Abstract
Abstract
Coronavirus disease 2019 (COVID-19) is caused by novel severe acute respiratory syndrome coronavirus (SARS-CoV-2). Its main protease, 3C-like protease (3CLpro), is an attractive target for drug design, due to its importance in virus replication. The analysis of the radial distribution function of 159 3CLpro structures reveals a high similarity index. A study of the catalytic pocket of 3CLpro with bound inhibitors reveals that the influence of the inhibitors is local, perturbing dominantly only residues in the active pocket. A machine learning based model with high predictive ability against SARS-CoV-2 3CLpro is designed and validated. The model is used to perform a drug-repurposing study, with the main aim to identify existing drugs with the highest 3CLpro inhibition power. Among antiviral agents, lopinavir, idoxuridine, paritaprevir, and favipiravir showed the highest inhibition potential.
Graphical abstract
Enzyme – ligand interactions as a key ingredient for successful drug design
Supplementary Information
The online version contains supplementary material available at 10.1007/s11030-021-10355-8.
Keywords: 3CLpro, COVID-19, Radial distribution function, QSAR, Drug repurposing, Paritaprevir
Introduction
Since the outbreak of severe acute respiratory syndrome in 2003 and Middle East respiratory syndrome in 2012 caused by SARS-CoV and MERS-CoV, respectively, viruses from the genus Coronavirus came into the focus of the scientific community [1–6]. Unfortunately, the appearance of a new member of the Coronaviridae family, SARS-CoV-2, in December 2019 in Wuhan, Hubei province, China, caused a global pandemic with significant effects on the health, social, economic, and environmental domains [7–10]. Recently, the first efficient vaccines against COVID-19, a disease caused by the SARS-CoV-2 virus, have become available [11–13]. Nevertheless, FDA has approved only one medicine, the antiviral drug remdesivir, for treatment of COVID-19 requiring hospitalization [14]. Design and development of SARS-CoV-2 drugs is ‘a hot potato’ in nowadays science and pharmaceutical industry. 3C-like protease (3CLpro), papain-like protease (PLpro), nonstructural protein 12 (nsp12) and RNA-dependent RNA polymerase (RdRp) have been selected as the main potential drug targets [15, 16].
The sequence of 3CLpro enzyme of SARS-CoV-2 and SARS-CoV has a high level of similarity (96%) [17]. 3CLpro is a homodimeric enzyme with an essential role in viral replication and transcription [18]. In a dimer, only one protomer demonstrates catalytic activity [19, 20]. With 306 residues, the protomer’s three-dimensional structure is usually divided into three domains (Fig. 1) [21, 22]. An antiparallel β-barrel is the main secondary structure motif of domains I (residues 8–101) and II (residues 102–184). On the other hand, five α-helices form compact antiparallel globular domain III (residues 201–303), connected to domain II by long linker group (residues 185–200). The first seven residues from the N-terminus side are forming the N-finger, which is believed to have a significant role both in dimerization and in establishing proteolytic activity [23, 24]. In the heart of the binding site, located in a cleft between domains I and II, is a conserved histidine-cysteine catalytic dyad. Here, Cys145 has a role of a nucleophile in the first step of the proteolysis, while His41 is the base catalyst [25]. The existence of allosteric binding sites in the groove between domains II and III has been proposed, with the possibility to stop the dimerization and prevent enzyme maturation [26, 27].
Fig. 1.
The structure of SARS-CoV-2 3CLpro in homodimeric form (left) and two perspectives of the monomer (right). The N-finger (residues 1–7) is depicted in dark blue, domain I (residues 8–101) in cornflower blue, domain II (residues 102–184) in orange, the loop region (residues 185–200) in green, domain III (residues 201–306) in red
The beginning of the global COVID-19 pandemic required immediate response—initial idea was to repurpose already approved drugs and use them to fight SARS-CoV-2. Chiou et al. [28] performed screening of 774 FDA-approved drugs against 3CLpro activity. Ethacrynic acid, naproxen, and allopurinol were shown as the most potent SARS-CoV-2 3CLpro inhibitors, with IC50 values below 5 μM. Combined docking and MM-PBSA study [29] of six available anti-HIV drugs, which act as HIV-1 protease inhibitors, identified indinavir and darunavir as potential anti-COVID-19 drugs. At the same time, clinical trials for lopinavir–ritonavir cocktail have shown the combination to be ineffective for the treatment of severe COVID-19 cases [30]. Alamri et al. [31] combined pharmacoinformatics with molecular dynamics studies to reveal potential covalent inhibitors capable of binding to the thiol group of Cys145. Additionally, they proposed paritaprevir and simeprevir, anti-hepatitis C virus drugs acting as NS3/4A serine protease inhibitors, as best hits from FDA-approved drugs list for clinical trials to fight COVID-19. Khan et al. [32] performed docking and molecular dynamics simulations to propose paritaprevir and raltegravir as lead candidates for inhibition of SARS-CoV-2 3CLpro, and dolutegravir and bictegravir for inhibition of 2′-O-ribose mathylotransferase. Alternative approach seeks inspiration from nature, from microbial natural products [26] to phytocompounds [33–36], to pinpoint active compounds useful for treatment of COVID-19 patients.
Structural and evolutionary analysis of SARS-CoV and SARS-CoV-2 main proteases indicated that the design of novel inhibitors or repurposing existing ones might be challenging [37]. Detailed molecular dynamics simulations of 3CLpro reveal several difficulties one has to be aware of in designing 3CLpro inhibitors. Although SARS-CoV and SARS-CoV-2 main proteases differ by 12 residues located mostly on the enzymes’ surface, both shape and size of the binding site experience significant changes, due to its flexibility and plasticity. The encouraging result of this study is the identification of a small number of residues with significant contribution to the protein stability—a potential target for a new class of inhibitors. Recently, Chen et al. [38] gave an overview of potential inhibitors of SARS-CoV, MERS-CoV, and SARS-CoV-2 main proteases in lower μM and sub-μM regimes. Because of the lack of antivirus activity of peptide-like 3CLpro inhibitors in animals due to interactions with host proteins, they suggested small molecular inhibitors, with higher solubility and lower cytotoxicity. In the same line is work by Zhang et al. [39] who reported a series of noncovalent 3CLpro inhibitors with 20 nM potency.
