Summary
Background
Synaptic proteins are increasingly studied as biomarkers for synaptic dysfunction and loss, which are early and central events in Alzheimer's disease (AD) and strongly correlate with the degree of cognitive decline. In this study, we specifically investigated the synaptic binding partners neurexin (NRXN) and neuroligin (Nlgn) proteins, to assess their biomarker's potential.
Methods
we developed a parallel reaction monitoring mass spectrometric method for the simultaneous quantification of NRXNs and Nlgns in cerebrospinal fluid (CSF) of neurodegenerative diseases, focusing on AD. Specifically, NRXN-1α, NRXN-1β, NRXN-2α, NRXN-3α and Nlgn1, Nlgn2, Nlgn3 and Nlgn4 proteins were targeted.
Findings
The proteins were investigated in a clinical cohort including CSF from controls (n=22), mild cognitive impairment (MCI) due to AD (n=44), MCI due to other conditions (n=46), AD (n=77) and a group of non-AD dementia (n=28). No difference in levels of NRXNs and Nlgns was found between AD (both at dementia and MCI stages) or controls or the non-AD dementia group for any of the targeted proteins. NRXN and Nlgn proteins correlated strongly with each other, but only a weak correlation with the AD core biomarkers and the synaptic biomarkers neurogranin and growth-associated protein 43, was found, possibly reflecting different pathogenic processing at the synapse.
Interpretation
we conclude that NRXN and Nlgn proteins do not represent suitable biomarkers for synaptic pathology in AD. The panel developed here could aid in future investigations of the potential involvement of NRXNs and Nlgns in synaptic dysfunction in other disorders of the central nervous system.
Funding
a full list of funding can be found under the acknowledgments section.
Keywords: synaptic proteins, neurexins, neuroligins, Alzheimer's disease, biomarkers, mass spectrometry
Abbreviations: ABC, Ammonium bicarbonate; Aβ, amyloid-beta; APP, amyloid precursor protein; Aβo, Aβ oligomers; Aβ1-42, Aβ peptide 1-42; AD, Alzheimer´s disease; CNS, central nervous system; CV, coefficient of variation; DLB, dementia with Lewy bodies; ELISA, enzyme-linked immunosorbent assays; F/T, freeze/thaw; bvFTD, behavioral variant frontotemporal dementia; GAP43, growth-associated protein 43; IS, internal standard; MRI, magnetic resonance imaging; MS, mass spectrometry; MCI, mild cognitive impairment; MMSE, mini-mental state examination; NRXNs, neurexins; NFTs, neurofibrillary tangles; Nlgns, neuroligins; PRM, parallel reaction monitoring; p-tau, phospho-tau; QC, quality control; SPE, solid-phase extraction; t-tau, total-tau; VaD, vascular dementia
Research in context.
Evidence before this study
The fluid biomarker field for neurodegenerative diseases is rapidly expanding and many investigations are directed towards the study of synaptic proteins as biomarkers for synaptic dysfunction and loss. Alterations of synaptic integrity is an early and central event in Alzheimer's disease (AD) and other neurodegenerative diseases, which also correlate with the degree of cognitive decline. Therefore, synaptic biomarkers are sought after as they might hold the ability of detecting early pathological changes and to follow disease progression. Neurexin (NRXN) and neuroligin (Nlgn) proteins are binding partners ubiquitously expressed at neuronal synapses, binding to each other at the synaptic cleft through their extracellular domains. NRXNs have been detected in cerebrospinal fluid (CSF), where several studies have reported altered levels of the proteins in AD, already at the mild cognitive impairment (MCI) stage, albeit with variable results. In our previous work, we investigated Nlgn1 in brain and CSF of AD patients, with promising results, but the other members of the Nlgn's family have never been explored as biomarkers. None of the previous studies investigated these proteins simultaneously. Given the importance of NRXNs and Nlgns at the synapse and the tight relationship these two families of proteins show, we decided with this study to investigate them simultaneously in a comprehensive clinical cohort, focusing on AD, but also including MCI patients, both due to AD and to other causes, and a non-AD dementia group.
Added value of this study
To our knowledge, this is the first study that simultaneously investigated NRXNs and Nlgns in CSF of AD patients. Through their extracellular domains, the presynaptic NRXNs bind the postsynaptic Nlgns across the synaptic cleft and their mechanisms appear to be interrelated. This study tried to answer the question whether these proteins change in AD patients and whether changes in the levels of NRXN proteins would affect Nlgns and vice versa. Moreover, the method developed here allows for their simultaneous quantification with high sensitivity and precision. In order to create this mass spectrometry panel, several different peptides per protein have been included, covering the extracellular part of NRXN and Nlgn proteins, of which soluble fragments are quantifiable in CSF. The study aims at reproducing previously published results and expand them, in order to increase our knowledge on NRXN and Nlgn proteins as possible synaptic biomarkers for AD.
Implications of all the available evidence
Despite previous studies showing NRXN proteins changing in CSF of AD patients, even at early stages, our results show no changes of these proteins in both AD dementia and MCI cases, as well as in the non-AD dementia group. Nlgns also do not show changes in their levels in any of the groups analysed. Thus, this study does not support the use of NRXNs and Nlgns as biomarkers for synaptic dysfunction in AD. If changes of these proteins happen at even earlier stages than those we investigated, or in different brain regions, or in other brain pathologies, are questions that remain to be investigated. Moreover, these results might suggest that synaptic dysfunction in AD is not a generalized disruption of synapses but it could entail more specific mechanisms, which affect a subset of proteins but not others. The work presented in this paper provides a novel methodology, which can be used for further specific studies of these proteins in order to elucidate their role at the synapse and implications in neurodegenerative and neuropsychiatric diseases.
Alt-text: Unlabelled box
Introduction
Synapses are central structures for memory function and information storage in the brain, and as such, their integrity and homeostasis are essential for proper cognitive function. Synapse health is disrupted in Alzheimer´s disease (AD), the most common cause of dementia, where abnormal depositions of proteins or peptides, such as amyloid-beta (Aβ) in plaques and tau protein in neurofibrillary tangles (NFTs), leads to synaptic degeneration, neuronal loss and clinical symptoms.1, 2, 3, 4 Additionally, synaptic dysfunction and synapse loss have also been shown in other types of dementia.5, 6, 7, 8, 9 However, the mechanisms leading to synaptic loss are not fully understood yet and further investigations are needed. Considering the high correlation between synaptic loss and cognitive symptom severity in AD,10,11 the study of synaptic proteins could increase our understanding of the pathophysiological processes underlying neurodegenerative changes and possibly yield valuable biomarkers to monitor them. The potential usefulness of synaptic biomarkers is also founded on the fact that synapses are the substrate of cognition and synaptic dysfunction is one of the earliest events in the course of neurodegenerative diseases. Therefore, it is suggested that pathological processing of synaptic proteins may reflect changes in cognition in neurodegenerative diseases at early stages.3,12, 13, 14 In AD, the Aβ peptide 1-42 (Aβ1-42) or the Aβ42/40 ratio, total-tau (t-tau) and tau phosphorylated at Thr181 (p-tau181) are well-established cerebrospinal fluid (CSF) biomarkers15 describing the main pathological events in the brain during the course of the disease. Several synaptic proteins have also been investigated as biomarkers.16 Among them neurogranin, growth-associated protein 43 (GAP43), synaptosomal-associated protein 25 (SNAP25) and synaptotagmin-1 are arguably the most investigated in AD.17,18 However, the complexity of the brain and the multiple mechanisms involved in synaptic regulation call for more investigations into processing of synaptic proteins to better understand different events in disease progression and to better discriminate between different dementias.
