Abstract
Background
The coronavirus disease 2019 (COVID-19) pandemic is still raging worldwide. Efficient, fast and low-cost severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nucleic acid detection methods are urgently needed.
Methods
A rapid PCR temperature change mode was explored by moving the reaction tube between the independent temperature modules with large temperature differences and a portable ultra-fast real-time PCR instrument were developed. We established a rapid SARS-CoV-2 test method using the ultra-fast real-time PCR instrument, a China Food and Drug Administration-certified SARS-CoV-2 reagent and optimized reaction condition. The analytical and clinical performances of the rapid tests were evaluated by comparing with the standard SARS-CoV-2 tests.
Results
The new temperature change mode can effectively shorten the amplification reaction time and be successfully used in the development of the ultra-fast real-time PCR instrument. The rapid SARS-CoV-2 test method was established and the time to yield results were greatly shortened from 81 min of the standard test to 31 min. Specificity of the rapid test was assessed and no non-specific amplification (0/63) was observed. The limits of detection of the rapid and standard tests were similar. Clinical performance was evaluated using 184 respiratory specimens from patients with suspected SARS-CoV-2 infection. The positive agreement between the rapid and standard tests was 100% (67/67), the negative agreement was 97.4% (114/117), and the kappa statistic was 0.965 (P<0.001). No significant differences in the Ct values for each target gene were observed between the rapid test and the standard test (P>0.05).
Conclusions
We had developed a 30-minute detection method for SARS-CoV-2 nucleic acid using a novel ultra-fast real-time PCR instrument. The rapid test method may impact on patient management.
Keywords: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), coronavirus disease 2019 (COVID-19), rapid test, ultra-fast real-time PCR instrument, pandemic prevention and control
Introduction
The coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), remains an enormous threat to human society (1-3). More than 199 million cases have been confirmed worldwide, and more than 4.2 million people had died from the infection as of August 4, 2021 according to the World Health Organization (https://covid19.who.int/). There are several kinds of SARS-CoV-2 vaccines being vaccinated globally, but it will still take a long time to achieve herd immunity. At the same time, there are still problems such as low vaccination rates in low- and middle-income countries, and the reduced vaccine protection due to virus mutation (4-7).
Decreasing viral transmission is always effective way to prevent and control the COVID-19 pandemic. Rapid diagnosis has been demonstrated to be very helpful in reducing the risk of SARS-CoV-2 transmission (8,9). Using conventional detection technologies, serum antibody against SARS-CoV-2 is often not detected in the first week to 10 days of symptoms. Therefore, antibody testing is generally unhelpful for acute diagnosis (10-12). Virus isolation in culture presents significant biosafety risks and has sensitivity limitations. Nucleic acid amplification tests (NAATs), especially Taqman real-time PCR tests of respiratory tract specimens are widely used worldwide to diagnose COVID-19 and are the gold standard diagnostic method (1,13). With the normalization of pandemic prevention and control, the need for rapid and low-cost detection equipment and methods is becoming more urgent.
Many factors may influence on the speed of real-time PCR tests. Under the constant reaction system, the temperature ramping speed of the PCR instrument and amplification program time setting are important factors. A conventional real-time PCR instrument performs the amplification reaction through the heating and cooling of the single thermal cycle module, and the temperature ramping speed is about 3.0-3.5oC/s. It usually takes more than 1h to complete the test. For Taqman real-time PCR test, the target nucleic acid length is about 60-150bp, detection time may be greatly reduced by optimizing settings.
In this work, we explored a rapid temperature rise and fall method using independent temperature modules with large temperature differences (ITM-LTD), and developed an ultra-fast real-time PCR instrument. Basing on this novel instrument and amplification condition optimization, we established and evaluated an approximately 30-min rapid SARS-CoV-2 test method using a China Food and Drug Administration (CFDA) approved reagent. The ability of the novel test to enable rapid, real-time detection of SARS-CoV-2 may increase the detection capabilities of primary medical units, expand the scope of detection, and improve patient management.
We present the following article in accordance with the STARD reporting checklist (available at https://dx.doi.org/10.21037/jtd-21-1288).
