Abstract
Sclerotinia sclerotiorum, the notorious necrotrophic phytopathogenic fungus with wide distribution, is responsible for sclerotium disease in more than 600 plant species, including many economic crops such as soybean, oilseed rape, and sunflower. The compound appressorium is a crucial multicellular infection structure that is a prerequisite for infecting healthy tissues. Previously, the Forkhead‐box family transcription factors (FOX TFs) SsFoxE2 and SsFKH1 were shown to play a key regulatory role in the hyphae growth, sexual reproduction, and pathogenicity of S. sclerotiorum. However, little is known about the roles of SsFoxE3 regulating growth and development and pathogenicity. Here, we report SsFoxE3 contributes to sclerotium formation and deletion of SsFoxE3 leads to reduced formation of compound appressoria and developmental delays. Transcripts of SsFoxE3 were greatly increased during the initial stage of infection and SsFoxE3 deficiency reduced virulence on the host, while stabbing inoculation could partially restore pathogenicity. The SsFoxE3 mutant showed sensitivity to H2O2, and the expression of reactive oxygen species detoxification and autophagy‐related genes were reduced. Moreover, expression of SsAtg8 was also decreased during the infection process of the SsFoxE3 mutant. Yeast 1‐hybrid tests suggested that SsFoxE3 interacted with the promoter of SsAtg8. Disruption of SsAtg8 resulted in a phenotype similar to that of the SsFoxE3 mutant. Comparative analysis of the level of autophagy in the wild type and SsFoxE3 mutant showed that N starvation‐induced autophagy was reduced in the SsFoxE3 mutant. Taken together, our findings indicate that SsFoxE3 plays an important role in compound appressorium formation and is involved in transcriptional activation of SsAtg8 during infection by S. sclerotiorum.
Keywords: autophagy, compound appressoria, pathogenicity, Sclerotinia sclerotiorum, SsFoxE3
Transcription factor SsFoxE3 plays an important role in compound appressorium formation and is involved in transcriptional activation of SsAtg8 during infection by Sclerotinia sclerotiorum.

1. INTRODUCTION
Sclerotinia sclerotiorum, a pathogen with a complex of pathogenicity factors (cell wall‐degrading enzymes, transcription factors [TFs], effectors, and oxalic acid), has two different lifestyles: biotrophic and necrotrophic (Bashi et al., 2016; Hegedus & Rimmer, 2005; Kabbage et al., 2015; Liang & Rollins, 2018; Lyu et al., 2016; Sang et al., 2019; Veluchamy et al., 2012). During infection, S. sclerotiorum saprotrophic hyphae differentiate to form a special infection structure named the multicellular appressorium (infection cushion) that penetrates the cuticle and cell walls of healthy plant issue (Li et al., 2012; Lumsden, 1973; Tariq & Jeffries, 1984). Several phytopathogenic fungi, such as Rhizoctonia solani, Fusarium graminearum, and Botrytis cinerea, use complex appressoria similar to infection cushions (Armentrout & Downer, 1987; Choquer et al., 2021; Jeffries, 1986; Mentges et al., 2020; Xu et al., 2018). SsGgt1, which encodes γ‐glutamyl transpeptidase involved in redox homeostasis, is known to functions in the regulation of infection cushion development (Li et al., 2012). In addition, transcriptomic studies of infection cushions in F. graminearum and B. cinerea have shown that infection cushions are arsenals: virulence‐associated factors including cell wall‐degrading enzymes, effector proteins and secondary metabolites are synthesized in infection cushions (Choquer et al., 2021; Mentges et al., 2020). Therefore, the formation and development of compound appressoria in S. sclerotiorum should be studied systematically.
Members of the Forkhead‐box (FOX) TF family contain a wing‐like helix structure in the DNA‐binding region, also called the winged helix TF (Postnikoff et al., 2012). FOX TFs show great functionality in growth, development, and cell division and participate in the control of the cell cycle process, growth, differentiation, and other biological processes (Bulmer et al., 2004; Postnikoff et al., 2012; Shimada et al., 2008). FOX TFs are conserved in animal and fungal genomes, but are not found in plants. Saccharomyces cerevisiae contains four FOX TFs: FKH1, FKH2, HCM1, and FHL1 (Arsenault et al., 2015; Pataki et al., 2017; Postnikoff et al., 2012). Fhl1 of Schizosaccharomyces pombe regulates the nitrogen starvation response and acts in the target of rapamycin signalling pathway (Pataki et al., 2017). CaFKH2, the FOX TF of Candida albicans, controls the morphogenesis of fungal hyphae and the toxicity of pseudohyphae (Bensen et al., 2002). In Magnaporthe oryzae, the absence of MoFKH1 affects mycelial growth, conidial germination, and its pathogenicity; the absence of MoHCM1 leads to defects in mycelial growth and conidial germination (Park et al., 2014). Fox1 is exclusively expressed during the biotrophic stage of Ustilago maydis infection; deficiency of fox1 can lead to reduced virulence, impaired tumour development, and induces the accumulation of H2O2 in and around infected cells (Zahiri et al., 2010). In S. sclerotiorum, there are four FOX TF members: SsFkh1, SsFoxE2, SsFoxE3, SsFox1 Silencing SsFkh1 resluts in significantly reduced pathogenicity and no formation of sclerotia (Fan et al., 2017). SsFoxE2 is required for apothecial development (Wang et al., 2016). However, the functions of SsFoxE3 and SsFox1 are still unknown.
Autophagy is a programmed cell degradation mechanism that is ubiquitous in eukaryotic cells. Organisms can remove and degrade excess biological macromolecules and self‐damaged cells that are produced in biological processes through autophagy and use the degradation products to provide energy, which is considered to be a strategy for obtaining nutrients by recycling their own cell components under starvation conditions, and is essential for maintaining cell homeostasis and the operation of life activities. Autophagy in filamentous fungi is not only related to nutrition balance but also to developmental processes such as cell differentiation, secondary metabolism, and pathogenicity, and is often a prerequisite for pathogenicity (Liu et al., 2012; Zhu et al., 2019). MoAtg8 is necessary for the differentiation of the M. oryzae infection structure (Liu et al., 2010). Deletion of ATG8 causes U. maydis autophagosomes to accumulate in vacuoles and reduces virulence (He et al., 2018). SmAtg8 is required for Sordaria macrospora fruiting body development and ascospore germination (Voigt et al., 2013). In Colletotrichum orbiculare, deficiency of coatg8 causes it to fail to form normal appressoria in the early steps of morphogenesis (Asakura et al., 2009). FgAtg8 is involved in the formation of fruiting bodies and the production of conidia of F. graminearum (Josefsen et al., 2012), which means that the autophagy process has a variety of functions in different species. However, the transcriptional regulation of autophagy‐related genes in S. sclerotiorum is not yet known, and the role of autophagy is still unclear.
In this study, we identified SsFoxE3 as a FOX TF and showed it is involved in polarized growth, pathogenicity, and the stress response in S. sclerotiorum. Moreover, we reveal that SsAtg8 may be the downstream target of SsFoxE3, which is also required for pathogenicity.