Correct interaction between the pre- and post-synaptic compartment is essential for proper function of synapses. Neurexin (NRXN) and neuroligin (Nlgn) families consist of important synaptic adhesion proteins taking part in this fundamental process.19, 20, 21 The presynaptic NRXNs and postsynaptic Nlgns22 comprise single-pass transmembrane proteins with short cytoplasmic domains and large extracellular domains, through which they bind each other in the synaptic cleft, stabilizing the two compartments of the synaptic bouton.20 Moreover, NRXNs and Nlgns cluster receptors and channels essential for synapse formation and differentiation of the synaptic compartment. How the different proteins exert their function is not entirely understood. However, NRXNs and Nlgns appear to have a tight relationship by which they regulate each other and downstream signalling through their interaction. To exert their function, both their extracellular and cytoplasmic domains seem to be important.19,23 NRXNs in humans are encoded by three genes which use two different promoters. Their transcription gives rise to α- and β-forms of NRXNs.19,24 NRXNs contain relatively well conserved cytoplasmic domains, but differ much more in their extracellular domain, where they can be spliced at five alternative positions, and theoretically produce more than a thousand different isoforms of the proteins.25 In humans, five NLGN genes have been described. Nlgn1, -2 and -3 proteins are predominantly expressed in the central nervous system (CNS) and appear to be the most abundant.24 Nlgn4 is designated as Nlgn4-X to distinguish it from the product of the fifth gene, Nlgn4-Y (occasionally referred to as Nlgn5), which is located on the Y chromosome and presents high sequence homology to Nlgn4-X.26 Although it is the least studied of the family, also Nlgn4 has been found expressed in the cerebral cortex, preferentially localized at dendritic spines, taking part in excitatory synaptic transmission.27
Genetic alterations of these proteins have been connected to synaptic dysfunction in mental disorders like schizophrenia and autism,25,28,29 and various studies have reported NRXNs and Nlgns to be altered in neurodegenerative diseases, like AD.30, 31, 32, 33, 34 Nlgn1 protein levels has been shown to be decreased in different brain regions of AD and a group of primary tauopathies.18,35 Moreover, the extracellular domains of NRXNs and Nlgns undergo proteolytic cleavage36, 37, 38, 39 which leads to the extracellular release of a soluble N-terminal ectodomain that can be detected in CSF. Independent studies have reported altered levels of NRXN-1α,40, 41, 42, 43 NRXN-2α, NRXN-3α and Nlgn2 in CSF of AD41,44,45 and from the earlier stage of mild cognitive impairment (MCI).46 However, these studies showed variable results and none of them investigated NRXNs and Nlgns simultaneously. Considering these investigations in brain and CSF and the tight relationship between NRXNs and Nlgns, the simultaneous monitoring of these proteins and the possibility to distinguish between the different isoforms would be highly valuable to increase our knowledge of the processes regulating these proteins at the synapse and evaluate the potential of using these proteins as biomarkers for synapse dysfunction in pathology. The heterogeneous nature of NRXN and Nlgn proteins complicates the use of antibody-based methods, whereas high-throughput and highly selective methods such as mass spectrometry (MS), offers an antibody-independent alternative, as well as the possibility of multiplexing. Indeed, MS is increasingly used to investigate biomarkers, showing good performance and reproducibility.47 Therefore, the aim of this study was to develop a targeted MS assay for the simultaneous quantification of NRXN and Nlgn proteins to study these proteins in CSF samples from patients of neurodegenerative diseases. Included in the parallel reaction monitoring (PRM) assay are the most commonly explored NRXN-2α, -3α, -1α and -1β and all the Nlgn proteins, Nlgn1 to 4. In a pilot study, we used the novel assay to explore the proteins in CSF having an AD and non-AD profile (defined by biochemical criteria). Subsequently, the study of the proteins was expanded to a clinical cohort including CSF from controls, AD at both MCI and dementia stages, and a group of non-AD MCI and non-AD dementia individuals.
Methods
CSF samples included in the study
Discovery cohort
The NRXNs-Nlgns MS panel was initially tested in a pilot study of CSF samples from the Neurochemistry Laboratory, Sahlgrenska University Hospital, Mölndal, Sweden, including AD CSF profile (n=21) and non-AD CSF profile (n=19). These samples were biochemically defined as AD and non-AD samples based on the analysis of CSF core biomarkers for AD (p-tau181, t-tau and Aβ1-42) following the respective diagnostic criteria.48
Clinical cohort
All included subjects were recruited at the Cognitive Neurology Centre, GHU APHP Nord Université de Paris Lariboisière Fernand Widal Paris, from 2015 to 2019. All methods and diagnosis processes have been extensively described in Tible et al.17 Briefly, recruited individuals were seen at the memory clinic and underwent CSF biomarkers analysis for a cognitive complaint. Consensus clinical diagnosis of the neurocognitive disorders was reached after review by a multidisciplinary team according to validated diagnostic criteria. Reference criteria were used for the inclusions of patients including those for AD dementia,49 MCI due to AD (MCI-AD),50 and for patients with other dementia (non-AD dementia) including vascular dementia (VaD),51 dementia with Lewy bodies (DLB),52 and frontotemporal dementia of behavioural variant type (bvFTD).53 MCI of non-neurodegenerative causes (non-AD MCI) were also included. The neurological control (NC) group included participants with subjective cognitive complain or non-neurological disorders. CSF core biomarker analysis was performed (see below) and, after analysis, AD and MCI due to AD had pathological amyloid ratio, high p-tau and t-tau, while neurological control subjects had normal CSF profile. Other dementia and neurological control groups had a CSF amyloid ratio in the range of normal.
CSF sampling and analysis of AD core biomarkers
CSF samples were obtained by lumbar puncture according to the European and French recommendations54,55 in the context of the diagnostic workup of patients with cognitive complaint or decline. CSF was collected in polypropylene tubes using a standardized procedure. Samples were centrifuged (2000 × g, 20 min, +4°C) and the supernatant stored at -80°C. For the discovery cohort, CSF AD core biomarker concentrations were quantified by commercially available INNOTEST enzyme-linked immunosorbent assay (ELISA) (Aβ1-42 cat.# 81583, t-tau cat.# 81572, p-tau181 cat.# 81581; Fujirebio, Ghent, Belgium). For analysis of CSF core AD biomarkers in the clinical cohort, a LUMIPULSE G1200 instrument (Fujirebio, Ghent, Belgium) was used according to the manufacturer's instructions. The normal ranges were defined as follows: Aβ42 > 620 ng/L, Aβ42/40 ratio > 0.61, p-tau181 < 61 ng/L, t-tau < 479 ng/L.56 Analysis of the synaptic biomarkers neurogranin and GAP43 was performed using in-house ELISA assays57,58 at the Neurochemistry Laboratory at Sahlgrenska University Hospital, Mölndal, Sweden. Quality controls (QC) used for assay validation consisted of pooled CSF samples, also obtained from the Neurochemistry Laboratory, which were aliquoted and stored in the same way as the individual patient samples. Demographics and biomarker characteristics of the patients are shown in Table 1.
Table 1.
Demographic characteristics, CSF AD core biomarkers and synaptic biomarkers concentration for the discovery and the validation cohorts
| Discovery cohort |
Clinical cohort from Paris |
||||||
|---|---|---|---|---|---|---|---|
| Patient group | Non-AD (n=19) | AD (n=21) | NC (n=22) | MCI-AD (n=44) | AD dementia (n=77) | Non-AD MCI (n=46) | Non-AD Dementia (n=28) |
| Sex (male/female) | 7/11¤ | 12/9 | 8/14 | 17/27 | 28/49 | 17/29 | 16/12 |
| Age (years) mean, [SD] | 54.2 [8.7] | 72.4 [9.7] | 64.4 [8.86] | 72 [7.7]*& | 72.2 [8.33]*& | 67 [10]# | 66 [7.5]# |
| MMSE, mean, [SD] | - | - | 27.14 [2.34] | 23.34[4.54]⁎# | 19.06 [5.63]* | 24.5 [3.65]# | 23.5 [4.95]# |
| Biomarkers | |||||||
| Aβ42 [pg/ml] mean, [SD] | 891 [173.32] | 346.9 [86.26] | 1095.9 [277.09] | 599 [332.04]*& | 508.33 [160.9]*& | 1080.5 [419.8]# | 974.9 [380.9]# |
| Aβ42/Aβ40 mean, [SD] | - | - | 0.093 [0.009] | 0.046 [0.012]*& | 0.042 [0.009]*& | 0.090 [0.010]# | 0.089 [0.014]# |
| t-Tau [pg/ml] mean, [SD] | 227.6 [65.56] | 530.19 [46.60] | 243.09 [66.09] | 594.63 [268.8]*& | 731.79 [384.9]*& | 296.35 [139.6]# | 329.1 [303.7]# |
| p-Tau [pg/ml] mean, [SD] | 37.28 [8.95] | 67.43 [3.89] | 32.84 [8.06] | 92.42 [46.06]*#& | 112.59 [55.86]*& | 37.07 [15.41]# | 34 [10.63]# |
| Neurogranin [pg/ml] mean, [SD] | - | - | 131.69 [40.19] | 259.8 [82.72]*& | 268.72 [78.61]*& | 172.4 [45.11]# | 163.3 [49.47]# |
| GAP43 [pg/ml] mean, [SD] | - | - | 2427.26 [707.16] | 4071.3 [1889.2]*& | 4497 [1828.5]* | 3071.58 [1263.7]# | 3012.9 [1422.6]# |
Abbreviations: AD= Alzheimer's disease, NC=neurological controls, MCI=mild cognitive impairment, GAP43=growth-associated protein 43, MMSE=Mini-Mental State Examination, SD=standard deviation
One missing data
Notes: Analysis of variance followed by Tukey's post hoc test (continuous variables) or contingency chi-square test (sex), were used to test differences between the groups. Significant differences compared to control (*) or AD (#) or non-AD dementia (&) in the same cohort are highlighted. Significance level set to p ≤ 0.05.
I will add the difference between the groups in the Paris cohort
Ethics
For the discovery cohort, the collection and storage of CSF samples were in accordance with the Swedish law of biobanks in healthcare (2002:297). The use of these patient samples was approved by the Ethics Committee at the University of Gothenburg (EPN 140811). For the clinical cohort, the study was approved by the Bichat Hospital Ethics Committee of Paris Diderot University. All patients signed an informed consent.
