Abstract
Objective
The mortality rate for critically ill COVID-19 cases was more than 80%. Nonetheless, research about the effect of common respiratory diseases on critically ill COVID-19 expression and outcomes is scarce.
Design
We performed proteomic analyses on airway mucus obtained by bronchoscopy from patients with severe COVID-19, or induced sputum from patients with chronic obstructive pulmonary disease (COPD), asthma, and healthy controls.
Results
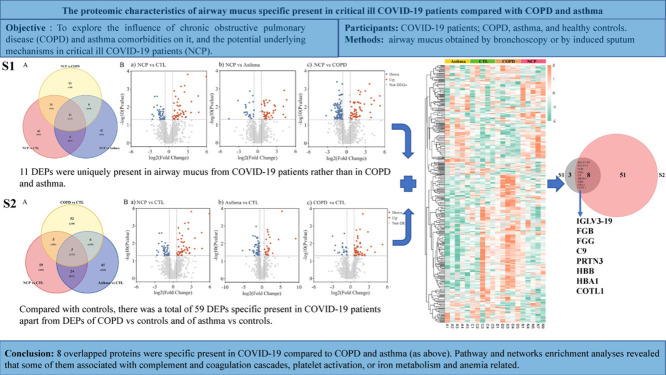
Of the total identified and quantified proteins, 445 differentially expressed proteins (DEPs) were found in different comparison groups. In comparison with COPD, asthma, and controls, 11 proteins were uniquely present in COVID-19 patients. Apart from DEPs associated with COPD versus controls and asthma versus controls, there was a total of 59 DEPs specific to COVID-19 patients. Finally, the findings revealed that there were 8 overlapping proteins in COVID-19 patients, including C9, FGB, FGG, PRTN3, HBB, HBA1, IGLV3-19, and COTL1. Functional analyses revealed that most of them were associated with complement and coagulation cascades, platelet activation, or iron metabolism, and anemia-related pathways.
Conclusions
This study provides fundamental data for identifying COVID-19–specific proteomic changes in comparison with COPD and asthma, which may suggest molecular targets for specialized therapy.
Keywords: Critically ill COVID-19, Proteomic sequencing, Airway mucus, COPD, Asthma
Graphical abstract
1. Introduction
COVID-19, which is caused by SARS-CoV-2, is a threat to global health and health care systems. Currently, the disease is spreading rapidly around the world. According to the World Health Organization's situation report for June 9, 2021, there had been more than 174,801,871 confirmed COVID-19 cases and approximately 3,756,350 COVID-19 related deaths worldwide. In addition, the report revealed that the global severity rate of COVID-19 ranges between 5% to 20%, with the rates varying from region to region. For example, in New York, 1,151 patients (20%) were diagnosed with severe COVID-19 and required mechanical ventilation (Richardson et al, 2020). In Italy, the proportion of intensive care unit (ICU) admissions was between 5% and 12% of the total COVID-19 cases (Livingston and Bucher, 2020). According to the Chinese Center for Disease Control and Prevention, 19% of COVID-19 patients developed severe or critical illness, in a study encompassing 44,415 COVID-19 cases (Wu and McGoogan, 2020). Surprisingly, the mortality rate of critically ill COVID-19 cases was more than 80% (Yang et al, 2020).
To date, there are still gaps in the mechanistic understanding of the disease process as reported by (Bhaskaran et al., 2022). For instance, data about the biochemical and molecular alterations associated with the severe form of COVID-19 are scarce. In addition, there is evidence that chronic respiratory diseases, including chronic obstructive pulmonary disease (COPD) and asthma, may predispose patients to SARS-CoV-2 infection. Nevertheless, the effects of COPD and asthma on disease expression and outcomes, as well as the potential underlying processes, are poorly investigated in COVID-19 patients.
The formation of mucus plugs has been observed in critically ill COVID-19 patients. Clinical findings show that the mucus plugs cause airway obstruction and respiratory failure in a significant proportion of affected patients (Lu et al, 2021, Zhang et al, 2021). In this study, it was speculated that this mucus is a mixture of secretions produced by airway and alveolar epithelial cells in response to viruses and inflammatory mediators, and the molecular changes may be indicative of the pathological changes of COVID-19. A previous study reported that COPD and asthma were associated with severe illness in COVID-19 patients (Gao et al, 2021). In this study, proteomic analyses of airway mucus from severe COVID-19, chronic obstructive pulmonary disease (COPD), and asthma patients were performed. The study contributes fundamental information to the understanding of the pathogenesis of critically ill COVID-19 patients and their associated comorbidities, which can be used to develop future targeted therapeutic approaches.
2. Material and Methods
2.1. Study design and clinical data collection
Five critically ill COVID-19 patients were diagnosed with laboratory-confirmed SARS-CoV-2 infection by the local health authorities. COVID-19 patients were classified into subgroups based on their different clinical manifestations using the Chinese Government Diagnosis and Treatment Guideline (Trial Seventh Version). Severe patients were characterized by respiratory distress and a respiratory rate ≥30 times/min, which corresponds to an oxygen saturation ≤93% in resting state or arterial blood oxygen partial pressure (PaO2)/oxygen concentration (FiO2) ≤300 mm Hg (1 mm Hg = 0.133 kPa). Patients classified as critically ill were those who had respiratory failure requiring mechanical ventilation, experienced shock, or required ICU care. The COPD inclusion and exclusion criteria were adapted as previously described (Lu et al, 2016). Asthma was defined according to Global Strategy for Asthma Management and Prevention 2018 (Bateman et al., 2018). The change in forced expiratory volume in 1 second (FEV1) was used as a diagnostic tool. An increase in FEV1, in response to bronchodilator reversibility (ΔFEV1BDR) following inhalation of 400 µg salbutamol, was considered significant if it was ≥12% and ≥200 mL compared with the initial FEV1.
