Abstract
Plants have evolved sophisticated defense systems to enhance drought tolerance. These include the microRNA (miRNA) group of small noncoding RNAs that act as post‐transcriptional regulators; however, details of the mechanisms by which they confer drought tolerance are not well understood. Here, we show that osa‐MIR171f, a member of osa‐MIR171 gene family, is mainly expressed in response to drought stress and regulates the transcript levels of SCARECROW‐LIKE6‐I (SCL6‐I) and SCL6‐II in rice ( Oryza sativa ). The SCL6 genes are known to be involved in shoot branching and flag leaf morphology. Osa‐MIR171f‐overexpressing (osa‐MIR171f‐OE) transgenic plants showed reduced drought symptoms compared with non‐transgenic (NT) control plants under both field drought and polyethylene glycol (PEG)‐mediated dehydration stress conditions. Transcriptome analysis of osa‐MIR171f‐OE plants and osa‐mir171f‐knockout (K/O) lines generated by clustered regularly interspaced short palindromic repeats (CRISPR/Cas9) revealed that osa‐mature‐miR171a‐f (osa‐miR171) regulates the expression of flavonoid biosynthesis genes, consequently leading to drought tolerance. This upregulation in the osa‐MIR171f‐OE plants, which did not occur in NT control plants, was observed under both normal and drought conditions. Our findings indicate that osa‐miR171 plays a role in drought tolerance by regulating SCL6‐I and SCL6‐II transcript levels.
Keywords: drought, flavonoids, miR171, Osa‐miR171f, Rice, SCL6
1. INTRODUCTION
Drought is a common environmental condition and can profoundly affect plant growth and development (Mittler, 2006; Peet & Kramer, 1980). Accordingly, plants have developed mechanisms to cope with drought stress (Devi et al., 2017; Fu & Dong, 2013; Heil & Bostock, 2002) that have been extensively studied due to the economic implications of reduced growth and yield on different crop species (Chung et al., 2016; Joshi et al., 2016; Vakilian, 2020). Yield reduction by drought is due to various factors, including reduction of photosynthesis efficiency, disturbance of assimilates allocation, and poor leaf development. Specially, drought stress at the pre‐anthesis stage is critical in grain filling and yield (Fahad et al., 2017).
One mechanism that is known to be involved in drought stress responses is the regulation of gene expression by microRNAs (miRNAs). A variety of miRNAs induced or repressed in response to drought stress and regulated its target genes to cope with drought stress (Sunkar et al., 2012; Wang et al., 2010; Yoshida et al., 2014). These are small (20–24 nucleotides) noncoding RNAs that act as negative post‐transcriptional regulators to modulate gene expression (Lee et al., 1993; Mallory & Vaucheret, 2004; Ramachandran & Chen, 2008; Reinhart et al., 2002). Mature‐miRNAs (miRNA) are processed from stem‐loop precursor miRNAs (pre‐miRNA), derived from primary miRNAs (pri‐miRNA) by RNase III‐type endonucleases, such as Dicer (Reinhart et al., 2002; Voinnet, 2009). This involves mRNA degradation where the mature‐miRNA is incorporated into the RNA‐induced silencing complex (RISC), which guides Argonaute (AGO) to cleave the target mRNA through base‐pairing (Bartel, 2009; Filipowicz et al., 2008; Jones‐Rhoades et al., 2006). The miRNA target sites are highly conserved (Axtell, 2008; Axtell & Bowman, 2008; Chorostecki et al., 2012) and so can be predicted, as has been reported in Arabidopsis thaliana and rice (O. sativa) (Jones‐Rhoades & Bartel, 2004; Rhoades et al., 2002; Wang et al., 2004).
In A. thaliana, ath‐miR171c regulates the GAI, RGA, and SCR (GRAS) transcription factor family members SCARECROW‐LIKE6‐II (SCL6‐II), SCL6‐III, and SCL6‐IV (also known as HAIRY MERISTEM [HAM] or LOST MERISTEMS [LOM]). Ath‐MIR171c overexpression leads to reduced shoot branching and increased plant height (Wang et al., 2010), consistent with the known roles of GRAS members in shoot, root, and chlorophyll development via regulation of gibberellic acid (GA)‐auxin signaling (Bolle, 2004; Huang et al., 2017). In rice, osa‐miR171c‐mediated regulation of OsHAM1 (SCL6‐II), OsHAM2, OsHAM3, and OsHAM4 (SCL6‐I) affects the heading date and leaf morphology (Fan et al., 2015), and in tomato (Solanum lycopersicum), Sly‐miR171 overexpressing plants were found to be taller and to show earlier phase transition and flowering time than wild‐type plants. Many studies have shown miRNA involvement in drought responses by transcriptional profiling (Chung et al., 2016; Ferdous et al., 2015; Zhou et al., 2010), and in rice, miR159, miR169, miR395, and miR474 transcripts are generally up‐regulated by drought stress, whereas miR156, miR168, miR170, miR172, miR396, miR397, and miR408 transcripts are generally down‐regulated (Khraiwesh et al., 2012; Zhou et al., 2010). However, within families of miRNAs, there can be different responses by individual members to drought stress. For example, in A. thaliana pre‐miR171b transcript levels were observed to increase in response to drought stress, while pre‐miR171a transcript levels decreased (Liu et al., 2008; Sunkar & Zhu, 2004). Similarly, in rice, pre‐miR171f transcript levels were reported to increase following drought stress, whereas the osa‐pre‐miR171a transcript abundance decreased (Chung et al., 2016). Thus, the exact miRNA‐based mechanisms controlling the drought response have yet to be clearly elucidated.
A diverse range of factors are involved in, and induced, by drought stress, including the content of flavonoid pigments. Flavonoids are secondary metabolites (Harborne & Williams, 2000) that mitigate against oxidative damage that can occur through accumulation of reactive oxygen species (ROS) caused by abiotic stress, such as drought (Catalá et al., 2011; Koops et al., 2011; Kusano et al., 2011; Saito et al., 2013; Stracke et al., 2010). The metabolic pathway of flavonoid has been well defined, and core biosynthetic enzymes are identified (Galland et al., 2014; Shih et al., 2008). The expression of the corresponding genes is induced by drought stress in plants (Gai et al., 2020; Sharma et al., 2019; Wang et al., 2020).
In this current study, we investigated the role of osa‐miR171 in SCL6‐I (OsHAM4) and SCL6‐II (OsHAM1) regulation of the drought response in rice, using phenotypic and transcriptome analysis of osa‐MIR171f‐overexpressing and knockout plants generated by Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9. We describe an association of this regulatory process with the expression of flavonoid biosynthesis genes and propose a model in which osa‐mature‐microRNA171a‐f (osa‐miR171) derived from osa‐pre‐miR171f regulates flavonoid biosynthesis as a response to drought using its SCL6‐I and SCL6‐II targets.
2. RESULTS
2.1. Drought stress induces expression of osa‐pre‐miR171f and osa‐miR171
Previous studies showed that the precursor miR171f (pre‐miR171f) accumulates under drought stress in rice (Chung et al., 2016). To investigate its function more broadly, we analyzed its expression pattern in response to a range of abiotic stresses (Figure 1a,b). We found that osa‐pre‐miR171f transcript levels increased in shoots following desiccation stress and abscisic acid (ABA) treatment (Figure 1a) and by high salt treatment, cold stress, and ABA treatment in roots (Figure 1b). We also analyzed osa‐mature‐microRNA171a‐f (osa‐miR171) expression in seedling roots and shoots and in the shoots of 4‐week‐old plants (Figure 1c,d). In the seedlings, osa‐miR171 transcript levels increased in roots during desiccation stress but were not detected in shoots under the same experimental conditions. However, osa‐miR171 transcript levels did increase in shoots in response to the drought treatment in the 4‐week‐old plants (Figure 1d). Previous our RNA seq data showed that osa‐MIR171e and f were expressed dominantly in rice and other osa‐MIR171 members were hard to detect. Besides, osa‐MIR171e expression was down‐regulated, and miR171f expression was up‐regulated response to drought treatment (Chung et al., 2016). Thus, the expression patterns of osa‐pre‐miR171f dominantly affected osa‐miR171 level under drought stress, suggesting that among members of the osa‐MIR171 gene family osa‐MIR171f shows primarily association with drought responses.
FIGURE 1.