Peptide selection
For each protein, peptides were chosen either based on previously published studies18,40,44 or selected based on sequence uniqueness, length, and amino acid composition after in silico tryptic digestion (Table 2). Exceptions to the uniqueness criteria were; peptide 3 for NRXN-1 (VDSSSGLGDYLELHIHQGK), which is common to the two forms NRXN-1α and NRXN-1β, and all peptides for Nlgn4, which are found in both Nlgn4-X and Nlgn4-Y. All the peptides belong to the extracellular domain of the proteins, which is released extracellularly after proteolytic processing.
Table 2.
Proteins and peptides included in the PRM-MS study
| Protein name | Protein abbreviation | Protein accession ID | Peptide sequence | Position |
|---|---|---|---|---|
| Neurexin-1α (3 peptides) | NRXN-1α | Q9ULB1 | EATVLSYDGSMFMK | [714-727] |
| LTVDDQQAMTGQMAGDHTR | [822-840] | |||
| VDSSSGLGDYLELHIHQGK c | [1167-1185] | |||
| Neurexin-2α (4 peptides) | NRXN-2α | Q9P2S2 | TALAVDGEAR | [123-132] |
| LSALTLSTVK | [160-169] | |||
| LGERPPALLGSQGLR | [183-197] | |||
| LQGDLSFR | [477-484] | |||
| Neurexin-3α (4 peptides) | NRXN-3α | Q9Y4C0 | SDLSFQFK | [48-55] |
| NGLILHTGK | [292-300] | |||
| ANDGEWYHVDIQR | [536-548] | |||
| FICDCTGTGYWGR | [664-676] | |||
| Neuroligin-1 (3 peptides) | Nlgn1 | Q8N2Q7 | LDDVDPLVATNFGK | [47-60] |
| WTSENIGFFGGDPLR | [279-293] | |||
| FEEVAWTR | [609-616] | |||
| Neuroligin-2 (3 peptides) | Nlgn2 | Q8NFZ4 | FQPPEAPASWPGVR | [83-96] |
| ELVDQDVQPAR | [335-345] | |||
| TLLALFTDHQWVAPAVATAK | [449-468] | |||
| Neuroligin-3 (1 peptide) | Nlgn3 | Q9NZ94 | VGCNVLDTVDMVDCLR | [337-352] |
| Neuroligin-4 (2 peptides) | Nlgn4 | Q8N0W4/ | WIEENVGAFGGDPK d | [232-245] |
| Q8NFZ3 | TGPEDTTVLIETK d | [655-667] |
aUnderlined cysteine (C) indicate the carbamidomethylation occuring through alkylation
bBlue colour-coded amino acids indicate heavy labelled [U-13C6,15N4]-arginine (R) and [U-13C6,15N2]-lysine (K)
Common peptide for NRXN-1α and NRXN-1β
Common peptides for the two Nlgn4 isoforms
Heavy-isotope-labelled standards
Thirty-one tryptic peptides, labelled at the C-terminal arginine or lysine with 13C and 15N, were purchased from JPT Peptide Technology (Berlin, Germany) and used as heavy-isotope-labelled internal standards (IS) for peptide quantification. Peptides (stated amount from the manufacturer ≈ 10 nmol) were reconstituted in 1 mL 10% acetonitrile solution in deionized water (v/v), aliquoted and stored at -20°C pending analysis. Aliquots of all peptides were pooled and diluted in 50 mM ammonium bicarbonate (ABC) to a final optimized concentration, in order to create an IS mix matching the respective protein levels in the CSF samples.
Sample preparation and SPE
For sample preparation, 100 µL of CSF samples were pipetted into Micronic 0.75-mL tubes (cat.# MP32069L), followed by the addition of 25 µL IS diluted in 50 mM ABC as mentioned above. Cysteine disulfide bridges in the samples were reduced by adding 25 µL of 30 mM dithiothreitol in 50 mM ABC and shaken for 30 min at 60°C. Samples were then cooled to room temperature and cysteines were blocked by alkylation with 25 µL 70 mM iodoacetamide in 50 mM ABC and shaken for 30 min in the dark. Next, 25 µL of trypsin/Lys-C (Promega, cat.# V5073), corresponding to 0.5 µg per sample, were added to every sample and digestion was performed at 37°C overnight (16 h) shaking at 90 rpm. The day after, samples were spun down, centrifuged and digestion was stopped by the addition of 25 µL 10% trifluoroacetic acid in deionized water (v/v). Subsequently, solid-phase extraction for sample clean-up from salt and detergents was performed using Oasis 30 μm HLB 96-well μElution Plates (Waters Co., Milford, MA, USA). The plates were first conditioned twice with 300 µL of methanol, then equilibrated twice with 300 µL deionized water using a rotary pump for controlled suction. Samples were then loaded into the plate wells, aspirated and washed twice with 300 µL of deionized water. Finally, elution of the samples was obtained by the addition of 2 × 100 µL of methanol, and eluates collected into Micronic 0.75-mL tubes. Subsequently, eluates were dried in a vacuum centrifuge and stored at -80°C pending analysis. For the analysis of the clinical cohort, samples were randomized in three 96-well plates, and eight QC samples (two different QCs in four replicates per plate) were added to account for variations and to allow performance check during MS analysis. Median of the total area ratio of QC1 was used for adjustment of plate variations.
PRM-MS and data analysis
The PRM-MS analysis was performed using a Q Exactive hybrid quadrupole-orbitrap high resolution mass spectrometer (Thermo Fisher Scientific), with electrospray ionization. On the day of analysis, samples were reconstituted in 100 µL of 50 mM ABC, shaken for 30 min at room temperature and 45 µL were loaded using a Vanquish UHPLC (Thermo Fisher Scientific). Sample peptides were separated on a Hypersil Gold reversed-phase column (particle size 1.9 μm, internal diameter 2.1 mm, length 100 mm, Thermo Fisher Scientific) operated at a flow rate of 300 μL/min by applying a broken gradient of 0-32% B for 24 min (total sample cycle time was 32 min). Mobile phases used were A: 0.1% formic acid in deionized water (v/v) and B: 0.1% formic acid and 84% acetonitrile in deionized water (v/v/v). A graphical representation of the gradient is shown in Supplementary Fig. 1. Electrospray conditions were set as follows; spray voltage at +4100 V, capillary temperature at 320°C, sheath gas setting of 25, aux gas setting of 10, sweep gas setting of 0, probe heater temperature at 300°C, and S-lens RF level setting of 55. Mass spectra were acquired using a scheduled PRM method with retention time windows of 2 min for each peptide and a toggle limit of four different peptide pairs. For data acquisition, isolation window was set to 3 m/z units, automatic gain control target value to 3 × 106 and maximum injection time to 250 ms with a matching resolution setting of 70,000.
For each peptide, collision energy was optimized in order to maximize the sensitivity of the PRM assay. Peak detection and area integration was performed using Skyline 20.2 (MacCoss Lab Software).59 Every peak was manually inspected and, when required, peak adjustment and removal of transitions affected by interference were applied. Relative peptide quantification was performed by dividing the sum of all measured fragment peak areas by the sum of the fragment peak areas of the corresponding IS. Representative peaks and corresponding transitions are shown in Supplementary Fig. 2.
Assay validation
To monitor intra- and inter-plate variations, peptide stability and method and dilution linearity, CSF pools were used as QC standards for the different tests. The variability was estimated by calculating the coefficient of variation (CV) for the QCs. In order to investigate the stability of NRXN and Nlgn peptides, different storage conditions and freeze/thaw (F/T) cycles were tested. Aliquots of CSF samples from six different individuals under different storage conditions and F/T cycles were analysed. F/T cycles were as follows: aliquot #1 was stored at -80°C (i.e., one F/T cycle), while aliquots #2, #3, #4 and #5 underwent a total of two, three, four and five F/T cycles (and stored at -80°C in between cycles), respectively. Storage conditions were as follows: aliquot #6 at 5-8°C for 24 h and then stored at -80°C, aliquot #7 at 5-8°C for one week and then stored at -80°C, aliquot #8 at room temperature for 24 h and then stored at -80°C, aliquot #9 stored at -20°C for one month, then at -80°C. To evaluate the linearity of the method, four-fold serial dilutions of the IS were prepared and spiked into QC samples, digested and analysed in triplicate by LC-MS/MS. To examine the possible matrix effects caused by the CSF amount, dilution linearity of the peptides was tested using different volumes of CSF; volumes tested were 120, 100, 80, 60 and 40 µL of QC samples. For both tests, the curve fits were created using weighted sum of squares (1/X2) to account for non-homogeneous distribution of the residuals.60
Statistical analysis
Data related to NRXNs and Nlgns were not normally distributed (even after logarithmic transformation) and, therefore, non-parametric tests were applied. Differences between groups were assessed using Mann-Whitney U test or Kruskal-Wallis test with Dunn's correction for multiple comparisons, when appropriate. Correlations were investigated using Spearman's rank correlation coefficient (rho). None of the peptides correlated with age and sex, therefore neither age nor sex were included as covariates, except for Nlgn1 and Nlgn4. For Nlgn1 data were adjusted for age and for Nlgn4 data were adjusted for age and sex (Supplementary table 1). The Nlgn1 and Nlgn4 data were log transformed to make it resemble a normal distribution, thus linear regression of analysis of covariance was used. The cut-off for Aβ+/Aβ- was defined by the Aβ42/40 ratio. For data normally distributed, differences between more than two groups were investigated using analysis of variance followed by Tukey's post hoc test (continuous variables) or contingency chi-square test (categorical variable). Data visualization and statistical analysis were performed using GraphPad Prism 7 (GraphPad Software, La Jolla, CA, USA). All tests were two-sided and p-values ≤ 0.05 were considered as the threshold level for significance. However, the p-values for correlations were adjusted using Bonferroni correction for multiple comparison (n=24) and consequently a probability of p ≤ 0.002 was considered statistically significant.
