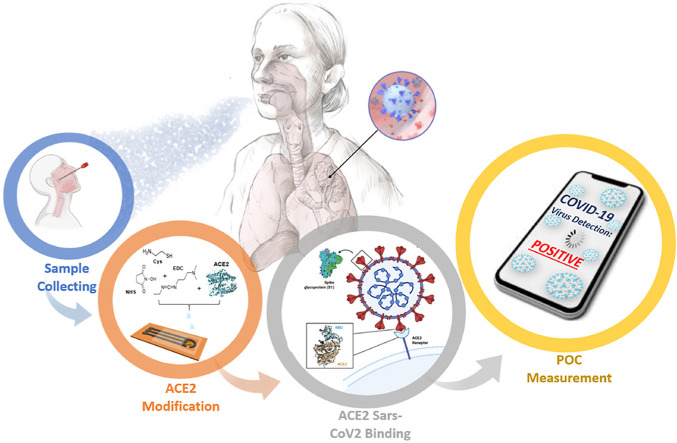
Abstract
Point of care (PoC) devices are highly demanding to control current pandemic, originated from severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2). Though nucleic acid-based methods such as RT-PCR are widely available, they require sample preparation and long processing time. PoC diagnostic devices provide relatively faster and stable results. However they require further investigation to provide high accuracy and be adaptable for the new variants. In this study, laser-scribed graphene (LSG) sensors are coupled with gold nanoparticles (AuNPs) as stable promising biosensing platforms. Angiotensin Converting Enzyme 2 (ACE2), an enzymatic receptor, is chosen to be the biorecognition unit due to its high binding affinity towards spike proteins as a key-lock model. The sensor was integrated to a homemade and portable potentistat device, wirelessly connected to a smartphone having a customized application for easy operation. LODs of 5.14 and 2.09 ng/mL was achieved for S1 and S2 protein in the linear range of 1.0–200 ng/mL, respectively. Clinical study has been conducted with nasopharyngeal swabs from 63 patients having alpha (B.1.1.7), beta (B.1.351), delta (B.1.617.2) variants, patients without mutation and negative patients. A machine learning model was developed with accuracy of 99.37% for the identification of the SARS-Cov-2 variants under 1 min. With the increasing need for rapid and improved disease diagnosis and monitoring, the PoC platform proved its potential for real time monitoring by providing accurate and fast variant identification without any expertise and pre sample preparation, which is exactly what societies need in this time of pandemic.
Keywords: COVID-19, SARS-CoV-2, Sensor, Machine learning, Point-of-care, Laser-scribed graphene
Graphical abstract
1. Introduction
Coronavirus disease 2019 (COVID-19), currently causing a global pandemic, originated from severe acute respiratory syndrome–coronavirus 2 (SARS-CoV-2) has been infectiously expanding around the globe and highly effecting health care and economy. The spike glycoproteins, protruding from SARS-CoV-2 membrane, are the most reachable protein units providing direct entry into host cells. Two subunits of the spike glycoprotein, spike 1 (S1) and (S2), contain the receptor binding domain, enabling the cell fusion of viral into cell membranes. As a crucial cellular receptor, Angiotensin-converting enzyme 2, known as ACE2, enables the direct entry the virus into the host cell(Idili et al., 2021). ACE2 is originally known as playing important role in angiotensin (Ang) mutation process. SARS-CoV-2 primarily cause infection by targeting ACE2 receptors, which are widespread in the lungs. Recent studies show that SARS-CoV-2 uses both subunits, S1 and S2, to enter the host cell(Davidson et al., 2020). During viral infection, S1 and S2 receptor recognition and membrane fusion of subunits is proven to be binding directly to the peptidase of ACE2 domain(Lee et al., 2021). While S1 binds to the ACE2 receptor directly, S2 is correlated with the membrane fusion(Crackower et al., 2002; Li et al., 2005).
The importance of monitoring the pandemic is remains critical to manage the situation in most efficient way(Pokhrel et al., 2020). Various detection methods including reverse transcription polymerase chain reaction (RT-PCR), widely known as the gold standard method, lateral flow immunoassays (LFIs) or CRISPR-Cas9-linked real time loop-mediated isothermal amplification (RT-LAMP) technologies, have been generated(Broughton et al., 2020; Chaibun et al., 2021; Ghorbanizamani et al., 2021a; Rahimi et al., 2021; Yu et al., 2020). However, viral particles may be affected during long processes of storage and transportation. As a result of insufficient, poor collection and management of clinical samples during testing, false-negative results may mislead the patients(Lee et al., 2021). Recently, electrochemical systems have been developed to detect both SARS-CoV-2 antibody and antigens(Beduk et al., 2021b; Guo et al., 2021; Li and Lillehoj, 2021; Yakoh et al., 2021). Commercial rapid tests are available in different sizes, shapes and configurations(Ghorbanizamani et al., 2021b; Yetisen et al., 2013). Developing practical systems for SARS-CoV-2 antigen detection in various media are much needed(Ghorbanizamani et al., 2021b). However, further improvements are required for current tastes due to the emerging variants of the virus(Jiang et al., 2021; Liu and Rusling, 2021; Peng et al., 2020). Thus, more specifications must be investigated for accurate testing for mutated variants of SARS-CoV-2. Yan et al. proved that the noncovalent binding of the S1 and S2 proteins happens on ACE2 modified surfaces simultaneously during the infection process(Yan et al., 2020). Moreover, the binding affinity between ACE2 and the S proteins of the coronaviruses is found to be enhanced in mutations(Chan et al., 2020; Lan et al., 2020).
