Abstract
A special-care neonatal unit from a large public hospital in Malawi was noted as having more frequent, difficult-to-treat infections, and a suspected outbreak of multi-drug-resistant Klebsiella pneumoniae was investigated using genomic characterisation. All K. pneumoniae bloodstream infections (BSIs) from patients in the neonatal ward (n=62), and a subset of K. pneumoniae BSI isolates (n=38) from other paediatric wards in the hospital, collected over a 4 year period were studied. After whole genome sequencing, the strain sequence types (STs), plasmid types, virulence and resistance genes were identified. One ST340 clone, part of clonal complex 258 (CC258) and an ST that drives hospital outbreaks worldwide, harbouring numerous resistance genes and plasmids, was implicated as the likely cause of the outbreak. This study contributes molecular information necessary for tracking and characterizing this important hospital pathogen in sub-Saharan Africa.
Keywords: Klebsiella pneumoniae, hospital outbreak, antimicrobial resistance, neonatal infection, sub-Saharan Africa, genome sequencing
Data Summary
The Klebsiella pneumoniae genome sequences completed using an Illumina platform from this study are deposited in GenBank under project no. PRJNA641987 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA641987) and the European Nucleotide Archive under project no. PRJEB19322 (https://www.ebi.ac.uk/ena/browser/view/PRJEB19322). The previously published K. pneumoniae sequences used as benchmarks of global context can be found in the European Nucleotide Archive under accession no. ERP000165.
Klebsiella pneumoniae is an opportunistic pathogen responsible for an increasing burden of hospital-acquired infections globally. It readily acquires antimicrobial resistance determinants via mobile genetic elements and plasmids [1], with particular sequence types (STs), such as those from clonal complex (CC) 258 (ST258, ST11), ST14/15 and ST405 reported to cause a large proportion of cephalosporin- and carbapenem-resistant infections with high mortality [2]. Critically, sepsis caused by multi-drug-resistant (MDR) K. pneumoniae is a growing challenge in neonatal care units; one systematic review of extended spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in neonatal care units reported that ESBL K. pneumoniae is the pathogen most frequently responsible for outbreaks in these settings, and is associated with a mortality rate of 31 % [3]. There are multiple reports detailing the prevalence and genetic epidemiology of MDR K. pneumoniae causing hospital outbreaks in high-income settings [2, 4, 5]. However, reports from low- and middle-income settings, where such data are of immense value in implementing appropriate infection prevention control and treatment regimes, are scarce.
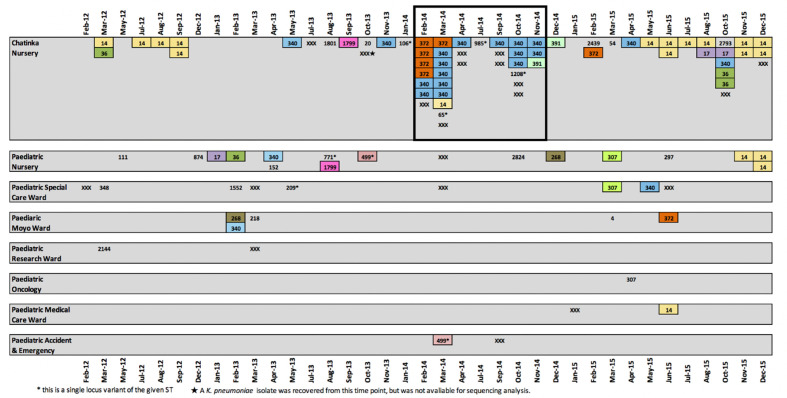
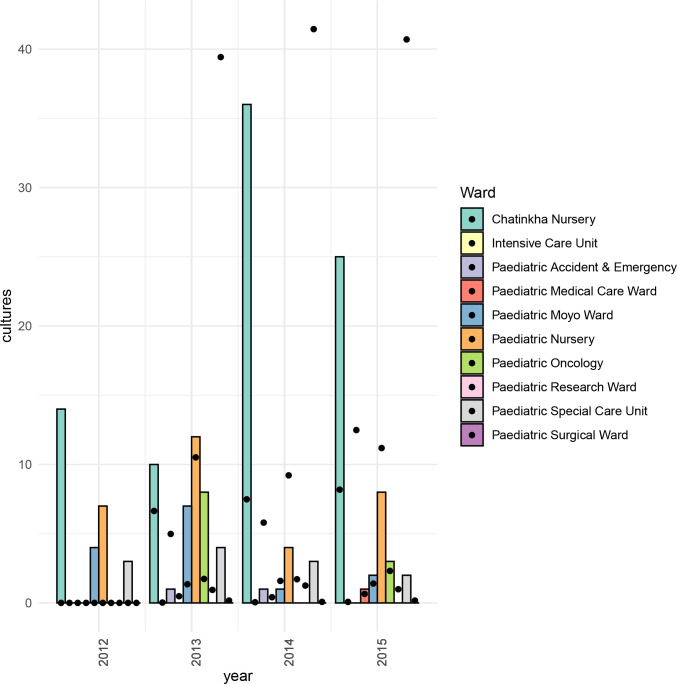
Queen Elizabeth Central Hospital (QECH) is the referral hospital in southern Malawi and care is free at the point of delivery. QECH serves both as district hospital for Blantyre (population 900 000) and is the tertiary referral hospital for the surrounding districts. QECH admits approximately 10 000 adult (aged ≥16 years) and 30 000 paediatric (aged <16 years) medical patients annually. Chatinkha is a 70-bed neonatal special care unit in QECH. Given that Chatinkha predominantly admits neonates born at QECH with complications, the majority of blood stream infections (BSIs) diagnosed on this unit are considered to be hospital-acquired. At QECH, all children with clinically suspected sepsis undergo a blood culture (BC) test as previously described [6]. The BC service typically identifies fewer than two K . pneumoniae BSI on Chatinkha each month, but a spike in K. pneumoniae isolates was observed in February (n=7) and March (n=9) 2014. The number of cases temporarily decreased, yet September to November 2014 showed a second peak in cases (Fig. 1). Despite Chatinkha having the smallest capacity of all of the paediatric wards at QECH, from February to November 2014 over 75 % of all paediatric K. pneumoniae BSI (n=33/43) were from neonates admitted to Chatinkha. This rise was not due to an increase in the number of blood cultures taken, which remained stable (Fig. 2). Furthermore, K. pneumoniae BSIs in Chatinkha during this period were untreatable with locally available antibiotics (most commonly penicillin, gentamicin, cotrimoxazole; less commonly ciprofloxacin, ceftriaxone; carbapenems unavailable); most displayed identical antimicrobial resistance patterns, suggesting the possibility of a clonal outbreak.