Methods
Independent temperature modules with large temperature differences (ITM-LTD)
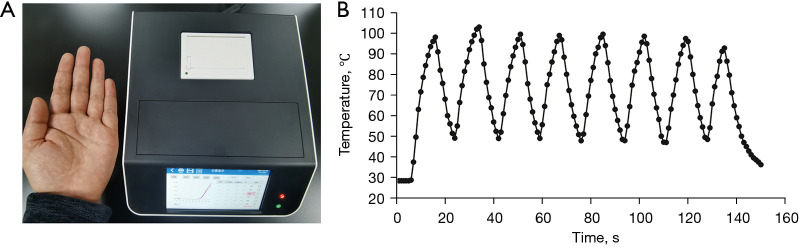
To achieve rapid temperature change, we set up two ITM which fixed at 120 and 40 °C using a melt bath (Shenhua Biotech, Guangzhou, China) and a PCR instrument (LongGene Scientific, Hangzhou, China), respectively. The PCR tube was moved manually between the high and low temperature modules (Figure 1). The digital thermometer probe (Lihuada, Guangdong, China) was put into the PCR reaction mixture to monitor temperature changes (Figure 1A). Paraffin oil was added to the tube to prevent the reaction buffer from evaporating.
Figure 1.
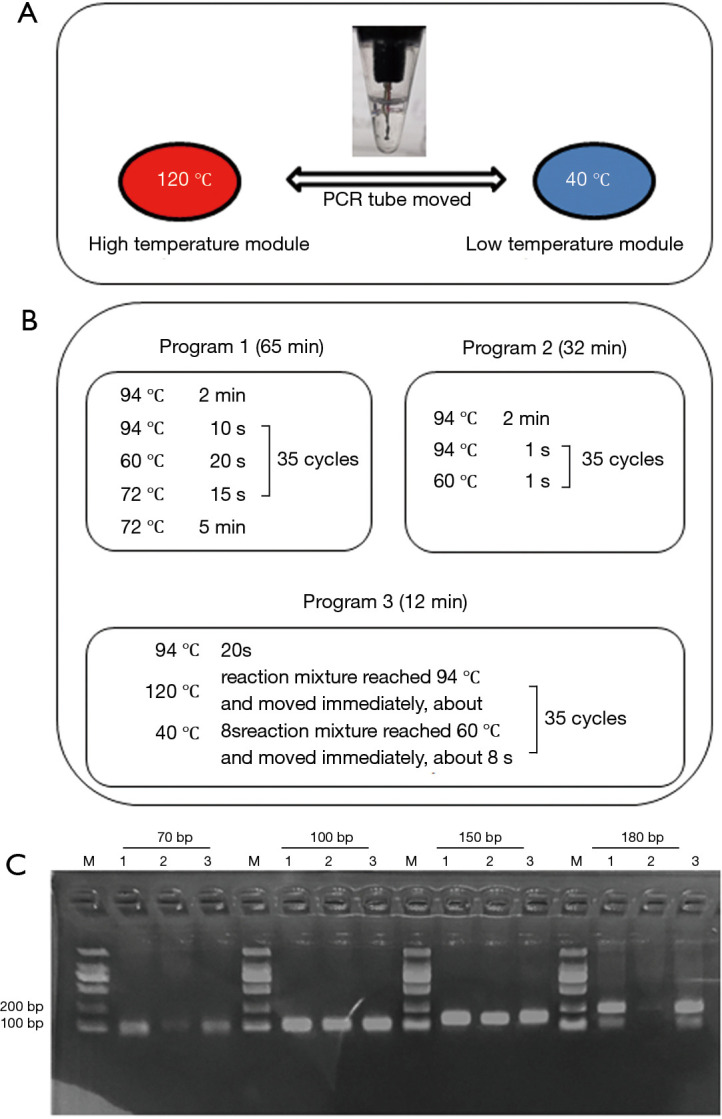
Rapid amplification of the short-length nucleic acid targets using independent temperature modules with large temperature differences and optimized programs. (A) The PCR tube was moved manually between the independent high (120 °C) and low (40 °C) temperature modules as the reaction mixture reached the setting temperature. (B) Program 1 and 2 were performed on the normal PCR instrument; Program 3 was performed on the independent temperature modules. C: 1, 2, 3 represented the product of Program 1, 2 and 3. M: 100 bp marker.
Short-length nucleic acids amplification using ITM-LTD
To evaluate the effect of ITM-LTD in the rapid amplification of short-length nucleic acids, a panel of primers for amplification targets about 70, 100, 150 and 180 bp of hepatitis B virus (HBV) were designed (Table S1). Promega Taq polymerase was used for amplification in total 25 µL reaction mixtures including 5 µL of template HBV-DNA according to the manufactory protocol. Three amplification programs were performed for each test simultaneously. Program 1 and 2 were performed on the normal PCR instrument (LongGene Scientific, Hangzhou, China), among which Program 1 was used as a control to set the normal condition; Program 2 had extreme short reaction time of 1 s. Program 3 was performed on ITM-LTD, and the set condition was to move the tube immediately once the predetermined temperature was reached (Figure 1B). Two µL amplification products were analyzed using 2.0% agarose gel electrophoresis.