2. RESULTS
2.1. Characterization of SsFoxE3 in S. sclerotiorum
The FOX family TFs are important regulators in controlling the cell cycle, primary metabolism, and morphogenesis. Four putative FOX TFs exist in S. sclerotiorum (SsFoxE2, SsFoxE3, SsFkh1, SsFox1). SsFoxE2 deletion mutants are deficient in apothecial development (Wang et al., 2016), SsFkh1 is involved in sclerotial formation and pathogenicity (Fan et al., 2017), suggesting that FOX TFs are transcriptional regulators that are shared by multiple pathways. We therefore analysed the function of SsFoxE3 in S. sclerotiorum. To distinguish between the TFs, we constructed a phylogenetic tree of the conserved domain (Forkhead domain IPR001766) of FOX TFs from the pathogenic fungiF. oxysporum, U. maydis, and M. oryzae, which revealed that SsFoxE3, Umfox1, and AfFox are in the same branch (Figure 1a,b). Alignment of Forkhead domains showed the most conserved residues to be Y707, L724, Y728, W746, R751, H752, N753, and L754 in SsFoxE3 (Figure 1c). The SsFoxE3 protein sequence contains 1232 amino acids including the forkhead‐associated (FHA) domain (IPR000253), which is a phosphopeptide recognition domain, suggesting that the protein has diverse functions (Figure 1b).
FIGURE 1.

Comparison of Forkhead (FH) domains and the phylogenetic tree of fungal Forkhead‐box family (FOX) transcription factors (TFs). (a) Dendrogram of FOX TFs. The phylogenetic tree was constructed by MEGA 7. GenBank accession numbers and sequence information are in Table S1. (b) Distribution map of Forkhead‐associated (FHA) and FH domains in FOX TFs. the information of the FHA (orange) and FH (purple) domains were analysed by Interpro (http://www.ebi.ac.uk/interpro) and drawn by GPS 2.0. (c) Alignments of the conserved FH domains of FOX TFs by ClustalW. Identical amino acids residues are marked as *
2.2. SsFoxE3 contributes to sclerotia formation
To investigate the role of SsFoxE3 in S. sclerotiorum, we constructed an SsFoxE3 mutant strain ∆SsFoxE3 by disruption with the hygromycin gene (Figure S1). First, we analysed the colony morphology of strain ∆SsFoxE3 by evaluating ∆SsFoxE3 and wild‐type UF‐1 strain growth on potato dextrose agar (PDA), minimal medium (MM), and complete medium (CM) for 48 h at 25°C. There was no significant difference in colony diameter between ∆SsFoxE3 and wild‐type UF‐1 (Figure 2a,b). To investigate whether SsFoxE3 interferes with cell wall synthesis related to hyphal development, the cell wall synthesis inhibitors Congo red, calcofluor white (CFW), and sodium dodecyl sulphate (SDS) were added to the medium. In each case, growth of ∆SsFoxE3 was significantly inhibited compared to the wild type (Figure 2a,b). When we used CFW to stain the cell wall, obvious blue fluorescence was seen at the growing tip of the wild‐type hyphae, while fluorescence in the mutant was diffuse, indicating that SsFoxE3 deficiency affected the normal development of the cell wall (Figure 2c,d). SsFoxE3 disruption also led to a decrease in the number of sclerotia (Figure 2e,f). The results indicate that SsFoxE3 may have an effect on the cell wall and affects the normal development of sclerotia.
FIGURE 2.
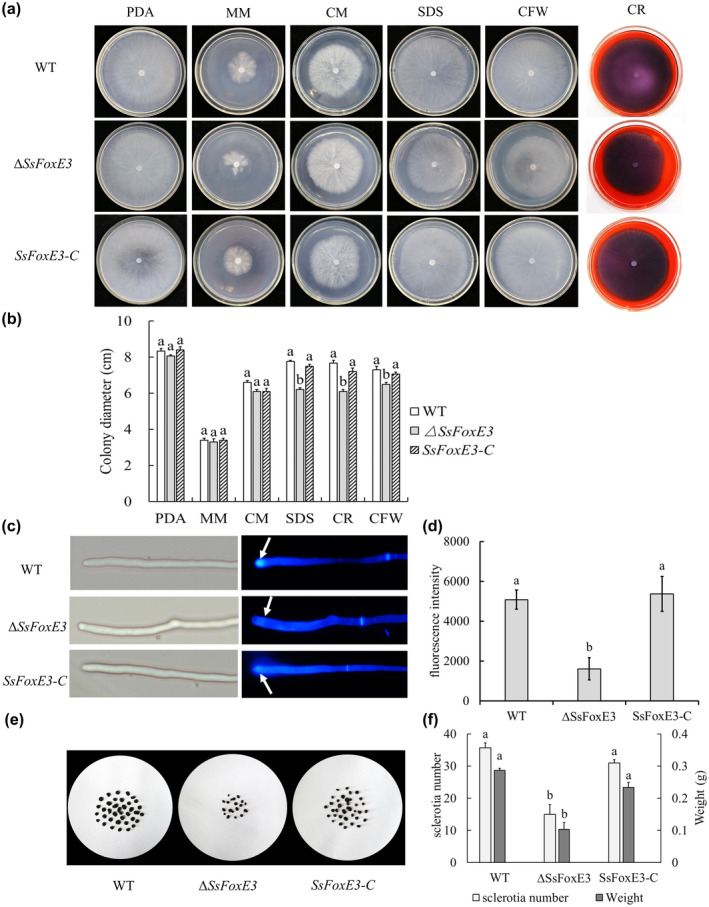
SsFoxE3 contributes to cell wall and sclerotia formation. (a) Mycelial growth is not affected by SsFoxE3. Strains were inoculated on potato dextrose agar (PDA), minimal medium (MM), and complete medium (CM), and cultured for 48 h at 25°C. (b) ΔSsFoxE3 is sensitive to cell wall synthesis inhibitors. Colony diameters of strains on medium amended with Congo red (CR), calcofluor white (CFW), or 0.001% sodium dodecyl sulphate (SDS) were measured by the cross method after 48 h. Error bars are the standard deviation, significant difference in data using SPSS software (p < 0.05). Each set of experiments was repeated three times. (c) SsFoxE3 is involved in the formation of the cell wall. The tips of hyphae were stained with CFW and observed under the fluorescence microscope. Arrows show the chitin content at the top of the mycelial growth. (d) The fluorescence intensity assay of strains. ImageJ was used to analyse the fluorescence intensity after staining by CFW. Different letters represent statistically significant differences (Student's t test, p < 0.05). (e) Sclerotial formation was reduced in ∆SsFoxE3. Strains were grown on PDA and cultured for 2 weeks at 25°C. (f) The number and the weight of sclerotia were measured and subjected to statistical analysis. Three repeats were performed and error bars represent the standard deviations. Different letters represent statistically significant differences (Student's t test, p < 0.05)
2.3. SsFoxE3 is required for compound appressoria formation and pathogenicity
Compound appressoria, play a vital role during the disease cycle of S. sclerotiorum (Fan et al., 2017; Kabbage et al., 2015). Compound appressoria formation of ∆SsFoxE3, the wild‐type UF‐1, and complemented strain SsFoxE3‐C was determined on a hydrophobic interface after incubation for 48 h. The formation of brown compound appresoria by the SsFoxE3 mutant was significantly reduced, and the number of compound appressoria was reduced almost 7‐fold (Figure 3a,b). The deficiency of SsFoxE3 also significantly reduced pathogenicity (Figure 3c). We ruled out the influence of oxalate (Figure S2). The pathogenicity of ∆SsFoxE3 onsoybean leaves was restored to a certain extent by wounding compared with that on unwounded leaves (Figure 3d,e). These results suggest that SsFoxE3 affects the development of the compound appressoria and the virulence of S. sclerotiorum.
FIGURE 3.