Great variety of artificial intelligence and machine learning methods were exploited with the aim to repurpose existing or to design new and effective anti-COVID-19 drugs [40–45]. Nand et al. [40] trained a decision stump machine learning predictive model to reduce a dataset of 1528 anti-HIV compounds to 356 compounds with strong bioactivity against 3CLpro. Then, in a series of steps, which included Lipinski’s rule of five filter, molecular docking, application of deep learning model, structural activity relationship analysis, and molecular dynamics simulations, two molecules—CID-230119 and CID-948801 were identified as hit compounds. Mohapatra et al. [45] designed a machine learning model based on the Naïve Bayes algorithm with 73% accuracy, and among FDA approved drugs found antiviral drug amprenavir as the most effective for the treatment of COVID-19. Except repurposing anti-HIV drugs against 3CLpro, Khan et al. [46] performed virtual screening of Traditional Chinese medicines database. For top hits compounds based on docking scores, they performed 100 ns molecular dynamics simulations. Based on RMSD, RMSF, and binding free energy (estimated using MM/GBSA approach) analysis, saquinavir and TCM5280805 emerged as compounds with the highest potential inhibitory role within the screened database. Kumar and Roy developed multiple linear regression (MLR) model and identified structural descriptors contributing to the increase and decrease of inhibitory potential [47]. Janairo et al. [48] build MLR, support vector regression (SVR), classification and regression trees (CART), random forest, and artificial neural networks (ANN) models predicting binding free energies and compared their performances. Exploiting five topological descriptors, the MLR model achieved the best score with R2 being 0.81. Among several developed machine learning models, Kumari and Subbarao pointed to the convolutional neural network (CNN) one, as being the most potent one in the binary classification of anti-SARS molecules [49].
In this paper, we calculated the recently introduced radial distribution function (RDF) weighted by the number of valence shell electrons [50–52] for 159 experimentally determined structures of SARS-CoV-2 3CLpro complexed with different ligands. The structural advantages of RDF, like it unambiguously describes 3D structures, its independence of the size of a molecule, and being invariant against translation and rotation of a molecule, are upgraded with electronic properties characteristic for each atom in a molecule. After design and careful validation of a model capable of predicting bioactivity against SARS-CoV-2 3CLpro, the activity was predicted for 6407 FDA approved and experimental compounds, revealing potential inhibitors of the main protease within the DrugBank database [53, 54].
Methods
Radial distribution function
The Protein Data Bank [55, 56] was queried for SARS-CoV-2 3CLpro (the main protease) structures with small molecules (ligands) bound in the active site. Our in-house software was used to manipulate downloaded pdb files. All water molecules, ions, additives, and small molecules outside the active site were removed. Since we are interested in estimating the similarity of the enzyme itself, we also deleted ligands. If the experimental structure was resolved for a homodimeric complex, only chain ‘A’ was retained. In the case when there were several structures with the same ligand, the structure with better resolution entered our dataset. Structures with two or more missing residues were not considered. For all main proteases from our dataset radial distribution function (RDF) weighted by the number of valence shell electrons (g[r]) were calculated.
Briefly, the RDF vector, whose size is defined by the distance of its two most distant atoms (rMAX), represents each structure [50, 57]. The elements of the vector are g(r) values, calculated in 0.1 Å intervals:
| 1 |
aij is the sum of atomic polarizabilities of atoms i and j, rij the distance between atoms i and j, N is the number of atoms in a molecule and the preexponential factors pi and pj account for the number of outer electrons of the i-th and j-th atoms, correspondingly. If we define average RDF, , as
| 2 |
where n is the number of structures in dataset, then the similarity index, σA, can be estimated
| 3 |
In that case, the similarity between RDFs of structure A and the averaged RDF is the overlapped area divided by the area under averaged RDF.
To test the hypothesis that perturbations induced by ligand bound into the catalytic pocket are local in nature, 109 structures with ligand in the proximity of Cys145 were selected. For each complex, all residues having at least one atom within 5.12 Å from the ligand were listed. 25 residues, referred from now on as catalytic pocket residues, fulfilled distance-based criteria for 7K40, complexed with ligand boceprevir. To treat all active sites on the same footing, from original pdb files catalytic pocket residues were extracted, followed by calculation and analysis of g(r). Ligands were excluded from calculations, to preserve consistency and for easier similarity index comparison. For more details see References [50, 52].
As an additional measure of similarity, the root mean square deviations of Cα atomic positions for all structures were evaluated. To do so, structures were superimposed by creating pairwise sequence alignments first, followed by fitting the aligned residues using the MatchMaker module of Chimera [58], and default parameters. Then, the pairwise root mean square deviations (RMSD) of CA atoms in the protein backbone for all structures were calculated.
QSAR model design and validation
A list of SARS-CoV 3CLpro inhibitors with experimentally determined IC50 values constituting our training set (see Table SI1 in the Supporting Information) is obtained from the ChEMBL database [59, 60]. The IC50 values were converted to pIC50, while 3D structures were generated using the GP_global module available at the chemosophia.com site [61]. Geometries were optimized by the MultiGen algorithm for global minimization with conserving initial stereochemistry [62, 63]. Those molecules were used to reconstruct the molecular field of the model receptor using 3D-QSAR Cinderella’s Shoe (CiS) algorithm introduced in References [64–66]. The molecular field in the CiS method is represented by Coulomb and van der Waals potential on the molecular surface of each m-th atom of the ligand molecule with j-th pseudo-atom of the modeled receptor [67, 68]. Those contributions are calculated using the MERA force field [67, 69]. The performance of the CiS algorithm was thoroughly tested using various small molecules datasets and for different kinds of bioactivities and proved as a high quality classification scheme, with cross-validation quality usually above 0.9 [62, 65, 70–72]. The neural network approach was used to model the relationship between bioactivity (pIC50) and CiS descriptors. The computed bioactivity was transformed to the desirability function. The desirability function [65], offers an alternative approach in the drug classification problem, defining the probability of the activity as a value between 0 (minimum probability of bioactivity) and 1 (maximum probability of bioactivity). As an external validation of the model, the desirability function for 38 molecules experimentally verified against the SARS-CoV-2 3CLpro target was predicted (see Table SI2 in Supporting Information for the list of the molecules and their desirability function). Based on the analysis of the confusion matrix, the desirability function’s threshold value was determined, discriminating active from inactive compounds. Technical details about QSAR model design and validation are summarized in Table 1. For specific implementation details see References [67, 68].
Table 1.
Parameters and validation of QSAR models for a prognosis of anti-SARS-CoV bioactivity
| QSAR model | SARS-CoV 3CLpro |
|---|---|
| Method | Neural network |
| Algorithm | CiS |
| Potential | Coulomb, van der Waals |
| Internal validation | Tenfold cross-validation with 80:20 set split |
| Cross-R2 | 0.91 |
| External validation | 38 molecules set from Mody et al. [73] |
Bioactivity prediction
A database of FDA approved and experimental drugs was obtained from DrugBank (version 5.1.7) [53, 54]. All mixtures, charged species, and compounds containing metals were excluded, and 6407 molecules remained in the final database, constituting the prediction set. In case the 3D structure of the drug was not part of the sdf file downloaded from DrugBank, the 3D molecular structure was generated by RDKit [74]. MM3 molecular mechanics force field was used for geometry optimization and global minimum search [75], with special attention paid to avoid inversion of chiral centers. For optimized structures, activity against SARS-CoV-2 3CLpro was predicted and transformed to desirability function using our newly developed model.
