Abstract
Objective
Familial intracranial aneurysms (FIAs) are found in approximately 6%–20% of patients with intracranial aneurysms (IAs), suggesting that genetic predisposition likely plays a role in its pathogenesis. The aim of this study was to identify possible IA-associated variants using whole exome sequencing (WES) in selected Korean families with FIA.
Materials and Methods
Among the 26 families in our institutional database with two or more IA-affected first-degree relatives, three families that were genetically enriched (multiple, early onset, or common site involvement within the families) for IA were selected for WES. Filtering strategies, including a family-based approach and knowledge-based prioritization, were applied to derive possible IA-associated variants from the families. A chromosomal microarray was performed to detect relatively large chromosomal abnormalities.
Results
Thirteen individuals from the three families were sequenced, of whom seven had IAs. We noted three rare, potentially deleterious variants (PLOD3 c.1315G>A, NTM c.968C>T, and CHST14 c.58C>T), which are the most promising candidates among the 11 potential IA-associated variants considering gene-phenotype relationships, gene function, co-segregation, and variant pathogenicity. Microarray analysis did not reveal any significant copy number variants in the families.
Conclusion
Using WES, we found that rare, potentially deleterious variants in PLOD3, NTM, and CHST14 genes are likely responsible for the subsets of FIAs in a cohort of Korean families.
Keywords: Whole exome sequencing, Genetics, Familial intracranial aneurysm
INTRODUCTION
The global prevalence of intracranial aneurysms (IAs) is estimated to be 3.2% [1]. In 6%–20% of patients with IA, one or more of their family members also have an IA [2]. These cases are defined as familial intracranial aneurysms (FIAs) and are reported to have a more severe phenotype in terms of a higher number of aneurysms and a higher risk of rupture than those without a familial history [3,4,5].
Several linkage studies and genome-wide association studies (GWAS) have identified a large number of candidate loci associated with FIAs [6,7,8]. However, the potential genetic defects in these loci have a relatively small effect on the risk of developing IA and can only explain a small fraction of heritability [6,7]. Recently, several studies using next-generation sequencing (NGS) have suggested rare variants in 10 candidate genes (ADAMTS15, THSD1, RNF213, ANGPTL6, LOXL2, ARHGEF17, C4orf6, SPDYE4, NFX1, and EDIL3) with larger effects related to FIA; however, some of these variants require further validation. Although the variants of these genes may explain some aneurysms in certain ethnic groups, they are rarely replicated across different studies.
Previous studies have mostly focused on the presence of an aneurysm and not on its phenotypic presentation, such as its location, shape, and size. We hypothesized that if a specific gene was associated with IA, the characteristics of the aneurysm would be shared among members of the family. Therefore, to increase the possibility of gene identification, we reasoned that detailed information on the aneurysm should be obtained and considered when recruiting the family. The purpose of this study was to use NGS to identify potential IA-associated variants in families that share a specific phenotype.
MATERIALS AND METHODS
Study Population
The Institutional Review Board of Asan Medical Center approved this prospective study (IRB No. 2018–1106). Informed written consent for blood sampling and magnetic resonance angiography screening was obtained from all study participants.
IA was defined as a saccular dilatation of any size occurring in the intracranial arteries; FIA was defined as when at least two first-degree relatives in a family were diagnosed with IA. A family history of IA was identified in 28 (4.4%) patients among the 638 patients with IA in a tertiary hospital's prospectively collected database from between January 2011 and August 2018. We then selected families with FIA for further genetic testing according to the following inclusion criteria: 1) demonstration of the pedigree of the disease status in the family, 2) two or more affected members and one or more non-affected members are available for genetic testing, 3) available angiographic data for the participants (both affected and unaffected), 4) genetically enriched samples where the family has a severe phenotype of IA (multiple, early onset, ruptured) and common site involvement among families [3,4,5]; and 5) consent to participated provided by the patient and family members.
We excluded patients who had 1) fusiform, mycotic, or dissecting aneurysms in the intracranial artery, 2) aneurysms associated with an arteriovenous malformation, or 3) aneurysms associated with syndromic disorders (e.g., polycystic kidney disease, Ehlers-Danlos syndrome, Marfan syndrome, fibromuscular dysplasia, and moyamoya disease). Physical examination, ultrasonography, and/or computed tomography angiography were performed to rule out any known or unknown syndromes associated with IA. Other first-degree relatives who had not been screened for IA underwent magnetic resonance angiography.
Whole Exome Sequencing (WES) Analysis
Genomic DNA was extracted from peripheral blood cells using the Chemagic Magnetic Separation Module I (Chemagic MSM I) extraction robot with a DNA Blood 200 µL Kit. SureSelect Human All Exon V5 (Agilent Technologies) was used for library preparation, and sequencing was performed on the Illumina NextSeq500 platform (Illumina Inc.), which generated 2 × 150 bp paired-end reads. The averages of the 30 × and 20 × coverage for the target regions were 89.18% and 94.34%, respectively. Trimmomatic v.0.36 was used to trim sequences of sequencing adapters and suffixes of low quality (i.e., Phred quality score < 10).
Variant Calling and Filtering Strategy
All reads were aligned to the human reference genome (GRCh37/hg19) using the Burrow-Wheeler Aligner (BWA version 0.7.12). The Picard tool (version 1.96, http://picard.sourceforge.net) was used to remove duplicate reads, and the Genome Analysis ToolKit (GATK version 3.30) was used for variant calling. The Annotation of Genetic Variants program (ANNOVAR, http://annovar.openbioinformatics.org) was used to annotate alterations using information from the following public databases: the Single Nucleotide Polymorphism database (dbSNP 147, https://www.ncbi.nlm.nih.gov/snp/), 1000 Genomes Project (https://www.internationalgenome.org/), Exome Aggregation Consortium (ExAC, http://exac.broadinstitute.org/), and Genome Aggregation Database (gnomAD, https://gnomad.broadinstitute.org/).