Graphene is a well-known promising material for various chemical or biosensing applications due to its highly advanced electrical and mechanical properties(Behrent et al., 2021; Kaidarova et al., 2020; Novoselov et al., 2012). Additional to various chemical synthesis methods, it was discovered that direct lasing of polymers forms high quality graphene by photothermal reactions(Beduk et al., 2020). The graphene produced by this straightforward and method is called laser scribed graphene (LSG)(Lahcen et al., 2020), used in many applications such as clinical diagnostics including SARS-CoV-2 S1 protein detection(Alhajji et al., 2020; Beduk et al., 2021a; Fenzl et al., 2017; Ghanam et al., 2020; Rauf et al., 2021b; Zhu et al., 2021).
In this study, we improved the previously reported affinity based, gold nanoparticle (AuNP) modified LSG sensor system for Alpha (B.1.1.7), Beta (B.1.351) and Delta (B.1.617.2) variants of SARS-CoV-2(Beduk et al., 2021a). To the best of our knowledge, this is the first LSG based electrochemical platform specifically structured for new variants by detecting SARS-CoV-2 S1 and S2 proteins using ACE2 receptor, which can form a matched pair with SARS-CoV-2 antigens. Since the validation of the sensor performance is very crucial for the self-diagnostic platform constructed in this study, a Dense Neural Network (DNN) architecture was established as a machine learning (ML) method. The final product was integrated to a home-made potentiostat to create a point-of-care (PoC) system including a smartphone attachment for a reliable, portable, and widely accessible SARS-CoV-2 detection for virus mutations. Clinical study carried out with 63 patients nasopharyngeal swabs collected from patients with different mutations and uninfected individuals to observe the affinity between ACE2 receptor and SARS-CoV-2 spike antigens, compared with clinically approved methods. The identification of the SARS-CoV-2 variants, B.1.1.7, B.1.351 and B.1.617.2, was achieved by the ML approach with 95.65% accuracy based on the direct current response of our sensor.
2. Experimental
2.1. Preparation of the electrochemical sensor
A CO2 laser was used for laser irradiation on polyimide substrate, then coated with gold (Rauf et al., 2021a). Further details about the materials, sensor fabrication and functionalization can be found in Supplementary Information (SI). The gold surface was functionalized by immobilizing 1 mg/mL of cysteamine to create amine functionalities. The surface then was to clean the excess unreacted cysteamine. Next, 10 μL of 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC): N- hydroxy succinimide (NHS) mixture with 50:50 mM concentration ratio was immobilized onto the amine functional AuNPs-LSG surface for 4 h. The excess reagent was rinsed with phosphate buffered saline (PBS) solution. Following the immobilization steps for the biomolecule binding, 10 μL from a human ACE2 receptor solution with concentration of 200 ng/mL was prepared in PBS solution and incubated for 16 h. All biofunctionalization steps were performed at 4 °C. After each incubation, working area of the sensor was rinsed with PBS solution to remove the excess solution. For the surface stabilization, 10 μL solution from a bovine serum albumin (BSA) solution prepared with 0.1 mg/mL concentration was immobilized on the sensor for 1 h. The aim of using BSA blocking as the last step is to eliminate the rest of the electroactive surface of the working area prior to analyte detection to avoid nonspecific interactions. The electrodeposition parameters, binder concentrations and incubation conditions etc., were validated in our previous work (Beduk et al., 2021a).
2.2. Preparation of nasopharyngeal swabs
Commercial RT-PCR test was performed for 63 clinical nasopharyngeal swabs by the hospital laboratory. Obtained COVID-19 positive swabs were classified according to the SARS-CoV-2 virus variants such as B.1.1.7 B.1.351 and B.1.617.2 by commercial RT-PCR. In particular, 10 samples of B.1.351 variant, 18 samples of B.1.1.7 variant, 23 samples of B.1.617.2 variant, 4 COVID-19 patients without any mutation and 8 negative control patients were obtained to test AuNPs-LSG sensor performance. Prior to electrochemical measurements, swabs were diluted at 3:4 ratio with PBS solution and incubated on working electrode for 2 h. After the incubation, the working surface was rinsed with PBS solution as the final step. For the interference study, Influenza B, Rhinovirus, Respiratory Syncytial Virus, Influenza A (H1N1), Human Coronavirus 229E/NL63 and Adenovirus/Rhinovirus samples were used in viral transport medium containing penicillin, streptomycin, and gentamicin with 1000-fold dilution by 0.1 M PBS.