Description of a novel ultra-fast real-time PCR instrument
An ultra-fast real-time PCR instrument was developed using ITM-LTD. The high and low temperature modules were designed at 120 and 40 °C, respectively. The reaction tubes were designed to move between the high and low temperature modules to achieve rapid temperature increases and decreases. Accurate temperature control of the reaction was maintained by moving the tubes up/down in the temperature modules according to calculations from the inner temperature sensor. The third adjustable temperature module was used for RNA reverse transcription and DNA amplification and was also the site where fluorescent signals were collected. The instrument had four fluorescence channels (1–4 channels) with a flux of 8. The size was 25 cm (L)×19 cm (W) ×12 cm (H). The weight was 3.2 kg (Figure 2A). The temperature rise and fall speeds of the reaction mixture in the instrument between 50–100 °C were 6.0±0.4 °C/s (mean ± standard deviation) and 6.1±0.4 °C/s, respectively. The fastest temperature change exceeded 10 °C/s (Figure 2B).
Figure 2.
Rapid temperature changes of reaction mixture in the ultra-fast real-time PCR instrument. (A) The instrument has four fluorescence channels with a flux of 8, a size of 25 cm (L) ×19 cm (W) ×12 cm (H), and a total weight of 3.2 kg; (B) the temperature changes of the reaction mixture tested in the instrument between 50–100 °C.
SARS-CoV-2 reagent
CFDA-certified SARS-CoV-2 detection reagents were purchased from Shanghai BioGerm Medical Technology Co., Ltd. (Shanghai, China). SARS-CoV-2 target genes (ORF1a/b and N) and a host cell housekeeping gene (RNase P) were amplified with FAM™, VIC™, and ROX™ labeled fluorescent probes, respectively. Reaction mixtures (25 µL total volumes) including 5 µL of template RNA were prepared using automated extraction methods (KeHua Bio-Engineering, Shanghai, China). The standard test used the ABI-7500 instrument or the QuantStudio6 (ABI-Q6) instrument (Life Technologies, Singapore). For standard tests, temperature was cycled as follows: 50 °C for 10 min, 94 °C for 5 min; 40 cycles of 94 °C for 10 s and 55 °C for 40 s. The standard test was used as a control for the establishment and evaluation of the rapid SARS-CoV-2 test.
Reaction condition optimization in the novel ultra-fast real-time PCR instrument
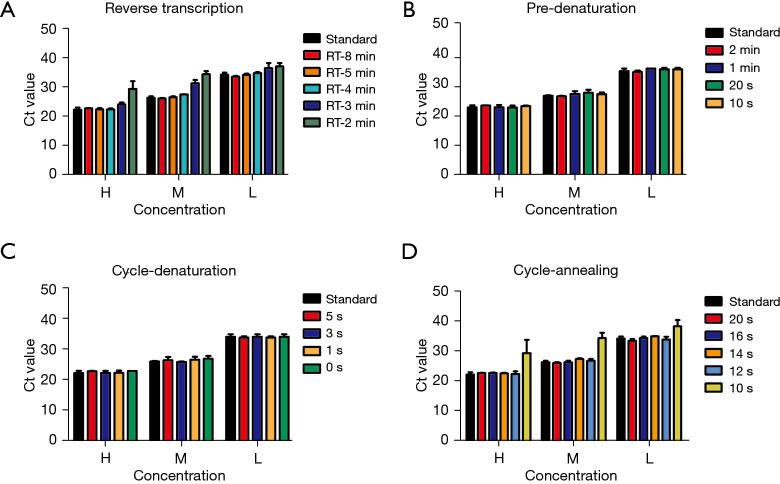
The rapid test conditions including the reaction time of reverse transcription, pre-denaturation and cycling conditions in the ultra-fast real-time PCR instrument were optimized to take advantage of fast scanning, temperature ramp speeds and short-length of the targets. Three SARS-CoV-2-positive samples which had high (Ct value 22.28), medium (Ct value 26.34) and low (Ct value 34.21) concentration were used for program optimization. Nucleic acids were extracted from samples using conventional column extraction or automated extraction methods, and then assayed simultaneously for rapid tests and standard tests.