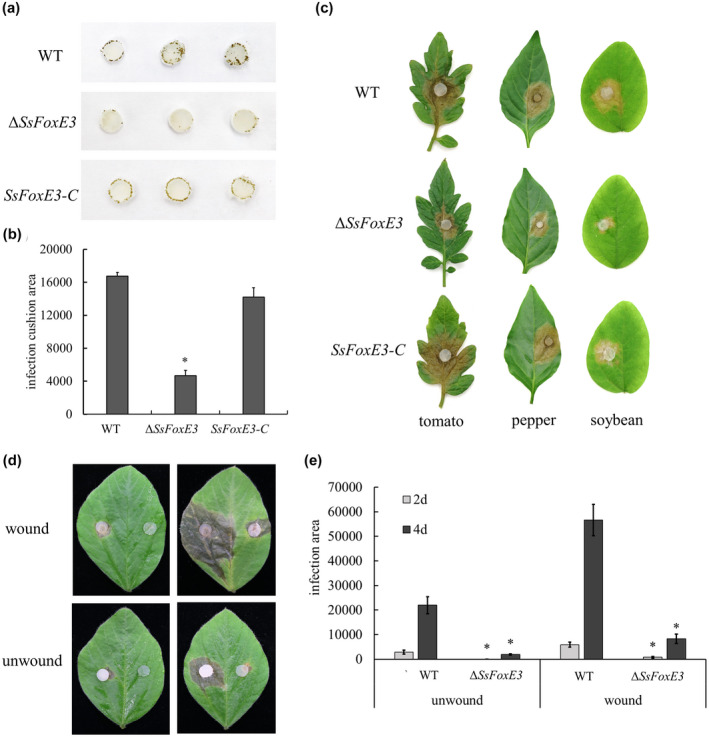
SsFoxE3 contributes to compound appressoria formation and pathogenicity. (a) Development of compound appressoria on a hydrophobic interface. Strains were grown on potato dextrose agar (PDA) for 48 h, then an agar disk containing mycelium (0.5 mm diameter) was inoculated on a glass slide, moisturized, then cultured for 48 h. (b) Statistical analysis of compound appressoria formation. The compound appressoria produced by strains were measured by ImageJ. Error bars represent standard deviations. *Significant differences from compound appressoria of wild type at α = 0.05 (Student's t test). (c) The SsFoxE3 deletion mutant showed reduced virulence on host leaves. Agar disks containing mycelium (0.5 mm diameter) of strains were inoculated on tomato, pepper, and soybean leaves. Leaves were harvested 48 h after inoculation. (d, e) Pathogenicity of ∆SsFoxE3 on unwounded and wounded soybean leaves. These experiments were performed three times with 10 pieces of soybean leaves. *Significant differences from wild type at α = 0.05 (Student's t test)
There was no obvious difference in morphology between the wild type and ∆SsFoxE3 compound appressoria on the glass slides (Figure S3); however, we found that SsFoxE3 disruption led to developmental retardation of compound appressoria (Figure 4a). To further clarify the influence of SsFoxE3 deletion on compound appressoria development during the infection process, we compared the ability to penetrate onion cells by ∆SsFoxE3 and the wild‐type strain. By staining with lactophenol blue 12 h after inoculation, it was found that the wild type formed a large group of compound appressoria (invasive hyphae) in onion cells but ∆SsFoxE3 did not (Figure 4b). These results indicate that the effect of SsFoxE3 deletion on the loss of pathogenicity is due to defects in the formation of compound appressoria.
FIGURE 4.
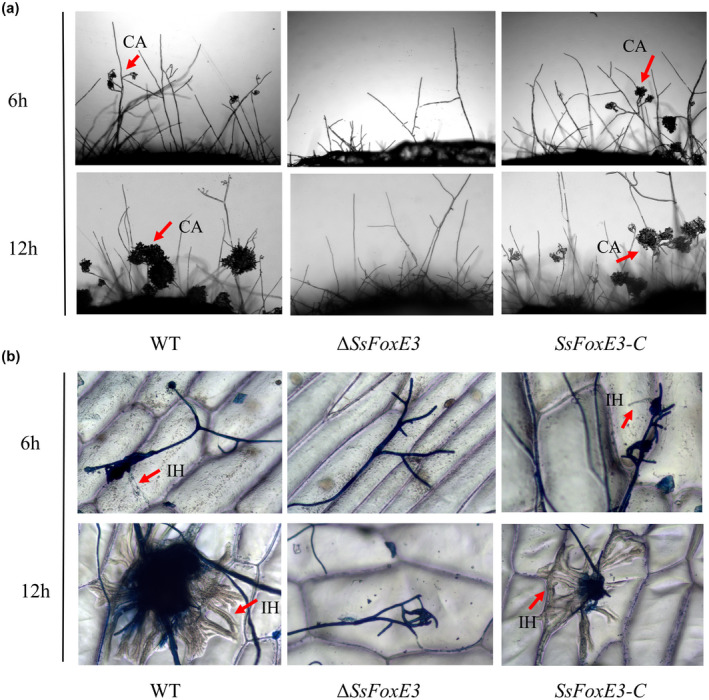
SsFoxE3 affects the formation of invasion hyphae. (a) Development of compound appressoria on a glass slide. The compound appressoria were induced by placing mycelial blocks on a glass slide for 6 and 12 h. (b) Penetration assay of the SsFoxE3 mutant on onion epidermis cell. The invasion hypha was stained with lactophenol blue and observed using differential interference contrast microscopy. Arrows indicate compound appressoria (CA) or invasive hyphae (IH) inside cells
2.4. SsFoxE3 disruption caused hypersensitivity to reactive oxygen species
For plant pathogens to successfully colonize and further develop in host plants, they need a tolerance mechanism to overcome plant defence mechanisms (Chi et al., 2009). Therefore, to explore whether SsFoxE3 plays an active role in tolerance to osmotic stress and exogenous H2O2, strains were inoculated on 5, 10, 15, 20, and 25 mM H2O2‐containing PDA. The hyphal growth of ∆SsFoxE3 was affected, the growth inhibition rate was higher than the wild type (Figure 5a,b). There was no significant difference in response to osmotic stress (Figure S4a). Subsequently, we used 3,3′‐diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) staining to measure the changes in H2O2 and O2− accumulation in the host after inoculation, respectively. The DAB staining of the leaf tissue of the mutant after inoculation was higher than that of the wild type, and the NBT staining was similar to that of wild type (Figure S4b). Because of this difference, we measured the expression of reactive oxygen metabolism‐related genes in the wild type and mutant, and found that SsSOD1 expression did not change significantly but Scat1 was significantly different. There were also significant differences in the expression of autophagy‐related genes in the mutant (Figure S4c). These results indicate that the weakened pathogenicity of the mutant may be because it lacks the mechanism to detoxify active oxygen.
FIGURE 5.