Variants with less than 10 × coverage and an allele frequency of more than 0.01 in public databases (1000 Genomes Project, Exome Aggregation Consortium, and gnomAD) were removed. Additionally, variants affecting protein-altering and splicing (e.g., non-synonymous amino acid changes, start codon alterations, stop loss changes, in-frame insertions/deletions, frameshifts, nonsense variants, and changes affecting consensus splice site sequences) were included.
In the family-based approach, we selected variants segregating as occurring only in affected members, and variants shared with unaffected members. For knowledge-based prioritization, the variants were screened among the 450 genes associated with aneurysm or vascular/connective tissue disorders in the Online Mendelian Inheritance in Man (OMIM) database (https://www.omim.org/), 16 IA candidate genes reported in PubMed-indexed studies (https://pubmed.ncbi.nlm.nih.gov/), and 77 genes from the GWAS catalog (https://www.ebi.ac.uk/gwas/) for brain aneurysms.
Variant Interpretation
The pathogenicity of the variants was predicted using Sorting Intolerant From Tolerant (SIFT) [9], Polyphen2[10], Genomic Evolutionary Rate Profiling (GERP) [11], Combined Annotation Dependent Depletion (CADD)[12], rare exome variant ensemble learner (REVEL) [13], Mendelian Clinically Applicable Pathogenicity (M-CAP) [14], pLI (the probability of being loss-of-function intolerant)[15], and % haploinsufficient (HI) [16]. Pathogenicity thresholds were chosen according to the respective authors' recommendations (SIFT, ≤ 0.05; Polyphen2, ≥ 0.9; GERP, ≥ 2; CADD, ≥ 20; REVEL, ≥ 0.5; M-CAP, ≥ 0.025; pLI, ≥ 0.9; and%HI, ≤ 10%).
Based on the standards and guidelines for the interpretation of sequence variants from the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) [17], the candidate variants were classified into five types: pathogenic variant (PV), likely pathogenic variant (LPV), variant of uncertain significance (VUS), likely benign variant (LBV), and benign variant (BV). We assigned PP3 (pathogenic supporting) as a variant if at least two out of three meta-predictors (CADD, REVEL, M-CAP) and SIFT or Polyphen2 calculated a pathogenicity score above their respective thresholds. All clinically significant and novel variants were confirmed using independent Sanger sequencing [18].
Chromosomal Microarray
To identify submicroscopic deletions or duplications that are difficult to assess using whole exome sequencing (WES), copy number analysis was performed using CytoScan HD (Affymetrix) according to the manufacturer's protocol. Regions of homozygosity and copy number variants (CNVs) shared between affected and unaffected siblings were eliminated as potential candidate regions. Thresholds for the detection of candidate pathogenic CNVs in affected subjects were set to 25 CNV markers for deletions and 50 CNV markers for duplications. CNVs were interpreted based on the technical standards of a joint consensus recommendation of the ACMG and the Clinical Genome Resource (ClinGen) [19].
RESULTS
Clinical Phenotypes of Three Families with FIA Used in This Study
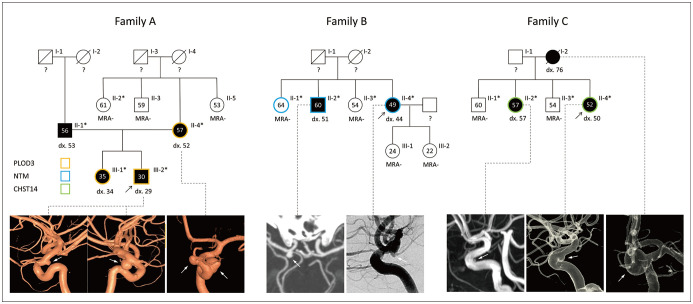
Thirteen individuals from three families were selected for WES. In all three families, two or more members had IAs at a common location (Fig. 1). In family A, the proband and his mother had paraclinoid aneurysms. The characteristics of this family included early onset (III-1, 2), presence of multiple aneurysms (average number of aneurysms ≥ 2) in a common location (II-4, III-1), and relatively few risk factors (Table 1). The father of the siblings also had an IA in the middle cerebral artery. Family B had two affected siblings and two unaffected siblings in the second generation. The proband (II-4) had two small unruptured aneurysms in the right internal carotid artery at the origin of the posterior communicating artery (P-COM) and at the top of the basilar artery, and her older brother (II-2) also had a small internal carotid artery aneurysm at the origin of the P-COM artery. Family C had three affected females (I-1, II-2, and II-4) whose IAs were commonly located in the paraclinoid region of the internal carotid artery.
Fig. 1. Pedigree of the families with genetic and phenotypical findings.
Asterisks indicate individuals that were subjected to whole-exome sequencing. black arrows = probands, black symbols = IA-affected cases, circles = females, diagonal line through a symbol = deceased, dx. number = age at IA diagnosis, IA = intracranial aneurysm, MRA- = negative result on magnetic resonance angiography, number in symbols = current age, Squares = males, white arrows = aneurysms on angiography, ? = unknown aneurysm status
Table 1. Baseline Characteristics of the Participants.
