Abstract
The modulation of protein-protein interactions (PPIs) has developed into a well-established field of drug discovery. Despite the advances achieved in the field, many PPIs are still deemed as ‘undruggable’ targets and the design of PPIs stabilizers remains a significant challenge. The application of fragment-based methods for the identification of drug leads and to evaluate the ‘tractability’ of the desired protein target has seen a remarkable development in recent years. In this study, we explore the molecular characteristics of the 14-3-3/Amot-p130 PPI and the conceptual possibility of targeting this interface using X-ray crystallography fragment-based screening. We report the first structural elucidation of the 14-3-3 binding motif of Amot-p130 and the characterization of the binding mode and affinities involved. We made use of fragments to probe the ‘ligandability’ of the 14-3-3/Amot-p130 composite binding pocket. Here we disclose initial hits with promising stabilizing activity and an early-stage selectivity toward the Amot-p130 motifs over other representatives 14-3-3 partners. Our findings highlight the potential of using fragments to characterize and explore proteins' surfaces and might provide a starting point toward the development of small molecules capable of acting as molecular glues.
Keywords: Amot-p130, 14-3-3 /protein-protein interactions stabilizers, Fragment-based drug discovery, X-ray crystallography, Ligandability
Abbreviations used: Amot, Angiomotin; AmotL1/2, Angiomotin-like 1/2; Lats 1/2, Large tumor suppressor 1/2; PTMs, post-translational modifications; YAP1, Yes-associated protein 1; AIP4, Atrophin-1 interacting protein 4; PPI, Protein-protein interaction; FBDD, Fragment-based drug discovery; FP, Fluorescence polarization; MST, Microscale thermophoresis
Graphical abstract
Highlights
-
•
Phosphorylation of Ser 175 mediates binding of Amot-p130 to 14-3-3.
-
•
The crystal structure of the 14-3-3σΔC/Amot-p130 peptide complex describes the interface.
-
•
A fragment-based exploration of the interface assesses ‘ligandability’.
-
•
Fragments binding at the 14-3-3/Amot-p130 interface display an initial stabilizing activity.
1. Introduction
Targeting the complex network of protein-protein interactions (PPIs) is an increasingly attractive challenge in the drug discovery of pharmacotherapeutics (Arkin and Wells, 2004; Scott et al., 2016). Most proteins do not function as single entities but are engaged in dynamic interactions with other proteins, constituting highly organized, responsive networks that are of pivotal importance in all biological processes and are often dysregulated in disease (Vidal et al., 2011). In particular, PPI stabilizers offer advantages over inhibition owing to their uncompetitive nature and their ability to exploit the equilibrium of the protein complexes formation, without artificially disrupting endogenous-ligand interactions. Additionally, orthosteric small-molecule stabilizers bind to composite pockets created at the interface of the protein complexes and hold great potential to achieve selectivity (Milroy et al., 2014; Andrei et al., 2017). Despite the advances achieved in the field, the design of small-molecule PPI stabilizers remains a challenging and largely unexplored area, with many PPIs still deemed as ‘undruggable’ targets (Mabonga and Kappo, 2019).
The application of fragment-based drug discovery (FBDD) methods, which allow probing a large chemical space using low molecular weight organic compounds, have given encouraging results in the development of PPI modulators (Valenti et al., 2019). Notably, one of these molecules – Venetoclax (Souers et al., 2013) – has received FDA approval for the treatment of chronic lymphocytic leukemia. The hits identified from a fragment screening campaign can be used as starting points for the development of drug leads, but also provide valuable tools to probe the chemical space and explore the ‘tractability’ of the desired protein target (Leach and Hann, 2011; Brown et al., 2018).
The 14-3-3 protein interactome represents an interesting platform for PPI modulation. This family of proteins consists of seven isoforms with highly related sequences (Bartel et al., 2014) that regulate the activity of many disease-related proteins through direct PPIs (Johnson et al., 2010). Depending on its interaction with specific protein partners, 14-3-3 participates in several regulatory processes, such as cell cycle control, survival signalling, enzymatic activity, cell adhesion, and neuronal plasticity (Ballone et al., 2018a). For example, 14-3-3 proteins have been implicated in the regulation of cytoplasmatic localization of the Hippo pathway transcription factors YAP1 and TAZ (Zhao et al., 2007; Kanai et al., 2000). The Hippo signalling pathway is a major controller of cell growth and organ size, mainly through its ability to regulate cell contact inhibition, proliferation, and apoptosis. It consists of a kinase cascade that prevents YAP1/TAZ to enter the nucleus and drive the expression of pro-proliferative genes (Yu et al., 2015). Probing the interface of the 14-3-3/TAZ complex using a fragment-based screening has recently led to the identification of non-covalent orthosteric binders as a starting point toward the development of small-molecule stabilizers of this PPI (Guillory et al., 2020).
