Abstract
Background
This study was to systematically test whether previously reported risk factors for chronic kidney disease (CKD) are causally related to CKD in European and East Asian ancestries using Mendelian randomization.
Methods
A total of 45 risk factors with genetic data in European ancestry and 17 risk factors in East Asian participants were identified as exposures from PubMed. We defined the CKD by clinical diagnosis or by estimated glomerular filtration rate of <60 ml/min/1.73 m2. Ultimately, 51 672 CKD cases and 958 102 controls of European ancestry from CKDGen, UK Biobank and HUNT, and 13 093 CKD cases and 238 118 controls of East Asian ancestry from Biobank Japan, China Kadoorie Biobank and Japan-Kidney-Biobank/ToMMo were included.
Results
Eight risk factors showed reliable evidence of causal effects on CKD in Europeans, including genetically predicted body mass index (BMI), hypertension, systolic blood pressure, high-density lipoprotein cholesterol, apolipoprotein A-I, lipoprotein(a), type 2 diabetes (T2D) and nephrolithiasis. In East Asians, BMI, T2D and nephrolithiasis showed evidence of causality on CKD. In two independent replication analyses, we observed that increased hypertension risk showed reliable evidence of a causal effect on increasing CKD risk in Europeans but in contrast showed a null effect in East Asians. Although liability to T2D showed consistent effects on CKD, the effects of glycaemic phenotypes on CKD were weak. Non-linear Mendelian randomization indicated a threshold relationship between genetically predicted BMI and CKD, with increased risk at BMI of >25 kg/m2.
Conclusions
Eight cardiometabolic risk factors showed causal effects on CKD in Europeans and three of them showed causality in East Asians, providing insights into the design of future interventions to reduce the burden of CKD.
Keywords: chronic kidney disease, cardiometabolic risk factors, Mendelian randomization, causality, trans-ethnic study
Key Messages.
This large-scale genetic study found robust evidence to support the causal roles of eight cardiometabolic risk factors on chronic kidney disease (CKD) among Europeans and three of these risk factors were causal among East Asians.
Trans-ethnic comparison suggested that hypertension showed a strong causal role on CKD in Europeans but no substantial role in East Asians.
The genetic evidence suggested that type 2 diabetes may have glucose-independent mechanisms to influence CKD.
This study highlighted importance of controlling the multimorbidity of cardiovascular disease and CKD as an intervention strategy to reduce the burden of both diseases.
Introduction
Chronic kidney disease (CKD) affects 10–15% of the population worldwide. It has a major effect on global health, both as a direct cause of morbidity and mortality, and as an important complication for cardiometabolic diseases.1–3 From 1990 to 2017, the global age-standardized mortality for many important non-communicable diseases has declined. For example, cardiovascular disease mortality has been reduced by 30.4%. However, the mortality decline for CKD has been just 2.8%.4 The majority of interventional trials have focused on disease treatment rather than primary prevention. In the literature, impaired fasting glucose, high blood pressure and high body mass index (BMI) are among the leading risk factors for CKD. However, even with existing interventions for these risk factors, the burden of CKD has not declined as expected.4 Moreover, CKD awareness is limited among the general public and healthcare authorities. Thus, a systematic assessment of the causal determinants of CKD is urgently needed to promote a shift from the treatment of CKD patients to the prevention of the disease in high-risk groups.
Well-designed randomized–controlled trials (RCTs) are usually the best approach to estimate a causal relationship between a risk factor and a disease. Whereas several studies have identified risk factors for CKD progression, there is a lack of reliable evidence to support their causal roles on CKD incidence. Mendelian randomization (MR) is an epidemiological method that can be used to obtain evidence about the causal effects of modifying intervention targets.5 MR exploits the random allocation of genetic variants at conception and is therefore less susceptible to confounding and reverse causality than traditional observational studies. The increasing availability of genetic-association resources provides a timely opportunity to test the causal effects of various risk factors on CKD.6,7
In this study, we aimed to investigate the causal effects of 45 previously reported risk factors on CKD using two-sample linear and non-linear MR approaches. We used the largest available genome-wide association studies (GWASs) for risk factors in European and East Asian ancestries. Summary data for CKD and estimated glomerular filtration rate (eGFR) were obtained from >1 million participants from the CKDGen consortium,8 UK Biobank,9 Trøndelag Health (HUNT) Study,10 Biobank Japan,11 China Kadoorie Biobank12 and Japan-Kidney-Biobank/ToMMo consortium.
Methods
The data, analytic methods and study materials will be made available to other researchers for purposes of reproducing the results. For more details, the genetic-association data of the selected risk factors are available in Supplementary Tables (available as Supplementary data at IJE online). The GWAS summary statistics for CKD and eGFR that were generated using UK Biobank and CKDGen data are available from the MRC-IEU OpenGWAS database (https://gwas.mrcieu.ac.uk/) and CKDGen website (http://ckdgen.imbi.uni-freiburg.de/) respectively. The GWAS results from HUNT, Biobank Japan, China Kadoorie Biobank and Japan-Kidney-Biobank/ToMMo can be accessed by request to the data holders. The analytical script of the MR analysis conducted in this study is available via the GitHub repository of the ‘TwoSampleMR’ R package (https://github.com/MRCIEU/TwoSampleMR/).
Study design
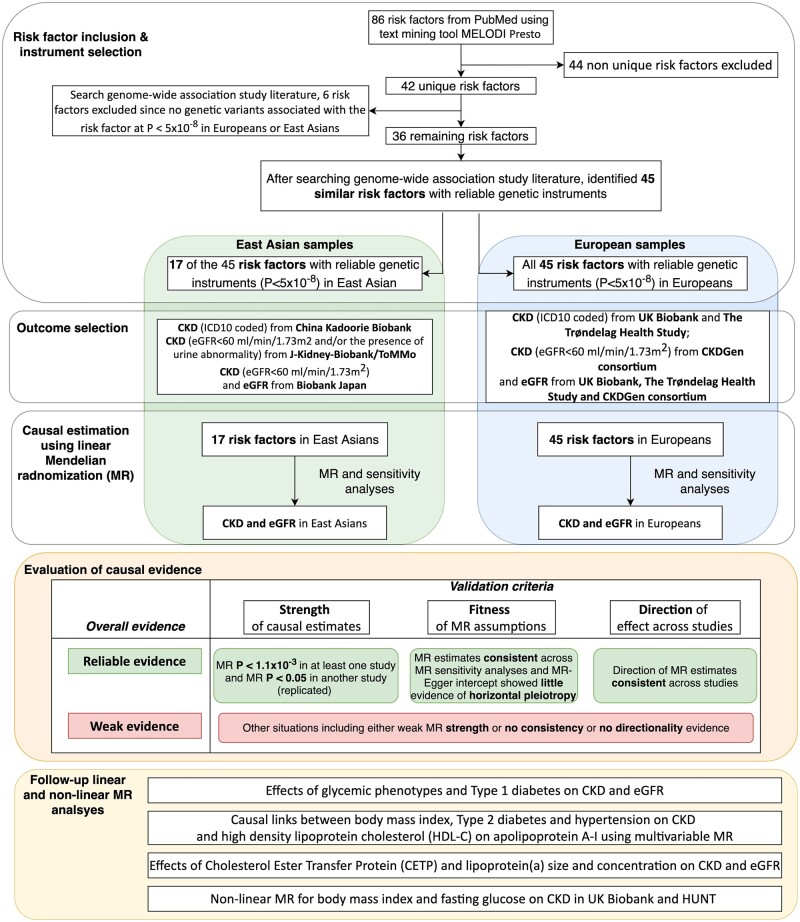
Our study consisted of four components (Figure 1). First, we identified 45 risk factors for CKD by mining PubMed. Second, we estimated the causal effects of these risk factors on CKD and eGFR in CKDGen,8 UK Biobank,9 HUNT Study,10 Biobank Japan,11 China Kadoorie Biobank12 and Japan-Kidney-Biobank/ToMMo consortium separately. Third, we evaluated the findings based on the strength and consistency of the evidence across MR methods and across individual studies. Finally, we conducted extensive follow-up analyses to confirm the findings for blood pressure, glycaemic and blood lipid phenotypes on CKD. Finally, non-linear MR was performed to estimate the optimal BMI and fasting glucose levels for reducing CKD risk in UK Biobank and the HUNT Study.
Figure 1.
Study design of the trans-ethnic Mendelian-randomization study of chronic kidney disease
CKD, chronic kidney disease; eGFR, estimated glomerular filtration rate.