3. Result and discussion
3.1. Characterization of the AuNPs-LSG sensor
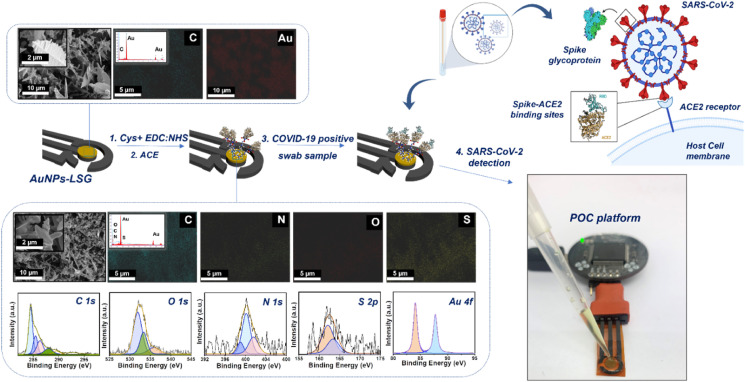
Fig. 1 summarizes the sensor production steps and portable potentiostat attachment. Scanning electron microscopy (SEM) data shows that the gold nanoparticle decorated surface is observed as highly rough and porous, while the surface structures have softer edges and different polarity after the binder and bioreceptor immobilizations. Further characterizations can be found in Figs. S1–S2 with details. The high resolution spectra of fully fabricated sensor (LSG:AuNPs:Cys:EDC:NHS:ACE2:BSA) shows that the surface is rich in C, N and S after functionalization steps due to the presence of ammino and carboxyl groups. The atomic percentages (%) given in Table S2 supports the presence of amino and carboxyl groups on the surface, while proving that S content exists only after the cysteamine deposition and N content increased after ACE2 receptor addition to the surface.
Fig. 1.
Representation of fabrication step of AuNPs-LSG sensor including (a–b) Scanning electron microscopy (SEM) images with corresponding elemental mapping images at a scale bar of 5 μm and 10 μm, (c) X-ray photoelectron spectroscopy (XPS) data containing C1s, O 1s, N 1s, S 2p, and Au 4f7 spectra of AuNPs-LSG sensor.
The high resolution spectra of fully fabricated sensor (LSG:AuNPs:Cys:EDC:NHS:ACE2:BSA) proves the presence of binders and ACE2 domain on the surface (Fig. 1c). The atomic percentages (%) show the elemental composition on the working area at each modification step are given in Table S1, resulting that S 2p content exists only after the cysteamine deposition. Moreover, N1s content has increased on the surface after ACE2 domain attachment. As the final step, the incubation of patient nasopharyngeal swab sample accelerates the increase in C1s and O1s content due to the presence of spike 1 and 2 proteins attached on the sensor surface.
3.2. Investigation of the sensitivity and selectivity of the AuNPs-LSG sensor
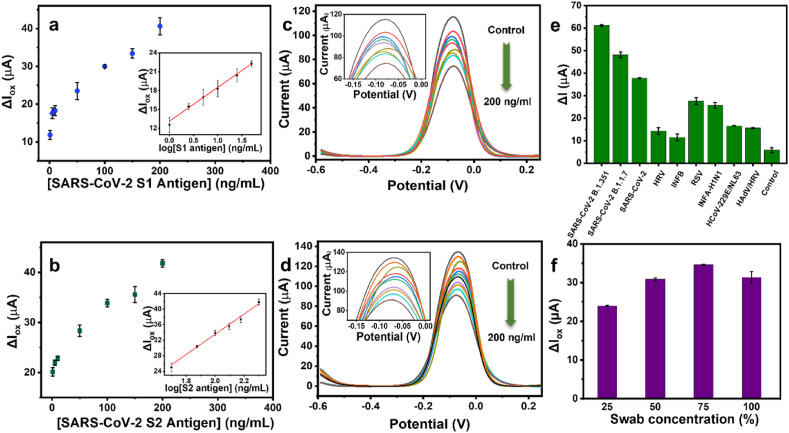
The experimental conditions, electrodeposition parameters, incubation time etc., were validated in our previous work(Beduk et al., 2021a). The difference in the oxidation current of the sensor (ΔIox) before and after spike protein incubation is considered as the sensor response, increasing depending on the spike protein concentration. Oxidation current difference (μA) between and ACE2-antigen attachment depending on the S1 and S2 protein concentration (ng/mL) is demonstrated in Fig. 2 . In particular, oxidation current decrease in the electrochemical response with increasing concentration of analyte. This trend has been validated by the calibration curves of S1 and S2 protein attachments in Fig. 2. Detailed explanation about the LOD calculations can be found in SI. In addition, selectivity test was performed in presence of various virus types mainly causing other respiratory diseases. Fig. 2e shows DPVs corresponding the oxidation current change of Influenza B, Rhinovirus, Respiratory Syncytial Virus, Influenza A (H1N1), Human Coronavirus 229E/NL63, Adenovirus/Rhinovirus, SARS-CoV-2 B.1.351, B.1.1.7 and B.1.617.2 variants, SARS-CoV-2 without mutation and control samples incubated onto the AuNPs-LSG sensor. CT values of the virus samples can be found in Table S2 Table S1. The relatively lower current responses were obtained from respiratory viruses compared to SARS-CoV-2 types, while slight attraction of ACE2 domain was observed towards influenza A and RSV, resulting a current response compared to other virus types. It is clearly observed that the current responses of respiratory viruses are lower compared to SARS-CoV-2 types. However, relatively high attraction of ACE2 domain towards influenza A and RSV has been observed in Fig. 2e, resulting relatively higher current response compared to other virus types. While the influenza viruses bind through the receptors called sialic acid, found in respiratory epithelial cells, other coronavirus types also have different −229E/NL63-receptors. Thus, only receptors of SARS-CoV and SARS-CoV-2 are observed as binding to ACE2 domain. is found to be ACE2 domain. As the binding receptors of other respiratory viruses are particularly different than SARS-CoV-2, their binding affinity towards ACE2 domain is naturally lower (Chang et al., 2020; Flerlage et al., 2021). Thus, the developed sensor has acceptable selectivity towards SARS-CoV-2 among other respiratory viruses.