Specificity assessment of rapid SARS-CoV-2 test
The specificity of the rapid test using optimized reaction conditions was evaluated. A total of 63 specimens containing 21 common non-SARS-CoV-2 respiratory pathogens were tested to assess specificity. Pathogens tested included common human coronaviruses (229E, OC43, NL63, and HKU1), influenza A and B viruses, respiratory syncytial virus, human parainfluenza virus types 1–4, adenovirus, enterovirus, human metapneumovirus, rhinovirus, human bocavirus, Mycoplasma pneumoniae, Chlamydophila pneumoniae, Streptococcus pneumonia, Staphylococcus aureus, and Haemophilus influenza. Specimens were stored at the State Key Laboratory of Respiratory Diseases and simultaneously used for the rapid and standard tests.
Limit of detection (LOD) of the rapid SARS-CoV-2 test
To evaluate the LOD of the rapid test, RNA was extracted from a SARS-CoV-2 positive sample. Ct values for SARS-CoV-2 genes (ORF1a/b and N) in the undiluted sample were determined using the standard test. The LOD of the rapid and standard tests were compared by assaying a 10-fold RNA dilution series in RNase-free water. Twenty-four replicates were tested for each dilution, including SARS-CoV-2-negative specimens.
Clinical performance of rapid SARS-CoV-2 test
The clinical study was conducted at two sites (Guangzhou Custom Technology Center and Kingmed Virology Diagnostic & Translational Center). Frozen respiratory specimens (nasopharyngeal swabs, alveolar lavage fluid, or sputum) from patients with suspected SARS-CoV-2 infections were collected and simultaneously assayed using the rapid and standard tests. Viral nucleic acids were extracted using automated extraction methods (KeHua Bio-Engineering and Liferiver, Shanghai, China). Data were collected by investigators at each study site and pooled for analysis.
Statistical analysis
Positive and negative percent agreements between the rapid and standard tests were calculated using two-by-two tables. Cohen’s kappa statistic was calculated. Box-and-whisker plots were used to show different Ct values for target SARS-CoV-2 genes. The Mann-Whitney U test was used to assess differences in Ct value distributions between two test conditions. Statistical analyses were performed using SPSS 19.0 (SPSS Inc., Chicago, IL, USA). Values of P<0.05 (two-tailed) were considered statistically significant.
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was reviewed and approved by The First Affiliated Hospital of Guangzhou Medical University Ethics Committee (No. 2020-77). Informed consent was taken from all individual participants. Parts of the study were considered to represent quality improvement activities and therefore were exempt from review.
Results
Rapid amplification of short-length nucleic acid targets using ITM-LTD
Four short-length nucleic acid targets (70, 100, 150 and 180 bp) were amplified with three programs simultaneously (Figure 1C). Rapid amplification was performed effectively in 12 min with Program 3. The reaction times of the Program 1 and 2 were 65 and 32 min, respectively. Program 1 and Program 3 had good and similar amplification effects, while the performance of Program 2 was relatively poor (Figure 1C).
Rapid reaction condition and duration of SARS-CoV-2 test in the ultra-fast real-time PCR instrument
The ultra-fast instrument monitors the temperature of the reaction mixture instead of the temperature of the heating module. We optimized the reaction time of reverse transcription, pre-denaturation and cycling conditions of the SARS-CoV-2 test. Effective reverse transcription reactions were carried out at a reaction time of 4 min or more at 50 °C (Figure 3A); the pre-denaturation reaction time could be shortened to 10 s without affecting the experimental effect (Figure 3B). As to the amplification reaction cycle, the denaturation reaction was set to 0 s (Figure 3C), and the annealing reaction was set to 12 s to achieve effective amplification (Figure 3D). In the end, in order to ensure the effective progress of the test, we set up conservative and rapid test condition as following: 50 °C for 5 min, 94 °C for 20 s; 40 cycles of 94 °C for 1 s and 55 °C for 14 s. Under the standard amplification reaction conditions, the reaction time using ABI-7500 instrument and ultra-fast instrument were 81 and 62 min, respectively; while under rapid amplification reaction conditions, the reaction time using ultra-fast instrument was greatly shortened to 31 min.
Figure 3.