| Family (Member) | Age (Year) | Sex | Risk Factors | Diagnosis (Age; Year) | Aneurysm Location (Size; mm) | Treatment (Age; Year) | WES |
|---|---|---|---|---|---|---|---|
| A (II-1)* | 56 | M | DM, HL, Smoking 90PY |
DSA (53) | Rt. MCA (3.8) | Clipping (53) | Yes |
| A (II-2) | 61 | F | None | MRA | Negative | N/A | Yes |
| A (II-3) | 59 | M | Smoking 30PY | MRA | Negative | N/A | No |
| A (II-4)* | 57 | F | HL | DSA (52) | Lt. paraclinoid (8.5, 5.1) | Coiling (52) | Yes |
| A (II-5) | 53 | F | None | MRA | Negative | N/A | No |
| A (III-1)* | 35 | F | None | MRA (34) | Rt. P-COM (1.7), Lt. P-COM (1.8) | No | Yes |
| A (III-2)* | 30 | M | Smoking 5PY | DSA (29) | Lt. paraclinoid (6.0), Rt. paraclinoid (3.2) Lt. AchA (2.0) |
Coiling (29) | Yes |
| B (II-1) | 64 | F | None | MRA | Negative | N/A | Yes |
| B (II-2)* | 60 | M | HTN, DM, HL, smoking 25PY | CTA (51) | P-COM (7.7) | Coiling (51) | Yes |
| B (II-3) | 54 | F | DM, HL | MRA | Negative | N/A | Yes |
| B (II-4)* | 49 | F | HTN, DM | DSA (44) | Basilar top (3.1), P-COM (2.1) |
Coiling (44) (basilar top) |
Yes |
| C (I-2)* | 82 | F | Cardiac (angina), HTN, HL | DSA (76) | Paraclinoid (14.3) | Coiling (76) | No |
| C (II-1) | 60 | M | Smoking 15PY, cardiac (MI) | MRA | Negative | N/A | Yes |
| C (II-2)* | 57 | F | Cardiac (arrhythmia) | MRA (57) | Paraclinoid (1.0) | No | Yes |
| C (II-3) | 54 | M | None | MRA | Negative | N/A | Yes |
| C (II-4)* | 52 | F | None | DSA (50) | Paraclinoid (4.5) | No | Yes |
*IA-affected subjects. AchA = anterior choroidal artery, An = aneurysm, CTA = computed tomography angiography, DM = diabetes mellitus, DSA = digital subtraction angiography, HL = hyperlipidemia, HTN = hypertension, Lt = left, MCA = middle cerebral artery, MI = myocardial infarction, MRA = magnetic resonance angiography, N/A = not applicable, P-COM = posterior communicating artery, PY = pack-year, Rt = right, WES = whole exome sequencing
WES Analysis and Variant Filtering
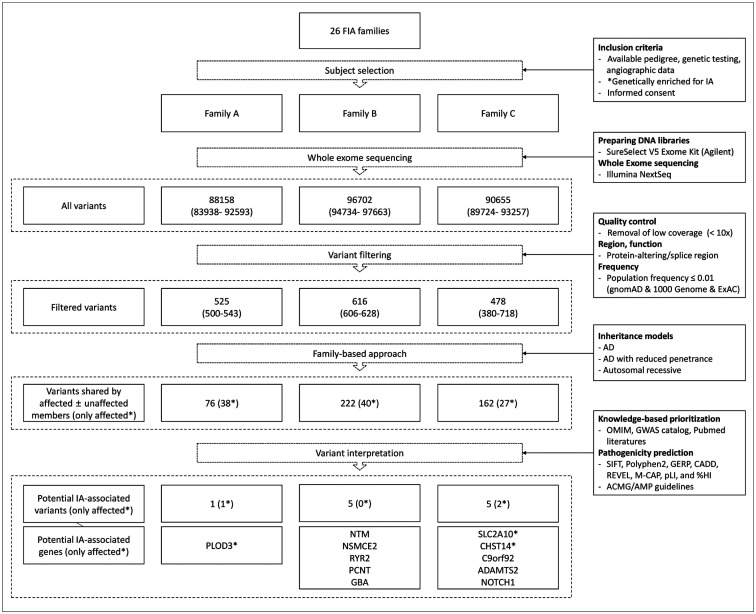
WES was performed in all living affected individuals and at least one unaffected first-degree relatives of the probands. Among the > 90000 variants initially discovered in the WES, an average of approximately 500 variants for each individual were selected after excluding those with insufficient coverage, a frequency of 0.01 or more in the population, and variants that did not affect the protein (Fig. 2). Through a family-based approach according to Mendelian inheritance patterns, 40, 38, and 27 variants (autosomal dominant) and 222, 76, 162 variants (autosomal dominant reduced penetrance) were selected in each family, respectively. There were no variants that showed segregation of autosomal recessive patterns among the three families. Finally, 11 pathogenic or damaging variants potentially associated with IA were derived through pathogenicity prediction algorithms and knowledge-based prioritization from previous genetic studies.
Fig. 2. Flowchart of variant filtering steps and results by family.
*Genetically enriched for IA: severe IA phenotype (multiple, early onset, or ruptured) and common site involvement within families. ACMG = American College of Medical Genetics and Genomics, AD = autosomal dominant, AMP = Association for Molecular Pathology, CADD = Combined Annotation Dependent Depletion, ExAC = exome aggregation consortium, FIA = familial intracranial aneurysm, GERP = Genomic Evolutionary Rate Profiling, GWAS = genome-wide association studies, HI = haploinsufficient, IA = intracranial aneurysm, M-CAP = Mendelian Clinically Applicable Pathogenicity, OMIM = Online Mendelian Inheritance in Man, pLI = the probability of being loss-of-function intolerant, REVEL = rare exome variant ensemble learner, SIFT = Sorting Intolerant From Tolerant
Potential IA-Associated Genes
All variants were heterozygous and missense variants, except for the nonsense mutation in the C9orf92 gene (Table 2). GBA and C9orf92 genes have been reported as susceptible genes associated with brain aneurysm in the GWAS catalog, and the remaining genes were reported to be associated with aneurysm or vascular/connective tissue disorders in the OMIM. The genes found in recent NGS studies were not identified in this study [8,20,21,22,23,24,25,26,27].