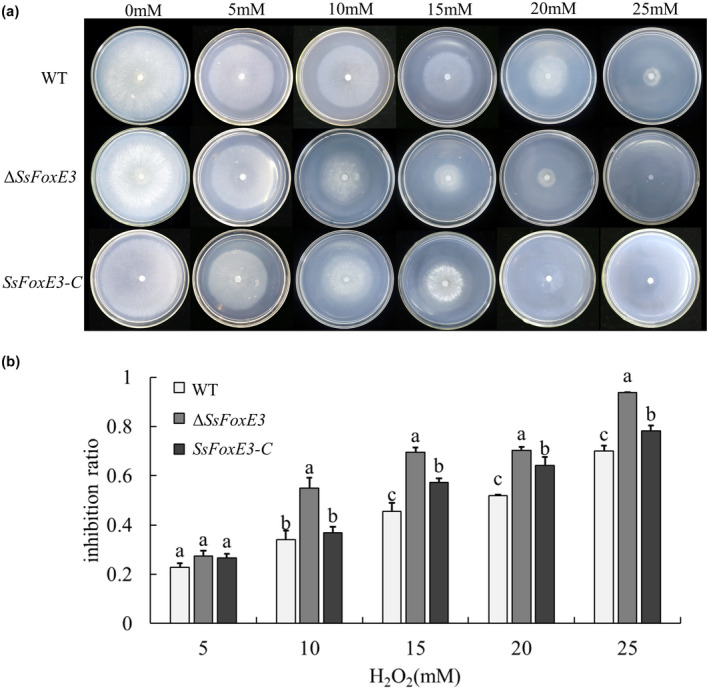
The SsFoxE3 deletion mutant is hypersensitive to oxidative stress. (a) Colonial morphology and mycelial growth of ∆SsFoxE3 under different concentrations of H2O2. (b) Growth inhibition rate of ∆SsFoxE3. Inhibition rate (%) = 100 × (colonial diameter of strain without H2O2 − colonial diameter of strain with H2O2)/(colonial diameter of strain without H2O2). Error bars are standard deviation, different letters represent statistically significant differences (Student's t test, p < 0.05)
2.5. SsFoxE3 transcriptional activation of autophagy‐related genes during invasion
To investigate the function of SsFoxE3 in S. sclerotiorum pathogenicity, we inspected its expression during the infection stage of S. sclerotiorum by reverse transcription quantitative PCR (RT‐qPCR). The expression of SsFoxE3 transcripts in invasive hyphae increased 2‐fold by 5 h postinoculation (hpi) on soybean leaves (Figure 6a). The results suggest that SsFoxE3 is highly activated during infection.
FIGURE 6.
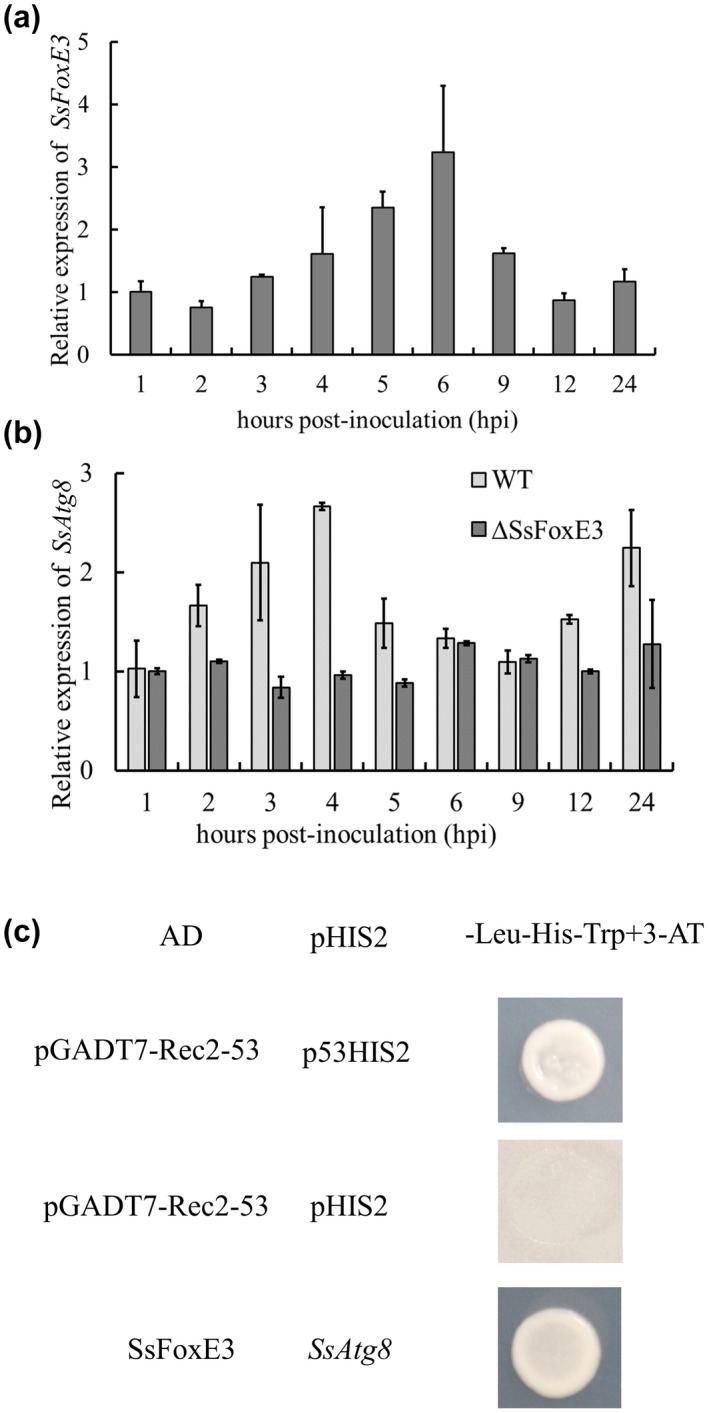
SsFoxE3 transcriptional activation of an autophagy‐related gene, SsAtg8, during invasion. (a) Expression of SsFoxE3 during invasion. (b) SsAtg8 gene expression analysis in the ΔSsFoxE3 mutant and wild type strain (WT) during invasion. (c) SsFoxE3 transcriptional activation of SsAtg8. Yeast 1‐hybrid analysis showed SsFoxE3 interacted with the promoter of SsAtg8. 3‐amino‐1,2,4‐triazole (3‐AT) was added to inhibit SsFoxE3 self‐activation
SsFoxE3 is a transcriptional activator that can self‐activate and make medium supplemented with X‐α‐gal appear blue (Figure S5). To clarify the effect of SsFoxE3 deletion on the expression of autophagy‐related genes (SsAtg1, SsAtg4, SsAtg6, SsAtg8, and SsAtg9), we analysed their expression levels in the wild type and ∆SsFoxE3 during the infection process (Figure S4). The expression of SsAtg8 increased in the early stages of wild‐type infection, but its expression in the ΔSsFoxE3 mutant was lower than that of the wild type, which indicates that SsFoxE3 deletion affected the normal expression of SsAtg8 (Figure 6b). Furthermore, yeast 1‐hybrid (Y1H) analysis showed that SsFoxE3 interacted with the promoter of SsAtg8 (Figure 6c). These results indicate that SsAtg8 may be down‐regulated in ∆SsFoxE3 during the infection process, which means SsFoxE3 could transcriptionally activate the expression of SsAtg8, enhancing autophagy to provide energy to form infectious hyphae.
2.6. SsAtg8 is involved in the formation of compound appressoria, sclerotia, and pathogenicity of S. sclerotiorum
To clarify the role of autophagy in the growth and development of S. sclerotiorum, SsAtg8 was knock out. Compared with the wild type, SsAtg8 deficiency reduced the formation of sclerotia (Figure S6). In ΔSsAtg8 and in ΔSsFoxE3, the number and the weight of sclerotia was significantly reduced on minimal medium, indicating that both of them were necessary for the development of sclerotia when growing under conditions of nutritional deficiency. The formation of compound appressoria decreased in ∆SsAtg8 compared to the wild type (Figure 7a–c). After inoculation of leaves, the pathogenicity of SsAtg8 mutant was significantly decreased, and was recovered by the complemented strain SsAtg8‐C (Figure 7d,e). The phenotype of the SsAtg8 mutant indicated that the effect of SsFoxE3 on sclerotia formation and infection may be related to the autophagy pathway.
FIGURE 7.