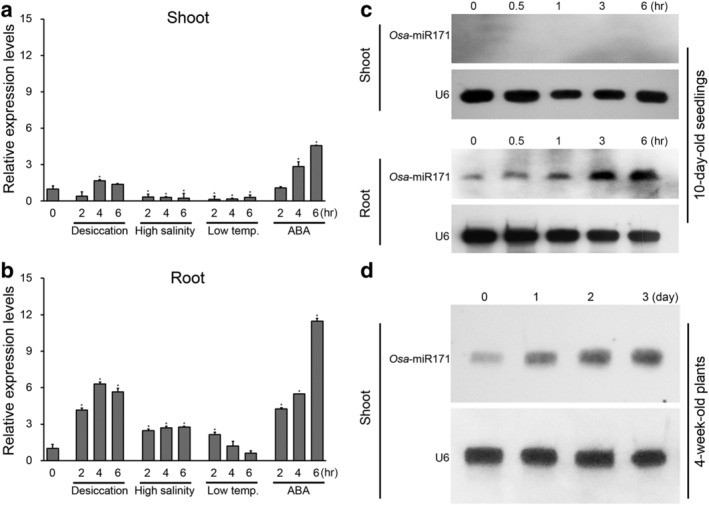
Analysis of osa‐pre‐miR171f and osa‐mature‐microRNA171a‐f (osa‐miR171) response to abiotic stress in rice. (a and b) Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analysis showing changes in osa‐pre‐miR171f expression levels in response to different stresses in shoots (a) and roots (b). Control plants were not treated with drought stress (0 h time). Error bars indicate SD of triplicate measurements. OsTubA2 and OsUbi1 were used as internal controls, and relative expression levels are shown as fold values. Asterisks indicate statistically significant differences between the corresponding samples and their control (p value < .01, Student's t test). ABA, abscisic acid. (c and d) osa‐miR171 expression levels in seedlings (c) and young plants (d) under drought stress. The expression level was assessed by Northern blot analysis. U6 transcript levels show equal loading of RNA. Total RNA was prepared from the 10‐day‐old and 4‐week‐old plants exposed to stresses for the indicated time periods
2.2. Overexpression of osa‐MIR171f promotes drought tolerance
To understand the function of osa‐MIR171f in drought stress, we generated transgenic rice lines overexpressing osa‐MIR171f under the control of the constitutive GOS2 promoter (Figure S1a) (Pater et al., 1992). We identified four independent transgenic lines (lines 2, 3, 5, and 8) with osa‐pre‐miR171f transcript levels using reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) assays (Figure S1b) and analyzed the expression levels of osa‐mature‐microRNA171a‐f (osa‐miR171) in these transgenic lines using Northern blot analysis (Figure 2a). To investigate the drought resistance of these osa‐MIR171f overexpressing (OE) plants, we compared their phenotypes to non‐transgenic plants (NT) in response to polyethylene glycol (PEG)‐mediated dehydration stress. The leaf‐rolling of 10‐day‐old osa‐MIR171f ‐OE plants clearly decreased compared to that of NT plants in the 22.5% PEG‐8000 (m/v) treatment (Figure S1c). This result indicated that osa‐MIR171f ‐OE plants are more tolerant of dehydration stress than NT plants. To investigate the effect of osa‐MIR171f loss of function during drought stress, we then measured osa‐miR171 expression levels in a osa‐mir171f ‐knockout mutant (line 2) generated using a CRISPR/Cas9 system (Chung et al., 2020) (Figures 2a and S2).
FIGURE 2.
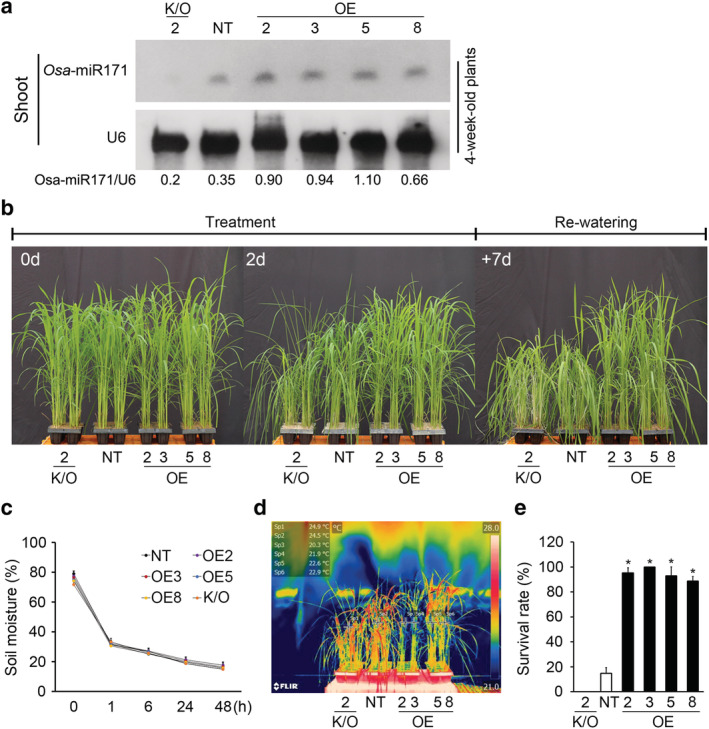
Analysis of osa‐MIR171f‐overexpressing transgenic plants and knockout mutants in response to drought stress. (a) Expression levels of osa‐mature‐microRNA171a‐f (osa‐miR171) in osa‐MIR171f‐overexpressing (OE) transgenic plants and knockout (K/O) mutants. The expression level was assessed by Northern blot analysis. U6 transcript levels show equal loading of RNA. Total RNA was prepared from the 4‐week‐old plants. Osa‐miR171/U6 value was calculated by band image analysis. (b) The phenotype of osa‐MIR171f‐OE plants and K/O mutants during drought stress. Four‐week‐old plants from four independent lines of osa‐MIR171f‐OE plants (2, 3, 5, 8), the K/O mutant (2), and non‐transgenic (NT) controls were exposed to drought stress for 2 days, followed by re‐watering for 7 days. (c) Measurement of soil moisture contents (mV). Data represent mean value ± SD of 10 measurements performed in different regions of the soil. (d), Thermal imaging of osa‐MIR171f‐OE plants (2, 3, 5, 8), K/O mutants (2), and NT plants. Different leaf temperatures are expressed as different colors: lower in blue and higher in red. (e) The survival rate of osa‐MIR171f‐OE plants (2, 3, 5, 8), K/O mutants (2), and NT plants 7 days after re‐watering. Data represent mean value + SD (n = 30)
We next characterized 4‐week‐old osa‐MIR171f‐OE plants and knockout (K/O) mutants grown under drought conditions in a greenhouse (Figure 2b–d). During the drought treatments, soil moisture content decreased uniformly over time (Figure 2c). Osa‐MIR171f‐OE, K/O, and non‐transgenic (NT) plants all showed induction of stress damage symptoms, such as wilting and leaf‐rolling (Figure 2b); however, the osa‐MIR171f‐OE plants recovered better during re‐watering for up to 7 days than did the K/O and NT plants. Notably, the survival rates of the osa‐MIR171f‐OE plants ranged from 85% to 98%, whereas the osa‐mir171f‐K/O and NT plants showed no signs of recovery (Figure 2b,e).
In general, plants with less water content have higher temperatures and emit more heat and infra‐red radiation, and this can be measured using infra‐red (IR) thermography, where a red color indicates a higher temperature, lower water content, and less stomatal conductance (Kim et al., 2020). Using this technique, leaves of osa‐MIR171f‐OE plants were observed to have lower temperatures (20.3–22.9°C) than those of NT and K/O plants, which had average temperature of 24.5°C and 24.9°C, respectively (Figure 2d). These results indicated that overexpressing osa‐MIR171f in rice enhances drought resistance during the vegetative stage compared to wild‐type plants.