Table 2. Variants Identified in Patients with Familial Intracranial Aneurysms according to the Application of Filtering Strategy and Segregation.
| Gene | Transcript ID | c. notation p. notation |
rs ID | MAF | Pathogenicity Predictions† | ACMG Interpretation‡ | Subjects | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gnomAD/ KRGDB | SIFT | Polyphen2 | GERP | CADD | REVEL | M-CAP | pLI | %HI | Classification | Fm | Member* | ||||
| PLOD3 | NM_001084.4 | c.1315G>A p.Ala439Thr |
rs138610113 | 0.000403/ 0.002105 | 0.011 | 0.646 | 5.13 | 21.9 | 0.087 | 0.06 | 0.06 | 0.65 | VUS PM1 + PP3 |
A | II-2*, III-1*, III-2* |
| NTM | NM_001048209.1 | c.968C>T p.Thr323Met |
rs755518659 | 0.000018/ 0.000000 | 0.223 | 0.958 | 5.21 | 23.6 | 0.247 | 0.138 | 1 | 14.03 | VUS PM2 +PP3 |
B | II-1, II-2*, II-4* |
| RYR2 | NM_001035.2 | c.3595G>A p.Asp1199Asn |
rs752376759 | 0.000065/ 0.000000 | 0.153 | 0.733 | 0.235 | 8.116 | 0.028 | 0.003 | 0 | 80.67 | LPV PM1 + PM2 + PP2 + PP3 |
B | II-2*, II-3, II-4* |
| PCNT | NM_006031.5 | c.545G>A p.Arg182His |
rs193268784 | 0.000113/ 0.003143 | 0.001 | 1 | 5.62 | 32 | 0.22 | 0.037 | 0.04 | 23.92 | LBV PM2 + BP1 + BP4 |
B | II-2*, II-3, II-4* |
| NSMCE2 | NM_173685.2 | c.217C>T p.Arg73Trp |
rs201903722 | 0.000191/ 0.007079 | 0.241 | 0.001 | 1.25 | 13.01 | 0.493 | 0.241 | 0 | 52.28 | VUS PM2 + PP3 + BP1 |
B | II-1, II-2*, II-3, II-4* |
| GBA | NM_000157.3 | c.680A>G p.Asn227Ser |
rs364897 | 0.000074/ 0.000262 | 0.001 | 0.979 | 4.3 | 23.6 | 0.244 | N/A | 0 | 51.13 | LPV PS3 + PM1 + PM2 + PP5 |
B | II-1, II-2*, II-3, II-4* |
| CHST14 | NM_130468.3 | c.58C>T p.Arg20Trp |
rs577809616 | 0.000216/ 0.003584 | 0 | 0.998 | 3.8 | 27.9 | 0.37 | 0.771 | 0.45 | 35.81 | VUS PM2 + PP2 + PP3 |
C | II-2*, II-4* |
| SLC2A10 | NM_030777.3 | c.931G>A p.Val311Ile |
rs139932041 | 0.000287/ 0.000786 | 0.283 | 0.084 | 3.88 | 14.58 | 0.14 | 0.018 | 0 | 71.81 | VUS PM1 + PM2 |
C | II-2*, II-4* |
| ADAMTS2 | NM_014244.4 | c.268G>A p.Ala90Thr |
rs776393146 | 0.000095/ 0.005500 | 0.595 | 0.212 | 4.18 | 19.61 | 0.044 | 0.008 | 0.97 | 25.76 | VUS PM1 + PM2 + BP1 |
C | II-2*, II-3, II-4* |
| NOTCH1 | NM_017617.4 | c.5422G>A p.Asp1808Asn |
rs571739078 | 0.000043/ 0.000786 | 0.091 | 0.676 | 4.55 | 23.7 | 0.381 | 0.512 | 1 | 0.15 | VUS PM2 + PP2 + PP3 |
C | II-2*, II-3, II-4* |
| C9orf92 | NM_001271829.1 | c.106C>T p.Gln36Ter | N/A | 0.000000/ 0.000000 | N/A | N/A | 1.98 | N/A | N/A | N/A | 0 | 0.52 | VUS PM2 |
C | II-2*, II-3, II-4* |
*IA-affected subject, †Pathogenicity thresholds: SIFT (≤ 0.05), Polyphen2 (≥ 0.9), GERP (≥ 2), CADD (≥ 20), REVEL (≥ 0.5), M-CAP (≥ 0.025), pLI (≥ 0.9), and %HI (≤ 10%), ‡ACMG standards and guidelines: VUS, LBV, LPV, each pathogenic criterion is weighted as very strong (PVS1), strong (PS1–4); moderate (PM1–6), or supporting (PP1–5), and each benign criterion is weighted as stand-alone (BA1), strong (BS1– 4), or supporting (BP1–6). ACMG = American College of Medical Genetics and Genomics, CADD = Combined Annotation Dependent Depletion, c.notation = nucleotide change, Fm = family, GERP = Genomic Evolutionary Rate Profiling, HI = haploinsufficient, KRGDB = Korean reference genome database, LBV = likely benign variant, LPV = likely pathogenic variant, MAF = minor allele frequency, M-CAP = Mendelian Clinically Applicable Pathogenicity, N/A = not available, pLI = the probability of being loss-of-function intolerant, p.notation = amino acid change, REVEL = rare exome variant ensemble learner, rs = reference, SIFT = Sorting Intolerant From Tolerant, SNP = single nucleotide polymorphism, VUS = variant of uncertain significance
Of the 11 genes, PLOD3, NTM, GBA, CHST14, SLC2A10, and C9orf92 genes have been reported to be related to IA or intracranial hemorrhage [28,29,30,31]. When assuming complete penetrance of the autosomal dominant variants, one variant of the PLOD3 gene in family A, no variants in family B, and two variants of the SLC2A10 and CHST14 genes in family C remained. Table 3 summarizes the function of all candidate genes and their related diseases.