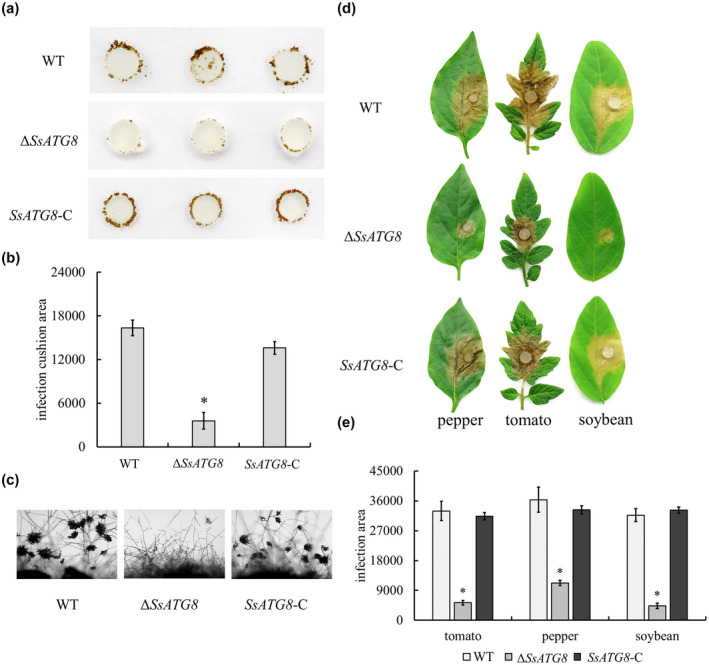
SsAtg8 contributes to compound appressoria formation and pathogenicity. (a) Development of compound appressoria on a hydrophobic interface. Strains were grown on potato dextrose agar for 48 h, then an agar disk containing mycelium (0.5 mm diameter) was inoculated on a sterilized glass slide, moisturized, and cultured for 48 h. (b) Statistical analysis of compound appressoria formation was measured by ImageJ. *Significant differences compared to compound appressoria of wild type (WT) at α = 0.05 (Student's t test). (c) Microscopic observation of compound appressoria formation. (d) Pathogenicity test of ∆SsAtg8 on pepper, tomato, and soybean leaves. (e) Statistical analysis of the lesion area. The experiments were performed three times with 10 pieces of host leaves for each strain. *Significant differences from the wild‐type strain at α = 0.05 (Student's t test)
2.7. SsFoxE3 response to N starvation‐activated autophagy
The generation and modulation of reactive oxygen species is essential for S. sclerotiorum during pathogenic development, successful colonization, and fungal development (Ding et al., 2020; Huang et al., 2021; Liang & Rollins, 2018). To examine whether SsAtg8 is involved in H2O2 tolerance, the wild type was treated with different H2O2 concentrations then changes in the expression level of SsAtg8 were measured. The growth of the ΔSsAtg8 mutant was significantly inhibited by 10 mM H2O2 (Figure S7). In addition to H2O2 induction, N starvation is the main condition for autophagy induction. Thus, we used minimal medium without nitrogen source (MM−N) medium for ∆SsAtg8 and ∆SsFoxE3 culture and found that the growth of ∆SsAtg8 and ∆SsFoxE3 was restricted (Figure 8a,b), indicating that H2O2 and nitrogen starvation could affect the autophagy of S. sclerotiorum.
FIGURE 8.
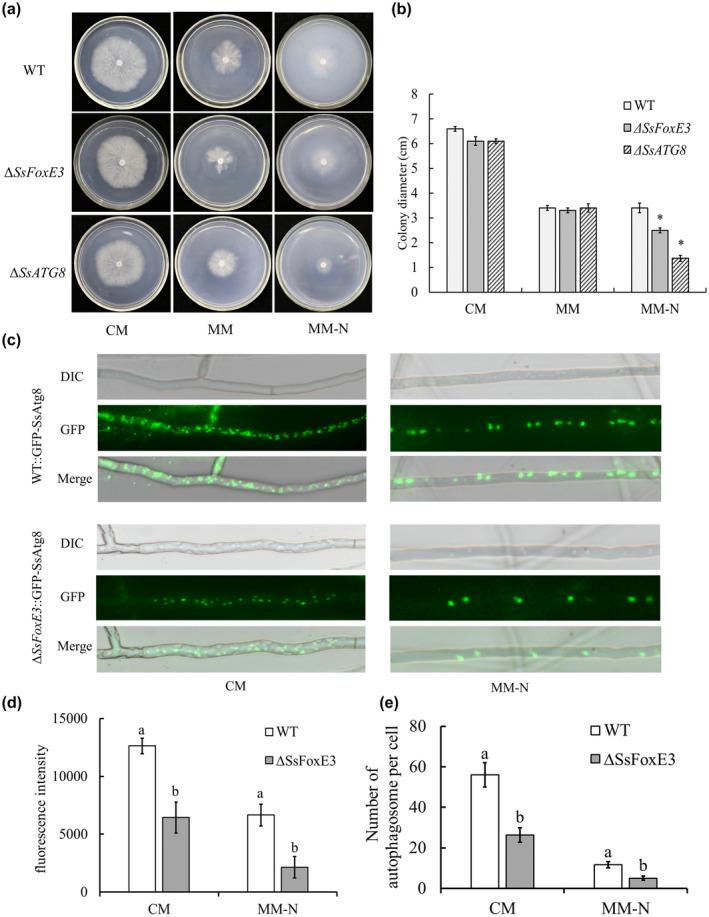
SsFoxE3 in response to N starvation and activated autophagy. (a) Colony morphology of ∆SsFoxE3 and ∆ SsAtg8 incubated on complete medium (CM), minimal medium (MM), and minimal medium without nitrogen source (MM−N at 25°C for 48 h. (b) Colony diameters of ∆SsFoxE3 and ∆ SsAtg8 incubated on CM, MM, and MM‐N for 48 h. Error bars are standard deviation and * indicates significant differences from the wild‐type strain at α = 0.05 (Student's t test). (c) Observation of the formation of autophagosomes in WT::GFP‐SsAtg8 and ∆SsFoxE3::GFP‐SsAtg8. WT::GFP‐SsAtg8 and ∆SsFoxE3::GFP‐SsAtg8 were first grown in liquid CM at 25°C for 24 hr, then transferred to liquid MM−N for 6 hr. (d) Fluorescence intensity assay of WT::GFP‐SsAtg8 and ∆SsFoxE3::GFP‐SsAtg8. ImageJ was used to analyse the fluorescence intensity by split channels. (e) The number of autophagosomes per cell. The experiments were performed three times with 20 cells of each strain. Different letters represent statistically significant differences (Student's t test, p < 0.05)
The autophagy process was observed using green fluorecent protein (GFP)‐tagged SsAtg8. Autophagosome formation of the wild type and ∆SsFoxE3 after N starvation‐induced autophagy was observed under the microscope. It was found that autophagosomes in ∆SsFoxE3 were decreased, and after induction on MM−N the number of autophagosomes in ∆SsFoxE3 were significantly lower than that of WT (Figure 8c–e). The results suggest that SsFoxE3 is involved in the autophagy pathway, and that the autophagy pathway is involved in the process of sclerotia formation and infection.