SARS-CoV-2 amplification reaction condition optimization in the novel ultra-fast real-time PCR instrument. The rapid test conditions including the reaction time of reverse transcription (A), pre-denaturation (B) and cycling conditions (C,D) in the ultra-fast real-time PCR instrument were optimized. H, M, and L represented high, medium and low concentration of SARS-CoV-2 RNA. Standard represented the standard test of SARS-CoV-2. Each experiment was repeated independently three times, and the mean values and standard deviations are shown. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Specificity, LOD of rapid SARS-CoV-2 test using the ultra-fast real-time PCR instrument
No non-specific amplification of common non-SARS-CoV-2 respiratory pathogens was observed using either the standard or rapid test. To evaluate the LOD of the rapid test, a SARS-CoV-2 positive specimen was amplified using the standard test, and the Ct values for amplification of ORF1a/b and N were 28.51 and 29.17, respectively. The LOD of the standard and rapid tests were compared using a 10-fold dilution series of specimen RNA. Both assays were positive for 100% of 24 replicates of the upper two dilutions. The rapid and standard tests showed similar LOD, amplifying 18/24 vs. 16/24 and 5/24 vs. 6/24 replicates of the lower two dilutions, respectively (Table 1).
Table 1. Limits of detection of the rapid tests of SARS-CoV-2 in the ultra-fast real-time PCR instrument.
| Specimen dilutiona | Rapid test, no. of positive/no. of replicates (%) | Standard test, no. of positive/no. of replicates (%) | Hit rate (rapid test/standard test), % |
|---|---|---|---|
| 1:10 | 24/24 (100.0) | 24/24 (100.0) | 100 |
| 1:100 | 24/24 (100.0) | 24/24 (100.0) | 100 |
| 1:1,000 | 18/24 (75.0) | 16/24 (66.7) | 112.5 |
| 1:10,000 | 5/24 (20.8) | 6/24 (25.0) | 83.3 |
| Negative | 0/24 (0) | 0/24 (0) | 100 |
a, the Ct values of the undiluted SARS-CoV-2-positive specimen were 28.51 (ORF1a/b) and 29.17 (N). SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Clinical performance
A total of 184 frozen clinical specimens from patients with suspected SARS-CoV-2 infections were simultaneously tested with the standard and rapid tests at two study sites. The performance of the SARS-CoV-2 rapid test was assessed by comparing with the standard test which has been verified.
Agreement between the rapid and the standard tests
Positive agreement between the rapid and standard tests was 100% (67/67), negative agreement was 97.4% (114/117), and total agreement was 98.4% (181/184) (Table 2). The kappa statistic was 0.965 (P<0.001).
Table 2. The performance of the SARS-CoV-2 rapid tests in 184 clinical specimens.
| Rapid test (no. of specimens) | Standard test (No. of specimens) | Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 67 | 3 | 70 |
| Negative | 0 | 114 | 114 |
| Total | 67 | 117 | 184 |
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Ct value distribution of clinical specimens
The Ct value distributions of the clinical specimens were analyzed. The median Ct values of ORF1a/b, N, RNase P genes in standard tests and rapid tests were 27.95, 29.94, 30.43 and 27.98, 29.41, 29.96, respectively (Figure 4). No significant differences were found in the Ct value distributions of each gene using the standard and rapid tests (P>0.05). The upper and lower quartiles of Ct values for ORF1a/b in the standard test were wider than those in the rapid test, but were similar for the other two genes (Figure 4, Table S2).
Figure 4.
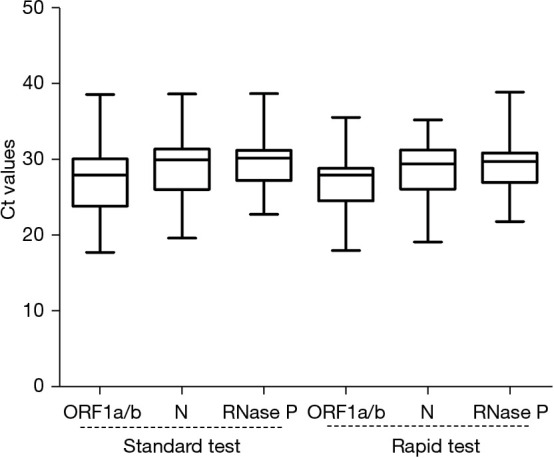
Ct value distributions of clinical specimens assayed using the rapid and the standard tests. Lines within boxes represent medians. The upper and lower boundaries of the boxes represent upper and lower quartiles. The bars represent minimum and maximum values.
Specimens with discrepant test results
Only three specimens showed discrepant results between the standard and rapid tests. All three specimens showed positive results for the N gene using the rapid test but negative results using the standard test (Table 3). The specimens were re-examined using two test conditions and similar results were obtained. The original records also showed that the specimens were borderline positive with Ct values >35 for the N gene.