Five participants who were negative for the SARS-CoV-2 nucleic acid test without any lung disease were included as healthy controls. Meanwhile, 5 COPD patients and 5 asthma patients were designated as disease controls. To aspirate the airway mucus, the critically ill COVID-19 patients presenting with expectoration difficulty and dyspnea underwent bronchoscopy using a PENTAX FB-15BS portable fiber bronchoscope (PENTAX Medical Shanghai Co, Ltd, Shanghai, China) via tracheal intubation. Airway mucus in COPD, asthma, and healthy control participants was induced using hypertonic (3%) saline solution inhalation administered via an ultrasonic nebulizer.
Clinical charts, nursing records, laboratory findings, and chest imaging of the COVID-19 patients were reviewed from January 26, 2020, to February 15, 2020. Electronic medical records were used to acquire epidemiological, clinical, laboratory, and radiological data. Two researchers independently reviewed the data collection forms to ensure that the collected data was accurate. All the procedures were approved by the Ethics Committee of the First Affiliated Hospital of Guangzhou Medical University (No.2020-65). Although informed consent was obtained from all participants, it was waived for COVID-19 patients because their family members were quarantined.
2.2. Airway mucus processing
The processing of airway mucus was conducted as previously described (Wang et al, 2019). Two independent physicians who were blind to clinical data performed the procedures. Supplementary Material 1 provides more information on airway mucus processing.
2.3. Protein extraction and trypsin digestion
Airway mucus processing was performed as previously described (Zhang et al, 2021). Supplementary Material 2 provides more information on protein extraction and trypsin digestion.
2.4. Quantification of proteomic data and liquid chromatography with tandem mass spectrometry analysis
Proteomic data were quantified and analyzed as previously described (Zhang et al, 2021). For label-free quantification, protein expression levels were estimated using the Intensity Based Absolute Quantification (iBAQ) algorithm embedded in MaxQuant (Schwanhausser et al, 2011). Detailed information is provided in Supplementary Material 3.
The peptides were subjected to the nanospray ionization (NSI) source followed by tandem mass spectrometry (MS/MS) in Q ExactiveTM Plus (Thermo Fisher Scientific), which was connected online to the Ultra-performance liquid chromatography (UPLC). Peptides were selected for MS/MS analysis using an normalized collision energy (NCE) setting of 28, and the fragments were detected in the Orbitrap at a resolution of 17,500. A principal components analysis (PCA) was performed to visualize the separation of COVID-19 patients, COPD, asthma, and healthy controls.
2.5. Differential expression/pathway analysis
Differential gene expression analysis was performed in R (v3.2.0) using the empirical Bayesian algorithm in the limma package. Up-regulated and down-regulated genes were defined using a fold-change of ≥1.5 or ≤0.67 and a P value <0.05. The cutoff value for fold-change was set at 1.2. The Gene Ontology (GO) annotation proteome was constructed using data from the UniProt-GOA database (http://www.ebi.ac. uk/GOA). The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to identify the enriched pathways. Further hierarchical clustering based on the functional classification of differentially expressed proteins (DEPs) was visualized using the “heatmap.2” function from the “gplots” in R-package. More information about pathway analysis is provided in Supplementary Material 4.