Fig. 2.
Oxidation response of the AuNPs-LSG sensor with magnified DPV peaks corresponding (a, c) SARS-CoV-2 S1 and (b, d) S2 protein various concentrations with respect to LSG RE. The insets indicate the logarithmic relation of the oxidation current difference (ΔIox) and S1 and S2 protein concentration (logCS1 and logCS2). Histograms showing (e) the response of the AuNPs-LSG sensor towards different viruses including Influenza B (INFB) (5.59 %RSD), Human Rhinovirus (HRV) (10.5 %RSD), Respiratory Syncytial Virus (RSV) (4.93 %RSD), Influenza A (H1N1) (INFA-H1N1) (0.85 %RSD), Human Coronavirus 229E/NL63 (HCoV-229E/NL63) (1.02 %RSD), Adenovirus/Rhinovirus, (HAdV/HRV) (1.97 %RSD), SARS-CoV-2 B.1.1.7 (2.59 %RSD), B.1.351 variants (0.46 %RSD), SARS-CoV-2 without mutation (0.58 %RSD) and control (negative) sample (18.7 %RSD),; (f) the change in oxidation current for nasopharyngeal swab dilution percentages 25% (0.65 %RSD), 50% (1.03 %RSD), 75% (0.47 %RSD), 100% (4.76 %RSD). (Error bars: ± SD for n = 3 for different sensors). 5.0 mM [Fe (CN)6]3–/4– containing 0.1 M PBS and 0.1 M KCl was used as redox buffer at 50 mV/s scan rate.
3.3. SARS-CoV-2 variant detection in human nasopharyngeal swabs
Prior to any patient sample measurements, dilution amount was optimized for patient samples. ΔIox values calculated from DPVs of corresponding 25, 50, 75 and 100% of swab dilutions were given in Fig. 2f, resulting the best response from 75% of dilution. Thus, patient samples were diluted by 75% with 0.05 M PBS and incubated onto AuNPs-LSG sensor without further purification and filtration in order to keep the virus concentration unaffected. The mean values and the corresponding standard deviation (n = 3) were considered for each measurement.
Table S2 S3 contains examples for the use of ACE2 domain in the sensor production for SARS-CoV-2 detection. Knowing that ACE2 domain and SARS-CoV-2 spike proteins have high binding affinity, various detection models including optical sensors, plasmonic sensors, LFIs and electrochemical sensors have been developed for COVID-19 diagnosis based on ACE2 domain. In particular, Lee et al. have developed a LFI platform based on pairing between ACE2 receptor and S1 antibody. Herein, LSG based electrochemical sensing platform offers an affordable solution for SARS-CoV-2 virus detection by pairing ACE2 receptor and spike antigens without the need of antibodies. Though our system requires further investigation to be able to perform fully quantitative identification, it has introduced as proof of concept for future diagnostics as the production processes relatively straightforward, keeping the time consumption minimum with high accuracy(Lee et al., 2021).
ACE2 receptor, a prior entry receptor for SARS-CoV-2 virus, was used as the functionalization since the binding affinity between ACE2 and the S1, S2 spike proteins enhanced in mutations. The interaction between ACE2 domain and spike antigen is directly correlated to the analytical performance and the stability of the sensor. This interaction is considered as a very promising for sensitive SARS-CoV-2 variant detection since this peptidase domain has high possibility to interact the glycoproteins via hydrogen bonding. Moreover, the combination of ACE2 domain and highly conductive gold nanoparticles on the surface leads to the possibility of enhanced H-bond formation(Mehranfar and Izadyar, 2020). However, existing methods often suffer from instrumental complexity and the long sample collection/preparation processes. In this work, the affinity of ACE2 domain towards SARS-CoV-2 spike proteins is used for the variant detection on LSG platform for the first time. Aiming to detect newly emerging SARS-CoV-2 variants, this study serves as an important starting point of understanding the need of updating current diagnostic methods. To increase the availability and practicality, the LSG platform is combined with our home-made portable potentiostat. Thus, compared to the existing COVID-19 diagnostic methods, our PoC platform provides high sensitivity and accuracy while offering high accessibility in both field and personal applications and identification of the virus specific variants.