Role of funders
A declaration of interest section, where funders are listed, can be found at the end of the manuscript. Funders did not have any role in the study design, data collection or analysis and interpretation of the results or manuscript writing. All authors had complete access to the data.
Results
Assay validation and performance
In total, 31 tryptic NRXN and Nlgn peptides were investigated in the CSF samples. Based on the repeatability analysis performed using eight QC replicates, 11 peptides could not be quantified (CSF level below limit of detection) or showed a CV higher than 20% (CSF level below limit of quantification); they were therefore disregarded from further analysis (excluded peptides are highlighted in Supplementary Table 2). Further, after peptide analysis and quantification in the clinical cohort, an additional four peptides were not taken into account for group comparison for equivalent reasons (highlighted in Supplementary Table 3). In total, 16 peptides (ten for NRXNs and six for Nlgns) were utilized for group comparison and final analysis in the clinical cohort (acquisition characteristics and repeatability analysis of the selected peptides are shown in Supplementary Table 3). The NRXN-1 peptide 3 sequence is common for both NRXN-1α and NRXN-1β proteins. However, from the initial screening, the unique peptide for NRXN-1β showed high variability, probably because of very low abundance of the protein in CSF. Indeed, α-forms of neurexins have been shown to be more abundant than the corresponding β-forms.61 Taking these pieces of evidence together, we believe NRXN-1 peptide 3 mainly reflects NRXN-1α. All the selected peptides for NRXNs and Nlgns showed analytical stability when the CSF samples underwent up to five F/T cycles, except for Nlgn2 peptide 1, Nlgn4 peptide 1, and NRXN-3α peptides 3 and 4. The same peptides showed high variation also upon different storage conditions, whereas the others were found to be stable (Supplementary Fig. 3). The method linearity test and the dilution linearity test, showed that the relative error corresponding to the back-calculated concentration were within the set limit (20%) for most of the peptides and at least for one peptide per protein (Supplementary Fig. 4 and 5). As mentioned above, several peptides showed suboptimal performance, especially among Nlgns, and were thus discarded from further analysis.
NRXNs-Nlgns CSF levels
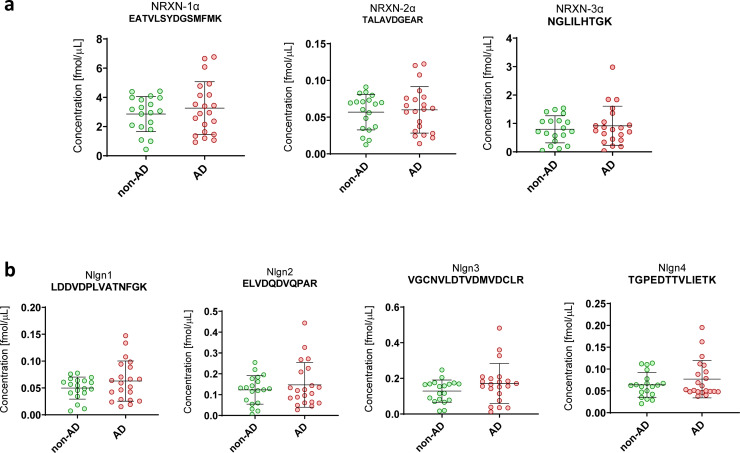
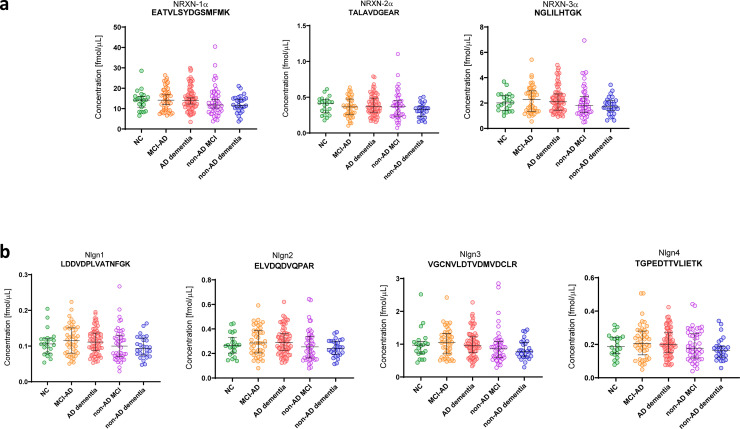
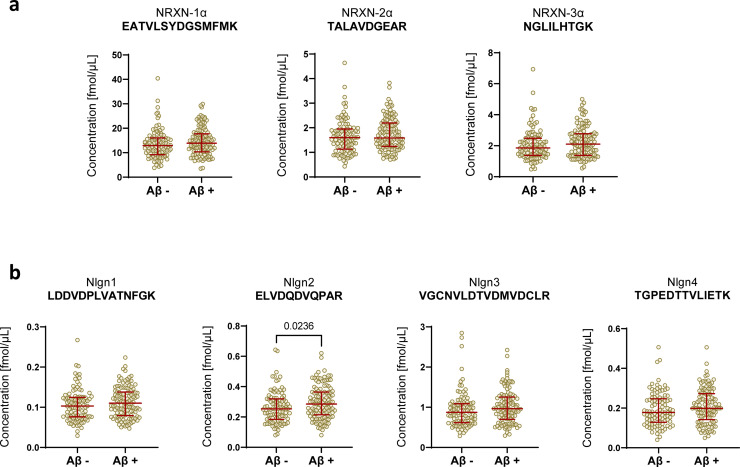
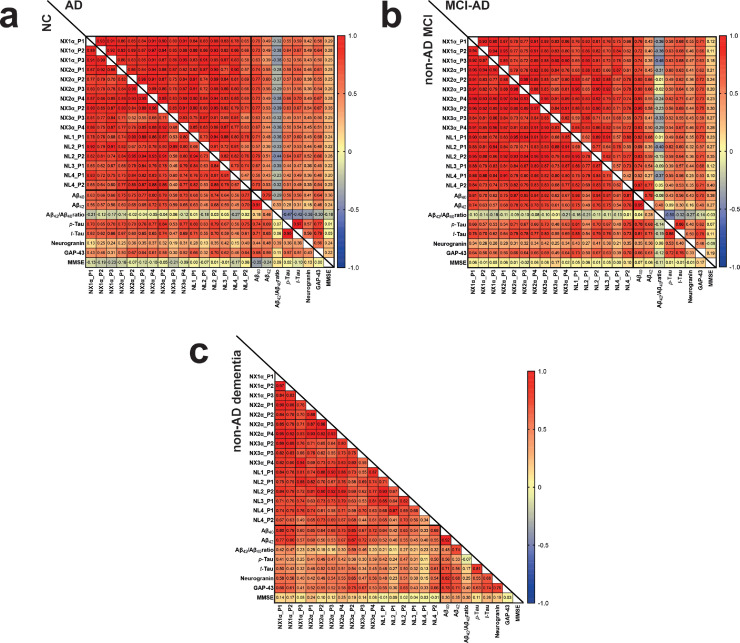
The NRXNs-Nlgns panel was initially evaluated in a pilot cohort, which included CSF samples from 21 AD and 19 non-AD patients. All the selected peptides were quantified, but no significant differences were found between the groups (Fig. 1). In order to extend these preliminary observations and further investigate the NRXN and Nlgn proteins in the AD continuum and in other neurodegenerative disorders, a larger clinical cohort including CSF from neurological controls (NC, n=22), AD (n=77), MCI due to AD (n=44), non-AD MCI (n=46) and non-AD dementia (n=28) was investigated. The CSF concentration and QC CV of all NRXN and Nlgn peptides are shown in Supplementary Table 3. Neither NRXNs nor Nlgns showed any significant change in AD, or in the MCI due to AD group when compared with neurological controls. Protein levels also did not change in the non-AD dementia and the non-AD related MCI groups, when compared with neurological controls or AD (Fig. 2). For a more comprehensive investigation, the non-AD dementia group was further divided into its constitutive subgroups, VaD (n=3), DLB (n=12), and bvFTD (n=13), and group differences in NRXNs and Nlgns levels were investigated. The proteins did not show any difference when the subgroups were compared to neurological controls (Supplementary Fig. 6). Protein levels were further investigated in relation to Aβ pathology, and groups were dichotomized into Aβ+ and Aβ- based on the Aβ42/40 ratio. Neither NRXNs nor Nlgns levels differed between the two groups, except for Nlgn2 (p=0.024, Mann-Whitney U test) although the difference was not deemed of importance because of the high overlap between the groups (Fig. 3). Nlgn1 peptide showed a significant correlation with age, and Nlgn4 peptide two, with age and sex (Supplementary Table 1), but correcting for it did not change the results. NRXN peptides were highly correlated with each other (rho=0.68-0.97, p≤0.002, Spearman's rank correlation coefficient test). The same strong correlation was also seen for all Nlgn peptides (rho=0.50-0.93, p≤0.002, Spearman's rank correlation coefficient test). NRXN and Nlgn proteins also significantly correlated with each other (Supplementary Fig. 7). A moderate to strong correlation between NRXN and Nlgn peptides was, with a few exceptions, found in all the groups (neurological control group rho=0.40-0.96, AD group rho=0.48-0.97, MCI due to AD rho=0.42-0.98, non-AD MCI rho=0.62-98, non-AD dementia rho=0.34-0.96, p≤0.002, Spearman's rank correlation coefficient test). Further, there was no change in correlations between the different groups (Fig. 4). NRXN and Nlgn proteins did not correlate with Aβ42/40 ratio (in any of the groups), whereas both correlated quite strongly with Aβ40 and moderately with Aβ42 across all groups. Generally, NRXNs and Nlgns moderately correlated also with t-tau and to a lesser degree with p-tau. When comparing NRXNs and Nlgns with other synaptic biomarkers, they correlated moderately with GAP43 but to a lesser extent with neurogranin (Fig. 4). Neurogranin and GAP43 moderately correlated with each other in the AD group (rho=0.56, p≤0.002, Spearman's rank correlation coefficient test) and in the non-AD dementia group (rho=0.76, p≤0.002, Spearman's rank correlation coefficient test), whereas a weak non-significant correlation was found in the other groups (neurological control group rho=0.43, MCI due to AD group rho=0.46, non-AD MCI rho=0.33). Neither NRXN nor Nlgn peptides correlated with the Mini-Mental State Examination (MMSE) score (Fig. 4). The correlations of the NRXNs and Nlgns and relative p-values are summarized in Supplementary Table 4.