Selection of risk factors
CKD risk factors were identified from a literature review using MELODI-Presto13,14 to search the PubMed database (Supplementary Note S1, available as Supplementary data at IJE online). We identified 45 risk factors for CKD, including blood-pressure phenotypes, glycaemic phenotypes, lipid phenotypes, obesity, smoking, alcohol intake, sleep disorders, nephrolithiasis, serum uric acid, coronary artery disease, bone mineral density, homocysteine, C-reactive protein, micro-nutrient phenotypes (serum metals and vitamins), dehydration and thyroid phenotypes. By searching the largest available GWASs (ensuring minimum sample overlap with the outcome samples), we extracted genetic variants associated with all 45 risk factors from European ancestry studies and extracted 17 of the 45 risk factors from East Asian ancestry studies (Supplementary Table S1 and Supplementary Note S1, available as Supplementary data at IJE online). To select the independent genetic variants, the genome-wide significant single-nucleotide polymorphism (SNPs) were grouped by linkage disequilibrium (LD) (r2 <0.001 for SNPs within 1 Mb genomic region) and the SNP with the lowest P-value per group was retained (Supplementary Tables S2 and S3, available as Supplementary data at IJE online).
Association of genetic variants with CKD and eGFR
In UK Biobank,9 HUNT Study10 and China Kadoorie Biobank,12 CKD was defined according to the International Classification of Diseases (ICD) 10th Revision. The CKD cases were defined as participants with ICD 10 code N18. The participants with any type of kidney conditions (N00 to N29) were excluded from the controls to reduce the possibility of including CKD cases in the control group. In Japan-Kidney-Biobank/ToMMo consortium, CKD was defined as eGFR of <60 ml/min/1.73 m2 and/or the presence of urine abnormality, which is similar to the clinical diagnosis for CKD. In CKDGen8 and Biobank Japan,11 CKD was defined as eGFR of <60 ml/min/1.73 m2. In all studies, eGFR was estimated from serum creatinine using the Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) formula.15 The genetic associations with CKD and eGFR were reported in three studies of European ancestry (CKDGen: 41 395 cases, 439 303 controls, 8.7% diabetes patients; UK Biobank: 6985 cases, 454 323 controls, 5.2% with diabetes; and HUNT Study: 3292 cases, 64 476 controls, 4.9% with diabetes). The genetic associations with CKD were reported in three East Asian studies (Biobank Japan: 8586 cases, 133 808 controls, 10.2% with diabetes; China Kadoorie Biobank: 461 cases, 94 887 controls, 6.7% with diabetes; Japan-Kidney-Biobank/ToMMo consortium: 4046 cases, 9423 controls, 7.3% with diabetes) and eGFR genetic associations were reported in Biobank Japan (Supplementary Table S4 and Supplementary Note S2, available as Supplementary data at IJE online). All participants included in the CKDGen,8 UK Biobank,9 HUNT,10 Biobank Japan,11 China Kadoorie Biobank12 and Japan-Kidney-Biobank/ToMMo provided written informed consent and studies were approved by their local research ethics committees and institutional review boards as applicable.
Statistical analysis
MR is an instrumental variable method that uses genetic variants as instruments to test the causal relationships between an exposure (e.g. BMI) and an outcome (e.g. CKD) and requires three core assumptions to be satisfied (Supplementary Figure S1 and Supplementary Note S3, available as Supplementary data at IJE online). For binary exposures [e.g. type 2 diabetes (T2D)], we converted the odds ratios (ORs) [multiplying log(ORs) by log (2) (equal to 0.693) and then exponentiating] in order to represent the OR of outcome per doubling of the odds of susceptibility to the exposure.16,17
The MR estimates for each risk factor were determined using inverse variance weighted (MR-IVW) analysis, which uses the random-effects meta-analysis approach to combine the Wald ratio estimates18 of the causal effect obtained from each of the tested SNPs. A set of sensitivity analyses, including MR–Egger,19 MR weighted median,20 MR mode estimator21 and a heterogeneity test,22 were conducted to test the underlying MR assumptions. We also examined the possibility of reverse causality using bidirectional MR23 and applied multivariable MR analyses of the correlated phenotypes (Supplementary Note S4, available as Supplementary data at IJE online). A conservative Bonferroni-corrected threshold (α = 1.11 × 10–3, as 45 risk factors were assessed) was used to account for multiple testing. Supplementary Note S5 (available as Supplementary data at IJE online) demonstrates the instrument strength estimation20 and power calculations.24,25 The MR and sensitivity analyses were conducted using the TwoSampleMR package.26
Follow-up MR analyses
To validate the different causal pattern of blood pressure across ancestries, we conducted a set of follow-up analyses: (i) to estimate the potential influence of instrument size and resulting power of the MR analyses, we conducted novel East Asian GWASs of hypertension (N cases = 40 318, N controls = 60 323), systolic blood pressure (SBP), diastolic blood pressure (DBP) and pulse pressure (PP) in 100 641 China Kadoorie Biobank participants. By extract genetic instruments from the well-powered GWASs, we further increased the instrument strength (F-statistics) of hypertension from 275.19 to 330.85 (Supplementary Table S3, available as Supplementary data at IJE online). Using these data as instruments, we conducted a validation MR between the blood-pressure phenotypes and CKD in the three East Asian studies; (ii) for the European SBP and DBP instruments, we checked whether their genetic associations were replicated in the East Asian GWASs.27 We then used the replicated SNPs of SBP and DBP (Supplementary Table S5, available as Supplementary data at IJE online) to conduct a second validation MR (noted as European variant + East Asian effect analysis); (iii) we compared the direction of effect and the heterogeneity of the genetic effects of hypertension in Europeans and East Asians using pair-wise Z test and ran sensitivity MR analysis to remove instruments with heterogeneity.
To better understand the causal mechanisms linking T2D with CKD, four additional MR analyses were conducted: (i) we validated the effects of eight glycaemic phenotypes on CKD using Steiger filtering28 and radial MR;29 (ii) we considered the influence of the genetic liability for type 1 diabetes (T1D)30 (Supplementary Table S2, available as Supplementary data at IJE online) on CKD; (iii) participants with eGFR measurements were stratified into diabetic (N = 11 529) and non-diabetic populations (N = 118 460)31 and we conducted MR analyses of T2D and five glycaemic phenotypes on eGFR in these two subpopulations; (iv) diabetic retinopathy was included as a positive control outcome to validate the analytical approach. The instruments for T2D and glycaemic phenotypes were used as exposures, whereas the CKD data from CKDGen, UK Biobank and HUNT as well as the diabetic retinopathy data from UK Biobank SAIGE release32 were used as outcomes (Supplementary Table S4, available as Supplementary data at IJE online).
To validate the MR findings of lipids on CKD, the following analyses were conducted: (i) to validate the high-density lipoprotein cholesterol (HDL-C) MR results in East Asians, we conducted the same European variant + East Asian effect analysis to boost the power of the MR findings (HDL-C data from Spracklen et al.33) (Supplementary Table S5, available as Supplementary data at IJE online); (ii) we tested the independent effect of HDL-C and apolipoprotein A-I on CKD using a multivariable MR model (Supplementary Note S4, available as Supplementary data at IJE online); (iii) we estimated the effect of circulating cholesteryl ester transfer protein levels34 on CKD (Supplementary Table S2, available as Supplementary data at IJE online); (iv) given that lipoprotein(a) levels for a fixed apolipoprotein(a) isoform size may vary, we estimated the effect of apolipoprotein(a) isoform size on CKD [lipoprotein(a) KIV2 repeats and apolipoprotein(a) protein isoform size data from Saleheen et al.35] (Supplementary Table S2, available as Supplementary data at IJE online).
Finally, for BMI and fasting glucose, a fractional polynomial approach36–38 was applied to estimate the non-linear shape of the association between these risk factors and CKD using data from UK Biobank and HUNT (Supplementary Note S6, available as Supplementary data at IJE online).
Evaluation of MR evidence
Previous studies have suggested that P-value thresholds should not be the only criteria to define ‘significance’.39,40,41 We therefore evaluated the MR evidence using three criteria: (i) MR evidence strength: whether the MR-IVW estimate of each risk factor passed the Bonferroni-corrected P-value threshold (P < 1.1 × 10–3) in at least one study and passed the replication threshold (P < 0.05) in at least one other study; (ii) fit of MR assumptions: whether the MR estimates for each risk factor showed the same direction of effect across MR sensitivity analyses and showed limited influence of horizontal pleiotropy using the MR–Egger intercept term and heterogeneity test; (iii) whether the direction of the MR effect of each risk factor on CKD was consistent across multiple studies. Figure 1 demonstrates how the MR evidence was evaluated in Europeans and East Asians separately: ‘Reliable evidence’ refers to risk factors that fulfilled all the three criteria, whereas ‘Weak evidence’ refers to risk factors that do not fulfil all the criteria (e.g. MR estimates with strong MR evidence but with inconsistent directionality).