Table 3. Summary of the Potential IA-associated Genes.
| Gene | Gene Full Name | Gene Function | OMIM Disease | OMIM Inheritance | Phenotype |
|---|---|---|---|---|---|
| PLOD3 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 | Forms hydroxylysine residues in -Xaa-Lys-Gly- sequences in collagens. These hydroxylysines serve as sites of attachment for carbohydrate units and are essential for the stability of the intermolecular collagen cross-links | Lysyl hydroxylase 3 deficiency, 612394 | AR | Stickler syndrome, variable features of Ehlers-Danlos syndrome, epidermolysis bullosa with vascular complications, intracranial arterial dilatation [34,35] |
| NTM | Neurotrimin | Neural cell adhesion molecule | Aneurysm, intracranial berry, 7, 612161 | AD | IA, cerebral hemorrhage, thoracic aortic aneurysm, aortic rupture [29] |
| RYR2 | Ryanodine receptor 2 (cardiac) | Calcium channel that mediates the release of Ca (2+) from the sarcoplasmic reticulum into the cytoplasm and thereby plays a key role in triggering cardiac muscle contraction. Aberrant channel activation can lead to cardiac arrhythmia. Required for embryonic heart development | Arrhythmogenic right ventricular dysplasia 2, 600996; Ventricular tachycardia, catecholaminergic polymorphic, 1, 604772 |
AD | Arrhythmogenic right ventricular dysplasia, ventricular tachycardia [47] |
| PCNT | Pericentrin | Integral component of the filamentous matrix of the centrosome involved in the initial establishment of organized microtubule arrays in both mitosis and meiosis. Plays a role, together with DISC1, in the microtubule network formation. Is an integral component of the PCM | Microcephalic osteodysplastic primordial dwarfism, type II, 210720 | AR | Extreme SS, dysmorphism |
| NSMCE2 | Non-SMC element 2, MMS21 homolog (S. cerevisiae) | E3 SUMO-protein ligase component of the SMC5-SMC6 complex, a complex involved in DNA double-strand break repair by homologous recombination. The complex is required for telomere maintenance via recombination in ALT (alternative lengthening of telomeres) cell lines | Seckel syndrome 10, 617253 | AR | Extreme SS, dysmorphism |
| GBA | Glucosidase, beta, acid | Lysosomal enzyme that catalyzes the breakdown of the glycolipid glucosylceramide to ceramide and glucose | Susceptibility to Parkinson disease, late-onset, 168600; Gaucher disease, 230800 |
AR, AD, Multifactorial | Susceptible to IA [28] and Parkinson’s disease [48], Gaucher disease |
| CHST14 | Carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14 | Catalyzes the transfer of sulfate to position 4 of the N-acetylgalactosamine (GalNAc) residue of dermatan sulfate. Plays a pivotal role in the formation of 4-0-sulfated IdoA blocks in dermatan sulfate. Appears to have an important role in the formation of the cerebellar neural network during postnatal brain development | Ehlers-Danlos syndrome, musculocontractural type 1, 601776 | AR | Intracranial hemorrhage [31], Craniofacial dysmorphism, musculoskeletal abnormality |
| SLC2A10 | Solute carrier family 2 (facilitated glucose transporter), member 10 | Facilitative glucose transporter glucose transporter 10 (GLUT10) | Arterial tortuosity syndrome, 208050 | AR | IA [30], arterial tortuosity, CNS stroke, dysmorphism, skin and joint abnormality |
| ADAMTS2 | ADAM metallopeptidase with thrombospondin type 1 motif, 2 | Cleaves the propeptides of type I and II collagen prior to fibril assembly. Does not act on type III collagen. May also play a role in development that is independent of its role in collagen biosynthesis | Ehlers-Danlos syndrome, type VIIC, 225410 | AR | Dermatosparaxis (tearing of skin), SS, dysmorphism |
| NOTCH1 | Notch 1 | Functions as a receptor for membrane-bound ligands Jagged1, Jagged2, and Delta1 to regulate cell-fate determination. May play an essential role in post-implantation development, probably in some aspect of cell specification and/or differentiation. May be involved in mesoderm development, somite formation, and neurogenesis | Adams-Oliver syndrome 5, 616028; Aortic valve disease 1, 109730 |
AD | Developmental disorder |
| C9orf92 | Chromosome 9 open reading frame 92 | No function information available | None | Unknown | Susceptible to IA in a Korean IA GWAS study [28] |
AR = autosomal recessive, AD = autosomal dominant, CNS = central nervous system, GWAS = genome-wide association study, IA = intracranial aneurysm, OMIM = Online Mendelian Inheritance in Man, PCM = pericentriolar material, SS = short stature, SMC = structural maintenance of chromosomes
Chromosomal Microarray
Several chromosomal losses or gains were found in each family, but most of the CNVs were benign or likely benign. One copy number gain of unknown significance was detected in family A, which did not segregate with the phenotype. In addition, no genes were potentially related to IA in the corresponding regions.
DISCUSSION
In this study, WES was performed in three selected FIA families to identify genetic variants associated with IAs. A total of 13 participants were sequenced, of whom 7 had IAs. Among the 11 potential IA-associated variants, we noted three rare, potentially deleterious variants (PLOD3 c.1315G>A, NTM c.968C>T, and CHST14 c.58C>T) after considering gene-phenotype relationships, gene function, co-segregation, and variant pathogenicity.
The PLOD3 gene encodes lysyl hydroxylase 3 (LH3), which is involved in post-translational modification of collagens, including type IV collagen [32,33]. As such, pathogenic variation of this gene can lead to complex connective tissue disorders resembling Stickler syndrome, Ehlers-Danlos syndrome, and epidermolysis bullosa [34,35,36]. Although vascular complications are rare manifestations of these syndromes, some cases of aneurysms or arterial dissection have been reported [34,35]. In addition, embryonic lethality with intracranial hemorrhage has been reported in LH3-knockout mice [33]. Although the PLOD3 mutation found in family A was a heterozygous variant, it could be a potential IA-associated variant considering the severe variability of the phenotype of PLOD3-related diseases [34].
Neurotrimin (NTM) belongs to the IgLON family of glycosylphosphatidylinisotol (GPI)-anchored cell adhesion molecules and has been implicated in the promotion of neurite outgrowth and adhesion [37]. Luukkonen et al.[29] reported that the NTM gene is associated with IA and thoracic aortic aneurysm and suggested that truncations in the NTM gene caused IA and thoracic aortic aneurysm in a family. The 11q25 chromosomal region has been suggested as a susceptibility locus for both IA and aortic aneurysms in several independent linkage studies [38,39]. Although the individual (II-1 in family B) unaffected by IA had a rare PV of the NTM gene, it is still considered a potential IA-associated variant when considering the reduced penetrance or late onset of the aneurysm phenotype.
