WRKY group I subfamily as it contains two WRKY domains. Although both the RepA and NbWRKY1 are nuclear‐targeting proteins, we observed no direct interaction between NbWRKY1 and RepA in yeast using a Y2H assay. Alternatively, an up‐regulated transcript level of NbWRKY1 was observed in N. benthamiana leaves infiltrated with RepA or MMDaV. Taken together with the fact that the repression of NbWRKY1 in N. benthamiana attenuated the cell death phenotype induced by RepA and MMDaV, we proposed a positive role of NbWRKY1 in MMDaV‐induced cell death. We deduced that MMDaV/RepA up‐regulated the transcript of NbWRKY1. The induced expression of NbWRKY1 participates in MMDaV/RepA‐triggered cell death possibly by modulating downstream components involved in plant antiviral defence, thus conferring plant resistance against MMDaV. Further characterization of the downstream components regulated by NbWRKY1 will help to elucidate the regulatory network of NbWRKY1 against geminiviruses, which could be potentially used to improve plant resistance against geminiviruses in crops.
4. EXPERIMENTAL PROCEDURES
4.1. Construction of the infectious clone of MMDaV
According to the complete nucleotide sequence of MMDaV (accession no. KP303687), primers MMDaV FL/SalI‐F (a SalI site was introduced) and MMDaV FL/KpnI‐R (containing a KpnI site) were designed to amplify the full‐length MMDaV genome, and the PCR product was inserted into pEasy‐T1 vector (Transgene) to produce T1‐MMDaV‐PL. T1‐MMDaV‐PL was then digested with SalI and KpnI and cloned into the binary plant transformation vector pBINPLUS to yield pBINPLUS‐MMDaV‐PL. The other full‐length fragment of MMDaV was amplified using MMDaV FL/KpnI‐F and MMDaV FL/KpnI‐R (containing a KpnI site) and cloned into pEasy‐T1 to obtain T1‐MMDaV‐FL. After that, the full‐length KpnI‐digested fragment of T1‐MMDaV‐FL was inserted into the unique KpnI restriction site of pBINPLUS‐MMDaV‐PL to generate a dimeric tandem repeat construct pBINPLUS‐MMDaV‐2A. Primer sequences are listed in Table S1. All the amplified fragments used for the construction of the MMDaV infectious clone were sequenced to ensure that no mutation was introduced into the clones by PCR. The resulting vector was mobilized into A. tumefaciens EHA105 by electroporation.
4.2. Construction of GFP‐fusion plasmids
The V1, V3, V4, V5, RepA, and Rep ORFs of MMDaV were cloned into pENTR/D‐TOPO Entry vector using standard protocols as per the manufacturer's instructions (Invitrogen) and then recombined into the Gateway‐compatible binary vector pEarleyGate 103 (N‐terminal GFP fusion) (Earley et al., 2006) to generate V1‐GFP, V3‐GFP, V3‐GFP, V5‐GFP, RepA‐GFP, and Rep‐GFP, respectively. GFP‐tagged V2 was described earlier (Yang et al., 2018).
To map the functional domains of the RepA protein, eight deletion mutations were introduced into the RepA gene of MMDaV. The DM1 and DM8 mutants of RepA were amplified from pENTR‐RepA using primer pairs DM1‐F/C1‐R and C1C2‐F/DM8‐R (Table S1), respectively. The resulting PCR fragments were inserted into pENTR/D‐TOPO and subsequently transferred to pEarleyGate 103 to yield RepADM1‐GFP and RepADM8‐GFP, respectively. For the construction of RepADM2‐GFP, RepADM3‐GFP, RepADM4‐GFP, RepADM5‐GFP, RepADM6‐GFP, and RepADM7‐GFP, mutations were introduced into pENTR‐RepA by inverse PCR using KOD‐Plus‐Mutagenesis Kit (Toyobo). The entry clones from DM2 to DM7 were selected and verified by DNA sequencing. The transfer of the mutated RepA fragments from the entry clone to pEarleyGate 103 was performed by LR reaction as per manufacturer's instructions (Invitrogen). To yield the NES‐ or nes‐tagged RepA mutant (NES‐RepA and nes‐RepA), an inverse PCR (Toyobo) was carried out using the plasmid pENTR‐RepA as a template and NES‐RepA‐F/R and nes‐RepA‐F/R as primers, respectively. The resulting pENTR‐NES‐RepA and pENTR‐nes‐RepA constructs were cloned to pEarleyGate 103 through LR reaction to obtain NES‐RepA‐GFP and nes‐RepA‐GFP, respectively. All the GFP‐tagged constructs were transformed into A. tumefaciens C58C1 by electroporation.