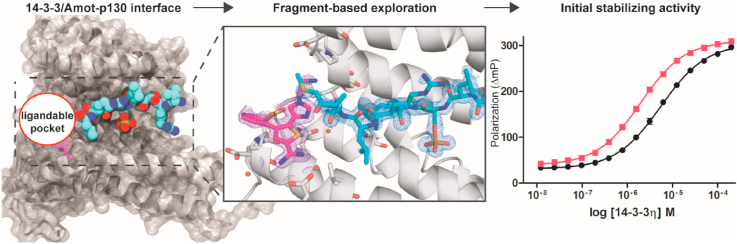
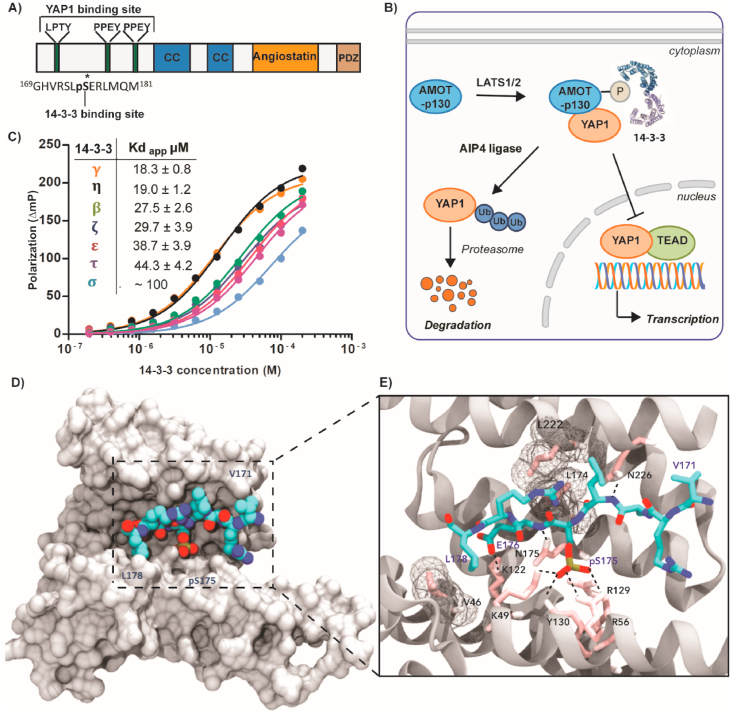
A previously unexplored PPI is the 14-3-3/Amot-p130 complex, which is a key interaction within the Hippo pathway. Amot-p130 belongs to the Motin protein family, which is represented by three members: Angiomotin (Amot), Angiomotin-like 1 (AmotL1), and Angiomotin-like 2 (AmotL2) (Bratt et al., 2002). Two isoforms of Amot are expressed: Amot-p130 and Amot-p80, which lacks the N-terminal 409 amino acids (Bratt et al., 2005). This family of adaptor proteins was originally recognized as angiostatin-binding proteins (Troyanovsky et al., 2001). The Amot proteins play an important role in angiogenesis, proliferation, migration of cancer and endothelial cells (Lv et al., 2017), the assembly of endothelial cell-cell junctions (Ernkvist et al., 2006), and were identified as major binding partners for YAP1 (Webb et al., 2011; Zhao et al., 2011). Taken together, the functional roles of the Amot proteins are highly correlated to the cellular context and the influence of post-translational modifications (PTMs) (Huang et al., 2018). Of particular interest is the phosphorylation of Amot-p130 at Ser175 by Lats1/2 kinase. This crucial PTM was found to repress YAP1-dependent cell proliferation and tumorigenesis by inducing the sequestration of the complex to the cytoplasm (Moleirinho et al., 2017). Additionally, phosphorylation of Ser175 enables Amot-p130 to bind 14-3-3 and, through Atrophin-1 Interacting Protein 4 (AIP4) mediated ubiquitination, can induce YAP1 degradation (Adler et al., 2013a, 2013b) (Fig. 1B). Targeted stabilization of the 14-3-3/Amot-p130 PPI could therefore be of interest to prevent YAP1's translocation to the nucleus and to promote the inhibition of YAP1-dependent signalling and tumor progression.
Fig. 1.
Characterization of the interaction between 14-3-3 and Amot-p130. (A) Domain organization of Amot-p130 including LPTY and PPEY binding motifs for YAP1 interaction, the conserved coiled-coil domains (CC), Angiostatin binding domain, and the C-terminal PDZ-binding domain. The 14-3-3 binding site and its sequence are indicated. (B) Schematic illustration of the interaction between 14-3-3, Amot-p130, and YAP1. The binding of 14-3-3 to Amot-p130 is induced by phosphorylation of S175 by the Lats 1/2 kinase. 14-3-3, Amot-p130, and YAP1 are recruited into a common complex that can interact with the AIP4 E3 ubiquitin ligase. The ubiquitination induced by AIP4 results in YAP1's degradation by the proteasome system. The sequestration of YAP1 to the cytosol and its ubiquitin-induced degradation prevents YAP1 from entering the nucleus to promote the transcription of pro-proliferative genes (Adler et al., 2013a). (C) Fluorescence Polarization data (mean ± SD; triplicates) of the 14-3-3 human isoforms titration against the FITC-labelled Amot-p130 peptide. Background polarization was subtracted from all values. SD error bars are smaller than data point symbols. (D) High resolution crystal structure of 13-mer Amot-p130 peptide in complex with 14-3-3σΔC (PDB ID: 7NMA). The crystals belong to the space group C2221 with one 14-3-3 monomer in the asymmetric unit. Front view of 14-3-3σ monomer (white surface) bound to the Amot-p130 phosphopeptide (cyan vdw spheres). (E) Close-up view of the binding groove. Polar contacts are indicated by dashed lines and hydrophobic interactions are depicted as a wireframe. The peptide sequence is 169GHVRSLpSERLMQM181. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In this work, we report the first structural elucidation of the 14-3-3σΔC/Amot-p130 interface and characterization of the binding mode and affinities involved. We explored the molecular characteristics of the PPI binding site and the conceptual possibility of targeting this interface using X-ray crystallography fragment-based screening. We identified how fragments can bind adjacent to the 14-3-3/Amot interface and engage with the complex. The hits displayed an initial stabilizing activity and an early-stage selectivity toward the Amot-p130 motifs over other representatives 14-3-3 partners. These findings represent an encouraging example of using fragments to characterize and target PPIs and might provide a starting point toward the development of small molecules able to establish contacts with both protein partners, acting as molecular glues.
2. Results and discussion
2.1. Characterization of the 14-3-3 binding site of Amot-p130
A critical step to assess the ‘ligandability’ of a PPI is the structural elucidation of its interface and the characterization of the binding mode and affinity of the binary complex. The 14-3-3 binding site is located in the N-terminal domain of the Amot-p130 protein (Adler et al., 2013a), and as with most 14-3-3 binding regions, the binding site is intrinsically disordered (Fig. 1A) (Huang et al., 2018). To study the interaction between 14-3-3 and Amot-p130, we made use of a peptide mimicking the disordered region of the binding motif. The peptide was composed of six amino acids of the protein's original sequence on each side of the phosphorylated Serine 175 (pSer175). To assess the affinity to different 14-3-3 isoforms, the peptide was labelled with FITC and tested using a Fluorescent Polarization (FP) assay. Analysis of the FP assay showed a significant increase in polarization upon titration of 14-3-3 proteins against the labelled peptide, although full saturation was not reached. The isoforms γ (Kd = 18 μM) and η (Kd = 19 μM) displayed the strongest binding. The affinities for the isoforms β, ζ, τ and ε ranged from 27 to 44 μM, and the σ isoform showed the weakest binding (Fig. 1C). This hierarchized affinity profile was similarly observed in previous studies including a wide range of different 14-3-3 binding motifs (Stevers et al., 2016; Kilisch et al., 2016; Centorrino et al., 2018; Ballone et al., 2018b) and represents a general feature of human 14-3-3 isoforms (Gogl et al., 2021).