2.6. Statistical analysis
Continuous variables were presented as median (IQR). Categorical variables were presented as a percentage (%) of the total sample (n). All analyses were performed using the GraphPad Prism 5 software, and 2-sided P values. Statistical significance was set at a P value <0.05.
3. Results
3.1. Clinical characteristics of participants
The clinical characteristics of COVID-19 patients, asthma, COPD, and healthy controls are shown in Table 1 . There was no significant difference in baseline characteristics (age, sex, and smoking status) between COVID-19, asthma, COPD, and healthy controls. In all COVID-19 patients, laboratory findings revealed characteristic clinical outcomes of SARS-CoV-2 infection, which were almost identical to those reported in previous studies.
Table 1.
Demographic, clinical, laboratory and radiographic findings of patients
| COVID-19 | Asthma | COPD | Healthy controls | |
|---|---|---|---|---|
| N = 5 | N = 5 | N = 5 | N = 5 | |
| Demographics and clinical characteristics | ||||
| Age, years | 70 (66-72) | 69.6 (65-79) | 68 (57-80) | 69 (63-75) |
| Male | 5 (100.0) | 5 (100.0) | 5 (100.0) | 5 (40.0) |
| Death | 0 | 0 | 0 | 0 |
| ICU admission | 5 (100.0) | 0 | 0 | 0 |
| ICU length of stay, days | 37 (10-43) | — | — | — |
| Hospital length of stay, days | 45 (41-48) | — | — | — |
| Time from illness onset to | 57 (53-68) | — | — | — |
| hospital admission, days | — | — | ||
| Severe | 5 (100.0) | 0 | 0 | 0 |
| Ever smoke | 4 (80.0) | 5 (100.0) | 5 (100.0) | 5 (40.0) |
| ARDS comorbidity | 5 (100.0) | 0 | 0 | 0 |
| Respiratory rate | 20 (14-20) | — | — | — |
| > 24 breaths per minute | 1 (20.0) | — | — | — |
| Pulse ≥100 beats per minute | 1 (20.0) | — | — | — |
| O2 pressure | 82.8 (69.0-110.0) | — | — | — |
| O2 concentration | 95.3 (93.3-95.4) | — | — | — |
| Fever (temperature ≥37.3°C) | 1 (20%) | 0 | 0 | 0 |
| Cough | 4 (80.0) | 5 (100.0) | 5 (100.0) | 0 |
| Sputum | 0 | 5 (100.0) | 5 (100.0) | 0 |
| Myalgia | 0 | 0 | 0 | 0 |
| Fatigue | 2 (40.0) | 0 | 0 | 0 |
| Diarrhea | 0 | 0 | 0 | 0 |
| Vomiting | 0 | 0 | 0 | 0 |
| Rhinobyon | 0 | 4 (80.0) | 2 (40.0) | 0 |
| Hemoptysis | 0 | 0 | 0 | 0 |
| Headache | 0 | 2 (40.0) | 1 (20.0) | 0 |
| Sore throat | 1 (20.0) | 4 (80.0) | 4 (80.0) | 0 |
| Polypnea | 5 (100.0) | 4 (80.0) | 4 (80.0) | 0 |
| Shiver | 0 | 0 | 0 | 0 |
| White blood cell count, × 10⁹/L | 11.1 (7.30-12.8) | 9.2±2.1 | 9.6±3.8 | — |
| Lymphocyte count, × 10⁹/L | 0.30 (0.25-0.55) | 1.5±0.67 | 1.6±0.83 | — |
| Monocyte count, × 10⁹/L | 0.40 (0.35-0.65) | 0.63±0.13 | 0.6±0.16 | — |
| Platelet count, × 10⁹/L | 117.0 (87.0-212.5) | 221±35 | 226±32 | — |
| Lactate dehydrogenase, U/L | 397 (356-535) | 191±23 | 183±19 | — |
| High-sensitivity cardiac | 0.01 (0.005-0.03) | 0.01±0.01 | 0.01±0.01 | — |
| troponin I, pg/mL | ||||
| Prothrombin time, s | 15.7 (13.6-18.1) | — | — | — |
| D-dimer, μg/mL | 1.390 (0.741-4.667) | — | — | — |
| IL-6, pg/mL | 22.2 (9.40-60.0) | — | — | — |
| Procalcitonin, ng/mL | 0.27 (0.09-0.43) | — | — | — |
| CRP, | 2.7 (1.5-12.9) | — | — | — |
| DBIL | 4.1 (3.0-8.7) | — | — | — |
| TBIL | 13.6 (11.9-20.4) | — | — | — |
| CK-MB | 11.0 (7.0-18.0) | — | — | — |
| Cr | 77.0 (69.1-91.7) | — | — | — |
| Imaging features | ||||
| Consolidation | 5 (100.0) | 0 | 0 | 0 |
| Ground-glass opacity | 5 (100.0) | 1 (20.0) | 1 (20.0) | 0 |
| Bilateral pulmonary infiltration | 5 (100.0) | 0 | 1 (20.0) | 0 |
Data are presented as median (IQR), mean ± SD, or n (%).
3.2. Proteomic profiling of airway mucus from all participants