4.3. Agrobacterium‐mediated infiltration
Agrobacterium‐mediated infiltration was performed as described previously (Yang et al., 2011). In brief, A. tumefaciens cultures harbouring the construct of MMDaV infectious clone or GFP‐tagged constructs were adjusted to an optical density OD600 = 1.0 and were incubated at room temperature for 2–3 h before infiltration. Fully expanded leaves of 4‐week‐old N. benthamiana or transgenic N. benthamiana plants expressing RFP‐H2B (Martin et al., 2009) were injected with Agrobacterium cultures using a needless syringe. The infiltrated plants were grown in an insect‐free growth chamber at 25°C under a 16 h light/8 h dark photoperiod.
4.4. Viral DNA detection
Nucleic acids from N. benthamiana leaves infiltrated with the MMDaV infectious clone were extracted at 3 dpi using the CTAB method as described previously (Xie et al., 2002). Specific primers V4/F and C1C2/F (Table S1) were used to detect viral DNA.
4.5. Measurement of electrolyte leakage
Ion leakage was determined from infiltrated N. benthamiana leaves as described previously (Kim et al., 2003). Briefly, 12 leaf discs (9 mm in diameter) from infiltrated N. benthamiana leaf areas were sampled and floated on distilled water for 2.5 h at room temperature. The conductivity in the solution was measured using an electrical conductivity meter (METTLER TOLEDO FE30). The leaf discs were put in the bathing solution and boiled for 20 min. Total conductivity was measured after the solution cooled to room temperature. The electrolyte leakage was expressed by the ratio of the conductivity to the total conductivity. All experiments were repeated four times, and the value for each treatment was calculated from the means of the four experiments.
4.6. DAB staining
DAB staining was carried out as described previously (Thordal‐Christensen et al., 1997). Briefly, N. benthamiana leaves were collected at 3 dpi and were immersed in the 1 mg/ml DAB‐HCl solution, pH 3.8 (Sigma). The samples were incubated in light for 8 h and were then destained in boiling 96% ethanol for 10 min. The intensity of DAB staining was assessed visually as a reddish‐brown colouration.
4.7. Laser scanning confocal microscopy
Epidermal cells in infiltrated leaves of transgenic N. benthamiana plants expressing RFP‐H2B (Martin et al., 2009) were imaged for fluorescence using a confocal microscope (LSM880; Carl Zeiss) at 36–48 hpi as described by Wang, Sun, et al. (2020). Excitations of GFP and RFP were set at 488 and 561 nm, respectively. Emissions of GFP and RFP were set at 500–530 nm and 600–630 nm, respectively. The red fluorescence derived from RFP‐H2B was used to mark the nucleus. Images were processed using Zeiss ZEN software.
4.8. Protein extraction and western blot analysis
Total soluble protein extraction and western blot analysis were performed as described by Yang et al. (2011). For detection of GFP, an anti‐GFP monoclonal antibody (Roche Diagnostic) and a secondary peroxidase‐conjugated goat anti‐mouse antibody (Cell Signaling Technology) was used. The signal of blotted proteins was developed and visualized using a chemiluminescence detection system (Tianneng).