Table 3. Specimens showing discrepant results using the rapid and standard tests of SARS-CoV-2.
| Specimen ID | Standard test (Ct value) | Rapid test (Ct value) | |||||
|---|---|---|---|---|---|---|---|
| ORF1a/b | N | RNase P | ORF1a/b | N | RNase P | ||
| 8075 | Negative | Negative | 28.05 | Negative | 34.93 | 27.53 | |
| 5994 | Negative | Negative | 24.16 | Negative | 35.05 | 24.08 | |
| 0981 | Negative | Negative | 23.57 | Negative | 35.05 | 23.08 | |
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Discussion
Prevention and control of the COVID-19 still faces huge challenges. Vaccines and active prevention and control measures are currently important means to defeat the pandemic, but vaccines will take a long time to vaccinate. Active pandemic prevention measures require a combination of early diagnosis and early intervention (14-21).
Many detection methods have been used for COVID-19 diagnosis, including NAATs (13), chest computed tomography (22,23), serological tests (24), antigens tests (25,26), and virus culture (27). Among NAATs, real-time reverse transcription PCR–based detection of SARS-CoV-2 RNA from respiratory samples is the gold standard for diagnosis. As the progress of the pandemic, low-cost, rapid test methods are urgently needed. Several rapid diagnostic methods have already been implemented in clinical practice (28,29), but additional methods are still needed to meet diverse application scenarios. Such methods could be conducive to the diagnosis of suspected infections in a wider range of individuals and regions.
In this study, we established and evaluated a rapid SARS-CoV-2 detection method that generates results in approximately 30 min using our novel ultra-fast real-time PCR instrument which based on ITM-LTD. The rapid test was mainly achieved from following aspects: (I) the PCR reaction temperature change mode was changed from the temperature rise and fall of a single module to the mechanical movement of PCR tubes between independent high and low temperature modules. (II) A large temperature difference between the temperature modules made further accelerating of the temperature change rate in the reaction mixture (Figure 1). In the SARS-CoV-2 tests, the same standard amplification conditions were performed in ABI7500 and the ultra-fast PCR instruments, the time-consuming was significantly decreased from 81 to 62 min, indicating the validity of the ITM-LTD. (III) Amplification of short fragments could be effectively done in the process of the temperature changes, which provides a basis for amplification reaction condition optimization. Amplification targets of Taqman real-time PCR are usually short-length. In this work, 70–180 bp short-length targets were effectively amplified in the temperature change process, as long as the temperature of the reaction mixture reached the required temperature (Program 1 vs. Program 3, Figure 1). The poor performance of Program 2 might be because the reaction mixture did not reach the predetermined temperature, because the PCR instrument monitored the temperature of the thermal cycler module instead of the reaction mixture. Basing on this, we optimized the rapid amplification conditions for SARS-COV-2 (Figure 3) and made the reaction time greatly shortened.
In this work, we developed an ultra-fast real-time PCR instrument using the principal of ITM-LTD. The instrument helped the PCR reaction mixture but not thermal cycle module to achieve the fastest temperature change >10 °C/s (Figure 2). The instrument which had a small size and four fluorescence channels with a flux of 8, provided a portable, low-cost equipment that could be used in a variety of test environments.
The rapid test of SARS-CoV-2 was established using the ultra-fast real-time PCR instrument with a CFDA certified-SARS-CoV-2 reagent and optimized reaction conditions. We evaluated the rapid test by comparing it with the standard test following the manufacturer’s protocols. The rapid test had excellent specificity and sensitivity compared with the standard test. No non-specific amplification of common non-SARS-CoV-2 respiratory pathogen-containing samples was observed. The rapid and standard tests showed similar sensitivity; in fact, the rapid test detected a higher proportion of 1:1,000-diluted SARS-CoV-2 RNA replicates (75%) than the standard test (66.7%) (Table 1). The rapid test had a significantly shorter reaction time than the standard test. Our results indicate that for amplification of short nucleic acid fragments, there is significant flexibility for optimization of reaction conditions without affecting specificity and sensitivity.
Our results showed excellent clinical performance of both the rapid and standard tests using 184 specimens from individuals with suspected SARS-CoV-2 infection (kappa =0.965) (Table 2). The Ct value distributions of the target genes were similar between the rapid and standard tests (P>0.05) (Figure 4). Three discrepant test specimens by the two methods were all positive for the N gene in the rapid test but negative in the standard test (Table 3).