To elucidate the structural basis of the 14-3-3/Amot-p130 interaction, co-crystallization trials were performed with 14-3-3σΔC and Amot-p130 peptide resulting in well-diffracting crystals. The structure was solved at a resolution of 1.75 Å (PDB ID: 7NMA) and interpretable electron density was found for 8 out of 13 residues of the Amot-p130 peptide. The phosphorylated peptide is bound to 14-3-3 in its conserved amphipathic groove (Fig. 1D and E). The Amot-p130 peptide binding is primarily mediated by the electrostatic interactions established by a positively charged sub-pocket shaped by Arg56, Arg129, Tyr130 and Lys49 of 14-3-3 and the negatively charged phosphate group of Ser175. The C-terminus of the peptide displays a polar amino acid (Glu176) after the phosphorylated Ser175 and gently curves out of the binding groove after the Leu178. The binding interface of the complex creates a composite PPI pocket at the peptide C-terminus that could be potentially exploited for small-molecule modulation (Fig. 2A and B). Direct polar contacts were observed between Asn175 and Asn226 of 14-3-3 and the backbone of the peptide, in addition to polar interactions between Lys122 of the protein and the Glu176 of Amot-p130. Residues Leu222, Leu174 and Val46 of 14-3-3 mediate hydrophobic interactions with the Leu178 and Leu174 of the Amot-p130 peptide (Fig. 1E).
Fig. 2.
Crystal structures of fragments binding to the 14-3-3σΔC/Amot-p130 peptide complex. (A–B) Front and side view of the pocket formed at the 14-3-3/Amot-p130 interface. 14-3-3σΔC protein is depicted with white cartoons and surface. The Amot-p130 peptide is represented with cyan sticks and surface. (C-D-E) Crystal structures of fragments binding to the 14-3-3σΔC protein/Amot-p130 complex. The fragments are depicted with colored sticks: 40 green (PDB ID: 7NMW), 12 pink (PDB ID: 7NMX), and 41 orange (PDB ID: 7NN2). The 2Fo-Fc electron density maps for both the peptide and fragments are displayed at 1σ. No direct contact with the peptide and the ligands were observed. (F–G–H) Close up of the interactions and distances in Å between the fragments and 14-3-3. 14-3-3 protein is depicted with white ribbons. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.2. Exploring ‘ligandability’ of the 14-3-3/Amot-p130 interface
The co-crystal structure of the Amot-p130 peptide bound to 14-3-3σΔC revealed a conformation of the peptide not entirely extended into the 14-3-3 binding groove, creating a potentially ‘ligandable’ pocket at the protein/peptide interface (Fig. 2A and B). Crystal structures of 14-3-3 bound to other interaction partners have shown how such a pocket can potentially accommodate the natural product Fusicoccin-A (FC-A) or its derivatives, increasing the stability of the ternary complex (Stevers et al., 2016; Würtele et al., 2003; Anders et al., 2013; Bier et al., 2016; Wolter et al., 2020; Lentini Santo et al., 2020). An initial exploration of the pocket was performed by using FC-A as a tool compound and a small selection of FC-like compounds, at single-dose in an FP assay. The Amot-p130 motif was not stabilized upon the addition of FC-A, the semisynthetic product ISIR 005, the aglycons of FC-A or FC-J. A decrease in polarization, and therefore inhibition of the protein-peptide interaction, was observed in presence of FC-THF (Würtele et al., 2003), which features an additional tetrahydrofuran ring, and in presence of FC-NAc, which has been shown to have a higher affinity for the apo 14-3-3 (Andrei et al., 2018) (Fig. S1). These findings pointed out that the fusicoccane compounds might possess an excessive steric hindrance to target the 14-3-3/Amot-p130 interface. This prompted us to address the pocket using a fragment-based approach to identify chemical entities that would not be as bulky as the tool compound FC-A, but still able to bind in the PPI pocket adjacent to the peptide.
To probe the pocket at the 14-3-3/Amot-p130 interface, we screened a subset of a non-covalent fragment library possessing a thiophene or benzothiophene scaffold (Fig. S2). Elements of this fragment library were previously described to bind Glu14 of 14-3-3 through an amidine moiety (Guillory et al., 2020). After visual inspection of the library, we selected a collection of fragments possessing varying substitutions of the common core, and the fragments were soaked into the 14-3-3σΔC/Amot-p130 crystals (Fig. 2). The crystal structures obtained for fragments 40 (PDB ID: 7NMW), 12 (PDB ID: 7NMX), and 41 (PDB ID: 7NN2) proved that these structures were able to bind in the ‘ligandable’ pocket adjacent to the Amot peptide (Fig. 2C, D, E). As expected, all the fragments interact with Glu14 of 14-3-3 through a salt bridge (Fig. 2G). Additionally, fragments 40 and 41 form non-canonical interactions with 14-3-3 via the side chain of Glu39 and their phenyl or pyridine ring (2.9–3.0 Å), respectively, via a CH-π interaction (Fig. 2F, H) (Platzer et al., 2020; Salonen et al., 2011). Furthermore, the 2-amino-pyridine forms an electrostatic interaction with the carboxylate of Glu39 (2.8 Å). From the analysis of the crystal structures no direct contacts with the peptide and the ligands were observed, as fragment binding was distal from the PPI interface.
To test if the fragments displayed activity on the complex, an FP assay was performed (Fig. S3). The fragments were titrated into a solution comprising a fixed concentration of 14-3-3/Amot-p130 complex that allowed 20% binding of the peptide initially (Polarization of ∼80 mP). The titrations were performed both in the presence and absence of 14-3-3 to exclude false-positive hits. Analysis of the FP assay showed that 41 and 12 did not induce ternary complex formation at a biophysically relevant concentration (≤1 mM). Notably, fragment 12 showed assay interference at high compound concentration, likely a result of aggregation or autofluorescence. Whilst fragment 40 also showed assay interference at high concentrations of the fragment, at lower concentrations of the fragment (above 31 μM) a significant increase in polarization was observed, independent of assay interference (Fig. S3A). This suggested that 40 does induce ternary complex formation, however accurate determination of the EC50 was not possible.