3. DISCUSSION
TFs are often the downstream effectors of signalling cascades and initiate the transcription and expression of specific genes, finally regulating responses to internal and external signals through the effect of gene products and playing an important regulatory role in environmental responses such as growth and development, morphogenesis, and biological and abiotic stress (Cho et al., 2012; van der Does et al., 2016; John et al., 2021). TFs have been widely used as novel antifungal drug targets (Bahn, 2015). FOX TF SsFkh1 is critical for sclerotia and pathogenicity in S. sclerotiorum (Fan et al., 2017). SsFoxE2 is involved in sexual development and affects the development of ascomycetes (Wang et al., 2016). In M. oryzae, deficiency of MoFKH1 affects mycelial growth, conidial germination, and its pathogenicity, while deficiency of MoHCM1 results in mycelial growth and conidial germination (Park et al., 2014). FKH1, FKH2, HCM1, and FHL1 play different roles in Saccharomyces cerevisiae (Arsenault et al., 2015; Martin et al., 2004; Pataki et al., 2017; Postnikoff et al., 2012). Our study of the function of SsFoxE3 is expected to further clarify the division and cooperation of the FOX TFs in S. sclerotiorum. First of all, we analysed the domain sequence of FOX TFs in S. sclerotiorum and other plant‐pathogenic fungi, and found that SsFoxE3 had the same domain distribution as SsFkh1 and MoFKH1 of M. oryzae, as well as FKH1 and FKH2 in yeast (Figure 1b). In addition, we conducted a cis‐acting regulatory element analysis on the promoter of SsFoxE3 and found that the promoter region contains light responsive (ATCT‐motif, Bo x 4, Box II, G‐Box, TCT‐motif, GT1‐motif), defence and stress responsive (TC‐rich repeats), methyl jasmonate (MeJA) responsive (TGACG‐motif and CGTCA‐motif), and GCN4 motifs, which provides ideas for the subsequent clarification of the expression response of SsFoxE3.
Further study on the function of SsFoxE3 showed that the number and weight of sclerotia of the mutant were significantly reduced, the formation of compound appressoria was abnormal, and the pathogenicity of the mutant was lower than that of the wild type.
The pathogenicity of the ΔSsFoxE3 mutant was reduced. Inoculation and staining revealed that the reduction of pathogenicity was related to the development of the compound appressoria and the formation of infectious hyphae, but was not related to the production of oxalic acid (Figure S2). Subsequently, the expression levels of SsFoxE3 and pathogenicity‐related genes were measured at different inoculation times, and it was found that the expression of SsAtg8 was reduced during the ∆SsFoxE3 infection process (Figure 6b). As an autophagy‐related protein, Atg8 is usually used as a marker protein to monitor the occurrence of autophagy (Liu et al., 2012, 2016). SsFoxE3 localizes in the nucleus (Figure S5a) and has transcriptional activation activity (Figure S5b). In RT‐qPCR and Y1H assays SsFoxE3 activated the transcription of SsAtg8 in response to starvation signals (Figures 6c and 8c). In addition, the SsAtg1, SsAtg8, and Scat1 genes in the SsFoxE3 mutant were still expressed, indicating that there are other TFs involved besides SsFoxE3 (Figure S4c).
Autophagy processes, which are highly conserved cellular processes, degrade cytoplasmic constituents in vacuoles and play important roles in filamentous fungal pathogenicity (Zhu et al., 2019). Subsequently, we analysed the biological characteristics of ∆SsAtg8. After deletion of SsAtg8, the number of sclerotia was reduced (Figure S6), the compound appressoria were defective, and the pathogenicity was reduced (Figure 7). The phenotype was similar to that of the ΔSsFoxE3 mutant. To further examine the relationship between SsFoxE3 and autophagy, we used a GFP‐SsAtg8 fusion protein as a marker. Under N starvation, the number of autophagosomes in the wild type was significantly higher than in ∆SsFoxE3, further indicating that SsFoxE3 is involved in the autophagy pathway (Figure 8c).
In the infection cycle of S. sclerotiorum, compound appressoria are important as an infection structure. They directly determine the parasitic relationship and the degree of disease. Autophagy of filamentous fungi is important not only to their nutritional balance, but also to developmental processes such as cell differentiation, secondary metabolism, and pathogenicity. Understanding the role of the SsFoxE3 TF in the regulation of S. sclerotiorum compound appressoria and autophagy is not only helpful for exploring the classic autophagy model, but also for the development of S. sclerotiorum compound appressoria and provides new clues for pathogenicity. However, the molecular mechanism of autophagy affecting the pathogenicity of S. sclerotiorum still needs further study, and the developmental mechanism of its sclerotia requires further analysis.
4. EXPERIMENTAL PROCEDURES
4.1. Strains and culture condition
S. sclerotiorum UF‐1 was used as the wild‐type strain in this study. ∆SsFoxE3 and SsFoxE3‐C (complemented strain), ∆SsAtg8 and ∆SsAtg8‐C, WT::GFP‐SsAtg8, and ∆SsFoxE3::GFP‐SsAtg8 were initially grown on PDA (200 g potato, 20 g glucose, 15 g agar per litre) at 25°C.
4.2. Identification and sequence analysis of SsFoxE3
The phylogenetic tree was inferred using the neighbour‐joining method. The associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches and the evolutionary analyses were conducted in MEGA 7. The domains contained in the sequence were analysed by Interpro (http://www.ebi.ac.uk/interpro) and visualized by GPS.
4.3. Cell integrity and stress treatment
Cell integrity of strains were determined by culture on PDA with Congo red (CR, 300 mg/ml), CFW (30 μg/ml), 0.001% SDS, 1 M glucose, 1 M sorbitol, 1 M KCl, and 1 M NaCl. For the analysis of oxidative stress, strains were inoculated on PDA with different concentrations of H2O2 (5, 10, 15, 20, 25 mM) (Huang et al., 2021). For testing nutrient uptake, strains were grown on CM (0.2 g KH2PO4, 0.25 g MgSO4.7H2O, 0.15 g NaCl, 1 g Ca(NO3)2.4H2O, 10 g glucose, 1 g yeast extract, 1 g casein hydrolysate, 15 g agar) and MM (without 1 g yeast extract and 1 g casein hydrolysate) and MM−N (i.e., MM without Ca(NO3)2.4H2O). To observe oxalic acid content, strains were grown on PDA with 100 μg/ml bromophenol blue (Li et al., 2018; Liu et al., 2018). The colony diameters were measured after 48 h and the inhibition of hyphal growth was calculated. Each experiment was repeated three times.
4.4. Analysis of virulence and the compound appressoria
To evaluate the virulence, mycelia‐colonized plugs (0.5 mm diameter) from wild type, ∆SsFoxE3, SsFoxE3‐C, ∆SsAtg8, and SsAtg8‐C were harvested from colony margins of 48 h‐old PDA cultures and then inoculated on tomato, pepper, and soybean leaves. The diameter of the lesions were measured after 48 h and each experiment was repeated three times. Compound appressoria of strains were observed on hydrophobic (glass slide) and onion surfaces, stained with lactophenol cotton blue (Sigma). The formation of compound appressoria was observed at 6 and 12 hpi under a microscope. The experiments were conducted three times.
4.5. Plasmid constructs and transformation
Plasmid pXEH‐SsFoxE3, which was used to generate the SsFoxE3 knock‐out transformants, was constructed as follows: plasmid pXEH, containing the HYG marker for hygromycin resistance, was used as the vector backbone. Oligonucleotides SsFoxE3F1EcoRI/SsFoxE3R1XhoI (5′‐GAATTCTGTATTGGATAGAGATGTTG‐3′/5′‐CTCGAGTCGTCCACCTTCTATTGT‐3′) and SsFoxE3F2 BamHI/SsFoxE3R2PstI (5′‐GGATCCCATTTCGTGGGAAGTAAG‐3′/5′‐CTGCAGGGATCCTATGGGCTGCGATGTGAT‐3′) were used to amplify the 5′ and 3′ fragments of SsFoxE3, which was digested with EcoRI/XhoI and PstI/BamHI, respectively, and cloned into pXEH to construct pXEH‐SsFoxE3. pXEH‐SsAtg8 was constructed as before with SsAtg8F1EcoRI/SsAtg8R1KpnI (5′‐GAATTCTCGGTCCTGGGTAACTAT‐3′/5′‐GGTACCGGGAAAGGGTATGGGAGT‐3′) and SsAtg8F1BamHI/SsAtg8R1PstI (5′‐GGATCCGAGGCGGATGATGGATAG‐3′/5′‐CTGCAGAGTGGAATGAGCGGAACA‐3′). These plasmids were used to transform S. sclerotiorum UF‐1 using the polyethylene glycol (PEG)‐mediated transformation method (Rollins, 2003) and protoplasts of S. sclerotiorum were prepared as described previously (Qu et al., 2014). Plasmids pYF11::RP27::eGFP::SsAtg8 and pYF11::RP27::eGFP::SsFoxE3 were used for the overexpression of mutants. Transformants were purified by using hygromycin (100 μg/ml) and G418 (100 μg/ml) selection of hyphal tips at least five times, respectively.