Molecular docking
The structure of the SARS-CoV-2 3CLpro was taken from Reference [26]. It was extracted from 900 ns molecular dynamics simulation, as a representative structure of the dominant conformation, with a population above 86%. Standard protocol for target preparation was followed—Gasteiger charges were added to each atom and nonpolar hydrogens were merged. After atom type determination, the structure was saved as pdbqt file using Chimera [76]. AutoDockTools 4 [77] were used to prepare fifteen FDA-approved drugs with the highest predicted activity against SARS-CoV-2 3CLpro for docking. The center of the grid box was at the position of Cys145 CA atom, with Cartesian coordinates 13.3, 58.2, and 45.4, and the size of the box 20 × 25 × 25 Å3. Exhaustiveness and the number of modes were set to 100. All ligand poses within 4 kcal mol−1 relative to the pose with the highest score were saved, and after visual inspection of the plausibility, the conformation with the lowest binding energy bound was kept. Docking experiments were performed using the AutoDock Vina [78] software. The appropriateness of our approach was validated in our previous study [27].
Results and discussion
Structural analysis
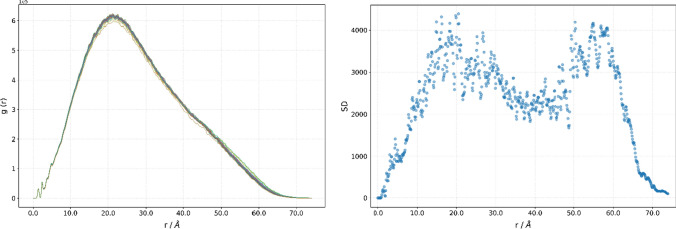
All SARS-CoV-2 3CLpro from our dataset share the same primary structure. 130 out of 159 structures have two residues whose positions have not been resolved. Those missing residues are Ser1 and Gln306, in two cases, and Phe305 and Gln306 in the rest of the cases. The role of a hydrogen atom and hydrogen bonds in the chemistry of life should not be underestimated [79–81]. However, since a hydrogen atom has only one electron, it is extremely hard to obtain its position accurately using an X-ray crystallography. To avoid introducing additional errors into the experimental structure by modelling protonation states and the site of protonation for residues’ side chains, g(r) was calculated only for non-hydrogen atoms [51]. Properties of g(r), like that it unambiguously describes a 3D arrangement of the atoms, and it is invariant against translation and rotation of a molecule, enable us to draw some conclusions about the proteases’ structures, just by comparing its g(r). As can be seen from Fig. 2, displaying g(r) of all 3CLpros from our dataset, RDF curves are very similar, sharing the same features. For example, two sharp spikes at 1.4 Å and 2.4 Å are followed by two less pronounced spikes at 3.8 Å and 4.9 Å. The fine structure is lost for distances above 5 Å. The global maximum of the function is in the 20.4 Å to 22.4 Å range, and the difference in the g(r) maximum value is below 4%. The analysis of standard deviations showed that the g(r) of proteases differs the most at 20.5 Å (Fig. 2, right). The standard deviation at that distance is 4386.5, being only 0.71% of the maximal value at that point.
Fig. 2.
Radial distribution function weighted by the number of valence shell electrons, g(r), for a series of SARS-CoV-2 3CLpro (left), and its standard deviations (right)
The maximum of the first spike, 1.4 Å, could be interpreted as the mean interatomic distance between two neighboring atoms, while a spike at 2.4 Å is a mean distance between two atoms separated by two bonds. Since only non-hydrogen atoms are considered, only carbon–carbon, carbon–oxygen, carbon–nitrogen, and carbon–sulphur interactions contribute to the total g(r). The similarity index, σ, varies between 0.9808 and 0.9998. Having in mind that there are no mutant proteases in our dataset, the differences in the structure can be explained by different experimental conditions or as a structural rearrangement due to perturbation introduced by bound ligands. In addition, in our dataset, both covalent and non-covalent inhibitors are present. High similarity between g(r) indicates that the structural changes experience the tertiary structure of the enzyme, most probably by reorientation of flexible loops and/or side chains.
Based on the analysis of the residues having at least one atom closer to a ligand then 5.12 Å, 25 catalytic pocket residues were identified (Thr25, Thr26, Leu27, His41, Met49, Tyr54, Phe140, Leu141, Asn142, Gln143, Ser144, Cys145, His163, His164, Met165, Glu166, Leu167, Pro168, Val186, Asp187, Arg188, Gln189, Thr190, Ala191, Gln192) (Fig. 3). As a template, a 7K40 structure was used, with boceprevir being the inhibitor having the most close contacts. On the other side, complexes 5RHB and 5RHC, having small methanimine derivatives, have only nine residues fulfilling the 5.12 Å criterion. To treat all structures on the same footing, in our analysis of the active pocket, we include all 25 catalytic pocket residues for all structures, and to reduce ‘noise’, inhibitors were excluded. g(r) of catalytic pocket share similar features for small distances with g(r) of all proteases, with resolved maxima at 1.4 Å, 2.4 Å, 3.7, Å and 4.8 Å. The similarity index, σ, is more spread, being in the range between 0.9485 and 0.9919. While the mean σ for the catalytic pocket is 0.9860 ± 0.0060, σ equals 0.9971 ± 0.0025 for the whole enzyme for the same data set. Two-sided Student’s t test showed that the difference is statistically significant (t = −17.9, p = 1 × 10–44). This finding corroborates our assumption that although large amplitude motions of domain III influence the geometry of the active site, perturbations introduced by bound ligands (even covalently bound) are local. One can see from the inset on Fig. 3 that g(r) of the active pocket has the biggest standard deviation at 8.8 Å. At this distance, we identify atoms with dominant contributions to the g(r = 8.8 Å). The dominant contribution is defined as a g(r) value larger than the mean g(r) contribution plus two standard deviations for a specific distance r [50]. Nitrogen atoms forming peptide bond from Glu166 and Met165, and peptide’s bond oxygen atom from Val186 are three atoms with a dominant contribution to 101, 99 and 94 complexes out of 109, respectively. While Met165 and Glu166 are part of the β sheet in domain II, Val186 is part of a nonstructured loop connecting domains II and III. In the 7K40 complex of 3CLpro with boceprevir, Glu166’s nitrogen forms a hydrogen bond with boceprevir’s carbonyl oxygen and the distance between the two atoms is 2.95 Å. Met165’s nitrogen and Val186’s oxygen are not in direct contact with the ligand, but those residues are neighbors through space and influence the depth of the active site. It is interesting to note a big difference in the occurrence of the atoms of two catalytic dyad residues as atoms with a dominant contribution. While the NE2 atom of His41 is dominant for 89 structures, the S atom of Cys145 is having the dominant contribution in only 11 complexes. Covalent inhibitors are binding to the S atom of Cys145, restricting its flexibility. At the same time, His41 has to adapt to bound ligands, and reorientation of the imidazole ring is the way to optimize interaction patterns.