The CHST14 gene encodes carbohydrate sulfotransferase 14/dermatan 4-O-sulfotransferase 1 (CHST14/D4ST1), which is required for the maturation of dermatan sulfate that is involved in collagen formation [40]. Along with variants in DSE, biallelic PVs in CHST14 are one of the causes of musculoskeletal Ehlers-Danlos syndrome [41]. Moreover, intracranial hemorrhage was reported in 9% of patients with mcEDS-CHST14 [31].
Among the other candidates, GBA and C9orf92 genes were suggested to be IA-susceptible genes in a recent GWAS study of the Korean population [28]. Biallelic PVs of the GBA gene cause Gaucher disease, and a heterozygous variant is a well-known risk factor for PD [42,43]. In a previous study, the rs75822236 in GBA gene showed the strongest association with the risk of IA formation (odds ratio = 161.46) with sufficient statistical power (1.1 × 10−19), whereas the SNP in the C9orf92 gene was underpowered because of the small sample size [28].
Another candidate gene in family C, SLC2A10, encodes the facilitative glucose transporter glucose transporter 10 (GLUT10). Homozygous or compound heterozygous PVs of this gene cause arterial tortuosity syndrome, which is characterized by tortuosity, elongation, stenosis, and aneurysm formation in major arteries [44]. In contrast, heterozygous carriers of this gene variant are asymptomatic and do not show any notable vascular anomalies [45]. The heterozygous carriers (II-2 and II-4 in family C) in our study also did not show any arterial abnormalities that indicated arterial tortuosity syndrome.
In our study, we selected families that would be most genetically enriched for IAs considering the phenotypes, which include common locations of the IA among family members, multiple IAs, early onset, and fewer risk factors. In particular, our study is distinct from other studies in terms of the selection criteria that the affected members in each family should share the same aneurysm location. We assume that the intuition of the physicians who diagnosed and treated the patients played an important role in identifying their genetic predisposition.
Many genetic studies have been performed on FIA, and several genetic variations have been identified through linkage studies, GWAS, and NGS; however, these can explain only a small proportion of the total IAs in certain ethnic groups [7]. The current literature suggests that marked genetic heterogeneity may exist in distinct populations, and only two genetic studies, a linkage study and a GWAS, have been performed on the Korean population to date [28,46]. Our study is the first FIA study using NGS in Korea and may serve as a basis for establishing a genetic database for Korean patients with aneurysms. If sufficient data on FIA is accumulated through genetic studies, simple genetic testing using an NGS panel could offer great clinical benefits in terms of risk stratification, treatment decisions, and the screening of unaffected family members.
Multiple factors are intricately involved in aneurysm development [3,4,5]. Gene-environment interaction and phenocopy hinder genetic studies on this matter, especially in patients with multiple risk factors such as hypertension, smoking, old age, and female sex. Therefore, it is difficult to determine whether the genetic variations in our study were entirely responsible for FIAs. Further validation using replication studies and expression or functional analyses are required to support our results.
The limitations of this study are as follows. First, this study suggested several candidate genes, but these have not been fully validated. Further validation studies, such as replication studies for sporadic IA groups or functional analysis of the corresponding genes are needed. Second, the basic assumption of this study was that there would be some rare variants with strong effects that could explain the IAs of each family. However, the IAs in the families may be caused by environmental factors or common genetic variants, rather than rare variants, even though we have selected the most genetically enriched families with FIA in our database. Third, there were no candidate variants that were only found in the affected members of family B, and we thus had to find the most probable candidate (NTM) by assuming reduced penetrance. In addition, although the variants in PLOD3 and CHST14 genes were segregated in families A and C, the number of affected members may not be sufficient to exclude the possibility of false-positive results. Lastly, some participants only underwent magnetic resonance angiography, which may have produced false-negative or false-positive findings, especially for tiny aneurysms. Despite these limitations, our study presented the use of a methodology for finding rare PVs using WES for IAs, a relatively common multifactorial disease. Further familial studies with more severe phenotypes and more affected members would be able to identify additional candidate genes with higher confidence.
In conclusion, we studied three families that were genetically enriched for IA and performed WES to identify possible IA-associated variants. We found that the rare, potentially deleterious variants in PLOD3, NTM, and CHST14 are likely responsible for a subset of FIAs. Our findings may contribute to the understanding of IA pathogenesis, the establishment of an FIA genetic database in Korea, and further validation of IA candidate genes.
Acknowledgments
We would like to thank Romain Bourcier, Gervaise Loirand, and Hubert Desal from INSERM, CNRS, UNIV Nantes, l'institut du thorax for sharing valuable information, meetings, and discussions on the topic of familial aneurysm.
Footnotes
Conflicts of Interest: The authors have no potential conflicts of interest to disclose.
- Conceptualization: Dae Chul Suh, Eul-Ju Seo.
- Data curation: Yunsun Song, Boseong Kwon, Jin-Ok Lee.
- Formal analysis: Yunsun Song, Eul-Ju Seo, Boseong Kwon, Jin-Ok Lee.
- Funding acquisition: Dae Chul Suh.
- Investigation: Dae Chul Suh, Eul-Ju Seo, Yunsun Song.
- Methodology: Dae Chul Suh, Eul-Ju Seo, Yunsun Song, Jong-Keuk Lee.
- Project administration: Dae Chul Suh, Eul-Ju Seo.
- Supervision: Dae Chul Suh, Eul-Ju Seo.
- Validation: all authors.
- Writing—original draft: Dae Chul Suh, Eul-Ju Seo, Yunsun Song.
- Writing—review & editing: Dae Chul Suh, Jong-Keuk Lee, Eul-Ju Seo.
Funding Statement: This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2018R1A2B6003143).