4.6. RT‐qPCR analysis
Gene expression during infection was observed in inoculated soybean leaves at hourly intervals. The RNA of test samples was extracted using a TransZol Up Plus RNA Kit and cDNA synthesis using EasyScript All‐in‐One First‐Strand cDNA Synthesis SuperMix for qPCR (One‐Step gDNA Removal) (TransGen Biotech). Quantitative expression assays were performed using TransStart Green qPCR SuperMix (TransGen Biotech). Primers used for qPCR are shown in Table S2 and actin was used as internal reference.
4.7. Yeast 1‐hybrid assays
The yeast 1‐hybrid (Y1H) assay is a method to analyse the interaction between DNA and proteins in vitro. The bait construct was generated by cloning the cDNA sequence of SsFoxE3 into pGADT7, the prey constructs were generated by cloning ProSsATG8 into pHIS2, and the pairs were cotransformed into yeast strain Y187 and grown on SD–Leu−His medium, then transferred to SD−Leu−His–Trp + 3‐amino‐1,2,4‐triazole. The positive and negative control strains were obtained from the BD library construction and screening kit.
4.8. Autophagy assay
To visualize the autophagic process in S. sclerotiorum, GFP‐SsAtg8 fusion protein plasmid pYF11::RP27:: eGFP::SsAtg8 was constructed and transformed into the wild‐type UF‐1 and ΔSsFoxE3. The autophagosomes were observed in the mycelia of WT::GFP‐SsAtg8 and ∆SsFoxE3::GFP‐SsAtg8, which were cultured in liquid CM at 25°C for 24 h then transferred to liquid MM−N for 6 h and viewed under a fluorescence microscope.
Supporting information
ACKNOWLEDGEMENTS
This work was financially supported by the National Natural Science Foundation of China (31772108, 31972978) and the Inter‐Governmental International Cooperation Special Project of National Key R&D Program of China (2019YFE0114200). All authors declare that there are no conflicts of interest.
Jiao, W. , Yu, H. , Cong, J. , Xiao, K. , Zhang, X. , Liu, J. , et al (2022) Transcription factor SsFoxE3 activating SsAtg8 is critical for sclerotia, compound appressoria formation, and pathogenicity in Sclerotinia sclerotiorum . Molecular Plant Pathology, 23, 204–217. 10.1111/mpp.13154
[Correction added on 9 November 2021, after first online publication: the article title has been updated in this version.]
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Armentrout, V.N. , Downer, A.J. , Grasmick, D.L. & Weinhold, A.R. (1987) Factors affecting infection cushion development by Rhizoctonia solani on cotton. Phytopathology, 77, 623–630. [Google Scholar]
- Arsenault, H.E. , Roy, J. , Mapa, C.E. , Cyert, M.S. & Benanti, J.A. (2015) Hcm1 integrates signals from Cdk1 and calcineurin to control cell proliferation. Molecular Biology of the Cell, 26, 3570–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura, M. , Ninomiya, S. , Sugimoto, M. , Oku, M. , Yamashita, S. , Okuno, T. et al. (2009) Atg26‐mediated pexophagy is required for host invasion by the plant pathogenic fungus Colletotrichum orbiculare . The Plant Cell, 21, 1291–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn, Y.S. (2015) Exploiting fungal virulence‐regulating transcription factors as novel antifungal drug targets. PLoS Pathogens, 11, e1004936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashi, Z.D. , Gyawali, S. , Bekkaoui, D. , Coutu, C. , Lee, L. , Poon, J. et al. (2016) The Sclerotinia sclerotiorum Slt2 mitogen‐activated protein kinase ortholog, SMK3, is required for infection initiation but not lesion expansion. Canadian Journal of Microbiology, 62, 836–850. [DOI] [PubMed] [Google Scholar]
- Bensen, E.S. , Filler, S.G. & Berman, J. (2002) A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans . Eukaryotic Cell, 1, 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer, R. , Pic‐Taylor, A. , Whitehall, S.K. , Martin, K.A. , Millar, J.B. , Quinn, J. et al. (2004) The forkhead transcription factor Fkh2 regulates the cell division cycle of Schizosaccharomyces pombe . Eukaryotic Cell, 3, 944–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, M.H. , Park, S.Y. , Kim, S. & Lee, Y.H. (2009) A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. PLoS Pathogens, 5, e1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, Y. , Srivastava, A. , Ohm, R.A. , Lawrence, C.B. , Wang, K.‐H. , Grigoriev, I.V. et al. (2012) Transcription factor Amr1 induces melanin biosynthesis and suppresses virulence in Alternaria brassicicola . PLoS Pathogens, 8, e1002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquer, M. , Rascle, C. , Goncalves, I.R. , de Vallee, A. , Ribot, C. , Loisel, E. et al. (2021) The infection cushion of Botrytis cinerea: a fungal “weapon” of plant‐biomass destruction. Environmental Microbiology, 23, 2293–2314. [DOI] [PubMed] [Google Scholar]
- Ding, Y. , Mei, J. , Chai, Y. , Yang, W. , Mao, Y. , Yan, B. et al. (2020) Sclerotinia sclerotiorum utilizes host‐derived copper for ROS detoxification and infection. PLoS Pathogens, 16, e1008919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Does, H.C. , Fokkens, L. , Yang, A. , Schmidt, S.M. , Langereis, L. , Lukasiewicz, J.M. et al. (2016) Transcription factors encoded on core and accessory chromosomes of Fusarium oxysporum induce expression of effector genes. PLoS Genetics, 12, e1006401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, H. , Gang, Y. , Liu, Y. , Zhang, X. , Liu, J. , Zhang, Y. et al. (2017) An atypical forkhead‐containing transcription factor SsFKH1 is involved in sclerotial formation and is essential for pathogenicity in Sclerotinia sclerotiorum . Molecular Plant Pathology, 18, 963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, M. , Xu, Y. , Chen, J. , Luo, Y. , Lv, Y. , Su, J. et al. (2018) MoSnt2‐dependent deacetylation of histone H3 mediates MoTor‐dependent autophagy and plant infection by the rice blast fungus Magnaporthe oryzae . Autophagy, 14, 1543–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus, D.D. & Rimmer, S.R. (2005) Sclerotinia sclerotiorum: when “to be or not to be” a pathogen? FEMS Microbiology Letters, 251, 177–184. [DOI] [PubMed] [Google Scholar]
- Huang, Z. , Lu, J. , Liu, R. , Wang, P. , Hu, Y. , Fang, A. et al. (2021) SsCat2 encodes a catalase that is critical for the antioxidant response, QoI fungicide sensitivity, and pathogenicity of Sclerotinia sclerotiorum . Fungal Genetics and Biology, 149, 103530. [DOI] [PubMed] [Google Scholar]
- Jeffries, V.T.P. (1986) Ultrastructure of penetration of Phaseolus spp. by Sclerotinia sclerotiorum . Canadian Journal of Botany, 64, 2909–2915. [Google Scholar]