Figure 1.
CSF concentrations obtained by PRM analysis of the NRXNs-Nlgns panel in the pilot cohort. In the figure, only one representative peptide for (a) NRXNs and (b) Nlgns is shown. The cohort consisted of biochemically defined non-Alzheimer's disease CSF profile (non-AD, n=19) and Alzheimer's disease (AD, n=21) CSF samples. Samples were analysed as singlicates. The bars indicate median with interquartile range.
Figure 2.
CSF concentrations obtained by PRM analysis of the NRXNs-Nlgns panel in the clinical cohort. In the figure, only one representative peptide for (a) NRXNs and (b) Nlgns is shown. The clinical cohort consisted of neurological controls (NC, n=22), Alzheimer's disease dementia (AD dementia, n=77), mild cognitive impairment due to AD (MCI-AD, n=44), MCI non-due to AD (non-AD MCI, n=46) and a group of non-AD dementia (n=28). Samples were analysed as singlicates. The bars indicate median with interquartile range.
Figure 3.
NRXNs and Nlgns levels in relation to Aβ pathology. In the figure, only one representative peptide for (a) NRXNs and (b) Nlgns is shown. Peptide levels are compared in dichotomized Aβ+ and Aβ- groups. Nlgn2 was slightly, although significantly increased, in the Aβ+ group (p≤0.05, Mann-Whitney U test). The bars indicate median with interquartile range.
Figure 4.
Correlation matrix between CSF correlation of NRXNs and Nlgns peptides. (a) Neurological controls (NC) and AD, (b) MCI-AD and non-AD MCI, (c) non-AD dementia, in the Paris clinical cohort. The correlation coefficients are presented as Spearman's rho. Abbreviations: NX=neurexins, NL=neuroligins, P=peptide number
Discussion
In this study, we have developed a targeted MS method for the simultaneous quantification of NRXNs and Nlgns in CSF of neurodegenerative diseases. The focus of the investigation was on AD, thus the panel was tested in a discovery cohort, comprising patients showing a typical AD CSF biomarkers profile and, subsequently, in a larger clinical cohort. The results obtained in both cohorts indicate no significant change in CSF levels of NRXNs and Nlgns, neither in the AD continuum (represented by the MCI due to AD cases and AD dementia patients), nor in the non-AD MCI group, when compared with neurological controls. The non-AD dementia group, including VaD, DLB, and bvFTD cases, also did not show any statistically significant difference in the levels of NRXNs and Nlgns. In our previous study on CSF samples from AD and neurological controls using western blot for quantification,18 we found that Nlgn1 levels showed weak or no difference between the two groups. Similarly, in the present study, the different levels of Nlgns between the AD and the neurological control groups did not reach significance. Several previous studies by other groups showed that levels of NRXNs are changed in CSF of AD or MCI patients, although in different directions. Decreased levels in CSF of AD cases have been reported for NRXN-1α and -3α but not for NRXN-2α.40,42 NRXN-1α, -2α and -3α were found to be elevated in MCI, with NRXN-3α showing the best performance in separating the groups, but no difference was found between controls and AD patients.46 In another study, NRXN-3α, -2α and also Nlgn2, showed what has been described as a biphasic profile, with decreased levels in the preclinical stage and elevated levels in the MCI and dementia stages which could separate these groups from controls.44 The high dynamics of these proteins at the synapse, and the variability in the time and disease stage of the CSF sampling could account for these contradictory results. Also, in some of the described studies, both sample size and effect size were small.40,42,43,45 Synapses are probably not the only source of NRXN and Nlgn peptides in the CSF, as more and more studies show that also astrocytes express these proteins.62, 63, 64 The astrocytic expression of NRXNs and Nlgns might be considered as an additional source of variation for the levels of these proteins in the CSF.
NRXN and Nlgn proteins did not correlate with the cognitive decline scored with the MMSE test, which tend to indicate that these proteins do not reflect the synaptic alteration seen in AD. NRXNs and Nlgns moderately correlated with p-tau, while there was no correlation with Aβ42/40 ratio. The high correlations found with Aβ40 and Aβ42 are probably reflecting the common proteolytic processing affecting both APP and NRXNs and Nlgns at the synapse.26,38 With the other synaptic proteins GAP43 and neurogranin, NRXNs and Nlgns showed a moderate to weak correlation, respectively. This might indicate that these proteins reflect different events at the synapse, with both neurogranin and GAP43 being located intracellularly, whereas NRXNs and Nlgns are transmembrane proteins with large extracellular domains communicating with the extracellular environment.
NRXNs and Nlgns have been connected to AD pathogenesis both genetically and at the protein level. The NRXN-3 gene has been suggested to have a role in sporadic AD susceptibility, with a protective effect in males,65 while altered expression of the NRXN-3 was shown in post-mortem frontal gyrus AD brain and has been associated with AD-related changes.34,66 A frameshift mutation for Nlgn1 has been described in a familial case of AD, which abolished the ability of the protein to exert its function in promoting glutamatergic synapse formation.67 At the protein level, NRXNs and Nlgns are proteolytically processed by metalloproteases, which cleave their respective extracellular domain leaving the C-terminal fragment as a substrate for γ-secretase, which catalytic units are mutated in familial cases of AD.68 Therefore, we can hypothesize that other substrates of the protease complex, including NRXNs and Nlgns, are affected by its altered activity.69 Amyloid-β aggregation in plaques is one of the major hallmarks of AD and according to the amyloid cascade hypothesis accumulation of Aβ peptides is the initial event leading to AD-related changes.70 Among the different Aβ forms, Aβ oligomers (Aβo) have been described as the most synaptotoxic.71 Both NRXNs72 and Nlgns33,73 have been shown to interact with Aβo at the synapse, an interaction that disrupts their function and is described as one of the possible mechanisms of Aβo toxicity at the synapse. Taken together, these studies make NRXNs and Nlgns interesting candidate biomarkers for synaptic pathology in AD. Moreover, the selective differential distribution of Nlgns in excitatory (Nlgn1) and inhibitory (Nlgn2) synapses74,75 would allow the simultaneous monitoring of related events in those different synapses. However, results from this study indicate otherwise, suggesting that these proteins do not show a CSF biomarker potential for synaptic impairment related to AD. Yet, this does not preclude their potential as biomarkers for synapse dysfunction and loss in other neurological diseases. Indeed, genetic alteration of NRXNs76 and Nlgn2,77,78 -3 and -428,79 has been identified in schizophrenia, autism and bipolar disorders and described as a common biological pathway for the synaptic dysfunction aetiology seen in these disorders.80,81
The strength of this study lies in the large sample size of the cohort analysed and in the high specificity offered by the PRM method. In addition, straightforward sample preparation and the possibility of multiplexing without the need for antibodies represent an advantage. Further studies can be directed toward the investigation of endogenous peptides (i.e., not digested in vitro), which might convey more valuable biomarker information. A more comprehensive characterization of NRXN and Nlgn species present in CSF would probably help in understanding how these proteins are processed outside the CNS and possibly yield a wider range of peptides to investigate as possible biomarkers. However, a major obstacle to the investigation of NRXN and Nlgn proteins is the limited choice of antibodies available and the large size of the proteins themselves, which are excessively long, and thus difficult to analyse by MS without prior proteolytic digestion.