References
- 1.Vlak MH, Algra A, Brandenburg R, Rinkel GJ. Prevalence of unruptured intracranial aneurysms, with emphasis on sex, age, comorbidity, country, and time period: a systematic review and meta-analysis. Lancet Neurol. 2011;10:626–636. doi: 10.1016/S1474-4422(11)70109-0. [DOI] [PubMed] [Google Scholar]
- 2.Ronkainen A, Hernesniemi J, Puranen M, Niemitukia L, Vanninen R, Ryynanen M, et al. Familial intracranial aneurysms. Lancet. 1997;349:380–384. doi: 10.1016/S0140-6736(97)80009-8. [DOI] [PubMed] [Google Scholar]
- 3.Broderick JP, Brown RD, Jr, Sauerbeck L, Hornung R, Huston J, 3rd, Woo D, et al. Greater rupture risk for familial as compared to sporadic unruptured intracranial aneurysms. Stroke. 2009;40:1952–1957. doi: 10.1161/STROKEAHA.108.542571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown RD, Jr, Broderick JP. Unruptured intracranial aneurysms: epidemiology, natural history, management options, and familial screening. Lancet Neurol. 2014;13:393–404. doi: 10.1016/S1474-4422(14)70015-8. [DOI] [PubMed] [Google Scholar]
- 5.Rinkel GJ, Djibuti M, Algra A, van Gijn J. Prevalence and risk of rupture of intracranial aneurysms: a systematic review. Stroke. 1998;29:251–256. doi: 10.1161/01.str.29.1.251. [DOI] [PubMed] [Google Scholar]
- 6.Hitchcock E, Gibson WT. A review of the genetics of intracranial berry aneurysms and implications for genetic counseling. J Genet Couns. 2017;26:21–31. doi: 10.1007/s10897-016-0029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou S, Dion PA, Rouleau GA. Genetics of intracranial aneurysms. Stroke. 2018;49:780–787. doi: 10.1161/STROKEAHA.117.018152. [DOI] [PubMed] [Google Scholar]
- 8.Sauvigny T, Alawi M, Krause L, Renner S, Spohn M, Busch A, et al. Exome sequencing in 38 patients with intracranial aneurysms and subarachnoid hemorrhage. J Neurol. 2020;267:2533–2545. doi: 10.1007/s00415-020-09865-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nat Protoc. 2016;11:1–9. doi: 10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- 10.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jagadeesh KA, Wenger AM, Berger MJ, Guturu H, Stenson PD, Cooper DN, et al. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat Genet. 2016;48:1581–1586. doi: 10.1038/ng.3703. [DOI] [PubMed] [Google Scholar]
- 15.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL, et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15:733–747. doi: 10.1038/gim.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen) Genet Med. 2020;22:245–257. doi: 10.1038/s41436-019-0686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourcier R, Le Scouarnec S, Bonnaud S, Karakachoff M, Bourcereau E, Heurtebise-Chretien S, et al. Rare coding variants in ANGPTL6 are associated with familial forms of intracranial aneurysm. Am J Hum Genet. 2018;102:133–141. doi: 10.1016/j.ajhg.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding X, Zhao S, Zhang Q, Yan Z, Wang Y, Wu Y, et al. Exome sequencing reveals a novel variant in NFX1 causing intracranial aneurysm in a Chinese family. J Neurointerv Surg. 2020;12:221–226. doi: 10.1136/neurintsurg-2019-014900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powell AE, Fernandez BA, Maroun F, Noble B, Woods MO. Familial intracranial aneurysm in newfoundland: clinical and genetic analysis. Can J Neurol Sci. 2019;46:518–526. doi: 10.1017/cjn.2019.230. [DOI] [PubMed] [Google Scholar]
- 23.Santiago-Sim T, Fang X, Hennessy ML, Nalbach SV, DePalma SR, Lee MS, et al. THSD1 (thrombospondin type 1 domain containing protein 1) mutation in the pathogenesis of intracranial aneurysm and subarachnoid hemorrhage. Stroke. 2016;47:3005–3013. doi: 10.1161/STROKEAHA.116.014161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Y, Li Z, Shi Y, Chen L, Tan H, Wang Z, et al. Exome sequencing identifies LOXL2 mutation as a cause of familial intracranial aneurysm. World Neurosurg. 2018;109:e812–e818. doi: 10.1016/j.wneu.2017.10.094. [DOI] [PubMed] [Google Scholar]
- 25.Yan J, Hitomi T, Takenaka K, Kato M, Kobayashi H, Okuda H, et al. Genetic study of intracranial aneurysms. Stroke. 2015;46:620–626. doi: 10.1161/STROKEAHA.114.007286. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, Li J, Fang Y, Zhang Z, Jin D, Chen X, et al. Rho guanine nucleotide exchange factor ARHGEF17 is a risk gene for intracranial aneurysms. Circ Genom Precis Med. 2018;11:e002099. doi: 10.1161/CIRCGEN.117.002099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou S, Ambalavanan A, Rochefort D, Xie P, Bourassa CV, Hince P, et al. RNF213 is associated with intracranial aneurysms in the French-Canadian population. Am J Hum Genet. 2016;99:1072–1085. doi: 10.1016/j.ajhg.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong EP, Kim BJ, Cho SS, Yang JS, Choi HJ, Kang SH, et al. Genomic variations in susceptibility to intracranial aneurysm in the Korean population. J Clin Med. 2019;8:275. doi: 10.3390/jcm8020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luukkonen TM, Poyhonen M, Palotie A, Ellonen P, Lagstrom S, Lee JH, et al. A balanced translocation truncates Neurotrimin in a family with intracranial and thoracic aortic aneurysm. J Med Genet. 2012;49:621–629. doi: 10.1136/jmedgenet-2012-100977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naunheim MR, Walcott BP, Nahed BV, MacRae CA, Levinson JR, Ogilvy CS. Arterial tortuosity syndrome with multiple intracranial aneurysms: a case report. Arch Neurol. 2011;68:369–371. doi: 10.1001/archneurol.2011.29. [DOI] [PubMed] [Google Scholar]