The rapid SARS-CoV-2 detection method developed in this study had the following advantages and limitations. First, the detection speed was fast. Rapid detection can greatly reduce waiting time for test results in cases of suspected infection and reduce the risk of disease transmission caused by the movement of individuals with early infection (8). Second, the miniaturization of the rapid test instrument may reduce the cost of testing, making the test conducive to a wider range of applications. Third, the rapid test can use existing technology and resource integration to achieve rapid implementation. These factors will directly or indirectly reduce the social and economic costs of pandemic prevention. However, we should also note that the rapid test established in this study also has problems such as low detection throughput, which is not suitable for large-scale screening work.
In Conclusion, we developed a test for rapid diagnosis of COVID-19 that yields results in approximately 30 min through the use of commercial SARS-CoV-2 reagents in combination with our novel ultra-fast real-time PCR instrument. The method has high sensitivity and specificity, and it showed excellent agreement with the standard test procedure using clinical specimens. Efficient, fast and low-cost diagnosis of COVID-19 or other emerging infectious diseases may enable real-time detection in fever clinics, airports, customs services, and primary medical units.
Supplementary
The article’s supplementary files as
Acknowledgments
We thank Changbing Wang and Bing Zhu (Guangzhou Women and Children’s Medical Center), and Yinhua Zhou (GIRM Biosafety (Guangzhou) Co., Ltd.) for technical assistance.
Funding: This study was supported by Emergency Key Program of Guangzhou Laboratory (EKPG21-13); the Guangzhou Science and Technology Program-Zhongnanshan Medical Foundation of Guangdong Province (202102010359-ZNSA-2020003); Special Project for COVID-19 Prevention and Control of Zhongnanshan Medical Foundation of Guangdong Province (ZNSA-2020012); Guangdong-Hong Kong-Macao Joint Laboratory of Respiratory Infectious Disease (GHMJLRID-Z-202109); the Research Project of Bioland Laboratory (1105101202) and the National Natural Science Foundation of China (81970003). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was reviewed and approved by The First Affiliated Hospital of Guangzhou Medical University Ethics Committee (No. 2020-77). Parts of the study were considered to represent quality improvement activities and therefore were exempt from review.
Footnotes
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://dx.doi.org/10.21037/jtd-21-1288
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jtd-21-1288
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jtd-21-1288). LL reports that she is an employee of GIRM Biosafety (Guangzhou) Co., Ltd.; LW, LXS and YL report that they are employees of Guangzhou Kingmed Center for Clinical Laboratory Co., Ltd. The companies had no role in study design, data collection and interpretation, or the decision to submit the work for publication. RZ reports Guangzhou Laboratory and Bioland Laboratory funding for this study; WKL reports Guangzhou Science and Technology Program-Zhongnanshan Medical Foundation of Guangdong Province, Zhongnanshan Medical Foundation of Guangdong Province, Guangdong-Hong Kong-Macao Joint Laboratory of Respiratory Infectious Disease and National Natural Science Foundation of China funding for this study. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The other authors have no conflicts of interest to declare.
References
- 1.Wiersinga WJ, Rhodes A, Cheng AC, et al. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020;324:782-93. 10.1001/jama.2020.12839 [DOI] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 2020;382:727-33. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan WJ, Ni ZY, Hu Y, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med 2020;382:1708-20. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fontanet A, Cauchemez S. COVID-19 herd immunity: where are we? Nat Rev Immunol 2020;20:583-4. 10.1038/s41577-020-00451-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burki TK. Herd immunity for COVID-19. Lancet Respir Med 2021;9:135-6. 10.1016/S2213-2600(20)30555-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plante JA, Liu Y, Liu J, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021;592:116-21. 10.1038/s41586-020-2895-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh PK, Kulsum U, Rufai SB, et al. Mutations in SARS-CoV-2 Leading to Antigenic Variations in Spike Protein: A Challenge in Vaccine Development. J Lab Physicians 2020;12:154-60. 10.1055/s-0040-1715790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rong XM, Yang L, Chu HD, et al. Effect of delay in diagnosis on transmission of COVID-19. Math Biosci Eng 2020;17:2725-40. 10.3934/mbe.2020149 [DOI] [PubMed] [Google Scholar]