2.3. Identification of osa‐miR171 target genes during drought stress
To further determine the basis of the enhanced drought tolerance in osa‐MIR171f overexpressing (OE) plants, we analyzed osa‐mature‐miR171a‐f (osa‐miR171) target mRNA in response to drought stress. To predict osa‐pre‐miR171f target genes and the associated mature miRNA‐target pair alignments, we used psRNATarget software (https://plantgrn.noble.org/psRNATarget/analysis) (Figure S3a). The SCL6‐I, SCL6‐II, ARC3, and DEPG10 transcripts were found to contain complementary sequences to osa‐miR171, and the sequences of SCL6‐I and SCL6‐II showed 100% base pairing. These results suggested that SCL6‐I and SCL6‐II are regulated by osa‐mature‐microRNA171a‐f (osa‐miR171) in rice, as was shown recently for SCL6‐II, SCL6‐III, and SCL6‐IV and pre‐miR171a and pre‐miR171c during shoot branching and elongation in A. thaliana (Llave et al., 2002; Wang et al., 2010).
We analyzed SCL6‐I, SCL6‐II, ARC3, and DEPG10 transcript levels in response to desiccation and drought stress by RT‐qPCR (Figures 3 and S2b–e). In 10‐day‐old plants, SCL6‐I expression decreased in roots in response to the desiccation stress but did not change in shoots (Figure S3b), while SCL6‐II expression decreased in both shoots and roots (Figure S3c). ARC3 and DEPG10 expression did not change significantly in response to desiccation stress (Figures 3e and S3d). In 4‐week‐old plants, ARC3 expression slightly decreased, while DEPG10 expression increased during the drought stress (Figure 3a,b). Unlike ARC3 and DEPG10 expression, SCL6‐I and SCL6‐II expression strongly decreased due to the drought stress (Figure 3c,d). In contrast, osa‐pre‐miR171f and osa‐miR171 expression increased (Figure 1). These results suggest that in rice, the SCL6‐I and SCL6‐II are putative osa‐miR171 target genes under drought stress conditions.
FIGURE 3.
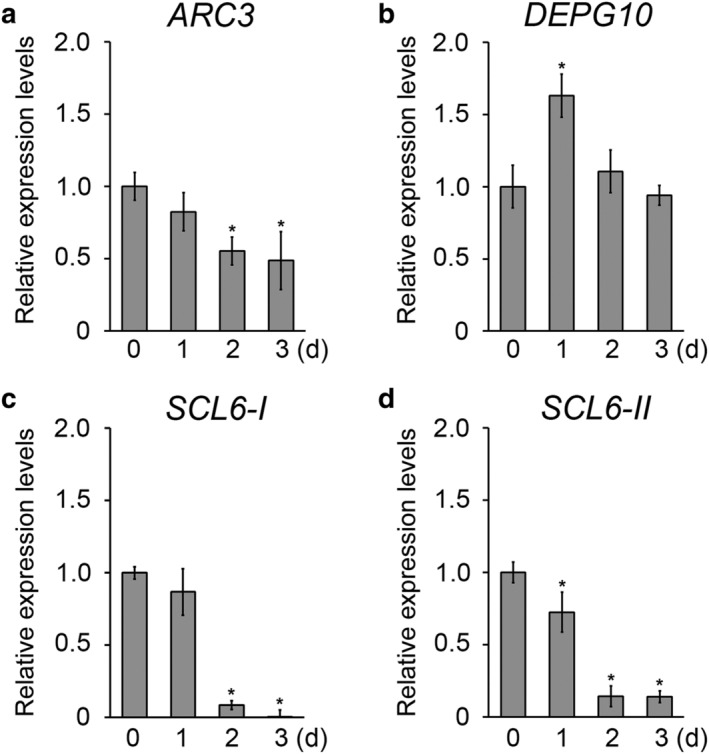
Expression of putative osa‐mature‐microRNA171a‐f (osa‐miR171) target genes in response to drought stress. (a–d) Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analysis shows changes in ARC3 (a), DEPG10 (b), SCL6‐I (c), and SCL6‐II (d) expression levels in response to drought stress. Total RNA was prepared from the 4‐week‐old plants exposed to drought stresses for the indicated time periods. Control plants were not treated with drought stress (0 days). X axis indicates days after drought treatment (d). Error bars indicate SD of triplicate measurements. OsTubA2 and OsUbi1 were used as internal controls, and relative expression levels are shown as fold values. Asterisks indicate statistically significant differences between the corresponding samples and the controls (p value < .01, Student's t test)
2.4. SCL6‐1 and SCL6‐II transcripts are regulated by osa‐MIR171f expression
If osa‐mature‐miR171f regulates both SCL6‐I and SCL6‐II transcript levels, it would be expected that they would have an opposite expression pattern to that of osa‐mature‐microRNA171a‐f (osa‐miR171) during rice development. To determine the spatial and temporal expression patterns of osa‐miR171, SCL6‐I, and SCL6‐II, we extracted total RNA from the coleoptile, roots, shoots, leaves, and flowers at different developmental stages and used it for RT‐qPCR analysis (Figure 4a). Osa‐miR171 expression was down‐regulated in coleoptiles, whereas SCL6‐II expression was up‐regulated in 4‐day‐old plants. In the roots, osa‐miR171 expression was up‐regulated, whereas SCL6‐II expression was down‐regulated in 10‐, 15‐, and 25‐day‐old plants. In leaves, osa‐miR171 expression was down‐regulated, whereas SCL6‐I expression was up‐regulated during development, and in flowers, osa‐miR171 expression increased, while SCL6‐I and SCL6‐II expression decreased at meiosis and before heading stages. These results showed that tissue‐specific expression of SCL6‐I or SCL6‐II correlates with osa‐miR171 transcript levels.
FIGURE 4.
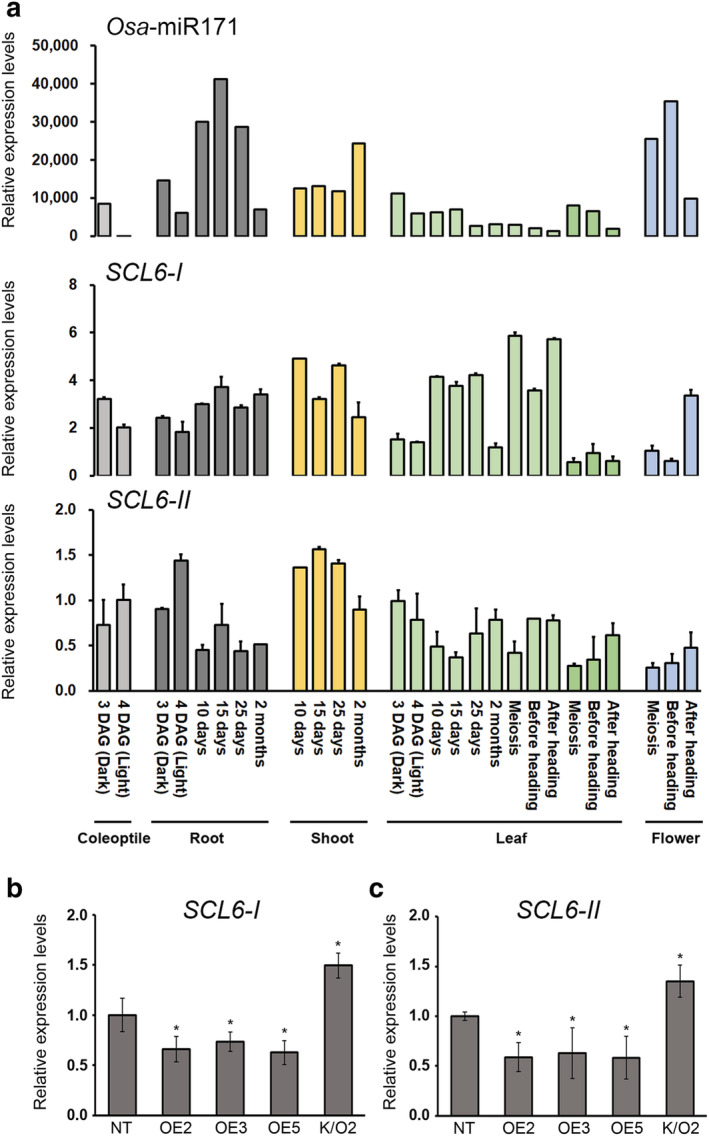
Expression patterns of SCL6 genes at various developmental stages and in osa‐MIR171f‐overexpressing (OE) plants and knockout (K/O) mutants. (a) Expression of osa‐mature‐microRNA171a‐f (osa‐miR171) (upper panel), SCL6‐I (middle panel), and SCL6‐II (lower panel) in various developmental stages and tissues. Total RNA was extracted, and the expression level was analyzed by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). (b) Expression level of SCL6‐I and SCL6‐II in osa‐MIR171f‐overexpressing plants and K/O mutants. The expression level was analyzed by RT‐qPCR. OsTubA2 and OsUbi1 were used as internal controls, and relative expression levels are shown as fold values. Total RNA was prepared from 4‐week‐old plants. Data represent the mean value of three biological replicates, and error bars indicate SD. Asterisks indicate statistically significant differences between the corresponding samples and their control (p value < .01, t test)
To validate the effect of osa‐MIR171f expression on SCL6‐1 and SCL6‐II expression, we performed RT‐qPCR on the OE, K/O, and NT plants (Figure 4b,c). We checked the SCL6‐1 and SCL6‐II expression in the leaf of miR171f OE and KO plants (4‐week‐old) because expression level of miR171f is significantly co‐related with SCL6‐1 and SCL6‐II in the leaf tissue at this developmental stage compared to other tissues, and it was expected that the miR171f overexpression effect on SCL6‐1 and SCL6‐II expression could be maximized. In osa‐MIR171f‐OE plants, SCL6‐1 and SCL6‐II transcript levels were down‐regulated compared to NT plants, while the opposite was true for the osa‐mir171f K/O mutants. These results indicated that SCL6‐1 and SCL6‐II transcript levels were regulated by the expression of osa‐MIR171f.