Fig. 3.
The catalytic pocket of 7K40, with inhibitor boceprevir (left). Radial distribution function weighted by the number of valence shell electrons, g(r), for a series of SARS-CoV-2 3CLpro catalytic pocket (right), and its standard deviations (inset)
Well-established procedures for getting insight into the structural differences include widely accepted protein overlay and root mean square deviation calculations. Pairwise RMSD of carbon atoms in the backbone was calculated. The resulting heat map of RMSD values is presented in Fig. 4. Although here side chains are neglected, and the analysis was performed only for the enzyme’s backbone, some valuable conclusions could be drawn. Only five structures have RMSD values above 1.0 Å—6LZE, 6M0K, 6W79, 7BUY, and 7JU7. 6M0K and 5RF9 differ the most, with the RMSD value being 1.78 Å. When those two structures are overlaid, one can see that the most significant difference is in the C-terminus. The last few residues, starting from Cys300, showed the greatest flexibility. In 5RF9, they are oriented toward domain II, while in 6M0K (and 6LZE, 6W79, 7BUY, and 7JU7) are pointing to the side of the domain III. Additionally, this position enables interaction between C- and N-terminus, with interatomic distance between CA atoms of Ser1 and Val303 being below 6.4 Å. The reason for the higher flexibility might lie in the fact that they do not participate in secondary structure formation and as terminal residues, their motion is restricted only from one side. Both N-finger and residues around the C-terminus’ last helix are known to have an important role in the enzyme dimerization [82, 83].
Fig. 4.
Structural analysis of SARS-CoV-2 3CLpro. Pairwise RMSD presented as a heat map (left). An overlay of two structures with the highest RMSD value (right) highlighting the area with the biggest structural difference (yellow rectangle). 5RF9 (blue), 6M0K (red)
Bioactivity prediction and docking
Reliable models with high predictive power are ‘must have’ tools for successful drug repurposing. Tenfold cross-validation of our model was performed as an internal validation method. The cross-Q2 equals to 0.91, indicating the model’s robustness and high predictive ability. Recently, Mody et al. [73] performed an in vitro enzymatic inhibitory assay study, testing enzymatic activity of 3CLpro against selected drugs (including viral protease inhibitors, viral non-protease inhibitors, and off-target drugs). We used those results as an external set to validate our model. The desirability function value of 0.82 is identified as the threshold for binary classification of compounds. Compounds are classified as being inactive when the desirability function predicted value is lower than 0.82, or active if equal or higher. The threshold is obtained by analyzing the confusion matrix, and statistical parameters derived from it, like accuracy and Matthews correlation coefficient (MCC). The elements of the confusion matrix, true positive (TP), false positive (FP), true negative (TN), and false negative (FN), were used to calculate both the MCC and the accuracy . For the desirability function being 0.82, the accuracy and the MCC are 0.84 and 0.41, respectively. Here is important to point out that according to Mody et al. [73] lopinavir is classified as non-active. But if lopinavir is classified as active, according to Zhang et al. [84], desirability function threshold is 0.87, with improved accuracy and MCC values of 0.89 and 0.62, respectively. Ivermectin, tipranavir and paritaprevir, with experimental IC50 values equal to 21.5 μM, 27.7 μM, and 73.4 μM [73], are also predicted to be active against 3CLpro enzyme by the model.
Since our model demonstrated predictive potential during validation, we predicted the activity of 6400 molecules from the DrugBank database against SARS-CoV-2 3CLpro. All results are compiled in Table SI3 in Supporting Information. 17 molecules with the highest activity are listed in Table 2. The highest predicted activity against 3CLpro has toremifene, a nonsteroidal selective estrogen receptor modulator, used in the treatment of advanced breast cancer. According to ClinicalTrials.gov (identifier NCT04531748), a randomized, double-blind, controlled clinical trial is trying to evaluate the effects of toremifene in adults with mild COVID-19 [85]. Martin and Cheng suggested toremifene’s mechanism of action as a potential blocker of the spike glycoprotein and methyltransferase nonstructural protein 14 (NSP14) inhibitor [86]. But, 500 ns long molecular dynamics simulation of 3CLpro complexed by toremifene showed that after 284 ns toremifene leaves the binding pocket.
Table 2.
Top 17 compounds from DrugBank with the highest predicted bioactivity against SARS-CoV-2 3CLpro, together with their 2D structures and docking score (in kcal mol−1)
The mechanism of cleavage of polyproteins by 3CLpro has been investigated for the SARS-CoV virus [18, 87]. Since the initial step includes deprotonation of Cys145 thiol and nucleophilic attack of anionic sulfur on the carbonyl carbon atom, a variety of peptidomimetics and small molecule covalent inhibitors have been proposed [88]. Independent of the nature of the potential inhibitor (covalent/non-covalent targeting active pocket), it has to interact with the target. Being aware of the drawback of the docking experiments [89–91], we performed docking of hit molecules against SARS-CoV-2 3CLpro, to get a general idea about interactions between potential inhibitor and target within the catalytic pocket. 900 ns molecular dynamics simulation of free, unbound 3CLpro performed by Novak et al. identified two dominant enzyme’s conformations, with predicted populations of 86.7% and 13.3% [26]. Although the main structural difference is the large amplitude motion of domain III, it affects the geometry of the catalytic pocket, being wider in the dominant conformer. Succinamide-CoA (DB03905) is a quite big compound, classified as experimental, with a molar mass of 850.6 g mol−1. Because of flexibility and numerous functional groups, it is capable to form a variety of interactions with residues forming the pocket (Fig. 5). For example, it forms hydrogen bonds with Thr26, Asn28, Asn119, Phe140, Asn142, Gly143, Cys145, Hie163, Hie164 residues. When it is bound, it completely blocks access to catalytic dyad residues, His41 and Cys145 (yellow in Fig. 5, left). 2D interaction networks between all hit molecules and 3CLpro are presented in Figure SI1 in Supporting Information. In most of the cases, hydrogen bonds, π-sulphur, π-alkylic, π–π, and van der Waals interactions are responsible for enzyme—ligand binding. More accurate calculations, like molecular dynamics simulations and free energy of binding calculations, are needed to provide a detailed description of complex interaction patterns.
Fig. 5.