- 31.D'hondt S, Van Damme T, Malfait F. Vascular phenotypes in nonvascular subtypes of the Ehlers-Danlos syndrome: a systematic review. Genet Med. 2018;20:562–573. doi: 10.1038/gim.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myllyharju J, Kivirikko KI. Collagens and collagen-related diseases. Ann Med. 2001;33:7–21. doi: 10.3109/07853890109002055. [DOI] [PubMed] [Google Scholar]
- 33.Ruotsalainen H, Sipila L, Vapola M, Sormunen R, Salo AM, Uitto L, et al. Glycosylation catalyzed by lysyl hydroxylase 3 is essential for basement membranes. J Cell Sci. 2006;119:625–635. doi: 10.1242/jcs.02780. [DOI] [PubMed] [Google Scholar]
- 34.Ewans LJ, Colley A, Gaston-Massuet C, Gualtieri A, Cowley MJ, McCabe MJ, et al. Pathogenic variants in PLOD3 result in a Stickler syndrome-like connective tissue disorder with vascular complications. J Med Genet. 2019;56:629–638. doi: 10.1136/jmedgenet-2019-106019. [DOI] [PubMed] [Google Scholar]
- 35.Salo AM, Cox H, Farndon P, Moss C, Grindulis H, Risteli M, et al. A connective tissue disorder caused by mutations of the lysyl hydroxylase 3 gene. Am J Hum Genet. 2008;83:495–503. doi: 10.1016/j.ajhg.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vahidnezhad H, Youssefian L, Saeidian AH, Touati A, Pajouhanfar S, Baghdadi T, et al. Mutations in PLOD3, encoding lysyl hydroxylase 3, cause a complex connective tissue disorder including recessive dystrophic epidermolysis bullosa-like blistering phenotype with abnormal anchoring fibrils and type VII collagen deficiency. Matrix Biol. 2019;81:91–106. doi: 10.1016/j.matbio.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 37.Sellar GC, Watt KP, Rabiasz GJ, Stronach EA, Li L, Miller EP, et al. OPCML at 11q25 is epigenetically inactivated and has tumor-suppressor function in epithelial ovarian cancer. Nat Genet. 2003;34:337–343. doi: 10.1038/ng1183. [DOI] [PubMed] [Google Scholar]
- 38.Worrall BB, Foroud T, Brown RD, Jr, Connolly ES, Hornung RW, Huston J, 3rd, et al. Genome screen to detect linkage to common susceptibility genes for intracranial and aortic aneurysms. Stroke. 2009;40:71–76. doi: 10.1161/STROKEAHA.108.522631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozturk AK, Nahed BV, Bydon M, Bilguvar K, Goksu E, Bademci G, et al. Molecular genetic analysis of two large kindreds with intracranial aneurysms demonstrates linkage to 11q24-25 and 14q23-31. Stroke. 2006;37:1021–1027. doi: 10.1161/01.STR.0000206153.92675.b9. [DOI] [PubMed] [Google Scholar]
- 40.Hirose T, Mizumoto S, Hashimoto A, Takahashi Y, Yoshizawa T, Nitahara-Kasahara Y, et al. Systematic investigation of the skin in Chst14-/-mice: a model for skin fragility in musculocontractural Ehlers-Danlos syndrome caused by CHST14 variants (mcEDS-CHST14) Glycobiology. 2020;31:137–150. doi: 10.1093/glycob/cwaa058. [DOI] [PubMed] [Google Scholar]
- 41.Kosho T, Mizumoto S, Watanabe T, Yoshizawa T, Miyake N, Yamada S. Recent advances in the pathophysiology of musculocontractural Ehlers-Danlos syndrome. Genes (Basel) 2019;11:43. doi: 10.3390/genes11010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balestrino R, Tunesi S, Tesei S, Lopiano L, Zecchinelli AL, Goldwurm S. Penetrance of glucocerebrosidase (GBA) mutations in Parkinson's disease: a kin cohort study. Mov Disord. 2020;35:2111–2114. doi: 10.1002/mds.28200. [DOI] [PubMed] [Google Scholar]
- 43.Pastores GM, Hughes DA. Gaucher disease. Ncbi.nlm.nih.gov Web site. [Accessed June 11th, 2021]. https://www.ncbi.nlm.nih.gov/books/NBK1269/
- 44.Coucke PJ, Willaert A, Wessels MW, Callewaert B, Zoppi N, De Backer J, et al. Mutations in the facilitative glucose transporter GLUT10 alter angiogenesis and cause arterial tortuosity syndrome. Nat Genet. 2006;38:452–457. doi: 10.1038/ng1764. [DOI] [PubMed] [Google Scholar]
- 45.Callewaert BL, Willaert A, Kerstjens-Frederikse WS, De Backer J, Devriendt K, Albrecht B, et al. Arterial tortuosity syndrome: clinical and molecular findings in 12 newly identified families. Hum Mutat. 2008;29:150–158. doi: 10.1002/humu.20623. [DOI] [PubMed] [Google Scholar]
- 46.Kim CJ, Park SS, Lee HS, Chung HJ, Choi W, Chung JH, et al. Identification of an autosomal dominant locus for intracranial aneurysm through a model-based family collection in a geographically limited area. J Hum Genet. 2011;56:464–466. doi: 10.1038/jhg.2011.27. [DOI] [PubMed] [Google Scholar]
- 47.Priori SG, Napolitano C, Tiso N, Memmi M, Vignati G, Bloise R, et al. Mutations in the cardiac ryanodine receptor gene (hRyR2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation. 2001;103:196–200. doi: 10.1161/01.cir.103.2.196. [DOI] [PubMed] [Google Scholar]
- 48.Malek N, Weil RS, Bresner C, Lawton MA, Grosset KA, Tan M, et al. Features of GBA-associated Parkinson's disease at presentation in the UK Tracking Parkinson's study. J Neurol Neurosurg Psychiatry. 2018;89:702–709. doi: 10.1136/jnnp-2017-317348. [DOI] [PMC free article] [PubMed] [Google Scholar]