4.9. VIGS assay in N. benthamiana
VIGS assays in N. benthamiana were performed using TRV‐based VIGS vector as described by Liu et al. (2002). About 350‐bp DNA fragments of NbNDR1, NbRAR1, NbCOI1, NbCTR1, NbNTF6, NbWRKY1, NbNPR1, and NbMEK2 were PCR amplified from N. benthamiana cDNA and cloned into the TRV2 vector to obtain TRV2‐NbNDR1, TRV2‐NbRAR1, TRV2‐NbCOI1, TRV2‐NbCTR1, TRV2‐NbNTF6, TRV2‐NbWRKY1, TRV2‐NbNPR1, and TRV2‐NbMEK2, respectively. N. benthamiana plants infiltrated with TRV‐phytoene desaturase (PDS) were used as a positive control and those with TRV‐GFP were used as a negative control. All constructs were confirmed by DNA sequencing. Primers used for plasmid construction are listed in Table S1.
For VIGS assays, TRV1 and the derivatives of TRV2 were individually introduced into A. tumefaciens EHA105 by electroporation. A. tumefaciens culture containing a TRV2 derivative was mixed with A. tumefaciens harbouring TRV1 at a 1:1 ratio to a final OD600 = 0.6. The mixture was subsequently infiltrated into the leaves of 4‐week‐old N. benthamiana plants as described above. Two weeks later, the systemic three or four leaves that showed stable silencing were used for further transient assays.
4.10. RNA extraction and quantitative real‐time PCR
To evaluate the silencing efficiency of TRV derivatives, total RNA was extracted from systemically infected N. benthamiana leaves at 10 dpi using TRIzol reagent as instructed (Invitrogen). To detect the expression of host factors responsive to MMDaV or RepA, total RNA was extracted from infiltrated N. benthamiana leaves at 3 dpi. First‐strand cDNA was synthesized from 1 μg of total RNA using SYBR PrimeScript RT‐PCR Kit (Takara). Real‐time quantitative PCR was performed using TransStart Green qPCR Supermix (Transgene) and the LightCycler 96 system (Roche). The stability of different reference genes in the context of TRV infection or the expression of MMDaV or RepA was validated as described by Liu et al. (2012). The expression of N. benthamiana GAPDH was further used as an internal control to normalize the expression value of target genes in each sample. Relative expression of target genes was determined using the comparative C t method (2−ΔΔ C t) (Livak & Schmittgen, 2001). Primers used for qPCR are also listed in Table S1.
4.11. Y2H assay
Interaction between RepA and NbWRKY1 was detected using a GAL4 Y2H system as described by Mei et al. (2018). RepA and WRKY1 were individually cloned into the GAL4 activation domain‐containing pGADT7 vector and the GAL4 DNA‐binding domain‐containing pGBKT7 vector, respectively. Constructs were cotransformed to the yeast Y2H GOLD strain using the Frozen‐EZ Yeast Transformation Kit (Zymo Research). Transformants were grown on yeast synthetic drop‐out media lacking leucine and tryptophan. Interaction between proteins was indicated by the growth of yeast cells on yeast synthetic drop‐out media lacking leucine, tryptophan, histidine, and adenine.
CONFLICT OF INTEREST
There is no conflict of interest to declare.
Supporting information
ACKNOWLEDGEMENTS
This work was financially supported by the National Natural Science Foundation of China (31972245 and 31720103914). The authors thank Michael Goodin (University of Kentucky) for providing RFP‐H2B transgenic N. benthamiana seeds.
Sun, S. , Ren, Y. , Wang, D. , Farooq, T. , He, Z. , Zhang, C. , et al (2022) A group I WRKY transcription factor regulates mulberry mosaic dwarf‐associated virus‐triggered cell death in Nicotiana benthamiana . Molecular Plant Pathology, 23, 237–253. 10.1111/mpp.13156
Contributor Information
Xiuling Yang, Email: yangxiuling@caas.cn.
Xueping Zhou, Email: zzhou@zju.edu.cn.
DATA AVAILABILITY STATEMENT
All data supporting the findings of this study are available within the paper and within its supplementary data published online.