Results
Causal effects of risk factors on CKD
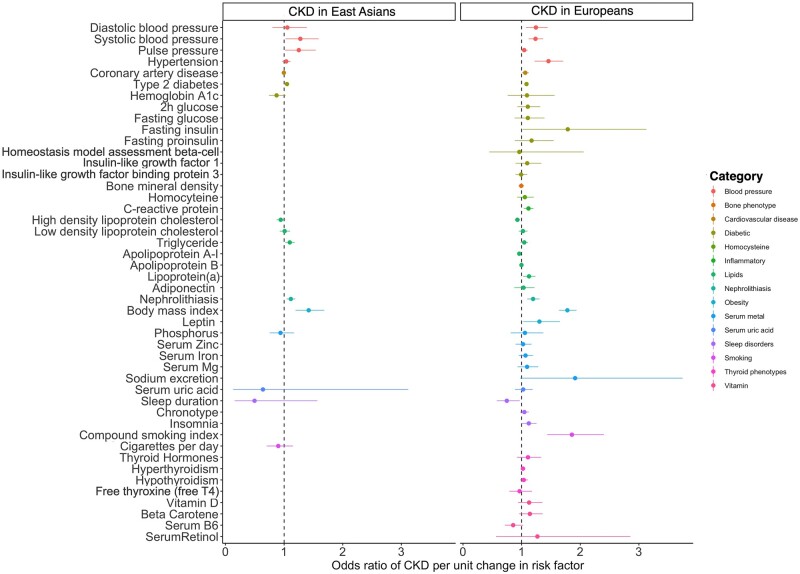
Most of the 45 risk factors had strong genetic instruments for both ancestries (F-statistics > 10 for 44 of the 45 risk factors in Europeans and 15 of the 17 risk factors in East Asians; Supplementary Tables S2 and S3, available as Supplementary data at IJE online). However, instruments tended to be stronger in Europeans compared with East Asians (Supplementary Table S6, available as Supplementary data at IJE online). Effect estimates for the 45 risk factors on CKD in Europeans and 17 risk factors in East Asians are presented in Figure 2. Effect estimates for the other sensitivity analyses can be found in Supplementary Tables S7–S12 (available as Supplementary data at IJE online). Detailed evaluations of the causal evidence in Europeans and East Asians are presented in Supplementary Tables S13A and S13B (available as Supplementary data at IJE online).
Figure 2.
Forest plot for causal effects of the 45 risk factors on chronic kidney disease in Europeans and the 17 risk factors on chronic kidney disease in Eastern Asians. (A) Causal estimates using European data; (B) causal estimates using Eastern Asian data. For binary exposures, the effect reported on the x-axis is the odds ratio of chronic kidney disease per doubling in the odds of the exposure. For continuous exposure, the effect on the x-axis is the odds ratio of chronic kidney disease per 1 standard deviation change in the exposure. CKD, chronic kidney disease.
Risk factors showing reliable MR evidence
In European ancestry, eight risk factors were associated with CKD. The OR [95% confidence intervals (CIs)] for CKD per 1-SD increase in continuous risk factors was 1.78 (1.64 to 1.94) for BMI, 1.24 (1.12 to 1.37) for SBP, 1.13 (1.07 to 1.19) for lipoprotein(a) levels, 0.93 (0.90 to 0.97) for HDL-C and 0.96 (0.94 to 0.98) for apolipoprotein A-I. The OR (95% CI) per doubling in the odds of genetic liability for the binary risk factors was 2.05 (1.59 to 2.64) for hypertension, 1.20 (1.09 to 1.31) for nephrolithiasis and 1.08 (1.05 to 1.12) for T2D. The effects of these eight risk factors on CKD were consistent across UK Biobank, CKDGen and HUNT (Supplementary Figure S2A and Supplementary Tables S7A, S8A and S9A, available as Supplementary data at IJE online).
In East Asian participants, genetically predicted higher BMI (OR = 1.42, 95% CI = 1.20 to 1.69, P = 6.49 × 10–5), increased nephrolithiasis risk (OR = 1.12, 95% CI = 1.04 to 1.19, P = 1.11 × 10–3) and increased T2D risk (OR = 1.07, 95% CI = 1.03 to 1.10, P = 1.66 × 10–4) were all associated with increased risk of CKD (Supplementary Figure S2B, available as Supplementary data at IJE online). The effect of T2D on CKD was consistent across the three East Asian studies. However, the effect of BMI and nephrolithiasis on CKD was not observed in the China Kadoorie Biobank—this is likely due to the limited number of CKD cases in this resource (Supplementary Tables S10A–S12, available as Supplementary data at IJE online).
We also conducted several sensitivity analyses. The bidirectional MR analysis found a consistent effect of increased CKD risk on increasing hypertension risk in European ancestry (Supplementary Table S14, available as Supplementary data at IJE online). The multivariable MR results for T2D, BMI and hypertension on CKD are included in Supplementary Table S15A–C, available as Supplementary data at IJE online. The MR analyses using eGFR as an outcome showed similar results to those for CKD (Supplementary Tables S7B–S10B, available as Supplementary data at IJE online).
Risk factors showing weak MR evidence
There was weak evidence to support a causal effect on CKD of the remaining 37 risk factors considered in the European ancestry analyses (Supplementary Tables S7–S9, available as Supplementary data at IJE online) and 14 risk factors considered in the East Asian ancestry analyses (Supplementary Tables S10–S12, available as Supplementary data at IJE online). Some established risk factors, such as smoking and serum uric acid, were among those with weak evidence. In addition, shorter sleep duration showed evidence to support an association with CKD in Japan-Kidney-Biobank/ToMMo (Supplementary Table S11, available as Supplementary data at IJE online) and in UK Biobank (Supplementary Table S7A, available as Supplementary data at IJE online), which was not replicated in other studies.
Follow-up MR analyses of key findings
Effect of blood-pressure phenotypes on CKD across populations
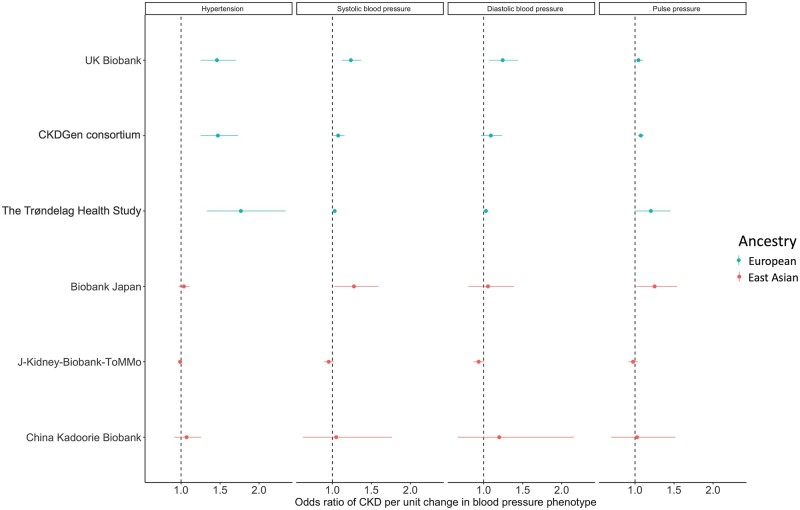
Figure 3 demonstrates that blood-pressure phenotypes, including genetic liability of hypertension and genetically predicted SBP and PP, showed strong causal effects on CKD in the European studies but appeared to show a null causal effect in the corresponding East Asian studies (ORs for liability of hypertension on CKD ranging from 1.46 to 1.77 in Europeans but only from 0.99 to 1.07 in East Asians). To validate these MR results, we first checked the strength of genetic instruments for the four blood-pressure phenotypes and observed that the instrument strengths were substantially above the F-statistics threshold of 10 for all four phenotypes in Europeans and East Asians (Supplementary Table S6, available as Supplementary data at IJE online). To further boost power, we used genetic instruments for hypertension, SBP, DBP and PP from 100 641 China Kadoorie Biobank participants [which obtained better instrument strength than the European hypertension data (F-statistics = 61.11 in Europeans vs 330.85 in East Asians); Supplementary Tables S2 and S3, available as Supplementary data at IJE online] and still observed null results in East Asians (Supplementary Table S16A, available as Supplementary data at IJE online). Third, we conducted an MR analysis using the European SBP and DBP instruments extracted from the East Asian studies (Supplementary Table S5, available as Supplementary data at IJE online), which showed similar null results (Supplementary Table S16B, available as Supplementary data at IJE online). Finally, we estimated the heterogeneity of genetic effects of hypertension across Europeans and East Asians and observed that 20.9% of the instruments showed distinguished effects across the two ancestries (Supplementary Table S17, available as Supplementary data at IJE online). Sensitivity MR analyses excluding the heterogenous instruments, controlling for different genetic architectures of BP across ancestries, showed similar MR results (Supplementary Table S18, available as Supplementary data at IJE online). These analyses provide additional evidence that blood pressure has a population-specific role in CKD aetiology.