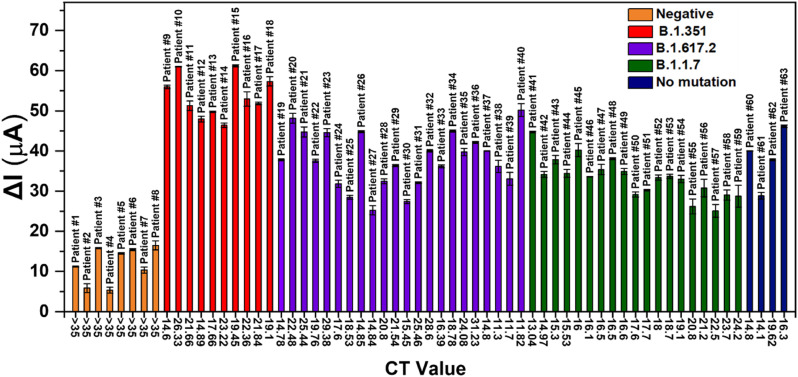
Fig. 3 shows the oxidation current response of 63 nasopharyngeal swabs collected from COVID-19 patients with alpha, beta and delta variant, patients having SARS-CoV-2 without any mutation and control patients (negative). The binding of S1 and S2 protein of viral components in patient samples inhibit the electrochemical response, eventually lowers the oxidation current in positive patient samples. Relatively high current response was obtained from samples with B.1.351 variant compared to the response of other variants. This behavior is most likely correlated with the high binding affinity of ACE2 domain towards the spike proteins on the surface as the active biological receptor, since the mutations on spike proteins vary in different variants of SARS-CoV-2. The response of ACE2-functionalized and swab-immobilized sensor were evaluated statistically relying on the t-test considering paired two sample for means, proving that there is no difference between the sensor response following ACE2 and swab immobilization statistically for the negative patient samples (Tables S4–S5). Moreover, the difference between the response of patients with B.1.351 variant and B1.1.7 variant was tested statistically By ANOVA test, resulting that the AuNPs-LSG sensor response towards two SARS-CoV-2 variants are statistically different.
Fig. 3.
Histogram showing the change in oxidation current values (ΔIox) of 63 nasopharyngeal swabs collected from control (negative) patients (orange), COVID-19 positive patients without any mutation (blue), with B.1.351 variant (red), B1.1.7 variant (purple) and B.1.617.2 variant (green). (Error bars: ± SD for n = 3 for different sensors). 5.0 mM [Fe (CN)6]3–/4– containing 0.1 M PBS and 0.1 M KCl was used as redox buffer at 50 mV/s scan rate. RSD values are given in Table S6 for n = 3. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Investigation of the AuNPs-LSG sensor into point of care platform
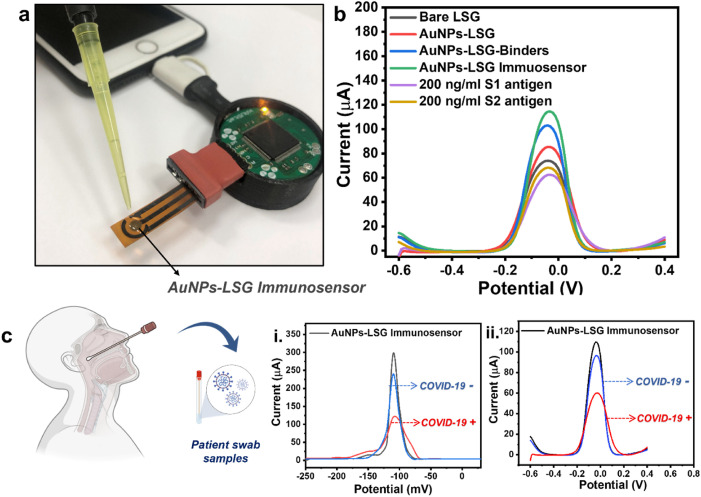
A home-made electrochemical analyzer, KAUSTat, developed in our previous study, is used for the COVID-19 diagnosis(Beduk et al., 2021a). The components of the device are shown in Fig. 4 a and Fig. S3 with the sensor attachment(Ahmad et al., 2019). The device enables a smartphone connection by a micro-USB port. The detailed explanation about the hardware can be found in SI including input/output peripherals (Fig. S4) and the circuitry (Fig. S5) explaining the main components as the Programmable System on Chip (PSoC) 5LP. KAUSTat connects directly to mobile application software consisting of multiple operation options such as connection, parameter, control, and data visualization windows, shown in Fig. S6.
Fig. 4.
(a) The portable PoC device with a smartphone connection by a USB-C connection including sensor attachment. (b) DPVs of the AuNPs-LSG sensor showing ΔIox (the oxidation current change) after each modification and after detecting 200 ng/mL of SARS-CoV-2 S1 and S2 antigens. (c) DPVs showing AuNPs-LSG sensor response for the swabs obtained from COVID-19 positive (+) and negative (−) patients by (i) the portable PoC device and (ii) the commercial potentiostat. (Error bars: ± SD for n = 3 for different sensors). 5.0 mM [Fe (CN)6]3–/4– containing 0.1 M PBS and 0.1 M KCl was used as redox buffer at 50 mV/s scan rate.