REFERENCES
- Al Rwahnih, M. , Dave, A. , Anderson, M.M. , Rowhani, A. , Uyemoto, J.K. & Sudarshana, M.R. (2013) Association of a DNA virus with grapevines affected by red blotch disease in California. Phytopathology, 103, 1069–1076. [DOI] [PubMed] [Google Scholar]
- Asai, S. & Yoshioka, H. (2009) Nitric oxide as a partner of reactive oxygen species participates in disease resistance to necrotrophic pathogen Botrytis cinerea in Nicotiana benthamiana . Molecular Plant‐Microbe Interactions, 22, 619–629. [DOI] [PubMed] [Google Scholar]
- Balint‐Kurti, P. (2019) The plant hypersensitive response: concepts, control and consequences. Molecular Plant Pathology, 20, 1163–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boualem, A. , Dogimont, C. & Bendahmane, A. (2016) The battle for survival between viruses and their host plants. Current Opinion in Virology, 17, 32–38. [DOI] [PubMed] [Google Scholar]
- Caplan, J.L. , Mamillapalli, P. , Burch‐Smith, T.M. , Czymmek, K. & Dinesh‐Kumar, S.P. (2008) Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell, 132, 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Zhang, L. , Li, D. , Wang, F. & Yu, D. (2013) WRKY8 transcription factor functions in the TMV‐cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 110, E1963–E1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Li, C. , Wang, H. & Guo, Z. (2019) WRKY transcription factors: evolution, binding, and action. Phytopathology Research, 1, 13. [Google Scholar]
- Chi, Y. , Yang, Y. , Zhou, Y. , Zhou, J. , Fan, B. , Yu, J.‐Q. et al. (2013) Protein–protein interactions in the regulation of WRKY transcription factors. Molecular Plant, 6, 287–300. [DOI] [PubMed] [Google Scholar]
- Choi, H.W. & Hwang, B.K. (2015) Molecular and cellular control of cell death and defense signaling in pepper. Planta, 241, 1–27. [DOI] [PubMed] [Google Scholar]
- Csorba, T. , Kontra, L. & Burgyan, J. (2015) Viral silencing suppressors: tools forged to fine‐tune host–pathogen coexistence. Virology, 479, 85–103. [DOI] [PubMed] [Google Scholar]
- Cui, X. , Tao, X. , Xie, Y. , Fauquet, C.M. & Zhou, X. (2004) A DNAβ associated with Tomato yellow leaf curl China virus is required for symptom induction. Journal of Virology, 78, 13966–13974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L. , Dietrich, R.A. & Richberg, M.H. (1996) Death don't have no mercy: cell death programs in plant‐microbe interactions. The Plant Cell, 8, 1793–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, J.D. , Goodwin, P.H. & Hsiang, T. (2005) Induction of glutathione S‐transferase genes of Nicotiana benthamiana following infection by Colletotrichum destructivum and C. orbiculare and involvement of one in resistance. Journal of Experimental Botany, 56, 1525–1533. [DOI] [PubMed] [Google Scholar]
- Diamos, A.G. & Mason, H.S. (2018) Modifying the replication of geminiviral vectors reduces cell death and enhances expression of biopharmaceutical proteins in Nicotiana benthamiana leaves. Frontiers in Plant Science, 9, 1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley, K.W. , Haag, J.R. , Pontes, O. , Opper, K. , Juehne, T. , Song, K. et al. (2006) Gateway‐compatible vectors for plant functional genomics and proteomics. The Plant Journal, 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Eulgem, T. & Somssich, I.E. (2007) Networks of WRKY transcription factors in defense signaling. Current Opinion in Plant Biology, 10, 366–371. [DOI] [PubMed] [Google Scholar]