Airway mucus samples were obtained from critically ill COVID-19 patients, asthma, COPD, and healthy control participants. Label-free quantification of proteomic (PTM Biolabs) was used to analyze airway mucus from each participant. The airway mucus from COVID-19 patients exhibited distinct proteomic patterns compared with asthma, COPD, and healthy controls. Of note, 91 DEPs were identified between COVID-19 and healthy controls, 78 between asthma and healthy controls, 66 between COPD and healthy controls, 69 between COVID-19 and asthma, and 143 between COVID-19 and COPD, as shown in Figure S1A. There were 2,257, 2,169, 2,093, and 2,175 proteins identified and quantified in the airway mucus of COVID-19 patients, asthmatic patients, COPD patients, and healthy controls, respectively (Figure S1B). The proteomics data sets (including fold-change and P values for the 2 groups’ comparisons) are provided in Table S1-S3. PCA, the median relative SD (RSD) of all internal standards in each sample, protein mass and coverage distribution, and protein sequence distribution were calculated as part of the quality control analysis (Figure S1C-F). The data of the current study were collected with a high degree of consistency and reproducibility. Figure S2-S3 depicts a heatmap, GO enrichment analysis, and KEGG pathway analysis for each proteomics data set.
3.3. Identification and enrichment analyses of COVID-19 specific proteins
3.3.1. Comparisons in COVID-19 versus controls, COVID-19 versus COPD, and COVID -19 versus asthma (method 1)
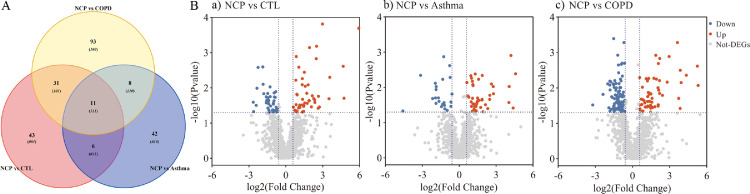
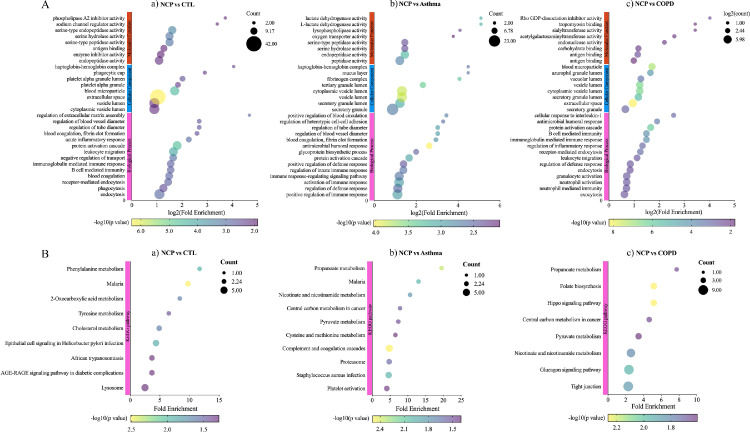
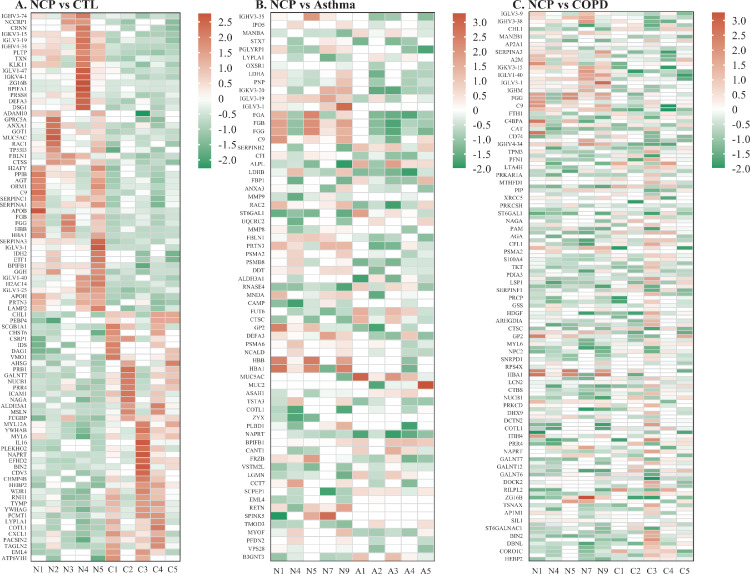
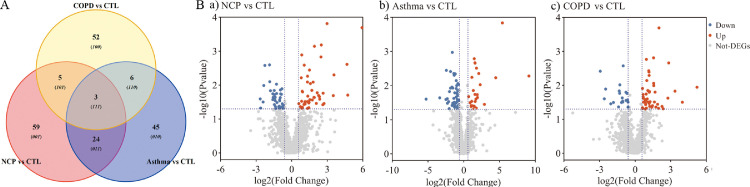
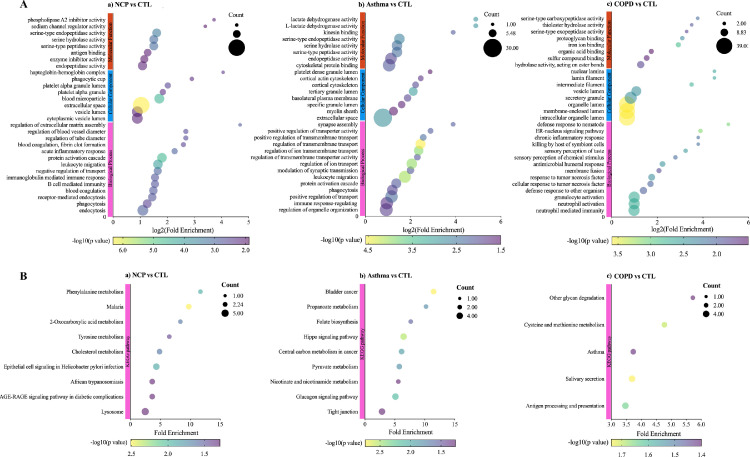
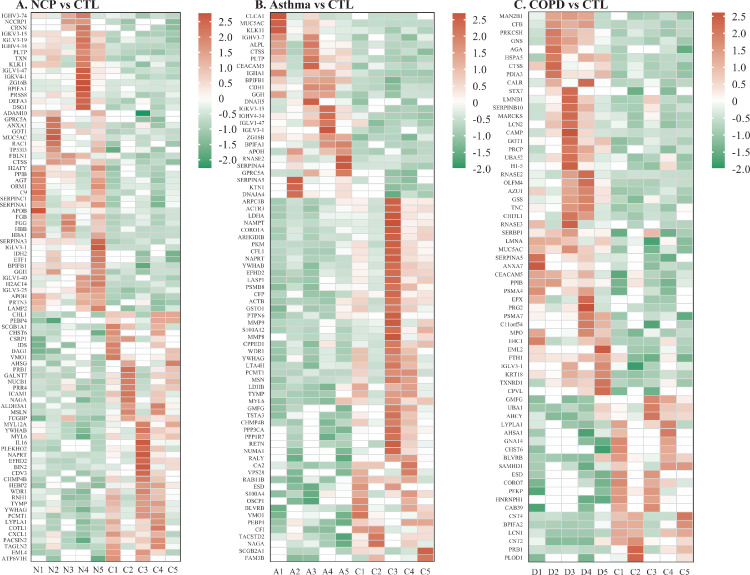
When COVID-19 was compared with healthy controls, Venn diagrams and volcano plots (Figure 1 ) indicated 91 dysregulated DEPs (50 up-regulated and 41 down-regulated). GO enrichment analysis showed that the significantly altered molecular function terms were enriched in serine-type peptidase activity, serine-type endopeptidase activity, and serine hydrolase activity. The biological process terms are mainly comprised protein activation cascade and leukocyte migration. Most of the proteins were in the extracellular space and blood microparticles (Figure 2 Aa). KEGG pathway analysis demonstrated that there were 2 pathways enriched in phenylalanine metabolism and 2-oxocarboxylic acid metabolism (Figure 2Ba), whereas all the DEPs are presented in a heatmap (Figure 3 A).
Figure 1.
Protein analysis shows proteins unique to COVID-19 patients (11 overlap proteins): A. Venn plot showing identification of the COVID-19 specific proteins among COVID-19 versus controls, COVID-19 versus asthma, and COVID-19 versus COPD; B. Volcano plot, a. COVID-19 versus controls; b. COVID-19 versus asthma; c. COVID-19 versus COPD. Blue: down-regulated proteins; red: up-regulated proteins. NCP;novel coronavirus pneumonia. COPD, chronic obstructive pulmonary disease.
Figure 2.
Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of differentiated expressed proteins (DEPs). A. GO annotation for biological processes, cellular compartments, and molecular function; B. KEGG enrichment analysis. a. COVID-19 versus controls; b. COVID-19 versus asthma; c. COVID-19 versus COPD. NCP; COPD, chronic obstructive pulmonary disease.
Figure 3.
Heatmap showing the differentiated expressed proteins (DEPs). A. COVID-19 versus controls; B. COVID-19 versus asthma; C. COVID-19 versus COPD. NCP; COPD, chronic obstructive pulmonary disease.