2.3. Fragments 09 and 22 display a weak stabilization effect
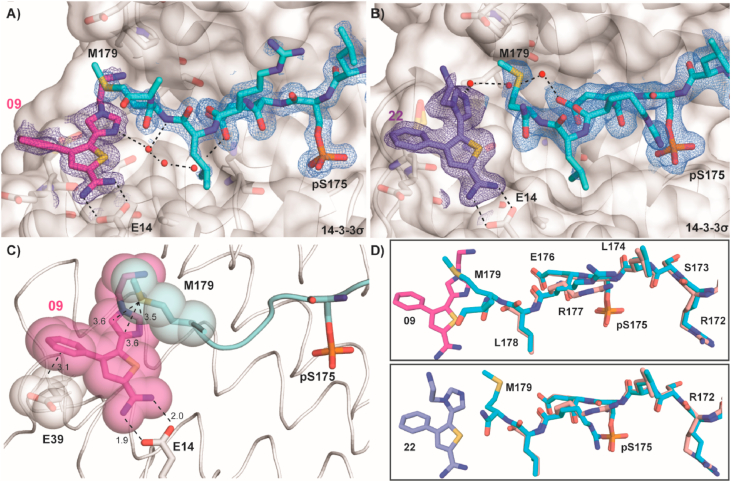
The solved crystal structures of the ternary complexes suggested that more extended substituents could reach into the composite PPI pocket towards the peptide. In combination with the promising activity of 40, this led us to explore fragments possessing additional functionality that extended from the C5 position of the thiophene ring into the composite binding pocket. As a result, we performed additional soaking experiments in the crystals of the binary complex, resulting in two hit compounds identified in crystal structures (Figs. 3A, B, and S8). Both fragments 09 and 22 were structural analogous of 40. Further, 09 and 22 were structural isomers. Analysis of the X-ray crystal structures shows that 09 and 22 bind to the protein complex in a pose similar to 40 and probe the composite pocket. Interestingly, the presence of the fragments allowed us to observe additional electron density at the Amot-p130 peptide C-terminus, specifically for Met179 (Fig. 3D). The lipophilic and flexible methionine of Amot-p130 is positioned in a co-planar fashion to the N-substituted imidazole ring engaging in van der Waals interactions, which is consistent with the interactions that methionine side chains typically establish with aryl rings (Bissantz et al., 2010) (Fig. 3C). The N-substituted imidazole ring extends into the water network between 14-3-3 and Amot-p130. The orientation of the imidazole ring of fragment 09 allows closer interaction with the methionine 179 and a water-mediated contact with the backbone of the peptide that could correlate to the slightly higher response of this fragment in the biophysical assays (vide infra).
Fig. 3.
Crystal structures of fragments 09 and 22 in complex with 14-3-3σΔC and the Amot-p130 peptide. (A–B) Crystal structures of fragments 09 (PDB ID: 7NND) and 22 (PDB ID: 7NNE) binding at the protein/peptide interface and stabilizing the C-terminus of the peptide allowing the fitting of Met179 in the electron density. The 2F0-FC electron density map for the Amot-p130 peptide and ligands is contoured at 1σ. Contacts between the fragments, Glu14, and the water network are depicted with black dashed lines. 14-3-3σΔC is shown with a white surface and cartoon. Amot peptide (cyan), fragment 09 (magenta), and fragment 22 (purple) are represented with colored sticks. (C) Close up of the interactions (dashed lines) established by fragment 09 with 14-3-3 and Amot-p130. Distances in Å are reported. (D) Overlay of the Amot-p130 peptide in presence of fragments 09 and 22 (cyan sticks) and absence of the fragments (pink sticks). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Additionally, the role of the length of the peptide on the fragments' binding mode was investigated. A 20-mer peptide with an extended C-terminus was co-crystallized with 14-3-3σΔC and the crystal structures in complex with fragments 09 (PDB ID: 7NPB) and 22 (PDB ID: 7NPG) were obtained, showing the same binding mode as for the shorter peptide (Fig. S7). The structures revealed that the ‘ligandable’ pocket is equally available with a longer peptide sequence and confirmed the methionine induced fit in presence of the fragments.
14-3-3η was used to quantify affinities as one of the isoforms that displayed the strongest binding. In FP assays, fragments titrated to a fixed protein/peptide concentration revealed a slight increase in polarization at the highest concentrations (Fig. S5). Microscale Thermophoresis (MST) was employed as an orthogonal assay. The assay's conditions were first optimized for the 14-3-3η/Amot-p130 complex. MST data of fragment 22 titration against 14-3-3η/Amot-p130 revealed an affinity for the complex in the micromolar range (Kd = 161 μM) (Fig. 4B).
Fig. 4.
Measurement of stabilization of fragments 09 and 22. (A) Chemical structures of fragments 22 and 09. (B) MST data (mean; duplicates) of fragment 22 titration (1:1 dilutions starting from 5 mM) against 14-3-3η/Amot-p130 complex. (C) MST data (mean; duplicates) of 14-3-3η titration against Cy5 labelled Amot-p130 peptide in presence of 1 mM 09 (red circles) or 22 (green circles). 1% DMSO control is represented with light blue circles. Because of the binding-dependent decrease in initial fluorescence upon protein titration, the raw fluorescence data were analyzed to generate binding curves (Fig. S6). (D) FP data (mean; triplicates) of 14-3-3η titration against FITC Amot-p130 peptide in presence of 1 mM 09 (pink squares) or 22 (purple triangle). 1% DMSO control is represented with black circles. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
14-3-3η protein titrations were performed in presence of fixed concentrations of fragments using both MST and FP assays (Fig. 4C and D). Analysis of the dose-response curves obtained suggested a 2-fold stabilization effect on the 14-3-3/Amot-p130 complex. Whilst the modulation on the complex is certainly modest, it is consistent with the weak stabilization effect that can be expected from a non-covalent fragment. The 14-3-3σ isoform used for the crystallography study was also tested in FP assay in presence of increasing fragments concentration, suggesting a similar stabilization profile to 14-3-3η (Fig. S4).
Finally, to validate the initial stabilization observed and to compare the modulation observed across different 14-3-3 binding modes, we tested fragment 22 in FP assay against the peptides Ataxin-pS776 and p53-pT387 as representative of different 14-3-3 binding modes (Fig. 5). The Ataxin-pS776 peptide is more extended into the 14-3-3 binding groove and directly interacts with Glu14 of 14-3-3, thereby making unavailable the main anchoring point for the amidine scaffolds (PDB ID: 6QIU) (Leysen et al., 2021) (Fig. 5D). The p53-pT387 peptide similarly to Amot-p130 presents a Glu at the +1 position after the phosphorylation site but displays a sharper turn at the C-terminus. The crystal structure of the fragment in complex with the p53-pT387 peptide was previously reported by Guillory et al. (PDB ID: 6RM7) (Guillory et al., 2020) and revealed a binding pose of the fragment distal from the peptide interface (Fig. 5E).