Figure 3.
Forest plot for causal effects of four blood-pressure phenotypes on chronic kidney disease risk. The subplots represent Mendelian-randomization results of different blood-pressure phenotypes. CKD, chronic kidney disease.
Effects of glycaemic phenotypes and CKD
Although the evidence for an effect of T2D on CKD was reliable, we detected little evidence to support the effects of eight glycaemic phenotypes [fasting insulin (FI), fasting glucose (FG), 2-hour glucose (2hGlu), fasting proinsulin (FP), haemoglobin A1c (HbA1c), HOMA-B, insulin-like growth factor binding protein 3 and insulin-like growth factor I] on CKD (Supplementary Figure S3, available as Supplementary data at IJE online) and eGFR (Supplementary Figure S4, available as Supplementary data at IJE online). Follow-up analyses showed that: (i) similar results were observed after controlling for possible reverse causation of instruments and potential outliers (Supplementary Table S19A, available as Supplementary data at IJE online); (ii) little evidence was observed that genetic liability to T1D was associated with CKD risk in any of the three outcome studies from European ancestry (Supplementary Table S19B, available as Supplementary data at IJE online), which further supported the weak effect of glucose on CKD; (iii) for the MR analysis using stratified eGFR in Europeans, little effect of glycaemic phenotypes on eGFR was observed in both diabetic and non-diabetic samples (Supplementary Table S19C, available as Supplementary data at IJE online), which suggested that the weak effect of glucose on CKD could be independent of diabetes; (iv) fasting glucose and genetic liability to T2D were associated with diabetic retinopathy (Supplementary Table S19D, available as Supplementary data at IJE online), suggesting that the genetic predictors of glycaemic phenotypes used for the main MR analyses were reliable.
Effects of blood lipids and CKD
For the MR findings of lipids, our follow-up analyses showed a few novel observations. First, we observed different MR evidence for genetically predicted HDL-C on CKD across Europeans and East Asians. In Europeans, good MR evidence was observed to support the effects of HDL-C and apolipoprotein A-I on CKD (Supplementary Tables S7–S9, available as Supplementary data at IJE online), whereas there was weaker MR evidence for the effect of HDL-C on CKD in East Asians (Supplementary Tables S10–S12, available as Supplementary data at IJE online). To test the potential influence of the power of the HDL-C effect on CKD in East Asians (OR = 0.94, 95% CI = 0.87 to 1.02), we conducted MR using better-powered European HDL-C instruments extracted from the East Asian studies (Supplementary Table S5, available as Supplementary data at IJE online). Using this approach, we found reliable MR evidence (OR = 0.89, 95% CI = 0.83 to 0.96; Supplementary Table S16C, available as Supplementary data at IJE online). This suggests that HDL-C may have an effect on CKD in both populations. Second, using European data, a multivariable MR considering both HDL-C and apolipoprotein A-I in the same model was conducted. This showed that the effect of HDL-C on CKD was independent of apolipoprotein A-I (Supplementary Table S15D, available as Supplementary data at IJE online). Third, following the HDL-C finding, we found an effect of the circulating cholesteryl ester transfer protein level on CKD in CKDGen (OR = 1.06, 95% CI = 1.01 to 1.11, P = 0.01; Supplementary Table S20, available as Supplementary data at IJE online). Finally, we investigated the potential influence of the apolipoprotein(a) size on CKD but found little evidence for a causal effect. This suggests that the effect of the lipoprotein(a) level on CKD may be independent of the apolipoprotein(a) size (Supplementary Table S21, available as Supplementary data at IJE online).
Non-linear effects of BMI and fasting glucose on CKD
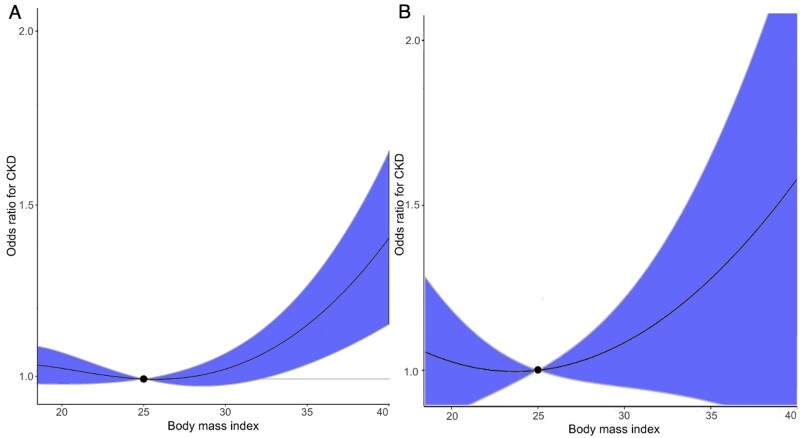
We observed a threshold relationship between genetically predicted BMI and CKD (Supplementary Table S22, available as Supplementary data at IJE online). The curved shape of this relationship suggests a higher risk of CKD in overweight or obese participants, with the optimal BMI threshold at ∼25 kg/m2 in both UK Biobank and HUNT (Figure 4). Stratified analyses split by sex (Supplementary Figure S5, available as Supplementary data at IJE online) and age (Supplementary Figure S6, available as Supplementary data at IJE online) suggested similar effects for genetically predicted BMI on CKD. Genetically predicted fasting glucose showed weak evidence for a non-linear relationship with CKD (Supplementary Figure S7 and Supplementary Table S22, available as Supplementary data at IJE online). This finding was consistent both among males and females (Supplementary Figure S8, available as Supplementary data at IJE online) and different age groups (Supplementary Figure S9, available as Supplementary data at IJE online).
Figure 4.
Non-linear Mendelian randomization of body mass index on chronic kidney disease risk. The dose–response curve between body mass index and chronic kidney disease risk for (A) UK Biobank and (B) the HUNT Study. The gradient at each point of the curve is the localized average causal effect. Shaded areas represent 95% confidence intervals. CKD, chronic kidney disease.
As a summary, we systematically compared the MR findings with existing clinical evidence in KDIGO guidelines and listed the potential clinical implications in Table 1.
Table 1.
Systematic evaluation of Mendelian-randomization evidence with existing clinical evidence
| Risk factors | Clinical evidence | MR evidence | Clinical implications |
|---|---|---|---|
| in KDIGO guideline | from this study | ||
| Blood pressure | Established | Strong evidence in Europeans | Suggest different prevention strategy for CKD across ancestries |
| Weak evidence in East Asians | |||
| BMI | Emerging | Strong in both ancestries | Suggest optimal control level of BMI as 25 |
| Nephrolithiasis | Emerging | Strong in both ancestries | Suggest assessment of kidney stones in high-risk groups |
| Diabetic phenotypes | Established | Strong for diabetes but weak evidence for other glycaemic traits | Imply glucose-independent effect of diabetes on CKD |
| Lipid phenotypes | Established | Strong evidence for HDL-C, cholesteryl ester transfer protein (CETP) and Lp(a) | Suggest cholesteryl ester transfer protein (CETP) and Lp(a) inhibition as intervention targets for CKD prevention |
| Low sleep duration | Emerging | Strong evidence in one study but lack of replication in other studies | Suggest future studies to confirm the effect of sleeping on CKD |
| Smoking, uric acid, CRP, bone, metal, vitamin, thyroid phenotypes | Emerging | Little evidence | – |
CKD, chronic kidney disease; BMI, body mass index; HDL, high-density lipoprotein; CETP, cholesteryl ester transfer protein; Lp(a), lipoprotein (a); CRP, C-reactive protein.