When COVID-19 was compared with asthma, Venn and volcano plots (Figure 1) showed that there were 46 up-regulated and 46 down-regulated DEPs. The GO enrichment analysis revealed significant changes in molecular function terms such as serine-type peptidase activity, serine hydrolase activity, and (serine-type) endopeptidase activity. Significantly altered biological process terms included protein activation cascade, antimicrobial humoral response, immune response, and regulation of defense response. Most of them were located in the vesicle lumen and granule lumen (Figure 2Ab). The KEGG pathway analysis showed that these DEPs were significantly enriched in complement and coagulation cascades as well as in propanoate metabolism (Figures 2Bb and 3B).
The comparison between COVID-19 and COPD groups showed the presence of 143 DEPs (Figure 1) in the mucus obtained from COVID-19 patients, including 56 up-regulated and 87 down-regulated proteins. The GO functional enrichment analysis revealed that protein activation cascade, antimicrobial humoral response, cellular response to interleukin-1 (IL-1), immunoglobulin mediated immune response, B cell-mediated immunity, regulation of inflammatory response, and receptor-mediated response were all enriched. Most of these proteins were in the extracellular space, vesicle lumen, and the vacuolar lumen. The molecular functions of these proteins were primarily distributed among 4-function processes: acetylgalactosaminyl transferase activity, endonuclease activity, carbohydrate-binding, and actin-binding (Figure 2Ac). According to the KEGG pathway analysis, these DEPs were significantly enriched in the folate biosynthesis, hippo signaling pathway, glucagon signaling pathway, and tight junction (Figures 2Bc and 3C).
3.3.2. Screening of COVID-19 specific proteins based on method 1
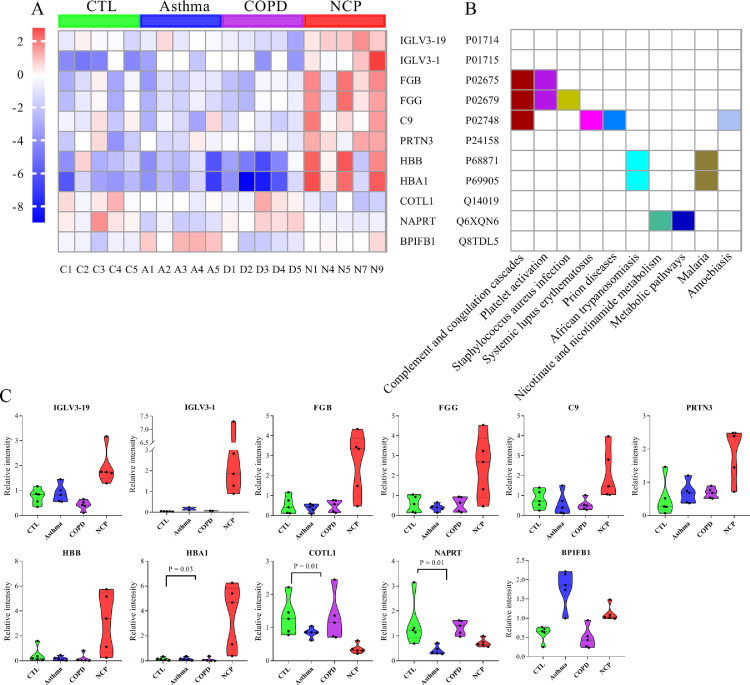
A total of 11 overlapped DEPs were identified in COVID-19 patients. They were discovered from the intersection of COVID-19 versus controls, COVID-19 versus asthma, and COVID-19 versus COPD. As illustrated in Figure 4 A-B, pathway and network enrichment analyses revealed that these intersecting DEPs were primarily associated with complement and coagulation cascades, platelet activation, Staphylococcus aureus infection, nicotinate, and nicotinamide metabolism, and metabolic pathways. According to the differential significance levels, the COVID-19 specific proteins were IGLV3-19, IGLV3-1, FGB, FGG, C9, PRTN3, HBB, HBA1, COTL1, NAPRT, and BPIFB1 (Figure 4C).
Figure 4.
Analysis of differentiated expressed proteins (DEPs) between different groups. A. Hierarchical clustering analysis of DEPs. A heatmap showing the top 11 DEPs. The red and the blue colors in the heatmap denote higher gene expression and lower gene expression, respectively. Target protein symbols for the top 11 DEPs are included; B. These dysregulated proteins are enriched in 2 pathways: complement and coagulation cascades pathways and platelet activation (both of which contain DEPs at a high frequency); C. The change in expression level (original value) of the 11 selected proteins with significance is indicated by the P value. COVID-19, NCP; COPD, chronic obstructive pulmonary disease.
3.3.3. Comparisons between COVID-19 versus controls, COPD versus controls, and asthma versus controls (method 2)
A comparison between COVID-19 patients and controls revealed 91 DEPs as previously reported (Figure 5 , Figure 6 Aa, 6Ba, and Figure 7 A). For asthma versus controls, 78 DEPs were significantly expressed, with 27 being up-regulated (Figure 5 and Figure 7B). GO enrichment analysis was performed to annotate the putative functional implications of these differently grouped DEPs. The results revealed that (L-) lactate dehydrogenase activity was enriched. In addition, most of these proteins were in the extracellular space and the tertiary granule lumen. The molecular function of these proteins was primarily distributed among 3 function processes: regulation of (ion) transmembrane transport, regulation of ion transport, and leukocyte migration (Figure 6Ab). KEGG pathway analysis revealed that these DEPs were significantly enriched in the hippo signaling pathway and glucagon signaling pathway (Figure 6Bb).
Figure 5.
Analysis of differentially expressed proteins (DEPs) between different groups. A. Venn plot, identification of the common proteins among COVID-19 versus healthy controls, COPD versus healthy controls, and asthma versus healthy controls; B. Volcano plot. a. COVID-19 versus healthy controls; b. COPD versus healthy controls; c. Asthma versus healthy controls. Blue: down-regulated proteins; red: up-regulated proteins. NCP; COPD, chronic obstructive pulmonary disease.
Figure 6.
Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of differentiated expressed proteins (DEPs). A. GO annotation for biological process, cellular compartment, and molecular function, respectively; B. KEGG enrichment analysis. a. COVID-19 versus healthy controls; b. COPD versus healthy controls; c. Asthma versus healthy controls. NCP; COPD, chronic obstructive pulmonary disease.
Figure 7.
Heatmap analysis of differentiated expressed proteins (DEPs). A. COVID-19 versus healthy controls; B. Asthma versus healthy controls; C. COPD versus healthy controls. NCP; COPD, chronic obstructive pulmonary disease.
There were 66 DEPs found in COPD versus controls, with 46 up-regulated and 20 down-regulated proteins (Figure 5 and Figure 7C). GO enrichment analysis showed that the significantly altered molecular function terms were enriched in iron ion binding and proteoglycan binding. The biological process terms comprised granulocyte/neutrophil activation, neutrophil-mediated immunity, response to tumor necrosis factor, and antimicrobial humoral response. Most of these proteins were found within the organelle/membrane-enclosed/intracellular organelle lumen (Figure 6Ac). KEGG pathway analysis revealed that there were 2 pathways enriched in salivary secretion, cysteine and methionine metabolism, antigen processing, and presentation (Figure 6Bc).
3.3.4. Screening of COVID-19 specific proteins according to method 2
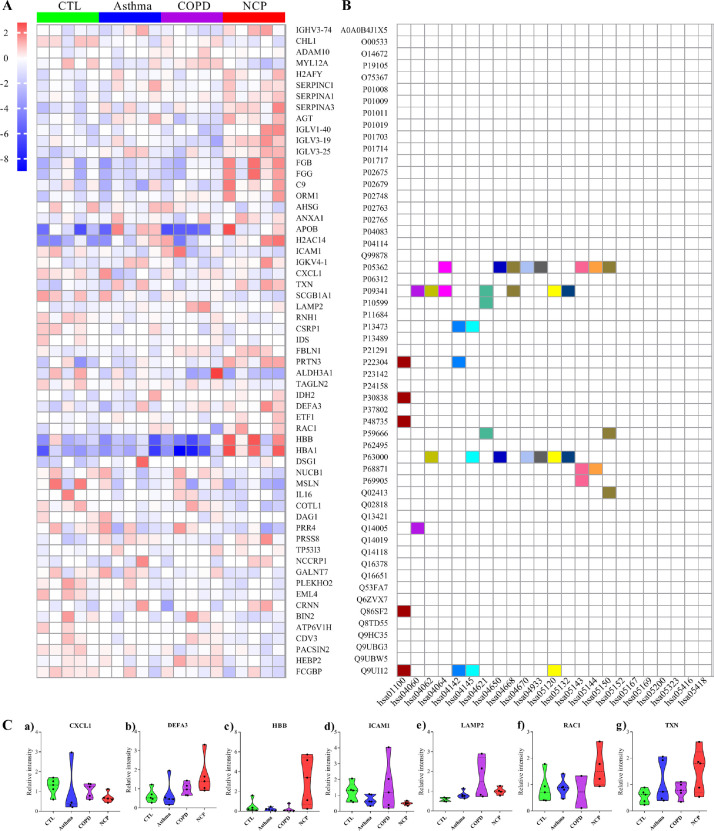
There were 59 DEPs detected in the mucus of COVID-19 patients compared with controls, excluding any DEPs detected in COPD versus controls or asthma versus controls. As indicated in Figure 8 A, pathway and network enrichment analysis revealed that the intersected DEPs were largely associated with metabolic pathways, lysosome, phagosome, and NOD-like receptor signaling pathways. The selected proteins included CXCL1, DEFA3, HBB, ICAM1, LAMP2, RAC1, and TXN, and were chosen because they were present in at least 2 pathways at a high frequency (Figure 8B).
Figure 8.
Analysis of differentiated expressed proteins (DEPs) between different groups. A. Hierarchical clustering analysis of DEPs. Heatmap of the top 59 DEPs. The red color in the heatmap denotes higher gene expression, and the blue color in the heatmap denotes lower gene expression. Target proteins symbols for the 59 DEPs are included; B. Pathways enrichment; C. The expression level change (original value) of the 7 selected proteins with significance is indicated by the P value. NCP; COPD, chronic obstructive pulmonary disease.
3.3.5. Screening of final COVID-19 specific proteins
COVID-19 patients’ specific proteins were defined as the intersection of specific DEPs in COVID-19 samples compared with healthy/disease controls (COVID-19 vs controls, COVID-19 vs COPD, and COVID-19 vs asthma). Simultaneously, any DEPs found in COPD versus controls or asthma versus controls were excluded from the analysis. For example, the filtered COVID-19 specific proteins were differentially expressed between COVID-19 and controls but not between COPD and controls or asthma and controls.
Finally, as determined by the 2 aforementioned techniques, the 8 overlapping differential proteins specific to COVID-19 patients were identified, including FGB, FGG, C9, PRTN3, HBB, HBA1, IGLV3-19, and COTL1 (Figure S4).
4. Discussion
The COVID-19 pandemic is a major threat to public health and the social-economic well-being of people globally. There is currently no effective treatment strategy to prevent the death of severely ill COVID-19 patients. Therefore, any lead to the discovery of therapeutic drug targets for critically ill COVID-19 patients is vital. In this study, compared with asthma and COPD, proteomic sequencing identified 8 key characteristics of the proteomic changes associated with hospitalized patients seriously infected with SARS-CoV-2.