2.5. Expression of osa‐MIR171f enhances drought tolerance
SCL6‐II, SCL6‐III, and SCL6‐IV are downregulated by ath‐miR171c and osa‐miR171a, and this affects shoot branching and plant height in A. thaliana and rice, respectively. Overexpression of either ath‐MIR171c and osa‐MIR171a results in reduced shoot branching and increased plant height compared with wild‐type (Wang et al., 2010). We, therefore, wanted to determine the phenotypic effect of osa‐MIR171f expression (Figure S4). The OE plants exhibited a reduced tiller number compared to NT plants. However, we observed no significant difference in the number of tillers between K/O mutants and NT plants (Figures 4b and S4a), and this was also true for plant height and culm length (Figure S4c). The flag leaf length of osa‐MIR171f ‐OE plants was greater than in NT plants (Figures 4e and S4d), while the flag leaf width was similar (Figure S4f), suggesting that osa‐MIR171f expression affects the number of tillers and flag leaf length but not plant height and flag leaf width.
The number of panicles with grains in osa‐MIR171f ‐OE plants was 11–19% less than in NT plants (Figure 5a and Table S1), and the number of spikelets per panicle was 3–34% greater in the transgenic plants. However, values for total grain weight, filling rate, and 1,000‐grain weight were not different between the transgenic and NT plants. In contrast, total grain weight and filling rate in the osa‐mir171f‐K/O mutants were reduced by 29% and 9%, respectively, compared with the NT plants. These results suggest that the expression of osa‐miR171 strongly affects the growth and productivity of rice under normal field conditions.
FIGURE 5.
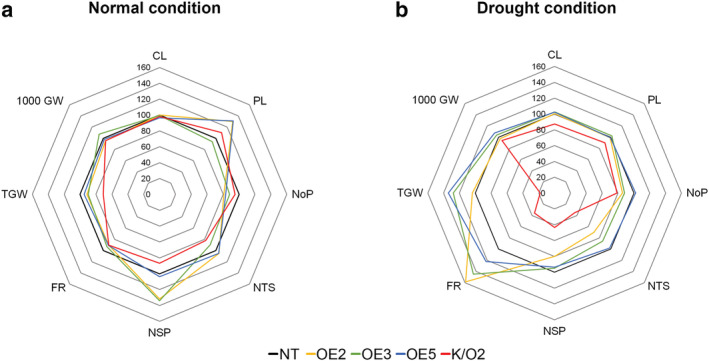
Agronomic traits of osa‐MIR171f‐overexpressing (OE) and ‐knockout (K/O) plants under normal growth conditions and drought stress conditions in the field. (a and b) Spider plots showing the agronomic traits of the osa‐MIR171f ‐OE and ‐K/O plants grown under normal field conditions (a) and under drought stress conditions (b). Osa‐MIR171f ‐OE and ‐K/O plants were analyzed together with non‐transgenic rice (NT) counterparts grown under the same conditions. The values represent the percentage of the mean values (n = 30). Mean values of controls were set at 100% as a reference. CL, culm length; PL, panicle length; NoP, number of panicles; NTS, total number of spikelets; NSP, the number of spikelets per panicle; FR, filling rate; TGW, total grain weight; 1,000 GW, 1,000 grain weight
We next investigated whether osa‐MIR171f expression correlates with grain yield under drought conditions in 14‐week‐old plants exposed to drought stress. After drought stress treatment, the growth and productivity of osa‐MIR171f‐OE, ‐K/O mutants, and NT plants were quantified at the reproductive stage (25‐week‐old) (Figure 5b and Table S2). The grain filling rate and total grain weight were higher in osa‐MIR171f–OE plants than in NT plants, consistent with enhanced drought resistance. In contrast, osa‐mir171f‐K/O mutants showed reduced values for all parameters compared to NT plants, indicating that osa‐mir171f‐K/O mutants are more susceptible to drought stress than NT plants. Collectively, these findings suggest that osa‐MIR171f expression promotes drought resistance in rice.
2.6. The expression of flavonoid biosynthesis genes is regulated by osa‐MIR171f
To better understand the mechanistic relationship between the expression of osa‐MIR171f and drought tolerance, we analyzed the transcriptome in osa‐MIR171f‐OE plants, osa‐mir171f‐K/O mutants, and NT plants using RNA‐sequencing analysis (Figure S5). We found that 45 of the differentially expressed genes (DEGs) overlapped between up‐regulated genes in osa‐MIR171f‐OE and the down‐regulated genes in osa‐mir171f‐K/O, compared to NT plants. In addition, 81 genes overlapped between down‐regulated genes in osa‐MIR171f‐OE and up‐regulated genes in osa‐mir171f ‐K/O, compared to NT plants (Figure S5a). Analysis of the DEG data using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.genome.jp/kegg/) indicated that flavonoid synthesis genes were regulated by osa‐MIR171f expression: CHALCONE SYNTHASE (OsCHS), CHALCONE ISOMERASE (OsCHI) FLAVANONE 3‐HYDROXYLASE (OsF3H), FLAVONOID 3'‐HYDROXYLASE (OsF3'H), FLAVONE SYNTHASE II (OsFNSII), 4‐COUMARATE‐COA LIGASE 2 (Os4CL2), and FLAVANONE 2‐HYDROXYLASE (OsF2H) were all up‐regulated in osa‐MIR171f‐OE and down‐regulated in osa‐mir171f‐K/O plants (Figure S5b). We validated the expression patterns of the flavonoid synthesis genes by RT‐qPCR (Figure 6). We also investigated whether osa‐MIR171f expression correlated with the expression of flavonoid synthesis genes under drought stress conditions (Figure 6). We observed that the transcript levels of all the investigated genes (OsCHS, OsCHI, OsF3H, OsF3'H, Os4CL2, and OsF2H) were up‐regulated by drought compared to normal conditions in osa‐MIR171f‐OE plants but were not significantly different in osa‐mir171f‐K/O mutants. These results indicated that osa‐MIR171f regulates the expression of flavonoid biosynthesis genes in the drought response, suggesting that osa‐MIR171f‐mediated drought tolerance involves flavonoid biosynthesis under the control of SCL6‐I and SCL6‐II expression.
FIGURE 6.