Discussion
In this trans-ethnic MR study, we comprehensively assessed the causality of 45 risk factors on CKD and eGFR in >1 million Europeans and 17 risk factors on CKD and eGFR in >250 000 East Asians. Using MR approaches, including five two-sample MR methods and multivariable MR, we found reliable evidence for the causal effects of eight cardiometabolic-related risk factors [BMI, SBP, hypertension, T2D, nephrolithiasis, HDL-C, apolipoprotein A-I and lipoprotein(a)] on CKD. The remaining 37 risk factors, including smoking and serum uric acid, had weak evidence to support causal effects on CKD using the currently available data. These findings are consistent with previous MR studies that analysed similar risk factors separately.40–44 In addition, the null finding of the serum uric acid agreed with the recent clinical trial investigating the effects of serum urate lowering (using Allopurinol) on CKD progression.45,46 Notably, our extensive MR and follow-up analyses suggested the possibility of glucose-independent pathways linking T2D with CKD. Using non-linear MR, we observed a threshold relationship between genetically predicted BMI and CKD risk, with increased CKD risk at a BMI of >25 kg/m2.
The causal patterns of 17 risk factors were compared across the two ancestries and we observed consistent effects of T2D, BMI and nephrolithiasis on CKD in Europeans and East Asians. In contrast, distinguishable causal patterns between ancestries were observed when examining the effect of hypertension on CKD, with a strong causal estimate in Europeans that was not replicated in the analysis of East Asians. These findings indicate that careful consideration is needed before implementing interventions for CKD risk factors in participants of one ancestry based on the evidence from another ancestry.
Among the prioritized risk factors, hypertension is one of the most common risk factors for kidney-function decline in patients with or without CKD.47–49 A recent bidirectional MR study in Europeans supported the causal effects of higher kidney function on lower blood pressure using eGFR instruments controlled by blood urea nitrogen. However, the same study suggested inconclusive evidence of an effect of blood pressure on eGFR.50 In our MR analysis, we found evidence of positive bidirectional causal effects between hypertension and CKD in Europeans. There are several potential explanations for the inconsistent MR findings across these studies. Yu et al. used genetic associations for blood pressure that were adjusted for BMI with genetic associations for eGFR and CKD that were not adjusted for BMI in their MR analysis. Given the causal role of BMI on both CKD and hypertension, only controlling for BMI in the exposure data may create unintended bias in the MR estimates, as described previously.51 An alternative explanation is the difference in CKD case ascertainment. Specifically, we used CKD cases that were clinically diagnosed, which may bring additional statistical power and provide more reliable evidence for the effect of blood pressure on CKD.
Given the difference in MR evidence across the ancestries that we observed, combined with previous evidence from the literature, it is possible that hypertension could have differential effects on CKD by ancestry. Ethnic disparities in relation to hypertension and CKD have previously been reported.52,53 For example, Chinese people with hypertension have a lower risk of CKD compared with European people with hypertension.52 Additionally, in 2019, hypertensive nephropathy accounted for 27% of the overall CKD cases in the USA but 20.8% of the overall CKD cases among Chinese.54,55 Further well-powered studies are needed to validate the causal effect of blood pressure on CKD across ancestries.
In addition, our MR analyses suggested substantial causal effects for BMI and nephrolithiasis on CKD. Previous observational studies have suggested that BMI is positively associated with CKD onset56 and end-stage renal disease57,58 and negatively associated with kidney function.59 The effect of weight loss on reducing the risk of diabetic nephropathy in patients with T2D60 and slowing kidney-function decline have also been reported.61 Using linear and non-linear MR approaches, we observed a threshold causal relationship between BMI and CKD. Moreover, nephrolithiasis is a common and serious health concern globally.62–64 There is increasing evidence to suggest that having kidney stones is a risk factor for CKD.62,65 For instance, people with kidney stones tend to have lower eGFR.63,66 A previous cohort study suggested that even a single kidney-stone episode was associated with an increased likelihood of adverse renal outcomes.67 A recent genetic study also suggested an inverse association between eGFR and kidney-stone formation.68 However, the causal relationship between nephrolithiasis and CKD had not been investigated previously. Our MR analysis supported the causal effect of increased nephrolithiasis risk on increasing CKD risk. This is of particular importance as obstructive nephropathy is among the leading causes of CKD in the general population. Specifically, it is the third leading cause of CKD among the Chinese population and has been estimated to be present in 15.6% of CKD cases.55
Notably, diabetic kidney disease is considered the most common type of CKD worldwide.69 A previous MR study of T2D on CKD in Chinese participants suggested a strong causal link between the two phenotypes,70 which aligned with our MR findings in both East Asians and Europeans. However, despite the reliable evidence for a causal effect of T2D on CKD, our linear and non-linear MR found limited evidence to support the causal effects of glucose- and insulin-related phenotypes on CKD. This is consistent with the findings from a previous MR study conducted in Europeans.71 It has also been observed that with increasing use of glucose-lowering medications, the prevalence of CKD in diabetics has not reduced as much as expected.72 This is supported by a meta-analysis of RCTs that found intensive glucose control to have an inconclusive effect on reducing the risk of end-stage renal disease.73 These findings, together with our MR results, suggest that glucose-independent pathways could play a role in the relationship between diabetes and CKD. Furthermore, it has consistently been suggested that the beneficial effects of SGLT2 inhibitors (antidiabetic medication) on renal outcomes may be mediated by glucose-independent pathways.74,75 One potential limitation of our analysis in relation to interrogating this finding is that the glucose GWAS that we used was conducted in a general population whose fasting glucose levels are <7 mmol/L. Existing MR studies using these data have made the assumption that the glucose change in the general population is similar to that in diabetic patients (i.e. individuals whose fasting glucose levels are typically >7 mmol/L), which may not necessarily be true. Although our stratified MR analysis showed little difference between diabetic and non-diabetic patients, we believe that better genetic instruments derived from a diabetic patient population and well-designed clinical trials are needed to evaluate the effect of glucose-dependent and -independent mechanisms on CKD prevention.
Hyperlipidaemia and dyslipidaemia have been widely documented to be associated with kidney disease.76,77 But the causal effects of lipid components on CKD are still unclear. A few recent MR studies have suggested a protective effect of higher HDL-C on CKD in Europeans,78 an adverse effect of higher triglycerides on CKD in Chinese79 and a nominal effect of lipoprotein(a) lowering on reducing CKD risk.80 In this study, we validated the HDL-C findings, confirmed the triglycerides effect in Biobank Japan and strengthened the evidence of the lipoprotein(a) finding in completely independent samples. Besides confirming these existing findings, our study also established novel causal evidence for the apolipoprotein A-I and non-apolipoprotein A-I properties of HDL-C on CKD risk in Europeans. Furthermore, our study extended the findings from recent studies of HDL-C81 and cholesteryl ester transfer protein inhibitors,82 which support the causal effect of circulating cholesteryl ester transfer protein levels on CKD in Europeans. The observed causal effect of HDL-C and the effect of cholesteryl ester transfer protein levels on CKD raises the possibility that increasing the HDL-C concentration may offer a potential intervention strategy for CKD prevention. Moreover, our study demonstrated that the causal effect of lipoprotein(a) levels on CKD was independent of the apolipoprotein(a) size. This finding, together with previous observational evidence,83–85 implies the possibility of lipoprotein(a)-reduction therapies, such as Pelacarsen [also known as IONIS-APO(a)-LRx], on reducing CKD risk.86 Overall, our findings have highlighted the potential for several lipid-management strategies in reducing CKD risk.
Strengths and limitations
Our study has some strengths compared with previous studies in this setting. We used clinically diagnosed CKD (instead of only using eGFR < 60 ml/min/1.73 m2 to define CKD) in two European (UK Biobank and HUNT) and two East Asian (China Kadoorie Biobank and Japan-Kidney-Biobank/ToMMo) studies. These four studies included participants with abnormal urine protein levels but with normal eGFR as CKD cases. This increased the robustness of the CKD definition. By comprehensively validating the MR findings in the six CKD studies, we also greatly enhanced the reliability of the causal atlas that we derived of risk factors for CKD.
Our study also has some potential limitations. First, we used the ICD 10 code to define CKD cases in three of the six studies. Such selection criteria excluded undiagnosed cases and diagnoses made in an outpatient setting. Considering the low disease awareness of CKD,87,88 such misclassification of the outcome may reduce the power of our study. However, as a trade-off, such an approach also excluded non-CKD samples from the case group (e.g. participants with a single eGFR measurement of <60 due to measurement error), which brought additional power to the statistical analysis. Second, we set up a stringent Bonferroni-corrected threshold together with other criteria (e.g. little evidence of pleiotropy) to select the top MR findings. Such a strategy could create some false-negative findings but minimize the possibility of identifying false-positive findings. With the aim of supporting the future clinical practice of CKD management, we decided to apply such a stringent strategy to provide the most reliable causal evidence using genetics. Second, in the MR analysis, genetic predictors for binary exposures (e.g. coronary artery disease) are not mimicking the exposure itself, but the predisposition to the exposure instead.89 Consequently, our results must be interpreted as the effect of removing the predisposition to the binary exposure (rather than treatment of the exposure) to reduce CKD risk. In addition, due to the relative lack of GWAS samples in East Asians, we could only examine causal effects for 17 of the 45 risk factors for this ancestry. For the same reason, the number of instruments for each risk factor in the analyses differed between the two ancestries. For risk factors with different MR evidence across ancestries, we conducted a comprehensive set of sensitivity analyses to minimize the influence of differences in power and instrument strength across ancestries. Other limitations of the study are listed in Supplementary Note S7 (available as Supplementary data at IJE online).