Around 20% to 51% of COVID-19 patients were associated with at least 1 comorbidity (Guan et al, 2020b, Huang et al, 2020). The 3 most prevalent comorbidities were hypertension, diabetes, and coronary heart disease, with frequencies ratios of 10%-30%, 10%-20%, and 7%-15%, respectively (Guan et al, 2020a, Wang et al, 2020, Zhou et al, 2020), which contributed to poorer clinical outcomes. It is reported that chronic respiratory disorders, including COPD and asthma, may predispose patients to SARS-CoV-2 infection (Guan et al, 2020b, Huang et al, 2020). Alternatively, the poor recognition by the general population and the lack of spirometric testing may result in the under-diagnosis of respiratory diseases (Guan et al, 2020a). For instance, it was reported that the frequencies of COVID-19 with COPD were 1.5% to 5% (Grasselli et al, 2020, Zhang et al, 2020) and for asthma 0% to 12.5%.18 Evidence suggests that the intrinsic pathophysiological features of COPD and asthma may modify the response to severe SARS-CoV-2 infection made possible by ACE2 expression (Song et al, 2021). Therefore, it is necessary to understand the effects of SARS-CoV-2 on unique proteomic changes compared with COPD and asthma, which may imply further research of molecular targets directed at specific therapy.
In this study, the 8 overlapped differential specific proteins were found in COVID-19 cases after intersecting. There was up-regulation of proteins, including FGB, FGG, C9, PRTN3, HBB, HBA1, and IGLV3-19, and down-regulation of COTL1 proteins in COVID-19 patients compared with the other groups. Pathway and network enrichment analysis revealed that the DEPs were mostly associated with complement and coagulation cascades, platelet activation pathways, or iron metabolism and anemia- related pathways. In the present study, an elevated complement system protein C9 was identified. It is reported that the complement system plays an important role in linking innate and adaptive immunity and that inflammation could further aggravate lung injury. Complement activation is detected cumulatively in conditions such as Acute respiratory distress syndrome (ARDS), pneumonia, asthma, pulmonary arterial hypertension, and COPD (Sarma et al, 2006). Evidence suggests that suppression of complement system protein C9 appears to be effective immunotherapy for the SARS-infected mouse model (Gralinski et al, 2018). In addition, FGB and FGG are crucial for blood clot formation (coagulation), and this study revealed that the 2 proteins were up-regulated. Previous proteomic study of plasma exosomes demonstrated that FGG and FGB levels were significantly higher in the malignant pulmonary nodules group than in the benign group (Kuang et al, 2019). FGB and FGG were 2 of the key epithelial-mesenchymal transition effectors associated with cell adhesion and cellular communication in lung cancer. Therefore, we indicate that critically ill COVID-19 patients may benefit from the suppression of the complement and coagulation systems.
Iron metabolism and anemia may play pivotal roles in multiple organ dysfunction syndromes in COVID-19. The hemoglobin proteins (HBB, HBA1, and HBA2) combine to form the adult hemoglobin molecule (HbA), which is a heterotetramer of 2 α and two β-globin chains. The dysregulated hemoglobin proteins result in an imbalanced globin chain synthesis and consequently impaired erythropoiesis. The severity of COVID-19 is heavily influenced by the degree of chain imbalance. Survival is dependent on regular blood transfusions in the worst-case scenario, which results in transfusional iron overload and secondary multi-organ damage due to iron toxicity. Understanding the relationship between HBB and HBA1 proteins and the severity of COVID-19 and whether these associations differ by age, sex, and the presence of chronic conditions is critical in the management of COVID-19.
Mucus is an integral part of respiratory physiology. It protects the respiratory tract by forming a physical barrier to inhaled allergens and pathogens. This study established that mucus accumulation contributed to recurrent airway infection, resulting in further obstruction. The inflammatory cytokine storm greatly contributes to the more serious clinical manifestations and worse outcomes in COVID-19 patients. It is particularly potent in accumulating mucus because it initiates many inflammatory cascades associated with mucus production. Numerous studies have demonstrated that the SARS-CoV-2 infection can result in an allergic reaction in the respiratory tract mucosa, which activates mucin secretion and modulates its chemical structure to enable the virus to enter the cells (Khan et al, 2021). Mucus accumulation can contribute to worse comorbidities indicated in COVID-19 patients, such as venous engorgement and pulmonary edema. Thus, it is important to understand the proteomic expression and functional changes of mucus to develop new therapeutic approaches.
In addition, this retrospective study identified several risk factors for COVID-19 patients. For example, increased levels of white blood cell count, D-dimer, blood IL-6, and lactate dehydrogenase, as well as lymphocytopenia, were all observed in severely ill COVID-19 patients. These risk factors were associated with COVID-19 outcomes and corroborated previously published studies (Zhang et al, 2021). In this study, there were no significant differences in age, gender, and smoking status among COVID-19, asthma, COPD, and healthy controls.
Our study has some limitations. First, the airway mucus obtained from COVID-19 patients using bronchoscopy may be a mixture of secretions produced by airway and alveolar epithelial cells in response to the virus and inflammatory mediators. In contrast, induced sputum was used for COPD, asthma, and control participants, all of whom may have variable content and sputum, cell count. Second, because the study design was retrospective, laboratory tests may have been underestimated in the medical records analyzed, making it difficult to investigate the effect on outcomes. Third, information on medications, disease control status, and phenotypes of diseases before admissions was incomplete. Furthermore, the effect of these factors on the risk of SASR-CoV-2 infection and disease expression needs further exploration. Finally, the sample size was relatively small. Prospect studies on a larger population should be conducted.
5. Conclusion
Airway mucus proteomic databases are highly valuable resources for elucidating the host proteomic changes associated with severe SARS-CoV-2 infection. This study analyzed proteins from COVID-19 patients, COPD, asthma, and controls to identify the unique proteomic molecular signatures associated with SARS-CoV-2 infection. This study contributes to our understanding of the pathological changes associated with COVID-19 and forms the basis for the development of potential therapeutic strategies.
Acknowledgments
Ethical approval and consent to participate
All the procedures were approved by the Ethics Committee of the First Affiliated Hospital of Guangzhou Medical University (No. 2020-65). Verbal informed consent was obtained from all participants because the family members were in quarantine.