- 9.Triggle CR, Bansal D, Farag EABA, et al. COVID-19: Learning from Lessons To Guide Treatment and Prevention Interventions. mSphere 2020;5:00317-20. 10.1128/mSphere.00317-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J, Yuan Q, Wang H, et al. Antibody Responses to SARS-CoV-2 in Patients With Novel Coronavirus Disease 2019. Clin Infect Dis 2020;71:2027-34. 10.1093/cid/ciaa344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haveri A, Smura T, Kuivanen S, et al. Serological and molecular findings during SARS-CoV-2 infection: the first case study in Finland, January to February 2020. Euro Surveill 2020;25:2000266. 10.2807/1560-7917.ES.2020.25.11.2000266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W, Liu L, Kou G, et al. Evaluation of Nucleocapsid and Spike Protein-Based Enzyme-Linked Immunosorbent Assays for Detecting Antibodies against SARS-CoV-2. J Clin Microbiol 2020;58:e00461-20. 10.1128/JCM.00461-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang YW, Schmitz JE, Persing DH, et al. Laboratory Diagnosis of COVID-19: Current Issues and Challenges. J Clin Microbiol 2020;58:e00512-20. 10.1128/JCM.00512-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zu J, Li ML, Li ZF, et al. Transmission patterns of COVID-19 in the mainland of China and the efficacy of different control strategies: a data- and model-driven study. Infect Dis Poverty 2020;9:83. 10.1186/s40249-020-00709-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou C, Chen J, Zhou Y, et al. The effectiveness of quarantine of Wuhan city against the Corona Virus Disease 2019 (COVID-19): A well-mixed SEIR model analysis. J Med Virol 2020;92:841-8. 10.1002/jmv.25827 [DOI] [PubMed] [Google Scholar]
- 16.Hellewell J, Abbott S, Gimma A, et al. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob Health 2020;8:e488-96. 10.1016/S2214-109X(20)30074-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matrajt L, Leung T. Evaluating the Effectiveness of Social Distancing Interventions to Delay or Flatten the Epidemic Curve of Coronavirus Disease. Emerg Infect Dis 2020;26:1740-8. 10.3201/eid2608.201093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kretzschmar ME, Rozhnova G, Bootsma MCJ, et al. Impact of delays on effectiveness of contact tracing strategies for COVID-19: a modelling study. Lancet Public Health 2020;5:e452-9. 10.1016/S2468-2667(20)30157-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S, Zhang Z, Yang J, et al. Fangcang shelter hospitals: a novel concept for responding to public health emergencies. Lancet 2020;395:1305-14. 10.1016/S0140-6736(20)30744-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang D, Pan S, Li Z, et al. Large-scale public venues as medical emergency sites in disasters: lessons from COVID-19 and the use of Fangcang shelter hospitals in Wuhan, China. BMJ Glob Health 2020;5:e002815. 10.1136/bmjgh-2020-002815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu P, Zhang H, Long X, et al. Management of COVID-19 patients in Fangcang shelter hospital: clinical practice and effectiveness analysis. Clin Respir J 2021;15:280-6. 10.1111/crj.13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hani C, Trieu NH, Saab I, et al. COVID-19 pneumonia: A review of typical CT findings and differential diagnosis. Diagn Interv Imaging 2020;101:263-8. 10.1016/j.diii.2020.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Xia L. Coronavirus Disease 2019 (COVID-19): Role of Chest CT in Diagnosis and Management. AJR Am J Roentgenol 2020;214:1280-6. 10.2214/AJR.20.22954 [DOI] [PubMed] [Google Scholar]
- 24.Lee CY, Lin RTP, Renia L, et al. Serological Approaches for COVID-19: Epidemiologic Perspective on Surveillance and Control. Front Immunol 2020;11:879. 10.3389/fimmu.2020.00879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baek YH, Um J, Antigua KJC, et al. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg Microbes Infect 2020;9:998-1007. 10.1080/22221751.2020.1756698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scohy A, Anantharajah A, Bodéus M, et al. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J Clin Virol 2020;129:104455. 10.1016/j.jcv.2020.104455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang CG, Lee KM, Hsiao MJ, et al. Culture-Based Virus Isolation To Evaluate Potential Infectivity of Clinical Specimens Tested for COVID-19. J Clin Microbiol 2020;58:e01068-20. 10.1128/JCM.01068-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basu A, Zinger T, Inglima K, et al. Performance of Abbott ID Now COVID-19 Rapid Nucleic Acid Amplification Test Using Nasopharyngeal Swabs Transported in Viral Transport Media and Dry Nasal Swabs in a New York City Academic Institution. J Clin Microbiol 2020;58:e01136-20. 10.1128/JCM.01136-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loeffelholz MJ, Alland D, Butler-Wu SM, et al. Multicenter Evaluation of the Cepheid Xpert Xpress SARS-CoV-2 Test. J Clin Microbiol 2020;58:e00926-20. 10.1128/JCM.00926-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as