Conclusions
By evaluating the causal evidence for 45 risk factors on CKD in >1 million individuals of European ancestry and 17 risk factors in >250 000 individuals of East Asian ancestry, we have shown that eight risk factors are reliably causal for CKD in Europeans and three of these are also causal in East Asians. These risk factors are predominantly related to cardiometabolic health, which supports the shared causal link between cardiometabolic health and kidney function. The different causal pattern between hypertension and CKD in Europeans compared with that in East Asians suggests that blood pressure might have an ancestry-specific role in CKD aetiology. Ultimately, our findings may have important clinical implications in terms of informing primary prevention in ‘at-risk’ individuals with normal renal function, which may in turn help to reduce the burden of CKD globally.
Supplementary data
Supplementary data are available at IJE online.
Ethics approval
The HUNT study was approved by the Central Norway Regional Committee for Medical and Health Research Ethics (REC Central no. 2015/1188) and written informed consent was given by all participants. The UK Biobank study has ethical approval from the North West Multicentre Research Ethics Committee (MREC). The China Kadoorie Biobank study obtained ethics approval from the Oxford University Tropical Research Ethics Committee (approval number: 025–04, 3.2.2005), the Chinese Centre for Disease Control and Prevention (CDC) Ethical Review Committee (approval number: 005/2004, 9.7.2004) and the local CDC of each study area.
Funding
This research has been conducted using the UK Biobank resource under Application Numbers ‘40135’ and ‘15825’. J.Z. is funded by a Vice-Chancellor Fellowship from the University of Bristol. This research was also funded by the UK Medical Research Council Integrative Epidemiology Unit [MC_UU_00011/1, MC_UU_00011/4 and MC_UU_00011/7]. J.Z. is supported by the Academy of Medical Sciences (AMS) Springboard Award, the Wellcome Trust, the Government Department of Business, Energy and Industrial Strategy (BEIS), the British Heart Foundation and Diabetes UK [SBF006\1117]. This study was funded/supported by the NIHR Biomedical Research Centre at University Hospitals Bristol NHS Foundation Trust and the University of Bristol (G.D.S., T.R.G. and R.E.W.). This study received funding from the UK Medical Research Council [MR/R013942/1]. J.Z., Y.M.Z. and T.R.G are funded by a BBSRC Innovation fellowship. J.Z. is supported by the Shanghai Thousand Talents Program. Y.M.Z. is supported by the National Natural Science Foundation of China [81800636]. H.Z. is supported by the Training Program of the Major Research Plan of the National Natural Science Foundation of China [91642120], a grant from the Science and Technology Project of Beijing, China [D18110700010000] and the University of Michigan Health System–Peking University Health Science Center Joint Institute for Translational and Clinical Research [BMU2017JI007]. N.F. is supported by the National Institutes of Health awards R01-MD012765, R01-DK117445 and R21-HL140385. R.C. is funded by a Wellcome Trust GW4 Clinical Academic Training Fellowship [WT 212557/Z/18/Z]. The Trøndelag Health Study (the HUNT Study) is a collaboration between HUNT Research Centre (Faculty of Medicine and Health Sciences, NTNU, Norwegian University of Science and Technology), Trøndelag County Council, Central Norway Regional Health Authority and the Norwegian Institute of Public Health. M.C.B. is supported by the UK Medical Research Council (MRC) Skills Development Fellowship [MR/P014054/1]. S.F. is supported by a Wellcome Trust PhD studentship [WT108902/Z/15/Z]. Q.Y. is funded by a China Scholarship Council PhD scholarship [CSC201808060273]. Y.C. was supported by the National Key R&D Program of China [2016YFC0900500, 2016YFC0900501 and 2016YFC0900504]. The China Kadoorie Biobank baseline survey and the first resurvey were supported by a grant from the Kadoorie Charitable Foundation in Hong Kong. The long-term follow-up is supported by grants from the UK Wellcome Trust [202922/Z/16/Z, 088158/Z/09/Z and 104085/Z/14/Z]. Japan-Kidney-Biobank was supported by AMED under Grant Number 20km0405210. P.C.H. is supported by Cancer Research UK [grant number: C18281/A19169]. A.K. was supported by DFG KO 3598/5–1. N.F. is supported by NIH awards R01-DK117445, R01-MD012765 and R21-HL140385. The views expressed in this publication are those of the author(s) and not necessarily those of the NHS, the National Institute for Health Research or the Department of Health.
Data availability
The genetic-association data of the selected risk factors are available in Supplementary Tables (available as Supplementary data at IJE online). The GWAS summary statistics for CKD and eGFR that were generated using UK Biobank and CKDGen data are available from the MRC-IEU OpenGWAS database (https://gwas.mrcieu.ac.uk/) and CKDGen website (http://ckdgen.imbi.uni-freiburg.de/), respectively. The GWAS results from HUNT, Biobank Japan, China Kadoorie Biobank and Japan-Kidney-Biobank/ToMMo can be accessed by request to the data holders. The raw China Kadoorie Biobank data underlying this article can be accessed via a formal data request to ckbaccess@ndph.ox.ac.uk, following the institution’s data-access policies. Preliminary event adjudication data are not publicly available. The data underlying this article will be shared on reasonable request to the corresponding author.
Author contributions
J.Z. is the guarantor; J.Z., Y.M.Z. and H.R. performed the linear MR analysis; J.Z. and H.R. performed the non-linear MR with support from S.F., Q.Y. and S.B.; J.Z., H.R. and L.F.T. performed the GWAS in UK Biobank and the HUNT Study; Y.S., M.Y. and N.K. conducted the GWAS in each cohort and performed the GWAS meta-analysis in the Japan-Kidney-Biobank/ToMMo study; M.A., M.K., K.M., Y.K. and Y.O. performed the GWAS in Biobank Japan; C.Q.Y. and J.C.L. conducted the GWAS in the China Kadoorie Biobank; J.Z., Y.M.Z. and B.E. performed the systematic review of CKD risk factors; R.E.W. performed the sensitivity analyses of smoking and CKD; P.C.H., A.H., J.R., B.M.B., L.F.T., K.H., S.H., A.K., C.P., M.W. and B.O.A. provided key data and supported the MR analyses; M.C.B., Y.C., R.C., S.H., N.F., A.P.M., G.D.S., S.B., C.Q.Y. and B.O.A. reviewed the paper and provided key comments; J.Z., Y.M.Z., H.R., V.W., Y.S., Y.L., G.D.S., S.B., B.O.A., H.Z. and T.R.G. wrote the manuscript; J.Z., Y.M.Z., H.Z. and T.R.G. conceived of and designed the study and oversaw all analyses.
Conflict of interest
T.R.G. and J.Z. receive funding from GlaxoSmithKline and Biogen for unrelated research. None declared by the other authors.
Supplementary Material
Acknowledgements
We thank all CKB participants, project staff, the China National Centre for Disease Control and Prevention and its regional offices (for access to death and disease registries) and the Chinese National Health Insurance scheme (for linkage to hospitalization data).