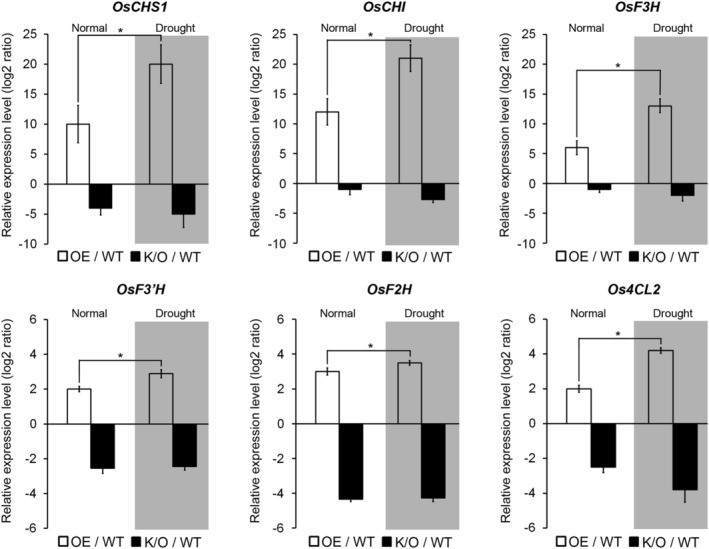
Expression patterns of flavonoid biosynthesis genes in osa‐MIR171f‐overexpressing (OE) and ‐knockout (K/O) plants. The reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analysis shows the expression levels of OsCHS1, OsCHI, OsF3H, OsF3'H, OsF2H, and Os4CL2 in non‐transgenic (NT), osa‐MIR171f‐OE, and K/O plants under normal and drought stress conditions (gray boxes). RT‐qPCR analysis shows changes in OsCHS1, OsCHI, OsF3H, OsF3'H, OsF2H, and Os4CL2 expression in response to drought stress. Total RNA was prepared from 4‐week‐old plants exposed to drought stress. Normal conditions mean plants were not treated with drought stress (0 h time). Error bars indicate SD of triplicate measurements. OsTubA2 and OsUbi1 were used as internal controls, and relative expression levels are shown as fold values. Asterisks indicate statistically significant differences between the corresponding samples and their control (p value < .01, Student's t test)
3. DISCUSSION
In both A. thaliana and rice, there are three miR171 precursors (pre‐miR171a to pre‐miR171c) and nine miR171 precursors (pre‐miR171a to pre‐miR171i), respectively. Despite a high degree of conservation of the mature miR171 sequences, the expression of MIR171 genes can be either down‐ or up‐regulated, depending on specific drought stress conditions and species (Chung et al., 2016; Ferdous et al., 2015; Jian et al., 2016). Osa‐MIR171f was previously identified as a drought stress‐responsive gene (Chung et al., 2016). In this study, we showed that osa‐pre‐miR171f and osa‐miR171 expression is increased by drought stress and ABA treatment (Figure 1). Previous studies reported that the osa‐MIR171f promoter contains both methyl‐jasmonate (MeJA) responsive elements and ABA responsive elements, as well as a MYB binding site (Zhu et al., 2015). At the vegetative, the osa‐MIR171f‐OE plants showed drought resistance phenotypes, while the osa‐mir171f‐K/O plants showed drought‐sensitive phenotypes (Figure 2a). Drought stress at the pre‐anthesis stage is critical in grain filling and yield (Fahad et al., 2017). Therefore, we applied the drought stress at the panicle heading stage and observed the significant yield difference between miR171f transgenic plants and NT plant (Figure 5). These findings suggest that rice osa‐MIR171f is involved in regulating the drought stress response across the development stage.
The miR171/GRAS modules are known to mediate meristem maintenance via regulation of the gibberellin and auxin signaling pathways (Bolle, 2004; Fan et al., 2015; Huang et al., 2017; Wang et al., 2010). In A. thaliana, plants overexpressing ath‐MIR171c, as well as triple scl6 mutant plants, are less branched and have narrower leaves than wild‐type plants. In addition, the triple scl6 mutants have increased chlorophyll accumulation compared to the wild‐type (Wang et al., 2010). In tomato, overexpression of sly‐miR171 increased plant height, whereas overexpression of the sly‐miR171 target gene, SlGRAS24, and sly‐miR171 knock‐down (short tandem target mimic construct, STTM171) plants showed increased branching and a dwarf phenotype (Huang et al., 2017; Kravchik et al., 2019). Here, we observed that osa‐MIR171f ‐OE plants had reduced panicle number and increased numbers of spikelets per panicle compared to NT plants at the reproductive stage (Figure S4). These results suggest that osa‐miR171 and its target genes SCL6‐I and SCL6‐II are important for tiller formation in rice, and that tiller formation enhances drought tolerance.
Drought stress induces reactive oxygen species (ROS), such as hydroxyl radicals (·OH), superoxide radicals (O2 •–), and hydrogen peroxide (H2O2), and is critical for initiating stress signaling. However, excess ROS accumulation causes cell death that can be attributed to a disturbance in cellular homeostasis (Agati et al., 2012; Bolouri‐Moghaddam et al., 2010; Nakabayashi et al., 2014; Vickers et al., 2009). Flavonoids can effectively scavenge ROS (Agati et al., 2020): for example, flavonoid over‐accumulating plants showed resistance to oxidative stress and drought stress. Moreover, cyanidin‐type anthocyanins and quercetin‐type flavonols exhibited strong radical‐scavenging activity in an 2,2‐diphenyl‐1‐picrylhydrazyl (DPPH) assay for analysis of flavonoids activity (Seyoum et al., 2006). Many studies show that the accumulation of flavonoids and expression of flavonoid biosynthesis genes increase under drought stress (Gai et al., 2020; Sharma et al., 2019; Wang et al., 2020). In a previous study in tobacco (Nicotiana tabacum), NtCHS‐overexpressing plants showed higher oxidative stress tolerance and drought tolerance (Hu et al., 2019), and in soybean (Glycine max), gma‐miR171o‐ and gma‐miR171q‐mediated expression of GmSCL‐6 and GmNSP2 were shown to regulate spatial and temporal aspects of nodulation (Hossain et al., 2019). Finally, in A. thaliana, miR156e‐3p‐overexpressing transgenic plants showed increased accumulation of anthocyanins in the lateral branches (Zhao et al., 2017). Here, we observed that key genes involved in flavonoid biosynthesis, such as OsCHS, OsCHI, OsF3H, OsF3'H, OsFNSII, Os4CL2, and OsF2H, were up‐regulated by osa‐MIR171f under drought stress (Figures 6 and S5b).
However, it is still unclear how osa‐MIR171f positively regulates the expression of the flavonoid biosynthesis genes via its repression of SCL6 gene expression. DELLA (GAI, RGA, and RGLs) proteins belonging to the GRAS family function as master negative regulators of GA signaling and are convergent integrators of multiple signaling pathways via direct interaction with various transcription and co‐transcription factors (Schwechheimer, 2011). It has been reported that DELLA proteins directly interact with MYB12/111 and positively regulate the expression of flavonoid biosynthesis genes, CHS and F3H (Tan et al., 2019), and other reports showed that SCL3 directly interacts with DELLA and plays an antagonistic role in GA signaling (Heo et al., 2011; Zhang et al., 2011). Based on these results, it can be speculated that DELLA and SCL have opposite regulatory effects on the transcription of flavonoid biosynthesis genes and hypothesized that an osa‐miR171/SCL6 module regulates the transcription of flavonoid biosynthesis genes via interaction with DELLA proteins.
Although many transcriptome analyses have suggested that osa‐MIR171 genes are involved in drought stress responses, the underlying mechanisms have not been identified. Based on the results presented here, we propose that rice osa‐MIR171f regulates the expression of flavonoid biosynthesis genes by regulating SCL6‐I and SCL6‐II expression under drought stress conditions, thereby conferring drought tolerance.
4. MATERIALS AND METHODS
4.1. Plant materials and growth conditions
O. sativa L. cv. Dongjin was used as non‐transgenic control plants. Rice seeds were germinated in Murashige and Skoog (MS) agar medium in a growth chamber in the dark at 28°C for 3 days. After 3 days of germination, plants were grown under long‐day conditions with 16‐h light/8‐h dark cycles at 28°C for 7 days.
4.2. Vector construction
For the construction of the GOS2::osa‐MIR171f plasmid, full‐length osa‐MIR171f cDNA was derived from total RNA extracted from Dongjin rice by reverse transcriptase polymerase chain reaction (RT‐PCR). The osa‐MIR171f cDNA was inserted into the p700 vector containing a promoter constitutively expressed in the whole body of the plant, GOS2 (Pater et al., 1992). The recombinant plasmid was verified by sequencing and then introduced into Agrobacterium tumefaciens LBA4404 for Agrobacterium‐mediated rice transformation. Primer sequences are listed in Table S3.