Fig. 4b shows the DPV response of the sensor at each fabrication step. A current increase observed in DPV response following the electrodeposition of gold nanoparticles on working electrode surface since the metal nanoparticle deposition enhances the conductivity and electroactivity at the electrode–electrolyte interface. The current continued to increase after cysteamine, EDC:NHS treatment and ACE2 domain attachment since the active ammonium groups are rich on the surface, attracting the negative ions at the electrode surface-redox solution interface, eventually leading to the high electron transfer. The incubation of non-electroactive S1 and S2 antigens on the surface leads to current decrease by the hindrance. Fig. 4c demonstrates the difference in DPV responses of the AuNPs-LSG sensor towards negative and positive patient samples measured by the commercial potentiostat and KAUSTat PoC device. Though the current response obtained from two potentiostats are different, we successfully showed that KAUSTat has the ability to identify COVID-19 positive and negative patient samples. Fig. S7 shows the boxplot analysis of patient swab samples showing threshold values of positive alpha, beta and delta variants. The plot is a statistical representation of the data, in which the colored part represents the distribution of 50% of all values, and whiskers represents the 25% of the values that are not close to the mean. The threshold (ΔIox) was defined as 19.49525 μA from the mean between the maximum negative and minimum positive averaged peak current. Thus, KAUSTat has proved that it has the potential of being an accurate and portable electrochemical detection system, compared to the large scale commercial electrochemical set ups.
3.5. Validation the SARS-CoV-2 identification in patient samples by a Tiny Machine Learning (TinyML) enabled PoC device
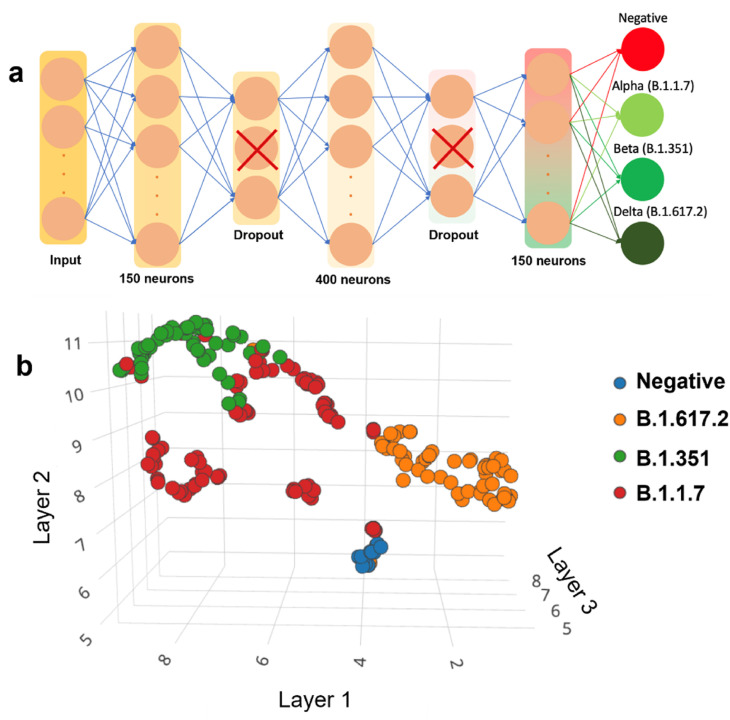
The COVID-19 diagnosis concept was coupled with the variant identification in this study. KAUSTat was improved with a ML concept to create a passible PoC diagnostic platform with high accuracy and availability. To improve and automate the results, a neural algorithm was developed to be able to run the KAUSTat device and detect the spike antigens directly from the oxidation current response. This method allows patients to self-diagnose themselves for COVID-19 from nasopharyngeal swabs. The dataset was then divided in 79% for training and 21% for validating the algorithm. A custom-made code in python was generated to subtract the patient sample response from the baseline, thus the resultant curve was feed to as input to the ML algorithm. A custom algorithm was generated to increase robustness against multiplicative and additive noise and improve performance of our algorithm, yielding in a dataset of 4224 DPV curves (Fig. S8). The details about the database created from the patient samples are given in SI. A block diagram with step by step of how we created our algorithm and the accuracy confusion matrix is shown in Fig. S9 and Table S7 . The spatial representation of the dataset is given in the Fig. 5 a. The dataset was then used to train a DNN using Edge Impulse IDE with EON Compiler. The Edge Impulse IDE combines different data processing tools to easily design and train neural networks. In this work, it was designed a DNN of 5 layers using Keras Python API running on Little TensorFlow platform. The architecture of the DNN proposed for KAUSTat is displayed in the Fig. 5b. For training this DNN, 200 training cycles were used with a learning rate of 0.0005 and a minimum confidence rating of 0.80, occupying a total space of 153.5 Kbytes. Edge Impulse IDE gave three options for the learning block, classification, Regression, and K-means. As K-means is optimized for recognizing unknown states and to find outliers, and Regression is optimized to predict numeric continuous values. Classification was the suitable choice for this project. The deployment of the DNN was done online, using live classification through data forwarder tool. The KAUSTat was linked to Edge impulse IDE via serial port using Command Line Interface (CLI) library. For offline use and replication, the DNN code was created in two different version, a C library, and an Arduino IDE compatible library.
Fig. 5.
(a) The Dense Neural Network (DNN) architecture prepared using Edge Impulse IDE with EON Compiler. (b) Spatial representation of the dataset collected by measuring nasopharyngeal swabs of COVID-19 positive and negative patients.
The DNN utilizes up to 58.5% of the flash memory and around 3.81% of the RAM memory. Added up to the potenstiostat firmware, the total flash occupancy is 70.45%. The DNN resulted in a 98.7% accuracy in inferring Beta variant, 99.5% accuracy in inferring Alpha variant, while 100% accuracy was obtained in inferring Delta and 99.37% accuracy in control patients. The total combined accuracy of the proposed neural algorithm was of 99.37%. Running at 80 MHz clock, KAUSTat takes around 1 min to read the prepared sensor after patient sample incubation for 2 h and approximately 20 ms to define the patient as positive or negative, and identify the specific type of variant.