- Fondong, V.N. (2013) Geminivirus protein structure and function. Molecular Plant Pathology, 14, 635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese, A. , Hann, D.R. , Gimenez‐Ibanez, S. , Jones, A.M.E. , He, K. , Li, J. et al. (2007) The receptor‐like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proceedings of the National Academy of Sciences of the United States of America, 104, 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. , Li, M.‐Y. , Wu, P. , Xu, Z.‐S. , Que, F. , Wang, F. et al. (2016) Members of WRKY Group III transcription factors are important in TYLCV defense signaling pathway in tomato (Solanum lycopersicum). BMC Genomics, 17, 788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J. , Ma, S. , Ye, N. , Jiang, M. , Cao, J. & Zhang, J. (2017) WRKY transcription factors in plant responses to stresses. Journal of Integrative Plant Biology, 59, 86–101. [DOI] [PubMed] [Google Scholar]
- Jin, M. , Li, C. , Shi, Y. , Ryabov, E. , Huang, J. , Wu, Z. et al. (2008) A single amino acid change in a geminiviral Rep protein differentiates between triggering a plant defence response and initiating viral DNA replication. Journal of General Virology, 89, 2636–2641. [DOI] [PubMed] [Google Scholar]
- Jin, Y. , Zhao, J.H. & Guo, H.S. (2020) Recent advances in understanding plant antiviral RNAi and viral suppressors of RNAi. Current Opinion in Virology, 46, 65–72. [DOI] [PubMed] [Google Scholar]
- Jones, R.A.C. & Naidu, R.A. (2019) Global dimensions of plant virus diseases: current status and future perspectives. Annual Review of Virology, 6, 387–409. [DOI] [PubMed] [Google Scholar]
- Kim, M. , Ahn, J.W. , Jin, U.H. , Choi, D. , Paek, K.H. & Pai, H.S. (2003) Activation of the programmed cell death pathway by inhibition of proteasome function in plants. Journal of Biological Chemistry, 278, 19406–19415. [DOI] [PubMed] [Google Scholar]
- Kumar, J. , Ramlal, A. , Kumar, K. , Rani, A. & Mishra, V. (2021) Signaling pathways and downstream effectors of host innate immunity in plants. International Journal of Molecular Sciences, 22, 9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D. , Shi, L. , Han, C. , Yu, J. , Li, D. & Zhang, Y. (2012) Validation of reference genes for gene expression studies in virus‐infected Nicotiana benthamiana using quantitative real‐time PCR. PLoS One, 7, e46451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. & Dinesh‐Kumar, S.P. (2004) Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N‐mediated resistance to tobacco mosaic virus. The Plant Journal, 38, 800–809. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. , Marathe, R. & Dinesh‐Kumar, S.P. (2002) Tobacco Rar1, EDS1 and NPR1/NIM1‐like genes are required for N‐mediated resistance to tobacco mosaic virus. The Plant Journal, 30, 415–429. [DOI] [PubMed] [Google Scholar]
- Livak, K. & Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔC T methods. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Loake, G. & Grant, M. (2007) Salicylic acid in plant defence – the players and protagonists. Current Opinion in Plant Biology, 10, 466–472. [DOI] [PubMed] [Google Scholar]
- Loconsole, G. , Saldarelli, P. , Doddapaneni, H. , Savino, V. , Martelli, G.P. & Saponari, M. (2012) Identification of a single‐stranded DNA virus associated with citrus chlorotic dwarf disease, a new member in the family Geminiviridae . Virology, 432, 162–172. [DOI] [PubMed] [Google Scholar]
- Luna, A.P. & Lozano‐Duran, R. (2020) Geminivirus‐encoded proteins: not all positional homologs are made equal. Frontiers in Microbiology, 11, 878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y. , Navarro, B. , Zhang, Z. , Lu, M. , Zhou, X. , Chi, S. et al. (2015) Identification and molecular characterization of a novel monopartite geminivirus associated with mulberry mosaic dwarf disease. Journal of General Virology, 96, 2421–2434. [DOI] [PubMed] [Google Scholar]
- Maio, F. , Arroyo‐Mateos, M. , Bobay, B.G. , Bejarano, E.R. , Prins, M. & van den Burg, H.A. (2019) A lysine residue essential for geminivirus replication also controls nuclear localization of the tomato yellow leaf curl virus Rep protein. Journal of Virology, 93, e01910‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, K. , Kopperud, K. , Chakrabarty, R. , Banerjee, R. , Brooks, R. & Goodin, M.M. (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. The Plant Journal, 59, 150–162. [DOI] [PubMed] [Google Scholar]