In conclusion, we developed a new robust and specific method for the simultaneous quantification of NRXN and Nlgn extracellular domain peptides in CSF. The possibility to measure multiple synaptic peptides in a panel assay provides a powerful tool to simplify the analysis of the complex biology of NRXNs and Nlgns in AD and other neurodegenerative diseases by detecting changes of those peptides in one assay, using one and the same sample. Further, longitudinal studies would be required to better describe if and how these proteins change in the CSF of patients with neurodegenerative diseases. Finally, this study provides the methodological groundwork to proceed on similar studies in mental disorders in which NRXNs and Nlgns have been shown to be involved (such as schizophrenia and autism), and possibly yield biomarkers for synapse pathology in those diseases.
Contributors
EC, JN, AV and GB designed the study. EC developed the method, performed the experiments, analysed the data and wrote the manuscript. JN and GB supervised during method development and the MS analysis. GB, KB and HZ were responsible for supervision, conceptualization, and verification of the underlying data. AV and CP provided the CSF samples of the clinical cohort. EC, CH participated in diagnosis of the patients and CSF samples collection. GB, BB, AB, HZ, KB contributed to the interpretation of the results and provided critical feedback of the manuscript. All authors have reviewed the manuscript.
Declaration of Competing Interest
HZ has served at scientific advisory boards for Eisai, Denali, Roche Diagnostics, Wave, Samumed, Siemens Healthineers, Pinteon Therapeutics, Nervgen, AZTherapies and CogRx, has given lectures in symposia sponsored by Cellectricon, Fujirebio, Alzecure and Biogen, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program. KB served as a consultant, at advisory boards, or at data monitoring committees for Abcam, Axon, Biogen, JOMDD/Shimadzu, Julius Clinical, Lilly, MagQu, Novartis, Prothena, Roche Diagnostics, and Siemens Healthineers. KB is a cofounder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program. CP is a member of the International Advisory Boards of Lilly; is a consultant for Fujiribio, Alzhois, Neuroimmune, Ads Neuroscience, Roche, AgenT, and Gilead; and is involved as an investigator in several clinical trials for Roche, Esai, Lilly, Biogen, Astra-Zeneca, Lundbeck, and Neuroimmune. The other authors declare no competing interests.
Acknowledgments
Acknowledgements
HZ is a Wallenberg Scholar supported by grants from the Swedish Research Council (#2018-02532), the European Research Council (#681712), Swedish State Support for Clinical Research (#ALFGBG-720931), the Alzheimer Drug Discovery Foundation (ADDF), USA (#201809-2016862), the AD Strategic Fund and the Alzheimer's Association (#ADSF-21-831376-C, #ADSF-21-831381-C and #ADSF-21-831377-C), the Olav Thon Foundation, the Erling-Persson Family Foundation, Stiftelsen för Gamla Tjänarinnor, Hjärnfonden, Sweden (#FO2019-0228), the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement (no.#860197) (MIRIADE), and the UK Dementia Research Institute at UCL. KB is supported by the Swedish Research Council (#2017-00915), the Alzheimer Drug Discovery Foundation (ADDF), USA (#RDAPB- 201809-2016615), the Swedish Alzheimer Foundation (#AF-742881), Hjärnfonden, Sweden (#FO2017-0243), the Swedish state under the agreement between the Swedish government and the County Councils, the ALF-agreement (#ALFGBG-715986), and European Union Joint Program for Neurodegenerative Disorders (JPND2019-466-236). GB is supported by the Swedish national infrastructure for biological mass spectrometry, BioMS. EC received support from Gamla tjänarinnor, Stohnes stiftelse, Demensfonden and Emil och Maria Palms stiftelse. AV is supported by Fondation Ophtalmologique Adolphe de Rothschild, Fondation Chatrier and AAIHP association. The funders had no role in data collection, analysis, or decision to publish.
Data sharing statement
Derived data supporting the findings of this study are available in the public, open access, Mendeley Data repository (https://data.mendeley.com/datasets/7sjrs6g6f9/1).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103793.
Appendix. Supplementary materials
References
- 1.Scheltens P, Blennow K, Breteler MMB, de Strooper B, Frisoni GB, Salloway S, et al. Alzheimer's disease. The Lancet. 2016;388(10043):505–517. doi: 10.1016/S0140-6736(15)01124-1. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe DJ. Alzheimer's Disease Is a Synaptic Failure. Science. 2002;298:789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- 3.Knobloch M, Mansuy IM. Dendritic spine loss and synaptic alterations in Alzheimer's disease. Mol Neurobiol. 2008;37(1):73–82. doi: 10.1007/s12035-008-8018-z. [DOI] [PubMed] [Google Scholar]
- 4.Vogels T, Murgoci AN, Hromadka T. Intersection of pathological tau and microglia at the synapse. Acta Neuropathol Commun. 2019;7(1):109. doi: 10.1186/s40478-019-0754-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardozo PL, de Lima IBQ, Maciel EMA, Silva NC, Dobransky T, Ribeiro FM. Synaptic Elimination in Neurological Disorders. Curr Neuropharmacol. 2019;17(11):1071–1095. doi: 10.2174/1570159X17666190603170511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taoufik E, Kouroupi G, Zygogianni O, Matsas R. Synaptic dysfunction in neurodegenerative and neurodevelopmental diseases: an overview of induced pluripotent stem-cell-based disease models. Open Biol. 2018;8(9) doi: 10.1098/rsob.180138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lepeta K, Lourenco MV, Schweitzer BC, Martino Adami PV, Banerjee P, Catuara-Solarz S, et al. Synaptopathies: synaptic dysfunction in neurological disorders - A review from students to students. J Neurochem. 2016;138(6):785–805. doi: 10.1111/jnc.13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheff SW, Neltner JH, Nelson PT. Is synaptic loss a unique hallmark of Alzheimer's disease? Biochem Pharmacol. 2014;88(4):517–528. doi: 10.1016/j.bcp.2013.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson JS, Witton J, Johnson JD, Ahmed Z, Ward M, Randall AD, et al. Altered Synapse Stability in the Early Stages of Tauopathy. Cell Rep. 2017;18(13):3063–3068. doi: 10.1016/j.celrep.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terry RD, Masliah E, Salmon DP, Butters N, DeTeresa R, Hill R, et al. Physical basis of cognitive alterations in Alzheimer's disease: synapse loss is the major correlate of cognitive impairment. Ann Neurol. 1991;30(4):572–580. doi: 10.1002/ana.410300410. [DOI] [PubMed] [Google Scholar]
- 11.Colom-Cadena M, Spires-Jones T, Zetterberg H, Blennow K, Caggiano A, DeKosky ST, et al. The clinical promise of biomarkers of synapse damage or loss in Alzheimer's disease. Alzheimers Res Ther. 2020;12(1):21. doi: 10.1186/s13195-020-00588-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheff SW, DA Price, Schmitt FA, Mufson EJ. Hippocampal synaptic loss in early Alzheimer's disease and mild cognitive impairment. Neurobiol Aging. 2006;27(10):1372–1384. doi: 10.1016/j.neurobiolaging.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Masliah E, Mallory M, Alford M, DeTeresa R, Hansen LA, McKeel DW, Jr., et al. Altered expression of synaptic proteins occurs early during progression of Alzheimer's disease. Neurology. 2001;56(1):127–129. doi: 10.1212/wnl.56.1.127. [DOI] [PubMed] [Google Scholar]
- 14.Hilton KJ, Cunningham C, Reynolds RA, Perry VH. Early Hippocampal Synaptic Loss Precedes Neuronal Loss and Associates with Early Behavioural Deficits in Three Distinct Strains of Prion Disease. PLoS One. 2013;8(6):e68062. doi: 10.1371/journal.pone.0068062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jack CR, Jr., Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer's disease. Alzheimers Dement. 2018;14(4):535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camporesi E, Nilsson J, Brinkmalm A, Becker B, Ashton NJ, Blennow K, et al. Fluid Biomarkers for Synaptic Dysfunction and Loss. Biomark Insights. 2020;15:1–17. doi: 10.1177/1177271920950319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tible M, Sandelius Å, Höglund K, Brinkmalm A, Cognat E, Dumurgier J, et al. Dissection of synaptic pathways through the CSF biomarkers for predicting Alzheimer disease. Neurology. 2020;95(8):e953–ee61. doi: 10.1212/WNL.0000000000010131. [DOI] [PubMed] [Google Scholar]