4. Conclusions
A versatile electrochemical sensor was developed for the identification of SARS-CoV-2 variants. Having ACE2 receptor on the sensor surface, successful attachment of spike proteins was obtained following the nasopharyngeal swab sample incubation. The device has been successfully applied to 63 nasopharyngeal swabs obtained from COVID-19 patients with B.1.351 variant, compared to other variants. LODs of ACE2-fuctionalized sensor was calculated as 5.14 and 2.09 ng/mL for S1 and S2 protein respectively. The sensor was successfully coupled with a portable and wireless potentiostat to obtain a PoC diagnostic platform. We developed a ML enabled diagnostic model which can be easily adapted to detection of emerging variants. KAUSTat takes 8 ms for the identification of the SARS-CoV-2 variant with 99.37% accuracy. Thus, we have created a portable and cheap diagnostic tool that has a potential to be widely accessible with further optimizations and database. We introduce the proposed device as a proof of concept and this model has potential to be a milestone for future diagnostic tools for further viruses and variants.
Author contributions
S.T., K.N.S., K.T., T.G., B.A., R.S., C.C., and F.Z. conceived and conceptualized the study, provided resources, and acquired funding. D.B., and T.B. performed the characterization and electrochemical measurements. J.I.O.F. developed the potentiostat device, wrote the software for the PoC measurements and conducted the Tiny Machine Learning process. D.B., T.B and D.H. wrote the manuscript, with the supervision of S.T. and K.N.S.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
Authors would like to express their acknowledgments to the financial support of funding from the Ege University, Research Foundation (Project number: TOA-2020-21862), Republic of Turkey, Ministry of Development (Project Grant No: 2016K121190) and King Abdullah University of Science and Technology (KAUST) Smart Health Initiative, Saudi Arabia. In addition, authors thank the laboratories of the Ege University Central Research Testing and Analysis Laboratory Research and Application Center (EGE-MATAL).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biosx.2022.100105.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ahmad R., Surya S.G., Sales J.B., Mkaouar H., Catunda S.Y.C., Belfort D.R., Lei Y., Wang Z.L., Baeumner A., Wolfbeis O.S., Alshareef H.N., Salama K.N. 2019. 2019 IEEE SENSORS; pp. 1–4. [Google Scholar]
- Alhajji E., Wang W., Zhang W., Alshareef H.N. ACS Appl. Mater. Interfaces. 2020;12(16):18833–18839. doi: 10.1021/acsami.0c01135. [DOI] [PubMed] [Google Scholar]
- Beduk T., Ait Lahcen A., Tashkandi N., Salama K.N. Sensor. Actuator. B Chem. 2020;314:128026. [Google Scholar]
- Beduk T., Beduk D., de Oliveira Filho J.I., Zihnioglu F., Cicek C., Sertoz R., Arda B., Goksel T., Turhan K., Salama K.N., Timur S. Anal. Chem. 2021;93(24):8585–8594. doi: 10.1021/acs.analchem.1c01444. [DOI] [PubMed] [Google Scholar]
- Beduk T., Durmus C., Hanoglu S.B., Beduk D., Salama K.N., Goksel T., Turhan K., Timur S. Trac. Trends Anal. Chem. 2021;143:116329. [Google Scholar]
- Behrent A., Griesche C., Sippel P., Baeumner A.J. Microchim. Acta. 2021;188(5):159. doi: 10.1007/s00604-021-04792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Nat. Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaibun T., Puenpa J., Ngamdee T., Boonapatcharoen N., Athamanolap P., O'Mullane A.P., Vongpunsawad S., Poovorawan Y., Lee S.Y., Lertanantawong B. Nat. Commun. 2021;12(1):802. doi: 10.1038/s41467-021-21121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., Procko E. Science. 2020;369(6508):1261. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang E.H., Willis A.L., Romanoski C.E., Cusanovich D.A., Pouladi N., Li J., Lussier Y.A., Martinez F.D. Am. J. Respir. Crit. Care Med. 2020;202(5):753–755. doi: 10.1164/rccm.202004-1343LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crackower M.A., Sarao R., Oudit G.Y., Yagil C., Kozieradzki I., Scanga S.E., Oliveira-dos-Santos A.J., da Costa J., Zhang L., Pei Y., Scholey J., Ferrario C.M., Manoukian A.S., Chappell M.C., Backx P.H., Yagil Y., Penninger J.M. Nature. 2002;417(6891):822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- Davidson A.M., Wysocki J., Batlle D. Hypertension. 2020;76(5):1339–1349. doi: 10.1161/HYPERTENSIONAHA.120.15256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenzl C., Nayak P., Hirsch T., Wolfbeis O.S., Alshareef H.N., Baeumner A.J. ACS Sens. 2017;2(5):616–620. doi: 10.1021/acssensors.7b00066. [DOI] [PubMed] [Google Scholar]