Molecular docking results—insight into the catalytic pocket (left) and interaction network (right) of Succinamide-CoA (DB03905) and SARS-CoV-2 3CLpro. Van der Waals surface of His41 and Cys145 is depicted in yellow, all other residues’ are in blue
Several drug repurposing studies suggested well-known antiviral drugs as potential SARS-CoV-2 3CLpro inhibitors. According to DrugBank [54], more than 70 compounds within our dataset are registered direct antiviral agents. Therefore, we were interested in the performance of our model and its possibility to point out promising antiviral drugs as 3CLpro inhibitors. Our predictive model identified eight currently used antiviral drugs as active against SARS-CoV-2 3CLpro (Table 3). In this section, we will put our results into a broader perspective, and see how the proposed molecules perform in in vitro, in vivo and in in silico studies. Lopinavir, a well-known HIV-1 protease inhibitor is predicted to have the highest activity against 3CLpro. It has been used to treat SARS and MERS patients but without proven efficacy [4]. Although lopinavir and its analogues were subjected to many studies [92–95], in clinical trials lopinavir-ritonavir combination have not been proven to be effective for the treatment of severe cases of COVID-19 [30]. Zhang et al. [84] demonstrated lopinavir inhibition potential against SARS-CoV-2 3CLpro in vitro, and by extrapolation to in vivo, they concluded that due to very low concentration of free, unbound to plasma proteins, lopinavir is not effective against SARS-CoV-2 in vivo. Paritaprevir [31, 32, 73, 96], favipiravir [34, 92], atazanavir [97–99], ganciclovir [95, 97], tipranavir [73, 98, 100–102], and bictegravir [32], were part of computational studies trying to repurpose existing drugs against COVID-19. Paritaprevir is a compound containing an acylsulfonamide moiety and is being used in treatment of hepatitis C. It is inhibiting viral NS3/4A serine protease, with Ser139, His57 and Asp81 constituting catalytic triad [103, 104]. Favipiravir, a pyrazinecarboxamide derivative, is a broad spectrum inhibitor of RNA viral replication, currently registered for influenza treatment [105, 106]. Clinical trials indicate its potential use on moderately to critically ill COVID-19 patients [107, 108]. Although computational studies of atazanavir and tipranavir, a HIV-1 protease inhibitors, suggested they might be good 3CLpro inhibitors, the careful analysis of its efficacy in cell culture and in vitro enzymatic assays revealed limited potential due to the requirement of high concentrations of the drugs to achieve significant inhibition [98]. Results of those studies show that mentioned antiviral drugs have potential to fight COVID-19 pandemic, and at the same time are an independent validation of our model. From the pool of more than 6400 different molecules, our approach enriched the final list of molecules with the compounds that were identified as active either by other theoretical methods or by experiments.
Table 3.
List of direct antiviral drugs that are predicted to show inhibition activity against SARS-CoV-2 3CLpro
| DB ID | Name | Activity | Drug descriptiona |
|---|---|---|---|
| DB01601 | Lopinavir | 0.991 | An HIV-1 protease inhibitor used in combination with ritonavir to treat human immunodeficiency virus (HIV) infection |
| DB00249 | Idoxuridine | 0.961 | A pyrimidine analog antiviral used for the treatment of viral eye infections, including herpes simplex keratitis |
| DB09297 | Paritaprevir | 0.894 | A direct acting antiviral agent used in combination with other antiviral agents for the treatment of Hepatitis C Virus (HCV) infections |
| DB12466 | Favipiravir | 0.881 | An antiviral used to manage influenza, and that has the potential to target other viral infections |
| DB01072 | Atazanavir | 0.865 | An antiviral protease inhibitor used in combination with other antiretrovirals for the treatment of HIV |
| DB10004 | Ganciclovir | 0.857 | A DNA polymerase inhibitor used to treat cytomegalovirus and herpetic keratitis of the eye |
| DB03312 | Brivudine | 0.844 | Brivudine is used in the treatment of herpes zoster |
| DB00932 | Tipranavir | 0.836 | A protease inhibitor used to treat HIV-1 resistant to more than 1 protease inhibitor |
ahttps://go.drugbank.com/categories/DBCAT002140 (Accessed 18 Mar 2021)
Conclusions
This study had three goals. First, structural similarity analysis based on radial distribution function weighted by the number of valence shell electrons of SARS-CoV-2 main protease obtained by X-ray crystallography was performed. Independent from different experimental conditions of crystallization, different space groups and different inhibitors bound into the enzyme’s catalytic pocket, the RDF-based similarity index is within the 0.9808 and 0.9998 range. This suggests that perturbations of the 3CLpro introduced by the ligand are local, concentrated in the vicinity of the active pocket. This finding is corroborated by independent analysis of the RMSD of CA atom type from protein’s backbone and additional analysis of the g(r) of the catalytic pocket.
The second goal was achieved by successful design and validation of the QSAR model capable of predicting activity against SARS-CoV-2 3CLpro. After reconstruction of the pseudo-receptor complementary to the external field of bioactive molecules using the CiS algorithm, the neural network was used to train the model. Internal predictive power was tested by tenfold cross-validation, giving the cross-Q2 equal to 0.91. Since a high value of cross-Q2 is a necessary condition for a model’s high predictive ability, but it is not a sufficient condition, additional external validation was performed. The value of R2 of 0.90 demonstrated the model’s high predictive ability for external molecules.
Finally, a newly developed predictive model was exploited for drug repurposing. From the list of FDA approved and experimental drugs, we identified molecules with the highest probability of being SARS-CoV-2 3CLpro inhibitors. Special attention was paid to existing antiviral drugs. Lopinavir, a HIV-1 protease inhibitor, is predicted to have the highest potential to inhibit SARS-CoV-2 3CLpro. Although it is confirmed by in vitro experiments that it inhibits SARS-CoV-2 3CLpro, its effectiveness in the treatment of severe COVID-19 cases is questionable due to the very low concentration of free lopinavir, unbound to plasma proteins. Other antiviral agents, like paritaprevir, identified by our model as prosperous are also under investigation by other groups or have already reached clinical trials. These independent results support the good performance of the model. Benefits of this research include short-term benefits, including fast drug repurposing possibilities, and on a long-term scale, reliable model for prediction of bioactivity against 3CLpro is developed and validated.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
J.N. would like to pay special gratitude and respect to the late colleague and supervisor Professor Vladimir A. Potemkin for the introduction to the exciting field of computer-aided drug design. Special thanks go to him for his great ideas, hard work, enthusiasm, and unique possibilities to explain complex concepts using everyday pictures and Russian proverbs.
Funding
This work was supported by the RFBR, DST, CNPq and SAMRCA under Grant 20-53-80002.
Declarations
Conflict of interest
The authors declare no competing financial interest.
Footnotes
This paper is dedicated to the memory of Professor Vladimir A. Potemkin, who passed away while conducting the research reported in this work.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Deceased: Vladimir A. Potemkin.