4.3. Northern blot analysis
To analyze osa‐mature‐microRNA171a‐f (osa‐miR171) transcripts, total RNA was extracted from the indicated samples and 15 μg was separated on a 12.5% urea‐polyacrylamide gel and electro‐transferred to a Hybond N+ membrane (GE Healthare). The miR171 probe was generated by labeling oligonucleotides complementary to the miR171 sequence (5′‐GATATTGGCGCGGCTCAATCA‐3′) using a DIG oligonucleotide tailing kit (Roche). U6 snRNA (5′‐ TTGCGTGTCATCCTTGCGCAGG‐3′) was used as equal loading indicator of RNA samples. An AP‐conjugated DIG antibody (Roche) was used for probe detection. Signals were visualized by treatment with an CSPD solution (Roche) and exposure to X‐ray film.
4.4. RT‐qPCR
RT‐qPCR analyses were performed using total RNA extracted from the indicated plants using TRIzol (Invitrogen). For cDNA synthesis, 20 μl reactions were performed using 2 μg of total RNA digested with RNAase‐free DNase I (Promega) and the RevertAid™ First Strand cDNA Synthesis Kit (Thermo Scientific™). To detect osa‐mature‐microRNA171a‐f (osa‐miR171), stem‐loop reverse transcription and RT‐PCR were performed as described in protocol (Varkonyi‐Gasic et al., 2007). Digested with RNAase‐free DNase I (Promega), 200 ng of total RNA was transcribed into cDNA using gene‐specific RT primers and a thermostable reverse transcriptase (Invitrogen). The miRNA was hybridized with the miRNA‐specific stem‐loop RT primer and reverse transcribed. A 20 μl mixture, containing 50 U Superscript III RT (Invitrogen), 4 U RNaseOUT (Invitrogen), and 1 μM stem‐loop RT primer, was incubated at 16°C for 45 min, and followed by 60 cycles of 30°C for 45 s, 42°C for 45 s, and 50°C for 1 s. For quantitative PCR, a Mx3000P Real‐Time PCR machine (Stratagene, CA, USA) with Solg™ 2× real‐time PCR smart mix and EvaGreen (SolGent, Deajeon, Korea) was used. PCR cycles included an initial denaturation at 95°C for 5 min followed by 45 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 10 s, and extension at 72°C for 10 s). OsUbi1 (Os06g0681400) and OsTubA2 (Os11g0247300) were used as internal controls. Relative expression level was represented by the mean value of the two independent experiments with OsUubA2 or OsUBi1. Primer sequences are listed in Table S3 and Figure S6.
4.5. Desiccation and drought stress
To analyze the effect of desiccation and drought stress on expressions of genes, 10‐day‐old plants grown on MS media or 4‐week‐old grown on soil were air‐dried on 3 M papers or drought for the indicated times. For the drought tolerance test at the vegetative stage, seeds were uniformly germinated on MS (Murashige and Skoog) media and placed in the dark (28°C) for 3 days. Seedlings were transferred into soil pots (4 × 4 × 6 cm; 3 plants per pot) and grown in a greenhouse (16 h‐light/8 h‐dark cycle) at 30°C. Four weeks after transplanting, drought stress was induced by withholding water for 3 days and then re‐watered for 7 days. The soil water content in each pot was adjusted to 80%. Soil moisture was monitored during the drought treatment using a Soil Moisture Sensor SM150 (Delta‐T Devices). For analysis of agronomic traits under drought stress, osa‐MIR171f‐overexpressing or ‐knockout plants planted in pots (15 cm diameter × 14 cm tall) were grown in a rain‐off shelter. At the panicle heading stage, the first round of drought stress treatments was applied by withholding water. After 7 days, plants were re‐watered and allowed to recover. Ten days after the first treatment, a second round of drought stress was applied in the same manner as before. For analysis of agronomic traits under normal conditions, three independent lines from the T4 (2020) generation of osa‐MIR171f‐overexpressing or osa‐mir171f‐knockout plants were grown in a paddy field located at Kyungpook National University, Korea (128:34E/36:15 N) together with corresponding NT plants.
4.6. Transcriptome analysis
Four‐week‐old NT, osa‐MIR171f‐OE and osa‐mir171f‐K/O plants were grown under normal conditions in a glasshouse. For the Illumina library preparation, total RNA was first extracted from NT, osa‐MIR171f‐OE and ‐K/O leaves using the Plant RNeasy mini kit (Qiagen, Valencia, USA). RNA from three independent lines of osa‐MIR171f‐OE, K/O, and NT plants was pooled for RNAseq according to the manufacturer's protocol and sequenced (Macrogen, Seoul, Korea) using the Illumina HiSeq2000 platform (Illumina, San Diego, USA). The transcripts were assembled and normalized by Fragment Per Kilobase of transcript per Killion mapped reads (FPKM), and the DEGs in each osa‐MIR171f‐OE versus NT and osa‐mir171f ‐K/O vs NT were selected by |log2 (fold change)| ≥ 1 and |log2 (fold change)| ≤ −1 using a P value <.05. The data set can be found at from GEO database with series accession number GSE180271 for RNA‐sequencing data.
4.7. Accession numbers
Sequence data from this article can be found in the in the Rice Annotation Project (RAP) database (https://rapdb.dna.affrc.go.jp/) or the microRNA database (miRBase) (http://www.mirbase.org/) under the following accession numbers: Osa‐MIR171f (MI0001137), ARC3 (Os09g0555600), DEPG10 (Os05g0417100), SCL6‐I (Os06g0105350), and SCL6‐II (Os02g0662700).
AUTHOR CONTRIBUTIONS
T. U. and J‐K K. designed this study; T. U., J. C., P. J. C., and J. S. S. (Jun Sung Seo) wrote the manuscript and prepared the figures; T. P., S. E. J., J. S. S. (Jae Sung Shim), Y. S. K., I‐Y. C., S. C. P., S‐J. O., and J‐K. K. performed phenotyping and contributed to data analysis. All authors have read and agreed to the published version of the manuscript.
CONFLICT OF INTEREST
The Authors did not report any conflict of interest.
Supporting information
Supplemental Figure S1. Generation of osa‐MIR171f‐overexpressing (OE) rice lines
Supplemental Figure S2. Generation of osa‐mir171f‐knockout (K/O) rice using the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system.
Supplemental Figure S3. Analysis of candidate osa‐miR171 targets in response to drying stress.
Supplemental Figure S4. Analysis of osa‐MIR171f‐overexpressing (OE) and ‐knockout (K/O) plant growth.
Supplemental Figure S5. Transcriptome analysis of osa‐MIR171f‐overexpressing (OE) and ‐knockout (K/O) plants.
Supplemental Table S1. Agronomic traits of in T4 generation of osa‐MIR171f overexpressing plants and of knockout mutants grown under normal field conditions (2020).
Supplemental Table S2. Agronomic traits of in T5 generation of osa‐MIR171f overexpressing plants and T4 generation of knockout mutants grown under drought
Supplemental Table S3. The sequences of oligonucleotides used in this study
ACKNOWLEDGMENTS
We thank the Rural Development Administration and Kyungpook National University for providing rice paddy fields. This work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ01597601 to J.‐K.K.)” Rural Development Administration, Republic of Korea.
Um, T. , Choi, J. , Park, T. , Chung, P. J. , Jung, S. E. , Shim, J. S. , Kim, Y. S. , Choi, I.‐Y. , Park, S. C. , Oh, S.‐J. , Seo, J. S. , & Kim, J.‐K. (2022). Rice microRNA171f/SCL6 module enhances drought tolerance by regulation of flavonoid biosynthesis genes. Plant Direct, 6(1), e374. 10.1002/pld3.374
DATA AVAILABILITY STATEMENT
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is Ju‐Kon Kim (jukon@snu.ac.kr).