References
- 1. Thomas R, Kanso A, Sedor JR.. Chronic kidney disease and its complications. Prim Care 2008;35:329–44, vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Webster AC, Nagler EV, Morton RL, Masson P.. Chronic kidney disease. Lancet 2017;389:1238–52. [DOI] [PubMed] [Google Scholar]
- 3. Dalrymple LS, Katz R, Kestenbaum B. et al. Chronic kidney disease and the risk of end-stage renal disease versus death. J Gen Intern Med 2011;26:379–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.GBD Chronic Kidney Disease Collaboration. Global, regional, and national burden of chronic kidney disease, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 2020;395:709–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Davies NM, Holmes MV, Davey Smith G.. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 2018;362:k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davey Smith G, Ebrahim S.. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 2003;32:1–22. [DOI] [PubMed] [Google Scholar]
- 7. Zheng J, Baird D, Borges M-C. et al. Recent developments in Mendelian randomization studies. Curr Epidemiol Rep 2017;4:330–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wuttke M, Li Y, Li M. et al. ; V. A. Million Veteran Program. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet 2019. ;51:957–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sudlow C, Gallacher J, Allen N. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Holmen J, Midthjell K, Krüger Ø. et al. The Nord-Trøndelag Health Study 1995-97 (HUNT 2). Norsk Epidemiol 2003;13:19–32. [Google Scholar]
- 11. Nagai A, Hirata M, Kamatani Y. et al. Overview of the Biobank Japan Project: study design and profile. J Epidemiol 2017;27:S2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Z, Chen J, Collins R. et al. ; China Kadoorie Biobank (CKB) orative group. China Kadoorie Biobank of 0.5 million people: survey methods, baseline characteristics and long-term follow-up. Int J Epidemiol 2011;40:1652–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Elsworth B, Dawe K, Vincent EE. et al. MELODI: mining enriched literature objects to derive intermediates. Int J Epidemiol 2018;47:369–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Elsworth B, Gaunt TR.. MELODI Presto: a fast and agile tool to explore semantic triples derived from biomedical literature. Bioinformatics 2021;37:583–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levey AS, Stevens LA, Schmid CH. et al. ; for the CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration). A new equation to estimate glomerular filtration rate. Ann Intern Med 2009;150:604–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gage SH, Jones HJ, Burgess S. et al. Assessing causality in associations between cannabis use and schizophrenia risk: a two-sample Mendelian randomization study. Psychol Med 2017;47:971–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sekula P, Del Greco M F, Pattaro C, Köttgen A.. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol 2016;27:3253–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lawlor DA, Harbord RM, Sterne JAC, Timpson N, Davey Smith G.. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Statist Med 2008;27:1133–63. [DOI] [PubMed] [Google Scholar]
- 19. Bowden J, Davey Smith G, Burgess S.. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan NA, Thompson JR.. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I 2 statistic. Int J Epidemiol 2016;45:1961–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hartwig FP, Davey Smith G, Bowden J.. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol 2017;46:1985–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bowden J, Hemani G, Davey Smith G.. Detecting individual and global horizontal pleiotropy in Mendelian randomization: a job for the humble heterogeneity statistic? Am J Epidemiol 2018;187:2681–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Timpson NJ, Nordestgaard BG, Harbord RM. et al. C-reactive protein levels and body mass index: elucidating direction of causation through reciprocal Mendelian randomization. Int J Obes 2011;35:300–08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Freeman G, Cowling BJ, Schooling CM.. Power and sample size calculations for Mendelian randomization studies using one genetic instrument. Int J Epidemiol 2013;42:1157–63. [DOI] [PubMed] [Google Scholar]
- 25. Burgess S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int J Epidemiol 2014;43:922–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hemani G, Zheng J, Elsworth B. et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7 e34408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takeuchi F, Akiyama M, Matoba N. et al. ; The International Genomics of Blood Pressure (iGEN-BP) Consortium. Interethnic analyses of blood pressure loci in populations of East Asian and European descent. Nat Commun 2018;9:5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hemani G, Tilling K, Davey Smith G.. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet 2017;13:e1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bowden J, Spiller W, Del Greco MF. et al. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int J Epidemiol 2018;47:1264–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Onengut-Gumuscu S, Chen W-M, Burren O. et al. ; Type 1 Diabetes Genetics Consortium. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet 2015;47:381–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pattaro C, Teumer A, Gorski M. et al. ; ECHOGen Consortium. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun 2016;7:10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou W, Nielsen JB, Fritsche LG, Wolford BN. et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet 2018;50:1335–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Spracklen CN, Horikoshi M, Kim YJ. et al. Identification of type 2 diabetes loci in 433,540 East Asian individuals. Nature 2020;582:240–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blauw LL, Li-Gao R, Noordam R. et al. CETP (Cholesteryl Ester Transfer Protein) concentration: a genome-wide association study followed by Mendelian randomization on coronary artery disease. Circ Genom Precis Med 2018;11:e002034. [DOI] [PubMed] [Google Scholar]
- 35. Saleheen D, Haycock PC, Zhao W. et al. Apolipoprotein(a) isoform size, lipoprotein(a) concentration, and coronary artery disease: a Mendelian randomisation analysis. Lancet Diabetes Endocrinol 2017;5:524–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun Y-Q, Burgess S, Staley JR. et al. Body mass index and all cause mortality in HUNT and UK Biobank studies: linear and non-linear Mendelian randomisation analyses. BMJ 2019;364:1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burgess S, Davies NM, Thompson SG; EPIC-InterAct Consortium. Instrumental variable analysis with a nonlinear exposure-outcome relationship. Epidemiology 2014;25:877–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Staley JR, Burgess S.. Semiparametric methods for estimation of a nonlinear exposure-outcome relationship using instrumental variables with application to Mendelian randomization. Genet Epidemiol 2017;41:341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sterne JA, Davey Smith G.. Sifting the evidence—what’s wrong with significance tests? BMJ 2001;322:226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li X, Meng X, Timofeeva M. et al. Serum uric acid levels and multiple health outcomes: umbrella review of evidence from observational studies, randomised controlled trials, and Mendelian randomisation studies. BMJ 2017;357:j2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen C, Xia F, Chen Y. et al. Association between thyroid-stimulating hormone and renal function: a Mendelian randomization study. Kidney Blood Press Res 2018;43:1121–30. [DOI] [PubMed] [Google Scholar]
- 42. Liu H-M, Hu Q, Zhang Q. et al. Causal effects of genetically predicted cardiovascular risk factors on chronic kidney disease: a two-sample Mendelian randomization study. Front Genet 2019;10:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Emanuelsson F, Nordestgaard BG, Tybjærg-Hansen A, Benn M.. Impact of LDL cholesterol on microvascular versus macrovascular disease: a Mendelian randomization study. J Am Coll Cardiol 2019;74:1465–76. [DOI] [PubMed] [Google Scholar]
- 44. Jordan DM, Choi HK, Verbanck M. et al. No causal effects of serum urate levels on the risk of chronic kidney disease: a Mendelian randomization study. PLoS Med 2019;16:e1002725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Badve SV, Pascoe EM, Tiku A. et al. ; CKD-FIX Study Investigators. Effects of allopurinol on the progression of chronic kidney disease. N Engl J Med 2020;382:2504–13. [DOI] [PubMed] [Google Scholar]
- 46. Doria A, Galecki AT, Spino C. et al. ; PERL Study Group. Serum urate lowering with allopurinol and kidney function in type 1 diabetes. N Engl J Med 2020;382:2493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Anderson AH, Yang W, Townsend RR. et al. ; Chronic Renal Insufficiency Cohort Study Investigators. Time-updated systolic blood pressure and the progression of chronic kidney disease: a cohort study. Ann Intern Med 2015;162:258–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu Z, Rebholz CM, Wong E. et al. Association between hypertension and kidney function decline: the Atherosclerosis Risk in Communities (ARIC) study. Am J Kidney Dis 2019;74:310–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Judson GL, Rubinsky AD, Shlipak MG. et al. Longitudinal blood pressure changes and kidney function decline in persons without chronic kidney disease: findings from the MESA study. Am J Hypertens 2018;31:600–08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yu Z, Coresh J, Qi GH. et al. A bidirectional Mendelian randomization study supports causal effects of kidney function on blood pressure. Kidney Int 2020;98:708–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hartwig FP, Tilling K, Smith GD, Lawlor DA, Borges MC.. Bias in two-sample Mendelian randomization by using covariable-adjusted summary associations. Int J Epidemiol 2021;50:1639–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu R, Zhang L, Zhang P, Wang F, Zuo L, Wang H.. Comparison of the prevalence of chronic kidney disease among different ethnicities: Beijing CKD survey and American NHANES. Nephrol Dial Transplant 2009;24:1220–26. [DOI] [PubMed] [Google Scholar]