Fig. 5.
Comparison of hit fragment 22 activity over different 14-3-3 binding epitopes. (A) Overview of the selected 14-3-3 partner and their binding epitopes observed in the crystal structures. (B) Overlay of the crystal structures of the 14-3-3 binding partners (cartoon representation). The phosphorylated site and C-terminus of Amot-p130 and Ataxin are highlighted with stick representations. Fragment 22 is depicted with purple lines and surface. 14-3-3σΔC protein is always depicted as a white surface. (C–E) Crystal structures of fragment 22 (purple sticks and surface) bound to 14-3-3σΔC in complex with Amot-p130 peptide (cyan sticks and surface) PDB ID: 7NNE or in complex with p53 peptide (green sticks and surface) PDB ID: 6RM7 (Guillory et al., 2020). (D) Binding mode of the Ataxin peptide (orange sticks and surface) PDB ID: 6QIU (Leysen et al., 2021). The polar contacts between Arg781 of the peptide and Glu14 of 14-3-3 are depicted as black dashed lines. Glu14 of 14-3-3 is highlighted with red sticks and surface. (F) FP data (mean; triplicates) of 14-3-3η titration in presence and absence of 1 mM 22 and TAMRA-labelled Amot-p130, Ataxin, or p53 peptides. Background polarization was subtracted from all values. DMSO in the assay was 1%. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Analysis of the FP assay data, showed a left shift upon 14-3-3η titration in presence of TAMRA (tetramethylrhodamine) labelled Amot-p130 peptide and fragment 22 (Fig. 5F). This shift is consistent with the 2-fold stabilizing activity detected in FP assay against the FITC-labelled Amot-p130 peptide and with the MST data. By contrast, the application of the same experimental settings did not result in a similar shift of the dose-response curves for Ataxin-pS776 and p53-pT387 peptides (Fig. 5G and H). These data indicate that the fragment is not able to engage with the partner peptides Ataxin-pS776 or p53-pT387 to increase the complex stability, suggesting an early stage selectivity that could be further explored with compound optimization.
The modulation of PPIs through the formation of ternary complexes can represent an alternative and novel strategy in drug discovery (Hughes and Ciulli, 2017). PPIs stabilizers such as molecular glues and bivalent inducers of protein degradation (PROTACs) can drive or enhance the association of two proteins, thus influencing regulatory or pathological mechanisms and have the potential to significantly expand the druggable proteome (Schreiber, 2021). Different from PPI inhibitors, which can be designed by mimicking one of the PPI partners, the identification of starting points for small-molecule stabilizers development is particularly arduous. In the early stages of this process, an important step is to assess whether the interfaces involved have a binding site that can be exploited for small molecules binding. In this regard, the structural and biophysical information gained by probing the proteins' interfaces with fragment-based approaches can be used to explore the target ‘tractability’ and provide starting points for future studies.
Our fragment-based exploration studies with the structural characterization of fragments in complex with 14-3-3/Amot-p130 provided evidence of the ‘ligandability’ of the interface. Moreover, the induced-fit of the C-terminus of the peptide in presence of the fragments contributed to obtaining some insights on the molecular characterization of the complex that can be used in future studies. Our binding assays suggested that a moderate stabilization and an early stage selectivity for the 14-3-3 binding modes that are more compatible with the fragments can be achieved and more importantly, that this effect could be further explored with the development of advanced compounds by rational design.
3. Materials and methods
3.1. Peptide sequences
Fluorescently labelled peptides (FITC or TAMRA) for fluorescence polarization assay, and 13-mer Amot-p130 peptide and 20-mer Amot-p130 for crystallography were purchased from GenScript Biotech BV (Netherlands) BV in purity higher than 95%. Sequences were: 13-mer Amot-p130 [169GHVRSL{pS}ERLMQM181], 20-mer Amot-p130 [169GHVRSL{pS}ERLMQMSLATSGV188], Ataxin [770TRKRRW{pS}APESRK782]. The 32-mer TAMRA labelled p53 peptide [362SRAHSSHLKSKKGQSTSRHKKLMFK{pT}EGPDSD393] was purchased from AnaSpec. The Cy5 labelled 13-mer Amot-p130 for MicroScale Thermophoresis was purchased from Life Tein LLC (USA) in purity higher than 95%. The fluorescent versions of the peptides were N-terminally labelled. Peptides used for crystallography were Acetylated at the C-terminus.
3.2. Protein expression and purification
His6-tagged 14-3-3 isoforms full-length and 14-3-3σΔC (C-terminally truncated after T231) were expressed in BL21 (DE3) competent cells via a pProEX HTb plasmid. Expression was induced with 0.4 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) overnight at 18 °C. After spinning down and lysis of the expression culture, the protein was purified on a Ni2+ -NTA column. The His6-tag was cleaved with Tobacco Etch Virus (TEV) protease in 1:0.05 mg ratio and a second nickel-affinity chromatography was performed followed by size exclusion chromatography (Superdex75) in 25 mM Hepes pH 7.5, 100 mM NaCl, 10 mM MgCl2, 2 mM β-mercaptoethanol.
3.3. Fragments
Fragments were obtained from a library of AstraZeneca Pharma as 100 mM DMSO stocks.
3.4. Fluorescence polarization assay
The Fluorescently labelled peptides, 14-3-3 protein and fragments (100 mM stock solution in DMSO) were diluted in FP buffer (10 mM Hepes pH 7.5, 150 mM NaCl, 0.1% Tween 20, 1.0 mg mL−1 BSA) to the desired concentrations. The final DMSO concentration was 1%. Fluorescently labelled peptides concentration was 100 nM. Dilution series of 14-3-3 protein or fragments were made on Corning black, round-bottom, low-binding 384-well plates (Product Number 4514), in a final sample volume of 10 μL. Polarization was measured with a Tecan Infinite F500 plate reader using appropriate excitation and emission wavelength for FITC (λex: 485 nm, λem: 535 nm) and TAMRA (λex: 535 nm, λem: 590 nm). FP data were fitted to a four-parameter dose-response curve using Graphpad Prism 5 software (GraphPad Software, San Diego, CA, USA).