Authors’ contributions
Wenju Lu, Lidong Liu, and Xiaoqing Liu conceived and designed the experiments. Fei Liu, Qiongqiong Li, and Yuanyuan Li conducted the sample preparation. Zili Zhang, Fanjie Lin, and Xinguang Wei conducted the data and bioinformatics analyses. Zili Zhang wrote the manuscripts. Zhanbei Zhu, Hua Guo, Wei Liu, Yaowei Fang, and Xinguang Wei collected and analyzed the clinical data. Wenju Lu oversaw the completion of this study and edited the manuscript.
Competing interests
The authors have no conflict of interest to declare.
Funding
This study was supported by grants 2016YFC0903700 and 2018YFC1311900 from the National Key R&D Program of China, grant 81520108001 from the National Natural Science Foundation of China, grant 1201620007 from the Guangzhou Department of Education, grant 2017BT01S155 from the Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program, grants 2020A1515010076 and 2021A1515011346 from the Guangdong Natural Science Foundation, and grants OP-201808 and OP-201912 from the Research Projects of State Key Laboratory of Respiratory Disease. It was also supported by grants ZNSA-2020001 and ZNSA-2021018 from the Zhongnanshan Medical Foundation of Guangdong Province and a grant specifically for COVID-19 study from the Guangzhou Institute of Respiratory Health.
Acknowledgements
We sincerely thank all the health care providers who are fighting this public health crisis and all the patients who participated in the study. We extend our heartfelt sympathy and condolences to the victims and bereaved families.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2022.01.008.
Appendix. Supplementary materials
References
- Bateman E.D., Hurd S.S., Barnes P.J., Bousquet J., Drazen J.M., FitzGerald J.M., et al. Global strategy for asthma management and prevention: GINA executive summary. Eur Respir J. 2018;31:143–178. doi: 10.1183/09031936.00138707. Eur Respir J 2018;51(2) [DOI] [PubMed] [Google Scholar]
- Bhaskaran K, Rentsch CT, Hickman G, Hulme WJ, Schultze A, Curtis HJ, et al. Overall and cause-specific hospitalisation and death after COVID-19 hospitalisation in England: A cohort study using linked primary care, secondary care, and death registration data in the OpenSAFELY platform. PLoS Med. 2022;19(1):e1003871. doi: 10.1371/journal.pmed.1003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YD, Ding M, Dong X, Zhang JJ, Kursat Azkur A, Azkur D, et al. Risk factors for severe and critically ill COVID-19 patients: A review. Allergy. 2021;76(2):428–455. doi: 10.1111/all.14657. [DOI] [PubMed] [Google Scholar]
- Gralinski LE, Sheahan TP, Morrison TE, Menachery VD, Jensen K, Leist SR, et al. Complement Activation Contributes to Severe Acute Respiratory Syndrome Coronavirus Pathogenesis. mBio. 2018;9(5) doi: 10.1128/mBio.01753-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasselli G, Zangrillo A, Zanella A, Antonelli M, Cabrini L, Castelli A, et al. Baseline Characteristics and Outcomes of 1591 Patients Infected With SARS-CoV-2 Admitted to ICUs of the Lombardy Region, Italy. JAMA. 2020;323(16):1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan WJ, Liang WH, Zhao Y, Liang HR, Chen ZS, Li YM, et al. Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur Respir J. 2020;55(5) doi: 10.1183/13993003.00547-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Khan ZA, Charles M, Pratap P, Naeem A, Siddiqui Z, et al. Cytokine Storm and Mucus Hypersecretion in COVID-19: Review of Mechanisms. J Inflamm Res. 2021;14:175–189. doi: 10.2147/JIR.S271292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang M, Peng Y, Tao X, Zhou Z, Mao H, Zhuge L, et al. FGB and FGG derived from plasma exosomes as potential biomarkers to distinguish benign from malignant pulmonary nodules. Clin Exp Med. 2019;19(4):557–564. doi: 10.1007/s10238-019-00581-8. [DOI] [PubMed] [Google Scholar]
- Livingston E, Bucher K. Coronavirus Disease 2019 (COVID-19) in Italy. JAMA. 2020;323(14):1335. doi: 10.1001/jama.2020.4344. [DOI] [PubMed] [Google Scholar]
- Lu W, Liu X, Wang T, Liu F, Zhu A, Lin Y, et al. Elevated MUC1 and MUC5AC mucin protein levels in airway mucus of critical ill COVID-19 patients. J Med Virol. 2021;93(2):582–584. doi: 10.1002/jmv.26406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Zheng Z, Chen X, Tan H, Wang J, Zhang Z, et al. Study Design and Interim Outcomes of Guangzhou Institute of Respiratory Disease COPD Biobank. COPD. 2016;13(2):203–213. doi: 10.3109/15412555.2015.1069807. [DOI] [PubMed] [Google Scholar]
- Richardson S, Hirsch JS, Narasimhan M, Crawford JM, McGinn T, Davidson KW, et al. Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA. 2020;323(20):2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma VJ, Huber-Lang M, Ward PA. Complement in lung disease. Autoimmunity. 2006;39(5):387–394. doi: 10.1080/08916930600739456. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Song J, Zeng M, Wang H, Qin C, Hou HY, Sun ZY, et al. Distinct effects of asthma and COPD comorbidity on disease expression and outcome in patients with COVID-19. Allergy. 2021;76(2):483–496. doi: 10.1111/all.14517. [DOI] [PubMed] [Google Scholar]
- Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Liang Z, Yang Y, Zhou L, Guan L, Wu W, et al. Reproducibility of fluid-phase measurements in PBS-treated sputum supernatant of healthy and stable COPD subjects. Int J Chron Obstruct Pulmon Dis. 2019;14:835–852. doi: 10.2147/COPD.S187661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, McGoogan JM. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JJ, Dong X, Cao YY, Yuan YD, Yang YB, Yan YQ, et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020;75(7):1730–1741. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wang T, Liu F, Zhu A, Gu G, Luo J, et al. The proteomic characteristics of airway mucus from critical ill COVID-19 patients. Life Sci. 2021;269 doi: 10.1016/j.lfs.2021.119046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.