References
- 1.Vijayanand P, Wilkins E, Woodhead M. Severe acute respiratory syndrome (SARS): a review. Clin Med (Northfield Il) 2004;4:152–160. doi: 10.7861/clinmedicine.4-2-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang H, Xie W, Xue X, et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3:e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng VCC, Lau SKP, Woo PCY, Kwok YY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fehr AR, Channappanavar R, Perlman S. Middle east respiratory syndrome: emergence of a pathogenic human coronavirus. Annu Rev Med. 2017;68:387–399. doi: 10.1146/annurev-med-051215-031152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song Z, Xu Y, Bao L, et al. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11:59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdelrahman Z, Li M, Wang X. Comparative review of SARS-CoV-2, SARS-CoV, MERS-CoV, and Influenza A respiratory viruses. Front Immunol. 2020 doi: 10.3389/fimmu.2020.552909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou P, Yang X-L, Wang X-G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicola M, Alsafi Z, Sohrabi C, et al. The socio-economic implications of the coronavirus pandemic (COVID-19): a review. Int J Surg. 2020;78:185–193. doi: 10.1016/j.ijsu.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mofijur M, Fattah IMR, Alam MA, et al. Impact of COVID-19 on the social, economic, environmental and energy domains: lessons learnt from a global pandemic. Sustain Prod Consum. 2021;26:343–359. doi: 10.1016/j.spc.2020.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voysey M, Costa Clemens SA, Madhi SA, et al. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet. 2021;397:881–891. doi: 10.1016/S0140-6736(21)00432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Logunov DY, Dolzhikova IV, Shcheblyakov DV, et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397:671–681. doi: 10.1016/S0140-6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.FDA Approves First Treatment for COVID-19 (2021) https://www.fda.gov/news-events/press-announcements/fda-approves-first-treatment-covid-19. Accessed 25 Feb 2021
- 15.Naqvi AAT, Fatima K, Mohammad T, et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165878. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang M-Y, Zhao R, Gao L-J, et al. SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol. 2020;10:1–17. doi: 10.3389/fcimb.2020.587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Zhao S, Teng T, et al. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12:244. doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anand K, Ziebuhr J, Wadhwani P, et al. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 19.Fan K, Wei P, Feng Q, et al. Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J Biol Chem. 2004;279:1637–1642. doi: 10.1074/jbc.M310875200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, Wei P, Huang C, et al. Only one protomer is active in the dimer of SARS 3C-like proteinase. J Biol Chem. 2006;281:13894–13898. doi: 10.1074/jbc.M510745200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suárez D, Díaz N. SARS-CoV-2 main protease: a molecular dynamics study. J Chem Inf Model. 2020;60:5815–5831. doi: 10.1021/acs.jcim.0c00575. [DOI] [PubMed] [Google Scholar]
- 22.Jin Z, Du X, Xu Y, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 23.Chen S, Chen L, Tan J, et al. Severe acute respiratory syndrome coronavirus 3C-like proteinase N terminus is indispensable for proteolytic activity but not for enzyme dimerization: biochemical and thermodynamic investigation in conjunction with molecular dynamics simulations. J Biol Chem. 2005;280:164–173. doi: 10.1074/jbc.M408211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu WC, Chang HC, Chou CY, et al. Critical assessment of important regions in the subunit association and catalytic action of the severe acute respiratory syndrome coronavirus main protease. J Biol Chem. 2005;280:22741–22748. doi: 10.1074/jbc.M502556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Lin D, Sun X, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novak J, Rimac H, Kandagalla S, et al. Can natural products stop the SARS-CoV-2 virus? A docking and molecular dynamics study of a natural product database. Future Med Chem. 2021;13:363–378. doi: 10.4155/fmc-2020-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novak J, Rimac H, Kandagalla S, et al. Proposition of a new allosteric binding site for potential SARS-CoV-2 3CL protease inhibitors by utilizing molecular dynamics simulations and ensemble docking. J Biomol Struct Dyn. 2021 doi: 10.1080/07391102.2021.1927845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiou WC, Hsu MS, Chen YT, et al. Repurposing existing drugs: identification of SARS-CoV-2 3C-like protease inhibitors. J Enzyme Inhib Med Chem. 2021;36:147–153. doi: 10.1080/14756366.2020.1850710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sang P, Tian SH, Meng ZH, Yang LQ. Anti-HIV drug repurposing against SARS-CoV-2. RSC Adv. 2020;10:15775–15783. doi: 10.1039/d0ra01899f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao B, Wang Y, Wen D, et al. A trial of Lopinavir-Ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. 2020;382:1787–1799. doi: 10.1056/nejmoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alamri MA, Tahir ul Qamar M, Mirza MU, et al. Pharmacoinformatics and molecular dynamics simulation studies reveal potential covalent and FDA-approved inhibitors of SARS-CoV-2 main protease 3CL pro. J Biomol Struct Dyn. 2021;39:4936–4948. doi: 10.1080/07391102.2020.1782768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan RJ, Jha RK, Amera GM, et al. Targeting SARS-CoV-2: a systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1753577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tahir ul Qamar M, Alqahtani SM, Alamri MA, Chen L-L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J Pharm Anal. 2020;10:313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choudhry N, Zhao X, Xu D, et al. Chinese therapeutic strategy for fighting COVID-19 and potential small-molecule inhibitors against severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) J Med Chem. 2020;63:13205–13227. doi: 10.1021/acs.jmedchem.0c00626. [DOI] [PubMed] [Google Scholar]
- 35.Jo S, Kim S, Kim DY, et al. Flavonoids with inhibitory activity against SARS-CoV-2 3CLpro. J Enzyme Inhib Med Chem. 2020;35:1539–1544. doi: 10.1080/14756366.2020.1801672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogidigo JO, Iwuchukwu EA, Ibeji CU, et al. Natural phyto, compounds as possible noncovalent inhibitors against SARS-CoV2 protease: computational approach. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1837681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bzówka M, Mitusińska K, Raczyńska A, et al. Structural and evolutionary analysis indicate that the SARS-CoV-2 Mpro is a challenging target for small-molecule inhibitor design. Int J Mol Sci. 2020;21:3099. doi: 10.3390/ijms21093099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen C, Yu X, Kuo C, et al. Overview of antiviral drug candidates targeting coronaviral 3C-like main proteases. FEBS J. 2021;288:5089–5121. doi: 10.1111/febs.15696. [DOI] [PubMed] [Google Scholar]