REFERENCES
- Agati, G. , Azzarello, E. , Pollastri, S. , & Tattini, M. (2012). Flavonoids as antioxidants in plants: Location and functional significance. Plant Science Journal, 196, 67–76. [DOI] [PubMed] [Google Scholar]
- Agati, G. , Brunetti, C. , Fini, A. , Gori, A. , Guidi, L. , Landi, M. , Sebastiani, F. , & Tattini, M. (2020). Are flavonoids effective antioxidants in plants? Twenty years of our Investigation. Antioxidants, 9, 1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M. J. (2008). Evolution of microRNAs and their targets: Are all microRNAs biologically relevant? Biochimica et Biophysica Acta ‐ Gene Regulatory Mechanisms, 1779, 725–734. [DOI] [PubMed] [Google Scholar]
- Axtell, M. J. , & Bowman, J. L. (2008). Evolution of plant microRNAs and their targets. Trends in Plant Science, 13, 343–349. [DOI] [PubMed] [Google Scholar]
- Bartel, D. P. (2009). MicroRNAs: Target recognition and regulatory functions. Cell, 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle, C. (2004). The role of GRAS proteins in plant signal transduction and development. Planta, 218, 683–692. [DOI] [PubMed] [Google Scholar]
- Bolouri‐Moghaddam, M. R. , le Roy, K. , Xiang, L. , Rolland, F. , & van den Ende, W. (2010). Sugar signalling and antioxidant network connections in plant cells. The FEBS Journal, 277, 2022–2037. [DOI] [PubMed] [Google Scholar]
- Catalá, R. , Medina, J. , & Salinas, J. (2011). Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 108(16475–16), 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorostecki, U. , Crosa, V. A. , Lodeyro, A. F. , Bologna, N. G. , Martin, A. P. , Carrillo, N. , Schommer, C. , & Palatnik, J. F. (2012). Identification of new microRNA‐regulated genes by conserved targeting in plant species. Nucleic Acids Research, 40(8893–8), 904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, P. J. , Chung, H. , Oh, N. , Choi, J. , Bang, S. W. , Jung, S. E. , Jung, H. , Shim, J. S. , & Kim, J.‐K. (2020). Efficiency of recombinant CRISPR/rCas9‐mediated miRNA gene editing in rice. International Journal of Molecular Sciences, 21, 9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, P. J. , Jung, H. , Jeong, D. H. , Ha, S. H. , Choi, Y. D. , & Kim, J. K. (2016). Transcriptome profiling of drought responsive noncoding RNAs and their target genes in rice. BMC Genomics, 17, 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi, E. L. , Kumar, S. , Singh, T. , Sharma, S. , Beemrote, A. , Devi, C. P. , Chongtham, S. K. , Singh, C. H. , Yumlembam, R. A. , Haribhushan, A. , Prakash, N. , & Wani, S. H. (2017). Adaptation strategies and defence mechanisms of plants during environmental stress. In Medicinal plants and environmental challenges (pp. 359–413). Springer, Cham. https://link.springer.com/book/10.1007/978-3-319-68717-9 [Google Scholar]
- Fahad, S. , Bajwa, A. A. , Nazir, U. , Anjum, S. A. , Farooq, A. , Zohaib, A. , Sadia, S. , Nasim, W. , Adkins, S. , Saud, S. , Ihsan, M. Z. , Alharby, H. , Wu, C. , Wang, D. , & Huang, J. (2017). Crop production under drought and heat stress: Plant responses and management options. Frontiers in Plant Science, 8(1), 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, T. , Li, X. , Yang, W. , Xia, K. , Ouyang, J. , & Zhang, M. (2015). Rice osa‐miR171c mediates phase change from vegetative to reproductive development and shoot apical meristem maintenance by repressing four OsHAM transcription factors. PLoS ONE, 10, e0125833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous, J. , Hussain, S. S. , & Shi, B. J. (2015). Role of microRNAs in plant drought tolerance. Plant Biotechnology Journal, 13, 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz, W. , Bhattacharyya, S. N. , & Sonenberg, N. (2008). Mechanisms of post‐transcriptional regulation by microRNAs: Are the answers in sight? Nature Reviews. Genetics, 9, 102–114. [DOI] [PubMed] [Google Scholar]
- Fu, Z. Q. , & Dong, X. (2013). Systemic acquired resistance: Turning local infection into global defense. Annual Review of Plant Biology, 64, 839–863. [DOI] [PubMed] [Google Scholar]
- Gai, Z. , Wang, Y. , Ding, Y. , Qian, W. , Qiu, C. , Xie, H. , Sun, L. , Jiang, Z. , Ma, Q. , & Wang, L. (2020). Exogenous abscisic acid induces the lipid and flavonoid metabolism of tea plants under drought stress. Scientific Reports, 10, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galland, M. , Boutet‐Mercey, S. , Lounifi, I. , Godin, B. , Balzergue, S. , Grandjean, O. , Morin, H. , Perreau, F. , Debeaujon, I. , & Rajjou, L. (2014). Compartmentation and dynamics of flavone metabolism in dry and germinated rice seeds. Plant & Cell Physiology, 55(1646‐1), 659. [DOI] [PubMed] [Google Scholar]
- Harborne, J. B. , & Williams, C. A. (2000). Advances in flavonoid research since 1992. Phytochemistry, 55, 481–504. [DOI] [PubMed] [Google Scholar]
- Heil, M. , & Bostock, R. M. (2002). Induced systemic resistance (ISR) against pathogens in the context of induced plant defences. Annals of Botany, 89, 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo, J. O. , Chang, K. S. , Kim, I. A. , Lee, M. H. , Lee, S. A. , Song, S. K. , Lee, M. M. , & Lim, J. (2011). Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow‐like 3 in the Arabidopsis root. Proceedings of the National Academy of Sciences of the United States of America, 108(2166–2), 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain, M. S. , Hoang, N. T. , Yan, Z. , Tóth, K. , Meyers, B. C. , & Stacey, G. (2019). Characterization of the spatial and temporal expression of two soybean miRNAs identifies SCL6 as a novel regulator of soybean nodulation. Frontiers in Plant Science, 10, 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, B. , Yao, H. , Gao, Y. , Wang, R. , Li, F. , Guo, J. , Li, K. , Zhao, M. , & Jin, L. (2019). Overexpression of chalcone synthase gene improves flavonoid accumulation and drought tolerance in tobacco. Preprints 2019060103 (doi: 10.20944/preprints201906.0103.v1). [DOI]
- Huang, W. , Peng, S. , Xian, Z. , Lin, D. , Hu, G. , Yang, L. , Ren, M. , & Li, Z. (2017). Overexpression of a tomato miR171 target gene Sl GRAS 24 impacts multiple agronomical traits via regulating gibberellin and auxin homeostasis. Plant Biotechnology Journal, 15, 472–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian, H. , Wang, J. , Wang, T. , Wei, L. , Li, J. , & Liu, L. (2016). Identification of rapeseed MicroRNAs involved in early stage seed germination under salt and drought stresses. Frontiers in Plant Science, 7, 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones‐Rhoades, M. W. , & Bartel, D. P. (2004). Computational identification of plant microRNAs and their targets, including a stress‐induced miRNA. Molecular Cell, 14, 787–799. [DOI] [PubMed] [Google Scholar]
- Jones‐Rhoades, M. W. , Bartel, D. P. , & Bartel, B. (2006). MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology, 57, 19–53. [DOI] [PubMed] [Google Scholar]
- Joshi, R. , Wani, S. H. , Singh, B. , Bohra, A. , Dar, Z. A. , Lone, A. A. , Pareek, A. , & Singla‐Pareek, S. L. (2016). Transcription factors and plants response to drought stress: Current understanding and future directions. Frontiers in Plant Science, 7, 1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh, B. , Zhu, J.‐K. , & Zhu, J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochimica et Biophysica Acta ‐ Gene Regulatory Mechanisms, 1819, 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. L. , Kim, N. , Lee, H. , Lee, E. , Cheon, K. S. , Kim, M. , Baek, J. , Choi, I. , Ji, H. , Yoon, I. S. , Jung, K. H. , Kwon, T. R. , & Kim, K. H. (2020). High‐throughput phenotyping platform for analyzing drought tolerance in rice. Planta, 252, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koops, P. , Pelser, S. , Ignatz, M. , Klose, C. , Marrocco‐Selden, K. , & Kretsch, T. (2011). EDL3 is an F‐box protein involved in the regulation of abscisic acid signalling in Arabidopsis thaliana . Journal of Experimental Botany, 62(5547–5), 560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchik, M. , Stav, R. , Belausov, E. , & Arazi, T. (2019). Functional characterization of microRNA171 family in tomato. Plants, 8, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano, M. , Tohge, T. , Fukushima, A. , Kobayashi, M. , Hayashi, N. , Otsuki, H. , Kondou, Y. , Goto, H. , Kawashima, M. , & Matsuda, F. (2011). Metabolomics reveals comprehensive reprogramming involving two independent metabolic responses of Arabidopsis to UV‐B light. The Plant Journal, 67, 354–369. [DOI] [PubMed] [Google Scholar]