- Flerlage T., Boyd D.F., Meliopoulos V., Thomas P.G., Schultz-Cherry S. Nat. Rev. Microbiol. 2021:1–17. doi: 10.1038/s41579-021-00542-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanam A., Lahcen A.A., Beduk T., Alshareef H.N., Amine A., Salama K.N. Biosens. Bioelectron. 2020;168:112509. doi: 10.1016/j.bios.2020.112509. [DOI] [PubMed] [Google Scholar]
- Ghorbanizamani F., Moulahoum H., Zihnioglu F., Evran S., Cicek C., Sertoz R., Arda B., Goksel T., Turhan K., Timur S. Biosens. Bioelectron. 2021;192:113484. doi: 10.1016/j.bios.2021.113484. [DOI] [PubMed] [Google Scholar]
- Ghorbanizamani F., Tok K., Moulahoum H., Harmanci D., Hanoglu S.B., Durmus C., Zihnioglu F., Evran S., Cicek C., Sertoz R., Arda B., Goksel T., Turhan K., Timur S. ACS Sens. 2021;6(8) doi: 10.1021/acssensors.1c00854. [DOI] [PubMed] [Google Scholar]
- Guo K., Wustoni S., Koklu A., Díaz-Galicia E., Moser M., Hama A., Alqahtani A.A., Ahmad A.N., Alhamlan F.S., Shuaib M., Pain A., McCulloch I., Arold S.T., Grünberg R., Inal S. Nat. Biomed. Eng. 2021;5(7):666–677. doi: 10.1038/s41551-021-00734-9. 2021 07. [DOI] [PubMed] [Google Scholar]
- Idili A., Parolo C., Alvarez-Diduk R., Merkoçi A. 2021. ACS Sensors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N., Tansukawat N.D., Gonzalez-Macia L., Ates H.C., Dincer C., Güder F., Tasoglu S., Yetisen A.K. ACS Sens. 2021;6(6):2108–2124. doi: 10.1021/acssensors.1c00669. [DOI] [PubMed] [Google Scholar]
- Kaidarova A., Alsharif N., Oliveira B.N.M., Marengo M., Geraldi N.R., Duarte C.M., Kosel J. Global Chall. 2020;4(4):2000001. doi: 10.1002/gch2.202000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahcen A.A., Rauf S., Beduk T., Durmus C., Aljedaibi A., Timur S., Alshareef H.N., Amine A., Wolfbeis O.S., Salama K.N. Biosens. Bioelectron. 2020;168:112565. doi: 10.1016/j.bios.2020.112565. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lee J.-H., Choi M., Jung Y., Lee S.K., Lee C.-S., Kim J., Kim J., Kim N.H., Kim B.-T., Kim H.G. Biosens. Bioelectron. 2021;171:112715. doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F., Li, W., Farzan, M., Harrison, S.C., 2005. 309(5742), 1864-1868. [DOI] [PubMed]
- Li J., Lillehoj P.B. ACS Sens. 2021;6(3):1270–1278. doi: 10.1021/acssensors.0c02561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Rusling J.F. ACS Sens. 2021;6(3):593–612. doi: 10.1021/acssensors.0c02621. [DOI] [PubMed] [Google Scholar]
- Mehranfar A., Izadyar M. J. Phys. Chem. 2020;11(24):10284–10289. doi: 10.1021/acs.jpclett.0c02677. [DOI] [PubMed] [Google Scholar]
- Novoselov K.S., Fal′ko V.I., Colombo L., Gellert P.R., Schwab M.G., Kim K. Nature. 2012;490(7419):192–200. doi: 10.1038/nature11458. [DOI] [PubMed] [Google Scholar]
- Peng L., Liu J., Xu W., Luo Q., Chen D., Lei Z., Huang Z., Li X., Deng K., Lin B., Gao Z. J. Med. Virol. 2020 doi: 10.1002/jmv.25936. 2020.2002.2021.20026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhrel P., Hu C., Mao H. ACS Sens. 2020;5(8):2283–2296. doi: 10.1021/acssensors.0c01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi H., Salehiabar M., Barsbay M., Ghaffarlou M., Kavetskyy T., Sharafi A., Davaran S., Chauhan S.C., Danafar H., Kaboli S., Nosrati H., Yallapu M.M., Conde J. ACS Sens. 2021;6(4):1430–1445. doi: 10.1021/acssensors.0c02312. [DOI] [PubMed] [Google Scholar]
- Rauf S., Lahcen A.A., Aljedaibi A., Beduk T., Ilton de Oliveira Filho J., Salama K.N. Biosens. Bioelectron. 2021;180:113116. doi: 10.1016/j.bios.2021.113116. [DOI] [PubMed] [Google Scholar]
- Rauf S., Mani V., Lahcen A.A., Yuvaraja S., Beduk T., Salama K.N. Electrochim. Acta. 2021;386:138489. [Google Scholar]
- Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Biosens. Bioelectron. 2021;176:112912. doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Science. 2020;367(6485):1444. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yetisen A.K., Akram M.S., Lowe C.R. Lab Chip. 2013;13(12):2210–2251. doi: 10.1039/c3lc50169h. 2013,13, 2210-2251. [DOI] [PubMed] [Google Scholar]
- Yu L., Wu S., Hao X., Dong X., Mao L., Pelechano V., Chen W.-H., Yin X. Clin. Chem. 2020;66(7):975–977. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Yu L., Beikzadeh S., Zhang S., Zhang P., Wang L., Travas-Sejdic J. Electrochim. Acta. 2021;378:138132. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.