- 18.Camporesi E, Lashley T, Gobom J, Lantero-Rodriguez J, Hansson O, Zetterberg H, et al. Neuroligin-1 in brain and CSF of neurodegenerative disorders: investigation for synaptic biomarkers. Acta Neuropathol Commun. 2021;9(1):19. doi: 10.1186/s40478-021-01119-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dean C, Scholl FG, Choih J, DeMaria S, Berger J, Isacoff E, et al. Neurexin mediates the assembly of presynaptic terminals. Nat Neurosci. 2003;6(7):708–716. doi: 10.1038/nn1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nam CI, Chen L. Postsynaptic assembly induced by neurexin-neuroligin interaction and neurotransmitter. Proc Natl Acad Sci U S A. 2005;102(17):6137–6142. doi: 10.1073/pnas.0502038102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Craig AM, Kang Y. Neurexin-neuroligin signaling in synapse development. Curr Opin Neurobiol. 2007;17(1):43–52. doi: 10.1016/j.conb.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bang ML, Owczarek S. A matter of balance: role of neurexin and neuroligin at the synapse. Neurochem Res. 2013;38(6):1174–1189. doi: 10.1007/s11064-013-1029-9. [DOI] [PubMed] [Google Scholar]
- 23.Luo F, Sclip A, Jiang M, Südhof TC. Neurexins cluster Ca(2+) channels within the presynaptic active zone. EMBO J. 2020;39(7) doi: 10.15252/embj.2019103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dean C, Dresbach T. Neuroligins and neurexins: linking cell adhesion, synapse formation and cognitive function. Trends Neurosci. 2006;29(1):21–29. doi: 10.1016/j.tins.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Südhof TC. Neuroligins and Neurexins Link Synaptic Function to Cognitive Disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bemben MA, Shipman SL, Nicoll RA, Roche KW. The cellular and molecular landscape of neuroligins. Trends Neurosci. 2015;38(8):496–505. doi: 10.1016/j.tins.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marro SG, Chanda S, Yang N, Janas JA, Valperga G, Trotter J, et al. Neuroligin-4 Regulates Excitatory Synaptic Transmission in Human Neurons. Neuron. 2019;103(4):617–626. doi: 10.1016/j.neuron.2019.05.043. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cast TP, Boesch DJ, Smyth K, Shaw AE, Ghebrial M, Chanda S. An Autism-Associated Mutation Impairs Neuroligin-4 Glycosylation and Enhances Excitatory Synaptic Transmission in Human Neurons. J Neurosci. 2021;41(3):392–407. doi: 10.1523/JNEUROSCI.0404-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasem E, Kurihara T, Tabuchi K. Neurexins and neuropsychiatric disorders. Neurosci Res. 2018;127:53–60. doi: 10.1016/j.neures.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Sindi IA, Tannenberg RK, Dodd PR. Role for the neurexin-neuroligin complex in Alzheimer's disease. Neurobiol Aging. 2014;35(4):746–756. doi: 10.1016/j.neurobiolaging.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 31.Dinamarca MC, Rios JA, Inestrosa NC. Postsynaptic Receptors for Amyloid-beta Oligomers as Mediators of Neuronal Damage in Alzheimer's Disease. Front Physiol. 2012;3:464. doi: 10.3389/fphys.2012.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sindi IA, Dodd PR. New insights into Alzheimer's disease pathogenesis: the involvement of neuroligins in synaptic malfunction. Neurodegener Dis Manag. 2015;5(2):137–145. doi: 10.2217/nmt.14.54. [DOI] [PubMed] [Google Scholar]
- 33.Brito-Moreira J, Lourenco MV, Oliveira MM, Ribeiro FC, Ledo JH, Diniz LP, et al. Interaction of amyloid-beta (Abeta) oligomers with neurexin 2alpha and neuroligin 1 mediates synapse damage and memory loss in mice. J Biol Chem. 2017;292(18):7327–7337. doi: 10.1074/jbc.M116.761189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng JJ, Li WX, Liu JQ, Guo YC, Wang Q, Li GH, et al. Low expression of aging-related NRXN3 is associated with Alzheimer disease: A systematic review and meta-analysis. Medicine (Baltimore) 2018;97(28):e11343. doi: 10.1097/MD.0000000000011343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dufort-Gervais J, Provost C, Charbonneau L, Norris CM, Calon F, Mongrain V, et al. Neuroligin-1 is altered in the hippocampus of Alzheimer's disease patients and mouse models, and modulates the toxicity of amyloid-beta oligomers. Sci Rep. 2020;10(1):6956. doi: 10.1038/s41598-020-63255-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bemben MA, Nguyen TA, Li Y, Wang T, Nicoll RA, Roche KW. Isoform-specific cleavage of neuroligin-3 reduces synapse strength. Mol Psychiatry. 2019;24(1):145–160. doi: 10.1038/s41380-018-0242-y. [DOI] [PubMed] [Google Scholar]
- 37.Restituito S, Khatri L, Ninan I, Mathews PM, Liu X, Weinberg RJ, et al. Synaptic autoregulation by metalloproteases and gamma-secretase. J Neurosci. 2011;31(34):12083–12093. doi: 10.1523/JNEUROSCI.2513-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Servián-Morilla E, Robles-Lanuza E, Sánchez-Hidalgo AC, Camacho-Garcia RJ, Paez-Gomez JA, Mavillard F, et al. Proteolytic Processing of Neurexins by Presenilins Sustains Synaptic Vesicle Release. J Neurosci. 2018;38(4):901–917. doi: 10.1523/JNEUROSCI.1357-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saura CA, Servián-Morilla E, Scholl FG. Presenilin/γ-secretase regulates neurexin processing at synapses. PLoS One. 2011;6(4):e19430. doi: 10.1371/journal.pone.0019430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brinkmalm G, Sjödin S, Simonsen AH, Hasselbalch SG, Zetterberg H, Brinkmalm A, et al. A Parallel Reaction Monitoring Mass Spectrometric Method for Analysis of Potential CSF Biomarkers for Alzheimer's Disease. Proteomics Clin Appl. 2018;12(1) doi: 10.1002/prca.201700131. [DOI] [PubMed] [Google Scholar]
- 41.Spellman DS, Wildsmith KR, Honigberg LA, Tuefferd M, Baker D, Raghavan N, et al. Development and evaluation of a multiplexed mass spectrometry based assay for measuring candidate peptide biomarkers in Alzheimer's Disease Neuroimaging Initiative (ADNI) CSF. Proteomics Clin Appl. 2015;9(7-8):715–731. doi: 10.1002/prca.201400178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hölttä M, Minthon L, Hansson O, Holmén-Larsson J, Pike I, Ward M, et al. An integrated workflow for multiplex CSF proteomics and peptidomics-identification of candidate cerebrospinal fluid biomarkers of Alzheimer's disease. J Proteome Res. 2015;14(2):654–663. doi: 10.1021/pr501076j. [DOI] [PubMed] [Google Scholar]
- 43.Abdi F, Quinn JF, Jankovic J, McIntosh M, Leverenz JB, Peskind E, et al. Detection of biomarkers with a multiplex quantitative proteomic platform in cerebrospinal fluid of patients with neurodegenerative disorders. J Alzheimers Dis. 2006;9(3):293–348. doi: 10.3233/jad-2006-9309. [DOI] [PubMed] [Google Scholar]
- 44.Lleo A, Nunez-Llaves R, Alcolea D, Chiva C, Balateu-Panos D, Colom-Cadena M, et al. Changes in Synaptic Proteins Precede Neurodegeneration Markers in Preclinical Alzheimer's Disease Cerebrospinal Fluid. Mol Cell Proteomics. 2019;18(3):546–560. doi: 10.1074/mcp.RA118.001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sathe G, Na CH, Renuse S, Madugundu AK, Albert M, Moghekar A, et al. Quantitative Proteomic Profiling of Cerebrospinal Fluid to Identify Candidate Biomarkers for Alzheimer's Disease. Proteomics Clin Appl. 2019;13(4) doi: 10.1002/prca.201800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duits FH, Brinkmalm G, Teunissen CE, Brinkmalm A, Scheltens P, Van der Flier WM, et al. Synaptic proteins in CSF as potential novel biomarkers for prognosis in prodromal Alzheimer's disease. Alzheimers Res Ther. 2018;10(1):5. doi: 10.1186/s13195-017-0335-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keshavan A, Pannee J, Karikari TK, Rodriguez JL, Ashton NJ, Nicholas JM, et al. Population-based blood screening for preclinical Alzheimer's disease in a British birth cohort at age 70. Brain. 2021 doi: 10.1093/brain/awaa403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dubois B, Feldman HH, Jacova C, Hampel H, Molinuevo JL, Blennow K, et al. Advancing research diagnostic criteria for Alzheimer's disease: the IWG-2 criteria. The Lancet Neurology. 2014;13(6):614–629. doi: 10.1016/S1474-4422(14)70090-0. [DOI] [PubMed] [Google Scholar]