- Lee, R. C. , Feinbaum, R. L. , & Ambros, V. (1993). The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14. Cell, 75, 843–854. [DOI] [PubMed] [Google Scholar]
- Liu, H.‐H. , Tian, X. , Li, Y.‐J. , Wu, C.‐A. , & Zheng, C.‐C. (2008). Microarray‐based analysis of stress‐regulated microRNAs in Arabidopsis thaliana . RNA, 14, 836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C. , Kasschau, K. D. , Rector, M. A. , & Carrington, J. C. (2002). Endogenous and silencing‐associated small RNAs in plants. Plant Cell, 14, 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A. C. , & Vaucheret, H. (2004). MicroRNAs: Something important between the genes. Current Opinion in Plant Biology, 7, 120–125. [DOI] [PubMed] [Google Scholar]
- Mittler, R. (2006). Abiotic stress, the field environment and stress combination. Trends in Plant Science, 11, 15–19. [DOI] [PubMed] [Google Scholar]
- Nakabayashi, R. , Yonekura‐Sakakibara, K. , Urano, K. , Suzuki, M. , Yamada, Y. , Nishizawa, T. , Matsuda, F. , Kojima, M. , Sakakibara, H. , & Shinozaki, K. (2014). Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. The Plant Journal, 77, 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pater, B. S. , van der Mark, F. , Rueb, S. , Katagiri, F. , Chua, N. H. , Schilperoort, R. A. , & Hensgens, L. A. (1992). The promoter of the rice gene GOS2 is active in various different monocot tissues and binds rice nuclear factor ASF‐1. The Plant Journal, 2, 837–844. [DOI] [PubMed] [Google Scholar]
- Peet, M. M. , & Kramer, P. J. (1980). Effects of decreasing source/sink ratio in soybeans on photosynthesis, photorespiration, transpiration and yield. Plant, Cell & Environment, 3, 201–206. [Google Scholar]
- Ramachandran, V. , & Chen, X. (2008). Small RNA metabolism in Arabidopsis. Trends in Plant Science, 13, 368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B. J. , Weinstein, E. G. , Rhoades, M. W. , Bartel, B. , & Bartel, D. P. (2002). MicroRNAs in plants. Genes & Development, 16(1616‐1), 626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M. W. , Reinhart, B. J. , Lim, L. P. , Burge, C. B. , Bartel, B. , & Bartel, D. P. (2002). Prediction of plant microRNA targets. Cell, 110, 513–520. [DOI] [PubMed] [Google Scholar]
- Saito, K. , Yonekura‐Sakakibara, K. , Nakabayashi, R. , Higashi, Y. , Yamazaki, M. , Tohge, T. , & Fernie, A. R. (2013). The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiology and Biochemistry, 72, 21–34. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C. (2011). Gibberellin signaling in plants—The extended version. Frontiers in Plant Science, 2, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyoum, A. , Asres, K. , & El‐Fiky, F. K. (2006). Structure–radical scavenging activity relationships of flavonoids. Phytochemistry, 67, 2058–2070. [DOI] [PubMed] [Google Scholar]
- Sharma, A. , Shahzad, B. , Rehman, A. , Bhardwaj, R. , Landi, M. , & Zheng, B. (2019). Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules, 24, 2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih, C. H. , Chu, H. , Tang, L. K. , Sakamoto, W. , Maekawa, M. , Chu, I. K. , Wang, M. , & Lo, C. (2008). Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta, 228(1043‐1), 054. [DOI] [PubMed] [Google Scholar]
- Stracke, R. , Favory, J. J. , Gruber, H. , Bartelniewoehner, L. , Bartels, S. , Binkert, M. , Funk, M. , Weisshaar, B. , & Ulm, R. (2010). The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet‐B radiation. Plant, Cell & Environment, 33, 88–103. [DOI] [PubMed] [Google Scholar]
- Sunkar, R. , Li, Y.‐F. , & Jagadeeswaran, G. (2012). Functions of microRNAs in plant stress responses. Trends in Plant Science, 17, 196–203. [DOI] [PubMed] [Google Scholar]
- Sunkar, R. , & Zhu, J.‐K. (2004). Novel and stress‐regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell, 16, 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, H. , Man, C. , Xie, Y. , Yan, J. , Chu, J. , & Huang, J. (2019). A crucial role of GA‐regulated flavonol biosynthesis in root growth of Arabidopsis. Molecular Plant, 12, 521–537. [DOI] [PubMed] [Google Scholar]
- Vakilian, K. A. (2020). Machine learning improves our knowledge about miRNA functions towards plant abiotic stresses. Scientific Reports, 10, 3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi‐Gasic, E. , Wu, R. , Wood, M. , Walton, E. F. , & Hellens, R. P. (2007). Protocol: A highly sensitive RT‐PCR method for detection and quantification of microRNAs. Plant Methods, 3, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers, C. E. , Gershenzon, J. , Lerdau, M. T. , & Loreto, F. (2009). A unified mechanism of action for volatile isoprenoids in plant abiotic stress. Nature Chemical Biology, 5, 283–291. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell, 136, 669–687. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Mai, Y. X. , Zhang, Y. C. , Luo, Q. , & Yang, H. Q. (2010). MicroRNA171c‐targeted SCL6‐II, SCL6‐III, and SCL6‐IV genes regulate shoot branching in Arabidopsis. Molecular Plant, 3, 794–806. [DOI] [PubMed] [Google Scholar]
- Wang, N. , Liu, W. , Yu, L. , Guo, Z. , Chen, Z. , Jiang, S. , Xu, H. , Fang, H. , Wang, Y. , & Zhang, Z. (2020). HEAT SHOCK FACTOR A8a modulates flavonoid synthesis and drought tolerance. Plant Physiology, 184(1273‐1), 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.‐J. , Reyes, J. L. , Chua, N.‐H. , & Gaasterland, T. (2004). Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biology, 5, R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, T. , Mogami, J. , & Yamaguchi‐Shinozaki, K. (2014). ABA‐dependent and ABA‐independent signaling in response to osmotic stress in plants. Current Opinion in Plant Biology, 21, 133–139. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. L. , Ogawa, M. , Fleet, C. M. , Zentella, R. , Hu, J. , Heo, J. O. , Lim, J. , Kamiya, Y. , Yamaguchi, S. , & Sun, T. P. (2011). Scarecrow‐like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 108(2160‐2), 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, D. , Xia, X. , Wei, M. , Sun, J. , Meng, J. , & Tao, J. (2017). Overexpression of herbaceous peony miR156e‐3p improves anthocyanin accumulation in transgenic Arabidopsis thaliana lateral branches. 3. Biotech, 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, L. , Liu, Y. , Liu, Z. , Kong, D. , Duan, M. , & Luo, L. (2010). Genome‐wide identification and analysis of drought‐responsive microRNAs in Oryza sativa . Journal of Experimental Botany, 61(4157‐4), 168. [DOI] [PubMed] [Google Scholar]
- Zhu, X. , Leng, X. , Sun, X. , Mu, Q. , Wang, B. , Li, X. , Wang, C. , & Fang, J. (2015). Discovery of conservation and diversification of miR171 genes by phylogenetic analysis based on global genomes. Plant Genome, 8, 1–11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Generation of osa‐MIR171f‐overexpressing (OE) rice lines
Supplemental Figure S2. Generation of osa‐mir171f‐knockout (K/O) rice using the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system.
Supplemental Figure S3. Analysis of candidate osa‐miR171 targets in response to drying stress.
Supplemental Figure S4. Analysis of osa‐MIR171f‐overexpressing (OE) and ‐knockout (K/O) plant growth.
Supplemental Figure S5. Transcriptome analysis of osa‐MIR171f‐overexpressing (OE) and ‐knockout (K/O) plants.
Supplemental Table S1. Agronomic traits of in T4 generation of osa‐MIR171f overexpressing plants and of knockout mutants grown under normal field conditions (2020).
Supplemental Table S2. Agronomic traits of in T5 generation of osa‐MIR171f overexpressing plants and T4 generation of knockout mutants grown under drought
Supplemental Table S3. The sequences of oligonucleotides used in this study
Data Availability Statement
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is Ju‐Kon Kim (jukon@snu.ac.kr).