- John, E. , Singh, K.B. , Oliver, R.P. & Tan, K.C. (2021) Transcription factor control of virulence in phytopathogenic fungi. Molecular Plant Pathology, 22, 858–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsen, L. , Droce, A. , Sondergaard, T.E. , Sørensen, J.L. , Bormann, J. , Schäfer, W. et al. (2012) Autophagy provides nutrients for nonassimilating fungal structures and is necessary for plant colonization but not for infection in the necrotrophic plant pathogen Fusarium graminearum . Autophagy, 8, 326–337. [DOI] [PubMed] [Google Scholar]
- Kabbage, M. , Yarden, O. & Dickman, M.B. (2015) Pathogenic attributes of Sclerotinia sclerotiorum: switching from a biotrophic to necrotrophic lifestyle. Plant Science, 233, 53–60. [DOI] [PubMed] [Google Scholar]
- Li, J. , Zhang, Y. , Zhang, Y. , Yu, P.L. & Rollins, J.A. (2018) Introduction of large sequence inserts by CRISPR‐Cas9 to create pathogenicity mutants in the multinucleate filamentous pathogen Sclerotinia sclerotiorum . mBio, 9, e00567‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Liang, X. & Rollins, J.A. (2012) Sclerotinia sclerotiorum γ‐glutamyl transpeptidase (Ss‐Ggt1) is required for regulating glutathione accumulation and development of sclerotia and compound appressoria. Molecular Plant‐Microbe Interactions, 25, 412–420. [DOI] [PubMed] [Google Scholar]
- Liang, X. & Rollins, J.A. (2018) Mechanisms of broad host range necrotrophic pathogenesis in Sclerotinia sclerotiorum . Phytopathology, 108, 1128–1140. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Wang, Q. , Zhang, X. , Liu, J. , Zhang, Y. & Pan, H. (2018) Ssams2, a gene encoding GATA transcription factor, is required for appressoria formation and chromosome segregation in Sclerotinia sclerotiorum . Frontiers in Microbiology, 9, 3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T.B. , Liu, X.H. , Lu, J.P. , Zhang, L. , Min, H. & Lin, F.C. (2010) The cysteine protease MoAtg4 interacts with MoAtg8 and is required for differentiation and pathogenesis in Magnaporthe oryzae . Autophagy, 6, 74–85. [DOI] [PubMed] [Google Scholar]
- Liu, X.‐H. , Gao, H.‐M. , Xu, F. , Lu, J.‐P. , Devenish, R.J. & Lin, F.‐C. (2012) Autophagy vitalizes the pathogenicity of pathogenic fungi. Autophagy, 8, 1415–1425. [DOI] [PubMed] [Google Scholar]
- Liu, X.H. , Xu, F. , Snyder, J.H. , Shi, H.B. , Lu, J.P. & Lin, F.C. (2016) Autophagy in plant pathogenic fungi. Seminars in Cellular and Developmental Biology, 57, 128–137. [DOI] [PubMed] [Google Scholar]
- Lumsden, R.D. (1973) Histopathology of Sclerotinia sclerotiorum infection of bean. Phytopathology, 63, 708–715. [Google Scholar]
- Lyu, X. , Shen, C. , Fu, Y. , Xie, J. , Jiang, D. , Li, G. et al. (2016) A small secreted virulence‐related protein is essential for the necrotrophic interactions of Sclerotinia sclerotiorum with its host plants. PLoS Pathogens, 12, e1005435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, D.E. , Soulard, A. & Hall, M.N. (2004) TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell, 119, 969–979. [DOI] [PubMed] [Google Scholar]
- Mentges, M. , Glasenapp, A. , Boenisch, M. , Malz, S. , Henrissat, B. , Frandsen, R.J.N. et al. (2020) Infection cushions of Fusarium graminearum are fungal arsenals for wheat infection. Molecular Plant Pathology, 21, 1070–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. , Kong, S. , Kim, S. , Kang, S. & Lee, Y.H. (2014) Roles of Forkhead‐box transcription factors in controlling development, pathogenicity, and stress response in Magnaporthe oryzae . Plant Pathology Journal, 30, 136–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pataki, E. , Weisman, R. , Sipiczki, M. & Miklos, I. (2017) fhl1 gene of the fission yeast regulates transcription of meiotic genes and nitrogen starvation response, downstream of the TORC1 pathway. Current Genetics, 63, 91–101. [DOI] [PubMed] [Google Scholar]
- Postnikoff, S.D. , Malo, M.E. , Wong, B. & Harkness, T.A. (2012) The yeast forkhead transcription factors fkh1 and fkh2 regulate lifespan and stress response together with the anaphase‐promoting complex. PLoS Genetics, 8, e1002583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, X. , Yu, B. , Liu, J. , Zhang, X. , Li, G. , Zhang, D. et al. (2014) MADS‐box transcription factor SsMADS is involved in regulating growth and virulence in Sclerotinia sclerotiorum . International Journal of Molecular Sciences, 15, 8049–8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins, J.A. (2003) The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Molecular Plant‐Microbe Interactions, 16, 785–795. [DOI] [PubMed] [Google Scholar]
- Sang, H. , Chang, H.X. & Chilvers, M.I. (2019) A Sclerotinia sclerotiorum transcription factor involved in sclerotial development and virulence on pea. mSphere, 4, e00615‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, M. , Yamada‐Namikawa, C. , Murakami‐Tonami, Y. , Yoshida, T. , Nakanishi, M. , Urano, T. et al. (2008) Cdc2p controls the forkhead transcription factor Fkh2p by phosphorylation during sexual differentiation in fission yeast. EMBO Journal, 27, 132–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq, V.N. & Jeffries, P. (1984) Appressorium formation by Sclerotinia sclerotiorum: Scanning electron microscopy. Transactions of the British Mycological Society, 82, 645–651. [Google Scholar]
- Veluchamy, S. , Williams, B. , Kim, K. & Dickman, M.B. (2012) The CuZn superoxide dismutase from Sclerotinia sclerotiorum is involved with oxidative stress tolerance, virulence, and oxalate production. Physiological and Molecular Plant Pathology, 78, 14–23. [Google Scholar]
- Voigt, O. , Herzog, B. , Jakobshagen, A. & Pöggeler, S. (2013) bZIP transcription factor SmJLB1 regulates autophagy‐related genes Smatg8 and Smatg4 and is required for fruiting‐body development and vegetative growth in Sordaria macrospora . Fungal Genetics & Biology, 61, 50–60. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Liu, Y. , Liu, J. , Zhang, Y. , Zhang, X. & Pan, H. (2016) The Sclerotinia sclerotiorum FoxE2 gene is required for apothecial development. Phytopathology, 106, 484–490. [DOI] [PubMed] [Google Scholar]
- Xu, L. , Li, G. , Jiang, D. & Chen, W. (2018) Sclerotinia sclerotiorum: An evaluation of virulence theories. Annual Review of Phytopathology, 56, 311–338. [DOI] [PubMed] [Google Scholar]
- Zahiri, A. , Heimel, K. , Wahl, R. , Rath, M. & Kaemper, J. (2010) The Ustilago maydis forkhead transcription factor Fox1 is involved in the regulation of genes required for the attenuation of plant defenses during pathogenic development. Molecular Plant‐Microbe Interactions, 23, 1118–1129. [DOI] [PubMed] [Google Scholar]
- Zhu, X.‐M. , Li, L. , Wu, M. , Liang, S. , Shi, H.‐B. , Liu, X.‐H. et al. (2019) Current opinions on autophagy in pathogenicity of fungi. Virulence, 10, 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