3.5. MicroScale thermophoresis
Measurements were carried out in PBS pH 7.4 and 0.05% Tween-20 buffer. Cy5 labelled peptide (λex: 650 nm, λem: 670 nm) was diluted to a final concentration of 50 nM. For the compound titration experiment, the fragment was diluted 1:1 from a starting concentration of 5 mM and titrated against a fixed concentration of protein-peptide complex corresponding to EC20. Thermophoresis was measured in a Nanotemper Monolith NT.115 using a red excitation laser with the settings 20% LED power and 80% MST power. The normalized fluorescence values were analyzed in Graphpad Prism 5 software. For protein titration experiments, the 14-3-3η protein was titrated in presence of the peptide and 1 mM fragments. The final DMSO concentration was 1%. Thermophoresis was measured at 20% LED power and 20% MST power. Because of binding-induced changes in the fluorescence signal upon protein titration (Fig. S6), the baseline fluorescence data were analyzed using MST analysis software (MO. Affinity Analysis v 2.1.3454) and the fraction bound (baseline-corrected normalized fluorescence/curve amplitude) was plotted against the protein concentration (Rainard et al., 2018). All measurements were performed at 25 °C using premium coated capillaries from Nanotemper (MO-K005). Default settings were as follow Fluo. Before 5 s, MST On 30s, Fluo. After 5 s and Delay 25 s.
3.6. Protein crystallization and structure elucidation
Crystals of the 14-3-3σΔC/13-mer Amot-p130 binary complex were co-crystallized by mixing 12.5 mg mL−1 14-3-3σΔC in a molar ratio of 1:2 with Amot-p130 peptide, in 20 mM Hepes pH 7.5, 2 mM MgCl2, and 2 mM β-mercaptoethanol, and incubated overnight at 4 °C. Crystals of the 14-3-3σΔC/20-mer Amot-p130 binary complex were obtained using 10 mg mL−1 14-3-3σΔC and 1:4 M ratio. The complexes were then set up for crystallization using the hanging drop method by mixing 1:1 with 0.095 M Hepes pH 7.5, 26% PEG 400, 0.19 M CaCl2, and 5% Glycerol. For the soakings, 100 mM stock solutions in DMSO of the fragments were added to the crystals of the binary complexes to a final concentration of 10 mM. Crystals were harvested after 2 weeks of incubation at 4 °C and directly flash cooled in liquid nitrogen. Data collection was performed either at the Diamond Light Source (DLS), Didcot, Oxfordshire, U.K. (beamlines I24, I03, I04, I04-1), or at the Deutsches Elektronen-Synchrotron (DESY) PETRA-III beamline PII, Hamburg, Germany, or in house on a Rigaku MicroMaz-003 (Rigaku Europe, Kemsing Sevenoaks, UK) equipped with a sealed tube X-ray source and a Rigaku Dectris PILATUS 200K detector (DECTRIS Ltd., Baden-Daettwil, Switzerland). Data were processed using Dials (Winter et al., 2018). Molecular replacement was carried out using Phaser (McCoy et al., 2007) and PDB ID:4JC3 as a model. The obtained model was subjected to reiterative rounds of model building and refinement using Coot (Emsley and Cowtan, 2004) and Phenix (Adams et al., 2010). Table S1 A-C provide further details of the crystallography data and refinement statistics. Figures were prepared using VMD (Humphrey et al., 1996) and Pymol (DeLano, 2002) software.
Accession numbers
The diffraction data and coordinates of the crystal structures have been deposited in the Protein Data Bank (http://www.pdb.org). The accession codes are PDB ID:7NMA (14-3-3σΔC/13-mer Amot), PDB ID: 7NMX (14-3-3σΔC/13-mer Amot/12), PDB ID: 7NMW (14-3-3σΔC/13-mer Amot/40), PDB ID: 7NN2 (14-3-3σΔC/13-mer Amot/41), PDB ID: 7NND (14-3-3σΔC/13-mer Amot/09), PDB ID: 7NNE (14-3-3σΔC/13-mer Amot/22), PDB ID: 7NP2 (14-3-3σΔC/20-mer Amot), PDB ID: 7NPB (14-3-3σΔC/20-mer Amot/09), PDB ID: 7NPG (14-3-3σΔC/20-mer Amot/22).
CRediT authorship contribution statement
Federica Centorrino: Formal analysis, Data curation, Investigation, Validation, Visualization, Writing – original draft, Writing – review & editing. Blaž Andlovic: Formal analysis, Investigation, Validation, Writing – review & editing. Peter Cossar: Conceptualization, Validation, Writing – review & editing. Luc Brunsveld: Conceptualization, Validation, Resources, Writing – review & editing. Christian Ottmann: Conceptualization, Supervision, Validation, Funding acquisition, Project administration, Resources, Writing – review & editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: LB and CO are co-founders and share-holders of Ambagon Therapeutics.
Acknowledgements
This research was supported through the Initial Training Network TASPPI, funded by the H2020 Marie Curie Actions of the European Commission under Grant Agreement 675179 and through a Eurotech Postdoctoral Fellow program (Marie Skłodowska-Curie Co-funded, grant number 754462). We would like to thank Dr. Hongming Chen and Helen Boyd (AstraZeneca) for providing the fragments for X-ray crystallography and initial FP validation. We thank Dr. Matthew Perry (AstraZeneca) for providing fragments 09 and 22 for MST assay and additional FP assays.