- McLellan, H. , Boevink, P.C. , Armstrong, M.R. , Pritchard, L. , Gomez, S. , Morales, J. et al. (2013) An RxLR effector from Phytophthora infestans prevents re‐localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathogens, 9, e1003670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei, Y. , Yang, X. , Huang, C. , Zhang, X. & Zhou, X. (2018) Tomato leaf curl Yunnan virus‐encoded C4 induces cell division through enhancing stability of Cyclin D 1.1 via impairing NbSKη ‐mediated phosphorylation in Nicotiana benthamiana . PLoS Pathogens, 14, e1006789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X. & Zhang, S. (2013) MAPK cascades in plant disease resistance signaling. Annual Review of Phytopathology, 51, 245–266. [DOI] [PubMed] [Google Scholar]
- Menke, F.L.H. , Kang, H.‐G. , Chen, Z. , Park, J.M. , Kumar, D. & Klessig, D.F. (2005) Tobacco transcription factor WRKY1 is phosphorylated by the MAP kinase SIPK and mediates HR‐like cell death in tobacco. Molecular Plant‐Microbe Interactions, 18, 1027–1034. [DOI] [PubMed] [Google Scholar]
- Ng, D.W. , Abeysinghe, J.K. & Kamali, M. (2018) Regulating the regulators: the control of transcription factors in plant defense signaling. International Journal of Molecular Sciences, 19, 3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise, V. (2014) Crop immunity against viruses: outcomes and future challenges. Frontiers in Plant Science, 5, 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Van der Does, D. , Zamioudis, C. , Leon‐Reyes, A. & Van Wees, S.C.M. (2012) Hormonal modulation of plant immunity. Annual Review of Cell and Developmental Biology, 28, 489–521. [DOI] [PubMed] [Google Scholar]
- del Pozo, O. , Pedley, K.F. & Martin, G.B. (2004) MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO Journal, 23, 3072–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, Y. , Hou, H. , Shen, Q. , Cai, X. , Sunter, G. & Zhou, X. (2015) RepA protein encoded by oat dwarf virus elicits a temperature‐sensitive hypersensitive response‐type cell death that involves jasmonic acid‐dependent signaling. Molecular Plant‐Microbe Interactions, 29, 5–21. [DOI] [PubMed] [Google Scholar]
- Qiu, Y. , Zhang, S. , Yu, H. , Xuan, Z. , Yang, L. , Zhan, B. et al. (2020) Identification and characterization of two novel geminiviruses associated with paper mulberry (Broussonetia papyrifera) leaf curl disease. Plant Disease, 104, 3010–3018. [DOI] [PubMed] [Google Scholar]
- Rodriguez, P.A. , Stam, R. , Warbroek, T. & Bos, J.I.B. (2014) Mp10 and Mp42 from the aphid Species Myzus persicae trigger plant defenses in Nicotiana benthamiana through different activities. Molecular Plant‐Microbe Interactions, 27, 30–39. [DOI] [PubMed] [Google Scholar]
- Rushton, P.J. , Somssich, I.E. , Ringler, P. & Shen, Q.J. (2010) WRKY transcription factors. Trends in Plant Science, 15, 247–258. [DOI] [PubMed] [Google Scholar]
- Thordal‐Christensen, H. , Zhang, Z. , Wei, Y. & Collinge, D.B. (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley—powdery mildew interaction. The Plant Journal, 11, 1187–1194. [Google Scholar]