- 39.Zhang C-H, Stone EA, Deshmukh M, et al. Potent noncovalent inhibitors of the main protease of SARS-CoV-2 from molecular sculpting of the drug perampanel guided by free energy perturbation calculations. ACS Cent Sci. 2021;7:467–475. doi: 10.1021/acscentsci.1c00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nand M, Maiti P, Joshi T, et al. Virtual screening of anti-HIV1 compounds against SARS-CoV-2: machine learning modeling, chemoinformatics and molecular dynamics simulation based analysis. Sci Rep. 2020;10:1–12. doi: 10.1038/s41598-020-77524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alimadadi A, Aryal S, Manandhar I, et al. Artificial intelligence and machine learning to fight covid-19. Physiol Genom. 2020;52:200–202. doi: 10.1152/physiolgenomics.00029.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Batra R, Chan H, Kamath G, et al. Screening of therapeutic agents for COVID-19 using machine learning and ensemble docking studies. J Phys Chem Lett. 2020;11:7058–7065. doi: 10.1021/acs.jpclett.0c02278. [DOI] [PubMed] [Google Scholar]
- 43.Kowalewski J, Ray A. Predicting novel drugs for SARS-CoV-2 using machine learning from a >10 million chemical space. Heliyon. 2020;6:e04639. doi: 10.1016/j.heliyon.2020.e04639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verma AK, Aggarwal R. Repurposing potential of FDA-approved and investigational drugs for COVID-19 targeting SARS-CoV-2 spike and main protease and validation by machine learning algorithm. Chem Biol Drug Des. 2021;97:836–853. doi: 10.1111/cbdd.13812. [DOI] [PubMed] [Google Scholar]
- 45.Mohapatra S, Nath P, Chatterjee M, et al. Repurposing therapeutics for COVID-19: rapid prediction of commercially available drugs through machine learning and docking. PLoS ONE. 2020;15:1–13. doi: 10.1371/journal.pone.0241543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khan A, Ali SS, Khan MT, et al. Combined drug repurposing and virtual screening strategies with molecular dynamics simulation identified potent inhibitors for SARS-CoV-2 main protease (3CLpro) J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1779128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar V, Roy K. Development of a simple, interpretable and easily transferable QSAR model for quick screening antiviral databases in search of novel 3C-like protease (3CLpro) enzyme inhibitors against SARS-CoV diseases. SAR QSAR Environ Res. 2020;31:511–526. doi: 10.1080/1062936X.2020.1776388. [DOI] [PubMed] [Google Scholar]
- 48.Janairo GIB, Yu DEC, Janairo JIB. A machine learning regression model for the screening and design of potential SARS-CoV-2 protease inhibitors. Netw Model Anal Heal Inform Bioinform. 2021;10:1–8. doi: 10.1007/s13721-021-00326-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumari M, Subbarao N. Deep learning model for virtual screening of novel 3C-like protease enzyme inhibitors against SARS coronavirus diseases. Comput Biol Med. 2021;132:104317. doi: 10.1016/j.compbiomed.2021.104317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novak J, Grishina MA, Potemkin VA, Gasteiger J. Performance of radial distribution function-based descriptors in the chemoinformatic studies of HIV-1 protease. Future Med Chem. 2020;12:299–309. doi: 10.4155/fmc-2019-0241. [DOI] [PubMed] [Google Scholar]
- 51.Novak J, Grishina MA, Potemkin VA. The influence of hydrogen atoms on the performance of radial distribution function-based descriptors in the chemoinformatic studies of HIV-1 protease complexes with inhibitors. Curr Drug Discov Technol. 2021;18:414–422. doi: 10.2174/1570163817666200102130415. [DOI] [PubMed] [Google Scholar]
- 52.Novak J, Grishina MA, Potemkin VA. Novel radial distribution function approach in the study of point mutations: the HIV-1 protease case study. Future Med Chem. 2020;12:1025–1036. doi: 10.4155/fmc-2020-0042. [DOI] [PubMed] [Google Scholar]
- 53.Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DrugBank. https://www.drugbank.ca/. Accessed 2 Feb 2021
- 55.Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.RCSB PDB. http://www.rcsb.org/. Accessed 4 Aug 2021
- 57.Hemmer MC, Steinhauer V, Gasteiger J. Deriving the 3D structure of organic molecules from their infrared spectra. Vib Spectrosc. 1999;19:151–164. doi: 10.1016/S0924-2031(99)00014-4. [DOI] [Google Scholar]
- 58.Meng EC, Pettersen EF, Couch GS, et al. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinform. 2006;7:1–10. doi: 10.1186/1471-2105-7-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davies M, Nowotka M, Papadatos G, et al. ChEMBL web services: streamlining access to drug discovery data and utilities. Nucleic Acids Res. 2015;43:W612–W620. doi: 10.1093/nar/gkv352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaulton A, Hersey A, Nowotka ML, et al. The ChEMBL database in 2017. Nucleic Acids Res. 2017;45:D945–D954. doi: 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Potemkin V, Grishina M. Grid-based technologies for in silico screening and drug design. Curr Med Chem. 2018;25:3526–3537. doi: 10.2174/0929867325666180309112454. [DOI] [PubMed] [Google Scholar]
- 62.Potemkin VA, Arslambekov RM, Bartashevich EV, et al. Multiconformational method for analyzing the biological activity of molecular structures. J Struct Chem. 2002;43:1126–1130. doi: 10.1023/A:1023615231976. [DOI] [Google Scholar]
- 63.Bartashevich EV, Potemkin VA, Grishina MA, Belik AV. A method for multiconformational modeling of the three-dimensional shape of a molecule. J Struct Chem. 2002;43:1033–1039. doi: 10.1023/A:1023611131068. [DOI] [Google Scholar]
- 64.Potemkin VA, Grishina MA, Bartashevich EV. Modeling of drug molecule orientation within a receptor cavity in the BiS algorithm framework. J Struct Chem. 2007;48:155–160. doi: 10.1007/s10947-007-0023-y. [DOI] [Google Scholar]
- 65.Potemkin VA, Grishina MA. A new paradigm for pattern recognition of drugs. J Comput Aided Mol Des. 2008;22:489–505. doi: 10.1007/s10822-008-9203-x. [DOI] [PubMed] [Google Scholar]
- 66.Potemkin V, Galimova O, Grishina M. Cinderella’s Shoe for virtual drug discovery screening and design. Drugs Future. 2010;35:14–15. [Google Scholar]
- 67.Potemkin VA, Pogrebnoy AA, Grishina MA. Technique for energy decomposition in the study of “receptor-ligand” complexes. J Chem Inf Model. 2009;49:1389–1406. doi: 10.1021/ci800405n. [DOI] [PubMed] [Google Scholar]
- 68.Potemkin V, Grishina M. Principles for 3D/4D QSAR classification of drugs. Drug Discov Today. 2008;13:952–959. doi: 10.1016/j.drudis.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 69.Potemkin VA, Bartashevich EV, Belik AV. A new approach to predicting the thermodynamic parameters of substances from molecular characteristics. Russ J Phys Chem. 1996;70:411–416. [Google Scholar]
- 70.Grishina MA, Pogrebnoi AA, Potemkin VA, Zrakova TY. Theoretical study of the substrate specificity of cytochrome P-450 isoforms. Pharm Chem J. 2005;39:509–513. doi: 10.1007/s11094-006-0011-0. [DOI] [Google Scholar]
- 71.Potemkin VA, Grishina MA, Fedorova OV, et al. Theoretical investigation of the antituberculous activity of membranotropic podands. Pharm Chem J. 2003;37:468–472. doi: 10.1023/B:PHAC.0000008246.07413.d9. [DOI] [Google Scholar]