Handling editor: Natalie Strynadka
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2021.12.003.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C., Richardson J.S., Terwilliger T.C., Zwart P.H. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler J.J., Johnson D.E., Heller B.L., Bringman L.R., Ranahan W.P., Conwell M.D., Sun Y., Hudmon A., Wells C.D. Serum deprivation inhibits the transcriptional Co-activator YAP and cell growth via phosphorylation of the 130-KDa isoform of Angiomotin by the LATS1/2 protein kinases. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17368–17373. doi: 10.1073/pnas.1308236110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler J.J., Heller B.L., Bringman L.R., Ranahan W.P., Cocklin R.R., Goebl M.G., Oh M., Lim H.-S., Ingham R.J., Wells C.D. Amot130 adapts Atrophin-1 interacting protein 4 to inhibit yes-associated protein signaling and cell growth. J. Biol. Chem. 2013;288:15181–15193. doi: 10.1074/jbc.M112.446534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders C., Higuchi Y., Koschinsky K., Bartel M., Schumacher B., Thiel P., Nitta H., Preisig-Müller R., Schlichthörl G., Renigunta V., Ohkanda J., Daut J., Kato N., Ottmann C. A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface. Chem. Biol. 2013;20:583–593. doi: 10.1016/j.chembiol.2013.03.015. [DOI] [PubMed] [Google Scholar]
- Andrei S.A., Sijbesma E., Hann M., Davis J., O'Mahony G., Perry M.W.D., Karawajczyk A., Eickhoff J., Brunsveld L., Doveston R.G., Milroy L.-G., Ottmann C. Stabilization of protein-protein interactions in drug discovery. Expet Opin. Drug Discov. 2017;12:925–940. doi: 10.1080/17460441.2017.1346608. [DOI] [PubMed] [Google Scholar]
- Andrei S.A., deVink P., Sijbesma E., Han L., Brunsveld L., Kato N., Ottmann C., Higuchi Y. Rationally designed semisynthetic natural product analogues for stabilization of 14 -3-3Protein –ProteinInteractions. Angew. Chem. Int. Ed. 2018;57:13470–13474. doi: 10.1002/anie.201806584. [DOI] [PubMed] [Google Scholar]
- Arkin M.R., Wells J.A. Small-molecule inhibitors of protein–protein interactions: progressing towards the dream. Nat. Rev. Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- Ballone A., Centorrino F., Ottmann C. 14-3-3: a case study in PPI modulation. Molecules. 2018;23:1386. doi: 10.3390/molecules23061386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballone A., Centorrino F., Wolter M., Ottmann C. Structural characterization of 14-3-3ζ in complex with the human Son of Sevenless Homolog 1 (SOS1) J. Struct. Biol. 2018;202:210–215. doi: 10.1016/j.jsb.2018.01.011. [DOI] [PubMed] [Google Scholar]
- Bartel M., Schäfer A., Stevers L.M., Ottmann C. Small molecules, peptides and natural products: getting a grip on 14-3-3 protein-protein modulation. Future Med. Chem. 2014;6:903–921. doi: 10.4155/fmc.14.47. [DOI] [PubMed] [Google Scholar]
- Bier D., Bartel M., Sies K., Halbach S., Higuchi Y., Haranosono Y., Brummer T., Kato N., Ottmann C. Small-molecule stabilization of the 14-3-3/Gab2 protein–protein interaction (PPI) interface. ChemMedChem. 2016;11:911–918. doi: 10.1002/cmdc.201500484. [DOI] [PubMed] [Google Scholar]
- Bissantz C., Kuhn B., Stahl M.A. Medicinal chemist's guide to molecular interactions. J. Med. Chem. 2010;53:5061–5084. doi: 10.1021/jm100112j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratt A., Wilson W.J., Troyanovsky B., Aase K., Kessler R., Meir E.G.V., Holmgren L. Angiomotin belongs to a novel protein family with conserved coiled-coil and PDZ binding domains. Gene. 2002;298:69–77. doi: 10.1016/s0378-1119(02)00928-9. [DOI] [PubMed] [Google Scholar]
- Bratt A., Birot O., Sinha I., Veitonmäki N., Aase K., Ernkvist M., Holmgren L. Angiomotin regulates endothelial cell-cell junctions and cell motility. J. Biol. Chem. 2005;280:34859–34869. doi: 10.1074/jbc.M503915200. [DOI] [PubMed] [Google Scholar]
- Brown K.K., Hann M.M., Lakdawala A.S., Santos R., Thomas P.J., Todd K. Approaches to target tractability assessment – a practical perspective. Med. Chem. Commun. 2018;9:606–613. doi: 10.1039/c7md00633k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centorrino F., Ballone A., Wolter M., Ottmann C. Biophysical and structural insight into the USP8/14-3-3 interaction. FEBS Lett. 2018;592:1211–1220. doi: 10.1002/1873-3468.13017. [DOI] [PubMed] [Google Scholar]
- DeLano W.L. CCP4 Newsletter on protein crystallography. Vol. 40. 2002. Pymol: an open-source molecular graphics tool; pp. 82–92. [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ernkvist M., Aase K., Ukomadu C., Wohlschlegel J., Blackman R., Veitonmäki N., Bratt A., Dutta A., Holmgren L. P130-Angiomotin associates to actin and controls endothelial cell shape. FEBS J. 2006;273:2000–2011. doi: 10.1111/j.1742-4658.2006.05216.x. [DOI] [PubMed] [Google Scholar]
- Gogl G., Tugaeva K.V., Eberling P., Kostmann C., Trave G., Sluchanko N.N. Hierarchized phosphotarget binding by the seven human 14-3-3 isoforms. Nat. Commun. 2021;12:1677. doi: 10.1038/s41467-021-21908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillory X., Wolter M., Leysen S., Neves J.F., Kuusk A., Genet S., Somsen B., Morrow J.K., Rivers E., van Beek L., Patel J., Goodnow R., Schoenherr H., Fuller N., Cao Q., Doveston R.G., Brunsveld L., Arkin M.R., Castaldi P., Boyd H., Landrieu I., Chen H., Ottmann C. Fragment-based differential targeting of PPI stabilizer interfaces. J. Med. Chem. 2020;63:6694–6707. doi: 10.1021/acs.jmedchem.9b01942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Zhou Y., Zhang J., Cheng A.S.L., Yu J., To K.F., Kang W. The physiological role of Motin family and its dysregulation in tumorigenesis. J. Transl. Med. 2018;16:98. doi: 10.1186/s12967-018-1466-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S.J., Ciulli A. Molecular recognition of ternary complexes: a new dimension in the structure-guided design of chemical degraders. Essays Biochem. 2017;61:505–516. doi: 10.1042/EBC20170041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Johnson C., Crowther S., Stafford M.J., Campbell D.G., Toth R., MacKintosh C. Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem. J. 2010;427:69–78. doi: 10.1042/BJ20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai F., Marignani P.A., Sarbassova D., Yagi R., Hall R.A., Donowitz M., Hisaminato A., Fujiwara T., Ito Y., Cantley L.C., Yaffe M.B. TAZ: a novel transcriptional Co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilisch M., Lytovchenko O., Arakel E.C., Bertinetti D., Schwappach B.A. Dual phosphorylation switch controls 14-3-3-dependent cell surface expression of TASK-1. J. Cell Sci. 2016;129:831–842. doi: 10.1242/jcs.180182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach A.R., Hann M.M. Molecular complexity and fragment-based drug discovery: ten years on. Curr. Opin. Chem. Biol. 2011;15:489–496. doi: 10.1016/j.cbpa.2011.05.008. [DOI] [PubMed] [Google Scholar]