- 49.Jack CR, Jr., Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, Research Framework NIA-AA, et al. Toward a biological definition of Alzheimer's disease. Alzheimers Dement. 2018;14(4):535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, et al. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7(3):270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sachdev P, Kalaria R, O'Brien J, Skoog I, Alladi S, Black SE, et al. Diagnostic criteria for vascular cognitive disorders: a VASCOG statement. Alzheimer Dis Assoc Disord. 2014;28(3):206–218. doi: 10.1097/WAD.0000000000000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKeith IG, Boeve BF, Dickson DW, Halliday G, Taylor JP, Weintraub D, et al. Diagnosis and management of dementia with Lewy bodies: Fourth consensus report of the DLB Consortium. Neurology. 2017;89(1):88–100. doi: 10.1212/WNL.0000000000004058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rascovsky K, Hodges JR, Knopman D, Mendez MF, Kramer JH, Neuhaus J, et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134(Pt 9):2456–2477. doi: 10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cognat E, Koehl B, Lilamand M, Goutagny S, Belbachir A, de Charentenay L, et al. Preventing Post-Lumbar Puncture Headache. Ann Emerg Med. 2021 doi: 10.1016/j.annemergmed.2021.02.019. [DOI] [PubMed] [Google Scholar]
- 55.Engelborghs S, Niemantsverdriet E, Struyfs H, Blennow K, Brouns R, Comabella M, et al. Consensus guidelines for lumbar puncture in patients with neurological diseases. Alzheimers Dement (Amst) 2017;8:111–126. doi: 10.1016/j.dadm.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leitão MJ, Silva-Spínola A, Santana I, Olmedo V, Nadal A, Le Bastard N, et al. Clinical validation of the Lumipulse G cerebrospinal fluid assays for routine diagnosis of Alzheimer's disease. Alzheimers Res Ther. 2019;11(1):91. doi: 10.1186/s13195-019-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sandelius Å, Portelius E, Källén Å, Zetterberg H, Rot U, Olsson B, et al. Elevated CSF GAP-43 is Alzheimer's disease specific and associated with tau and amyloid pathology. Alzheimers Dement. 2019;15(1):55–64. doi: 10.1016/j.jalz.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Portelius E, Olsson B, Hoglund K, Cullen NC, Kvartsberg H, Andreasson U, et al. Cerebrospinal fluid neurogranin concentration in neurodegeneration: relation to clinical phenotypes and neuropathology. Acta Neuropathol. 2018;136(3):363–376. doi: 10.1007/s00401-018-1851-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pino LK, Searle BC, Bollinger JG, Nunn B, MacLean B, MacCoss MJ. The Skyline ecosystem: Informatics for quantitative mass spectrometry proteomics. Mass Spectrom Rev. 2020;39(3):229–244. doi: 10.1002/mas.21540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gu H, Liu G, Wang J, Aubry A-F, Arnold ME. Selecting the Correct Weighting Factors for Linear and Quadratic Calibration Curves with Least-Squares Regression Algorithm in Bioanalytical LC-MS/MS Assays and Impacts of Using Incorrect Weighting Factors on Curve Stability, Data Quality, and Assay Performance. Anal Chem. 2014;86(18):8959–8966. doi: 10.1021/ac5018265. [DOI] [PubMed] [Google Scholar]
- 61.Schreiner D, Simicevic J, Ahrné E, Schmidt A, Scheiffele P. Quantitative isoform-profiling of highly diversified recognition molecules. eLife. 2015;4:e07794. doi: 10.7554/eLife.07794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stogsdill JA, Ramirez J, Liu D, Kim YH, Baldwin KT, Enustun E, et al. Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature. 2017;551(7679):192–197. doi: 10.1038/nature24638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sakers K, Eroglu C. Control of neural development and function by glial neuroligins. Curr Opin Neurobiol. 2019;57:163–170. doi: 10.1016/j.conb.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trotter JH, Dargaei Z, Wöhr M, Liakath-Ali K, Raju K, Essayan-Perez S, et al. Astrocytic Neurexin-1 Orchestrates Functional Synapse Assembly. bioRxiv. 2020:2020.08.21.262097.
- 65.Martinez-Mir A, González-Pérez A, Gayán J, Antúnez C, Marín J, Boada M, et al. Genetic study of neurexin and neuroligin genes in Alzheimer's disease. J Alzheimers Dis. 2013;35(2):403–412. doi: 10.3233/JAD-122257. [DOI] [PubMed] [Google Scholar]
- 66.Hishimoto A, Pletnikova O, Lang DL, Troncoso JC, Egan JM, Liu QR. Neurexin 3 transmembrane and soluble isoform expression and splicing haplotype are associated with neuron inflammasome and Alzheimer's disease. Alzheimers Res Ther. 2019;11(1):28. doi: 10.1186/s13195-019-0475-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tristan-Clavijo E, Camacho-Garcia RJ, Robles-Lanuza E, Ruiz A, van der Zee J, Van Broeckhoven C, et al. A truncating mutation in Alzheimer's disease inactivates neuroligin-1 synaptic function. Neurobiol Aging. 2015;36(12):3171–3175. doi: 10.1016/j.neurobiolaging.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 68.Dorszewska J, Prendecki M, Oczkowska A, Dezor M, Kozubski W. Molecular Basis of Familial and Sporadic Alzheimer's Disease. Curr Alzheimer Res. 2016;13(9):952–963. doi: 10.2174/1567205013666160314150501. [DOI] [PubMed] [Google Scholar]
- 69.Bot N, Schweizer C, Ben Halima S, Fraering PC. Processing of the synaptic cell adhesion molecule neurexin-3beta by Alzheimer disease alpha- and gamma-secretases. J Biol Chem. 2011;286(4):2762–2773. doi: 10.1074/jbc.M110.142521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer's disease at 25 years. EMBO Mol Med. 2016;8(6):595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ding Y, Zhao J, Zhang X, Wang S, Viola KL, Chow FE, et al. Amyloid Beta Oligomers Target to Extracellular and Intracellular Neuronal Synaptic Proteins in Alzheimer's Disease. Front Neurol. 2019;10:1140. doi: 10.3389/fneur.2019.01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Naito Y, Tanabe Y, Lee AK, Hamel E, Takahashi H. Amyloid-β Oligomers Interact with Neurexin and Diminish Neurexin-mediated Excitatory Presynaptic Organization. Sci Rep. 2017;7:42548. doi: 10.1038/srep42548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dinamarca MC, Di Luca M, Godoy JA, Inestrosa NC. The soluble extracellular fragment of neuroligin-1 targets Abeta oligomers to the postsynaptic region of excitatory synapses. Biochem Biophys Res Commun. 2015;466(1):66–71. doi: 10.1016/j.bbrc.2015.08.107. [DOI] [PubMed] [Google Scholar]
- 74.Chih B, Engelman H, Scheiffele P. Control of excitatory and inhibitory synapse formation by neuroligins. Science. 2005;307(5713):1324–1328. doi: 10.1126/science.1107470. [DOI] [PubMed] [Google Scholar]
- 75.Poulopoulos A, Aramuni G, Meyer G, Soykan T, Hoon M, Papadopoulos T, et al. Neuroligin 2 drives postsynaptic assembly at perisomatic inhibitory synapses through gephyrin and collybistin. Neuron. 2009;63(5):628–642. doi: 10.1016/j.neuron.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 76.Tromp A, Mowry B, Giacomotto J. Neurexins in autism and schizophrenia-a review of patient mutations, mouse models and potential future directions. Mol Psychiatry. 2020 doi: 10.1038/s41380-020-00944-8. [DOI] [PubMed] [Google Scholar]
- 77.Jiang DY, Wu Z, Forsyth CT, Hu Y, Yee SP, Chen G. GABAergic deficits and schizophrenia-like behaviors in a mouse model carrying patient-derived neuroligin-2 R215H mutation. Mol Brain. 2018;11(1):31. doi: 10.1186/s13041-018-0375-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kathuria A, Lopez-Lengowski K, Watmuff B, McPhie D, Cohen BM, Karmacharya R. Synaptic deficits in iPSC-derived cortical interneurons in schizophrenia are mediated by NLGN2 and rescued by N-acetylcysteine. Transl Psychiatry. 2019;9(1):321. doi: 10.1038/s41398-019-0660-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maćkowiak M, Mordalska P, Wędzony K. Neuroligins, synapse balance and neuropsychiatric disorders. Pharmacol Rep. 2014;66(5):830–835. doi: 10.1016/j.pharep.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 80.Calabrese F, Riva MA, Molteni R. Synaptic alterations associated with depression and schizophrenia: potential as a therapeutic target. Expert Opin Ther Targets. 2016;20(10):1195–1207. doi: 10.1080/14728222.2016.1188080. [DOI] [PubMed] [Google Scholar]
- 81.Carroll LS, Owen MJ. Genetic overlap between autism, schizophrenia and bipolar disorder. Genome Med. 2009;1(10):102. doi: 10.1186/gm102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.