- 72.Potemkin VA, Grishina MA, Belik AV, Chupakhin ON. Quantitative relationship between structure and antibacterial activity of quinolone derivatives. Pharm Chem J. 2002;36:22–25. doi: 10.1023/A:1015744707357. [DOI] [Google Scholar]
- 73.Mody V, Ho J, Wills S, et al. Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents. Commun Biol. 2021 doi: 10.1038/s42003-020-01577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.RDKit: Open-source cheminformatics
- 75.Grishina MA, Bartashevich EV, Pereyaslavskaya ES, Potemkin VA. Novel techniques for virtual discovery for study of multistage bioprocesses. Drugs Future. 2007;32:27. [Google Scholar]
- 76.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 77.Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kool ET. Hydrogen bonding, base stacking, and steric effects in DNA replication. Annu Rev Biophys Biomol Struct. 2001;30:1–22. doi: 10.1146/annurev.biophys.30.1.1. [DOI] [PubMed] [Google Scholar]
- 80.Hubbard RE, Kamran Haider M. Encyclopedia of life sciences. Chichester: Wiley; 2010. Hydrogen bonds in proteins: role and strength. [Google Scholar]
- 81.Bowie JU. Membrane protein folding: how important are hydrogen bonds? Curr Opin Struct Biol. 2011;21:42–49. doi: 10.1016/j.sbi.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anand K. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shi J, Song J. The catalysis of the SARS 3C-like protease is under extensive regulation by its extra domain. FEBS J. 2006;273:1035–1045. doi: 10.1111/j.1742-4658.2006.05130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang L, Liu J, Cao R, et al. Comparative antiviral efficacy of viral protease inhibitors against the novel SARS-CoV-2 in vitro. Virol Sin. 2020;35:776–784. doi: 10.1007/s12250-020-00288-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.ClinicalTrials.gov. https://clinicaltrials.gov/ct2/home. Accessed 15 Mar 2021
- 86.Martin WR, Cheng F. Repurposing of FDA-approved toremifene to treat COVID-19 by blocking the spike glycoprotein and NSP14 of SARS-CoV-2. J Proteome Res. 2020;19:4670–4677. doi: 10.1021/acs.jproteome.0c00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hsu M-F, Kuo C-J, Chang K-T, et al. Mechanism of the maturation process of SARS-CoV 3CL protease. J Biol Chem. 2005;280:31257–31266. doi: 10.1074/jbc.M502577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pillaiyar T, Manickam M, Namasivayam V, et al. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J Med Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 90.Chen YC. Beware of docking! Trends Pharmacol Sci. 2015;36:78–95. doi: 10.1016/j.tips.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 91.Ramírez D, Caballero J. Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data? Molecules. 2018;23:1–17. doi: 10.3390/molecules23051038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rafi MO, Bhattacharje G, Al-Khafaji K, et al. Combination of QSAR, molecular docking, molecular dynamic simulation and MM-PBSA: analogues of lopinavir and favipiravir as potential drug candidates against COVID-19. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1850355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bolcato G, Bissaro M, Pavan M, et al. Targeting the coronavirus SARS-CoV-2: computational insights into the mechanism of action of the protease inhibitors lopinavir, ritonavir and nelfinavir. Sci Rep. 2020;10:20927. doi: 10.1038/s41598-020-77700-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu J, Zhai Y, Liang L, et al. Molecular modeling evaluation of the binding effect of five protease inhibitors to COVID-19 main protease. Chem Phys. 2021;542:111080. doi: 10.1016/j.chemphys.2020.111080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Feng Z, Chen M, Xue Y, et al. MCCS: a novel recognition pattern-based method for fast track discovery of anti-SARS-CoV-2 drugs. Brief Bioinform. 2021;22:946–962. doi: 10.1093/bib/bbaa260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bahadur Gurung A, Ajmal Ali M, Lee J, et al. Structure-based virtual screening of phytochemicals and repurposing of FDA approved antiviral drugs unravels lead molecules as potential inhibitors of coronavirus 3C-like protease enzyme. J King Saud Univ Sci. 2020;32:2845–2853. doi: 10.1016/j.jksus.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Beck BR, Shin B, Choi Y, et al. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput Struct Biotechnol J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mahdi M, Mótyán JA, Szojka ZI, et al. Analysis of the efficacy of HIV protease inhibitors against SARS-CoV-2’s main protease. Virol J. 2020;17:1–8. doi: 10.1186/s12985-020-01457-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maffucci I, Contini A. In silico drug repurposing for SARS-CoV-2 main proteinase and spike proteins. J Proteome Res. 2020;19:4637–4648. doi: 10.1021/acs.jproteome.0c00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bello M, Martínez-Muñoz A, Balbuena-Rebolledo I. Identification of saquinavir as a potent inhibitor of dimeric SARS-CoV2 main protease through MM/GBSA. J Mol Model. 2020;26:340. doi: 10.1007/s00894-020-04600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Komatsu TS, Okimoto N, Koyama YM, et al. Drug binding dynamics of the dimeric SARS-CoV-2 main protease, determined by molecular dynamics simulation. Sci Rep. 2020;10:1–11. doi: 10.1038/s41598-020-74099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Indu P, Rameshkumar MR, Arunagirinathan N, et al. Raltegravir, Indinavir, Tipranavir, Dolutegravir, and Etravirine against main protease and RNA-dependent RNA polymerase of SARS-CoV-2: a molecular docking and drug repurposing approach. J Infect Public Health. 2020;13:1856–1861. doi: 10.1016/j.jiph.2020.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Swanstrom R, Anderson J, Schiffer C, Lee SK. Viral protease inhibitors. Handb Exp Pharmacol. 2009;189:85–110. doi: 10.1007/978-3-540-79086-0_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sharma A, Gupta SP, Siddiqui AA, Sharma N. HCV NS3/4A protease and its emerging inhibitors. J Anal Pharm Res. 2017;4:1–9. doi: 10.15406/japlr.2017.04.00108. [DOI] [Google Scholar]
- 105.Furuta Y, Takahashi K, Shiraki K, et al. T-705 (favipiravir) and related compounds: novel broad-spectrum inhibitors of RNA viral infections. Antiviral Res. 2009;82:95–102. doi: 10.1016/j.antiviral.2009.02.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Furuta Y, Komeno T, Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc Jpn Acad Ser B. 2017;93:449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Alamer A, Alrashed AA, Alfaifi M, et al. Effectiveness and safety of favipiravir compared to supportive care in moderately to critically ill COVID-19 patients: a retrospective study with propensity score matching sensitivity analysis. Curr Med Res Opin. 2021;37:1085–1097. doi: 10.1080/03007995.2021.1920900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ivashchenko AA, Dmitriev KA, Vostokova NV, et al. AVIFAVIR for treatment of patients with moderate coronavirus disease 2019 (COVID-19): interim results of a phase II/III multicenter randomized clinical trial. Clin Infect Dis. 2021;73:531–534. doi: 10.1093/cid/ciaa1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.