- Lentini Santo D., Petrvalska O., Obsilova V., Ottmann C., Obsil T. Stabilization of protein-protein interactions between CaMKK2 and 14-3-3 by fusicoccins. ACS Chem. Biol. 2020;15:3060–3071. doi: 10.1021/acschembio.0c00821. [DOI] [PubMed] [Google Scholar]
- Leysen S., Burnley R.J., Rodriguez E., Milroy L., Soini L., Adamski C.J., Davis R., Obsil T., Brunsveld L., Crabbe T., Zoghbi H.Y., Ottmann C., Davis J.M. Structural insights into the cytoplasmic chaperone effect of 14-3-3 proteins on Ataxin-1. J. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2021.167174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv M., Shen Y., Yang J., Li S., Wang B., Chen Z., Li P., Liu P., Yang J. Angiomotin family members: oncogenes or tumor suppressors? Int. J. Biol. Sci. 2017;13:772–781. doi: 10.7150/ijbs.19603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabonga L., Kappo A.P. Protein-protein interaction modulators: advances, successes and remaining challenges. Biophys. Rev. 2019;11:559–581. doi: 10.1007/s12551-019-00570-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milroy L.-G., Grossmann T.N., Hennig S., Brunsveld L., Ottmann C. Modulators of protein–protein interactions. Chem. Rev. 2014;114:4695–4748. doi: 10.1021/cr400698c. [DOI] [PubMed] [Google Scholar]
- Moleirinho S., Hoxha S., Mandati V., Curtale G., Troutman S., Ehmer U., Kissil J.L. Regulation of localization and function of the transcriptional Co-activator YAP by Angiomotin. Elife. 2017;6 doi: 10.7554/eLife.23966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platzer G., Mayer M., Beier A., Brüschweiler S., Fuchs J.E., Engelhardt H., Geist L., Bader G., Schörghuber J., Lichtenecker R., Wolkerstorfer B., Kessler D., McConnell D.B., Konrat R. PI by NMR: probing CH–π interactions in protein–ligand complexes by NMR spectroscopy. Angew. Chem. Int. Ed. 2020;59:14861–14868. doi: 10.1002/anie.202003732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainard J.M., Pandarakalam G.C., McElroy S.P. Using microscale Thermophoresis to characterize hits from high-throughput screening: a European lead factory perspective. SLAS Discov. 2018;23:225–241. doi: 10.1177/2472555217744728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen L.M., Ellermann M., Diederich F. Aromatic rings in chemical and biological recognition: energetics and structures. Angew. Chem. Int. Ed. 2011;50:4808–4842. doi: 10.1002/anie.201007560. [DOI] [PubMed] [Google Scholar]
- Schreiber S.L. The rise of molecular glues. Cell. 2021;184:3–9. doi: 10.1016/j.cell.2020.12.020. [DOI] [PubMed] [Google Scholar]
- Scott D.E., Bayly A.R., Abell C., Skidmore J. Small molecules, big targets: drug discovery faces the protein–protein interaction challenge. Nat. Rev. Drug Discov. 2016;15:533–550. doi: 10.1038/nrd.2016.29. [DOI] [PubMed] [Google Scholar]
- Souers A.J., Leverson J.D., Boghaert E.R., Ackler S.L., Catron N.D., Chen J., Dayton B.D., Ding H., Enschede S.H., Fairbrother W.J., et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- Stevers L.M., Lam C.V., Leysen S.F.R., Meijer F.A., van Scheppingen D.S., de Vries R.M.J.M., Carlile G.W., Milroy L.G., Thomas D.Y., Brunsveld L., Ottmann C. Characterization and small-molecule stabilization of the multisite Tandem binding between 14-3-3 and the R domain of CFTR. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E1152–E1161. doi: 10.1073/pnas.1516631113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanovsky B., Levchenko T., Månsson G., Matvijenko O., Holmgren L. Angiomotin: an angiostatin binding protein that regulates endothelial cell migration and tube formation. J. Cell Biol. 2001;152:1247–1254. doi: 10.1083/jcb.152.6.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenti D., Hristeva S., Tzalis D., Ottmann C. Clinical candidates modulating protein-protein interactions: the fragment-based experience. Eur. J. Med. Chem. 2019;167:76–95. doi: 10.1016/j.ejmech.2019.01.084. [DOI] [PubMed] [Google Scholar]
- Vidal M., Cusick M.E., Barabási A.-L. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb C., Upadhyay A., Giuntini F., Eggleston I., Furutani-Seiki M., Ishima R., Bagby S. Structural features and ligand binding properties of Tandem WW domains from YAP and TAZ, nuclear effectors of the Hippo pathway. Biochemistry. 2011;50:3300–3309. doi: 10.1021/bi2001888. [DOI] [PubMed] [Google Scholar]
- Winter G., Waterman D.G., Parkhurst J.M., Brewster A.S., Gildea R.J., Gerstel M., Fuentes-Montero L., Vollmar M., Michels-Clark T., Young I.D., Sauter N.K., Evans G. DIALS: implementation and evaluation of a new integration package. Acta Crystallogr. D Struct. Biol. 2018;74:85–97. doi: 10.1107/S2059798317017235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter M., de Vink P., Neves J.F., Srdanović S., Higuchi Y., Kato N., Wilson A., Landrieu I., Brunsveld L., Ottmann C. Selectivity via cooperativity: preferential stabilization of the P65/14-3-3 interaction with semisynthetic natural products. J. Am. Chem. Soc. 2020;142:11772–11783. doi: 10.1021/jacs.0c02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würtele M., Jelich-Ottmann C., Wittinghofer A., Oecking C. Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J. 2003;22:987–994. doi: 10.1093/emboj/cdg104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.-X., Zhao B., Guan K.-L. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Wei X., Li W., Udan R.S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., Zheng P., Ye K., Chinnaiyan A., Halder G., Lai Z.-C., Guan K.-L. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li L., Lu Q., Wang L.H., Liu C.-Y., Lei Q., Guan K.-L. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.