- 53. Hull S, Dreyer G, Badrick E, Chesser A, Yaqoob MM.. The relationship of ethnicity to the prevalence and management of hypertension and associated chronic kidney disease. BMC Nephrol 2011;12:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Saran R, Robinson B, Abbott KC. et al. US Renal Data System 2019 annual data report: epidemiology of kidney disease in the United States. Am J Kidney Dis 2020;75(1 Suppl 1):A6–7. [DOI] [PubMed] [Google Scholar]
- 55. Zhang L, Zhao M-H, Zuo L. et al. China Kidney Disease Network (CK-NET) 2015 annual data report. Kidney Int Suppl 2019;9:e1–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Garofalo C, Borrelli S, Minutolo R, Chiodini P, De Nicola L, Conte G.. A systematic review and meta-analysis suggests obesity predicts onset of chronic kidney disease in the general population. Kidney Int 2017;91:1224–35. [DOI] [PubMed] [Google Scholar]
- 57. Iseki K, Ikemiya Y, Kinjo K, Inoue T, Iseki C, Takishita S.. Body mass index and the risk of development of end-stage renal disease in a screened cohort. Kidney Int 2004;65:1870–76. [DOI] [PubMed] [Google Scholar]
- 58. Hsu C-Y, McCulloch CE, Iribarren C, Darbinian J, Go AS.. Body mass index and risk for end-stage renal disease. Ann Intern Med 2006;144:21–28. [DOI] [PubMed] [Google Scholar]
- 59. Tonelli M, Wiebe N, Kovesdy CP. et al. ; for the Alberta Kidney Disease Network. Joint associations of obesity and estimated GFR with clinical outcomes: a population-based cohort study. BMC Nephrol 2019;20:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. O'Brien R, Johnson E, Haneuse S. et al. Microvascular outcomes in patients with diabetes after bariatric surgery versus usual care: a matched cohort study. Ann Intern Med 2018;169:300–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Navaneethan SD, Yehnert H, Moustarah F, Schreiber MJ, Schauer PR, Beddhu S.. Weight loss interventions in chronic kidney disease: a systematic review and meta-analysis. Clin J Am Soc Nephrol 2009;4:1565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Moe OW. Kidney stones: pathophysiology and medical management. Lancet 2006;367:333–44. [DOI] [PubMed] [Google Scholar]
- 63. Scales CD Jr, Smith AC, Hanley JM, Saigal CS.. Urologic Diseases in America Project. Prevalence of kidney stones in the United States. Eur Urol 2012;62:160–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ward JB, Feinstein L, Pierce C. et al. ; NIDDK Urologic Diseases in America Project. Pediatric urinary stone disease in the United States: the urologic diseases in America Project. Urology 2019;129:180–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Uribarri J. Chronic kidney disease and kidney stones. Curr Opin Nephrol Hypertens 2020;29:237–42. [DOI] [PubMed] [Google Scholar]
- 66. Gillen DL, Worcester EM, Coe FL.. Decreased renal function among adults with a history of nephrolithiasis: a study of NHANES III. Kidney Int 2005;67:685–90. [DOI] [PubMed] [Google Scholar]
- 67. Alexander RT, Hemmelgarn BR, Wiebe N. et al. ; for the Alberta Kidney Disease Network. Kidney stones and kidney function loss: a cohort study. BMJ 2012;345:e5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Morris AP, Le TH, Wu H. et al. Trans-ethnic kidney function association study reveals putative causal genes and effects on kidney-specific disease aetiologies. Nat Commun 2019;10:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jha V, Garcia-Garcia G, Iseki K. et al. Chronic kidney disease: global dimension and perspectives. Lancet 2013;382:260–72. [DOI] [PubMed] [Google Scholar]
- 70. Xu M, Bi Y, Huang Y. et al. Type 2 diabetes, diabetes genetic score and risk of decreased renal function and albuminuria: a Mendelian randomization study. EBioMedicine 2016;6:162–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Emanuelsson F, Marott S, Tybjærg-Hansen A, Nordestgaard BG, Benn M.. Impact of glucose level on micro- and macrovascular disease in the general population: a Mendelian randomization study. Diabetes Care 2020;43:894–902. [DOI] [PubMed] [Google Scholar]
- 72. de Boer IH, Rue TC, Hall YN, Heagerty PJ, Weiss NS, Himmelfarb J.. Temporal trends in the prevalence of diabetic kidney disease in the United States. JAMA 2011;305:2532–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Coca SG, Ismail-Beigi F, Haq N, Krumholz HM, Parikh CR.. Role of intensive glucose control in development of renal end points in type 2 diabetes mellitus: systematic review and meta-analysis intensive glucose control in type 2 diabetes. Arch Intern Med 2012;172:761–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Perkovic V, Jardine MJ, Neal B. et al. Canagliflozin and renal outcomes in type 2 diabetes and nephropathy. N Engl J Med 2019;380:2295–306. [DOI] [PubMed] [Google Scholar]
- 75. Cannon CP, Perkovic V, Agarwal R. et al. Evaluating the effects of canagliflozin on cardiovascular and renal events in patients with type 2 diabetes mellitus and chronic kidney disease according to baseline HbA1c, including those with HbA1c <7%: results from the CREDENCE trial. Circulation 2020;141:407–10. [DOI] [PubMed] [Google Scholar]
- 76. Florens N, Calzada C, Lyasko E, Juillard L, Soulage CO.. Modified lipids and lipoproteins in chronic kidney disease: a new class of uremic toxins. Toxins 2016;8:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Visconti L, Benvenga S, Lacquaniti A. et al. Lipid disorders in patients with renal failure: role in cardiovascular events and progression of chronic kidney disease. J Clin Transl Endocrinol 2016;6:8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lanktree MB, Thériault S, Walsh M, Paré G.. HDL cholesterol, LDL cholesterol, and triglycerides as risk factors for CKD: a Mendelian randomization study. Am J Kidney Dis 2018;71:166–72. [DOI] [PubMed] [Google Scholar]
- 79. Zhang Y-B, Sheng L-T, Wei W. et al. Association of blood lipid profile with incident chronic kidney disease: a Mendelian randomization study. Atherosclerosis 2020;300:19–25. [DOI] [PubMed] [Google Scholar]
- 80. Emdin CA, Khera AV, Natarajan P. et al. ; CARDIoGRAM Exome Consortium. Phenotypic characterization of genetically lowered human lipoprotein(a) levels. J Am Coll Cardiol 2016;68:2761–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Holmes MV, Davey Smith G.. Challenges in interpreting multivariable Mendelian randomization: might ‘good cholesterol’ be good after all? Am J Kidney Dis 2018;71:149–53. [DOI] [PubMed] [Google Scholar]
- 82. Bowman L, Hopewell JC, Chen F. et al. ; HPS3/TIMI55–REVEAL Collaborative Group. Effects of anacetrapib in patients with atherosclerotic vascular disease. N Engl J Med 2017;377:1217–27. [DOI] [PubMed] [Google Scholar]
- 83. Lin J, Reilly MP, Terembula K, Wilson FP.. Plasma lipoprotein(a) levels are associated with mild renal impairment in type 2 diabetics independent of albuminuria. PLoS One 2014;9:e114397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kronenberg F, Kuen E, Ritz E. et al. Lipoprotein(a) serum concentrations and apolipoprotein(a) phenotypes in mild and moderate renal failure. J Am Soc Nephrol 2000;11:105–15. [DOI] [PubMed] [Google Scholar]
- 85. Kovesdy CP, Astor BC, Longenecker JC, Coresh J.. Association of kidney function with serum lipoprotein(a) level: the third National Health and Nutrition Examination Survey (1991-1994). Am J Kidney Dis 2002;40:899–908. [DOI] [PubMed] [Google Scholar]
- 86. Tsimikas S, Viney NJ, Hughes SG. et al. Antisense therapy targeting apolipoprotein(a): a randomised, double-blind, placebo-controlled phase 1 study. Lancet 2015;386:1472–83. [DOI] [PubMed] [Google Scholar]
- 87. Wang F, Zhang L, Wang H; China National Survey of CKD Working Group. Awareness of CKD in China: a national cross-sectional survey. Am J Kidney Dis 2014;63:1068–70. [DOI] [PubMed] [Google Scholar]
- 88. Coresh J, Byrd-Holt D, Astor BC. et al. Chronic kidney disease awareness, prevalence, and trends among U.S. adults, 1999 to 2000. J Am Soc Nephrol 200n;16:180–88. [DOI] [PubMed] [Google Scholar]
- 89. Munafò MR, Tilling K, Taylor AE. et al. Collider scope: when selection bias can substantially influence observed associations. Int J Epidemiol 2018;47:226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genetic-association data of the selected risk factors are available in Supplementary Tables (available as Supplementary data at IJE online). The GWAS summary statistics for CKD and eGFR that were generated using UK Biobank and CKDGen data are available from the MRC-IEU OpenGWAS database (https://gwas.mrcieu.ac.uk/) and CKDGen website (http://ckdgen.imbi.uni-freiburg.de/), respectively. The GWAS results from HUNT, Biobank Japan, China Kadoorie Biobank and Japan-Kidney-Biobank/ToMMo can be accessed by request to the data holders. The raw China Kadoorie Biobank data underlying this article can be accessed via a formal data request to ckbaccess@ndph.ox.ac.uk, following the institution’s data-access policies. Preliminary event adjudication data are not publicly available. The data underlying this article will be shared on reasonable request to the corresponding author.