- Varsani, A. , Roumagnac, P. , Fuchs, M. , Navas‐Castillo, J. , Moriones, E. , Idris, A. et al. (2017) Capulavirus and Grablovirus: two new genera in the family Geminiviridae . Archives of Virology, 162, 1–13. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Sun, S. , Ren, Y. , Li, S. , Yang, X. & Zhou, X. (2020) RepA promotes the nucleolar exclusion of the V2 protein of mulberry mosaic dwarf‐associated virus. Frontiers in Microbiology, 11, 1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. , Zhang, X. , Yao, X. , Zhang, P. , Fang, R. & Ye, J. (2020) A 7‐amino‐acid motif of Rep protein essential for virulence is critical for triggering host defense against Sri Lankan cassava mosaic virus. Molecular Plant‐Microbe Interactions, 33, 78–86. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Ren, Y. , Zhou, J. , Du, J. , Hou, J. , Jiang, R. et al. (2017) The cell death triggered by the nuclear localized RxLR effector PITG_22798 from Phytophthora infestans is suppressed by the effector Avr3b. International Journal of Molecular Sciences, 18, 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani, S.H. , Anand, S. , Singh, B. , Bohra, A. & Joshi, R. (2021) WRKY transcription factors and plant defense responses: latest discoveries and future prospects. Plant Cell Reports, 40, 1071–1085. [DOI] [PubMed] [Google Scholar]
- Xie, Y. , Zhou, X.P. , Zhang, Z.K. & Qi, Y.J. (2002) Tobacco curly shoot virus isolated in Yunnan is a distinct species of Begomovirus . Chinese Science Bulletin, 47, 197–200. [Google Scholar]
- Yang, B. , Wang, Q. , Jing, M. , Guo, B. , Wu, J. , Wang, H. et al. (2017) Distinct regions of the Phytophthora essential effector Avh238 determine its function in cell death activation and plant immunity suppression. New Phytologist, 214, 361–375. [DOI] [PubMed] [Google Scholar]
- Yang, X. , Guo, W. , Li, F. , Sunter, G. & Zhou, X. (2019) Geminivirus‐associated betasatellites: exploiting chinks in the antiviral arsenal of plants. Trends in Plant Science, 24, 519–529. [DOI] [PubMed] [Google Scholar]
- Yang, X. , Guo, W. , Ma, X. , An, Q. & Zhou, X. (2011) Molecular characterization of tomato leaf curl China virus, infecting tomato plants in China, and functional analyses of its associated betasatellite. Applied and Environment Microbiology, 77, 3092–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Ren, Y. , Sun, S. , Wang, D. , Zhang, F. , Li, D. et al. (2018) Identification of the potential virulence factors and RNA silencing suppressors of mulberry mosaic dwarf‐associated geminivirus. Viruses, 10, 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G. , Xian, L. , Sang, Y. & Macho, A.P. (2019) Cautionary notes on the use of Agrobacterium‐mediated transient gene expression upon SGT1 silencing in Nicotiana benthamiana . New Phytologist, 222, 14–17. [DOI] [PubMed] [Google Scholar]
- Zerbini, F.M. , Briddon, R.W. , Idris, A. , Martin, D.P. , Moriones, E. , Navas‐Castillo, J. et al. (2017) ICTV virus taxonomy profile: Geminiviridae . Journal of General Virology, 98, 131–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Gong, H. & Zhou, X. (2009) Molecular characterization and pathogenicity of tomato yellow leaf curl virus in China. Virus Genes, 39, 249–255. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Li, D. , Wang, M. , Liu, J. , Teng, W. , Cheng, B. et al. (2012) The Nicotiana benthamiana mitogen‐activated protein kinase cascade and WRKY transcription factor participate in Nep1(Mo)‐triggered plant responses. Molecular Plant‐Microbe Interactions, 25, 1639–1653. [DOI] [PubMed] [Google Scholar]
- Zhao, L. , Rosario, K. , Breitbart, M. & Duffy, S. (2019) Eukaryotic circular Rep‐encoding single‐stranded DNA (CRESS DNA) viruses: ubiquitous viruses with small genomes and a diverse host range. Advances in Virus Research, 103, 71–133. [DOI] [PubMed] [Google Scholar]
- Zhao, P. , Yao, X. , Cai, C. , Li, R. , Du, J. , Sun, Y. , et al. (2019). Viruses mobilize plant immunity to deter nonvector insect herbivores. Science Advances, 5, eaav9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, S. & Li, Y. (2021) Current understanding of the interplays between host hormones and plant viral infections. PLoS Pathogens, 17, e1009242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and within its supplementary data published online.