Fig. 1.
Schematic showing the location, sequence type (ST) and date of isolation of the K. pneumoniae isolates that were subjected to whole genome sequencing (n=86). STs that were identified more than once in the dataset are highlighted with a coloured box reflecting the STs. Singleton STs are not highlighted. Non-viable isolates (n=4) and those that failed sequencing (n=14) but were earlier confirmed as K. pneumoniae by the diagnostic laboratory are marked on the schematic as ‘XXX’ in order to give a complete picture of the number of BSI K. pneumoniae cases reported from Chatinkha each month during the study period. Months where no cases were reported are omitted from the schematic. The suspected outbreak period is highlighted with a black outline.
Fig. 2.
Bar chart showing the number of K. pneumoniae -positive blood cultures for Chatinkha ward and other paediatric wards at QECH. The chart is overlaid with a scatter plot showing the total number of all blood cultures (including those positive for any bacteria and those that were culture-negative) taken from the paediatric wards at QECH, Malawi, from 2012 to 2015 divided by 100. Data on the total number of blood cultures taken were not available for 2012 and are not shown.
Here, we performed a genomic investigation into the suspected K. pneumoniae outbreak, sequencing genomes from February to November 2014 (boxed in Fig. 1) and all viable, archived K. pneumoniae BSI isolates from Chatinkha (n=62) and a subset of K. pneumoniae BSI isolates (n=38) from other paediatric wards, isolated from January 2012 to December 2015. Patient records were retrospectively analysed to establish mortality outcomes. Blood culture collection, processing and antimicrobial susceptibility testing methods, based on the disc diffusion method and BSAC guidelines, for isolates at QECH have been previously described [6].
All DNA samples (n=100) were extacted with a Wizard Genomic DNA Purification Kit (Promega) and subjected to whole genome sequencing using the Illumina HiSeq X-Ten platform at the Wellcome Sanger Centre, UK, and the Oxford Genomics Centre. Fourteen sequenced genomes failed initial quality control using Kraken (more than 5 % of a species not part of the K. pneumoniae species complex assigned) [7]; the remaining 86 samples (Chatinkha n=56, other wards n=30) yielded, on average, 2.3 million reads per sample and an average of 77 contigs (range 22–259). The sequences were deposited in the European Nucleotide Archive (ENA) under project number PRJNA641987 and NCBI under project number PRJEB19322.
Multilocus sequence (MLST) typing was performed in silico as described elsewhere [8]. Resistance genes were identified using SRST2 v0.2.0[9] (https://github.com/katholt/srst2), plasmids using in silico PCR-based replicon typing [10] and PlasmidFinder version 2.0.1[11], capsular and O-antigen types using Kaptive (version 0.5.1; https://github.com/katholt/Kaptive) [12] and virulence genes using Kleborate v0.3.0 (https://github.com/katholt/Kleborate) [13, 14]. In order to place the study dataset within a global context, we compared our genomes to a previously published dataset from an international K. pneumoniae study [2]. A core gene alignment of the combined genomic dataset was generated using roary [15] and, from this, single nucleotide variants were used to generate a phylogeny with RaxML v.7.8.6 [16].
Whole genome sequencing analysis identified two lineages, ST340 and ST14, as the dominant K. pneumoniae STs recovered from neonates admitted to Chatinkha (Fig. 1).
Impact Statement.
Multi-drug-resistant Klebsiella pneumoniae is an important nosocomial pathogen that has been flagged by the World Health Organization and the Centre for Disease Control as an urgent threat to human health. Despite the importance of this pathogen and its worldwide distribution, the reporting and tracking of K. pneumoniae across sub-Saharan Africa is limited, and even less so from Malawi, especially using genomic data. In this study, we investigate an outbreak of extremely multiply resistant K. pneumoniae from a neonatal ward in a large public hospital in southern Malawi, using whole genome sequencing, across a 4 year period (2012–2015). We analysed the genomes of the isolates using various approaches to understand the phylogenetic typing features, antibiotic resistance and virulence properties and compared these with sequenced strains from other paedeatric wards in the hospital and put them in a global context. This study helps to understand the optimal local treatment options available and establish the virulent strain types of this important pathogen that are circulating in Malawi as well as adding information to the critical global network of K. pneumoniae genomic data.
There was a discrete outbreak of MDR ST340
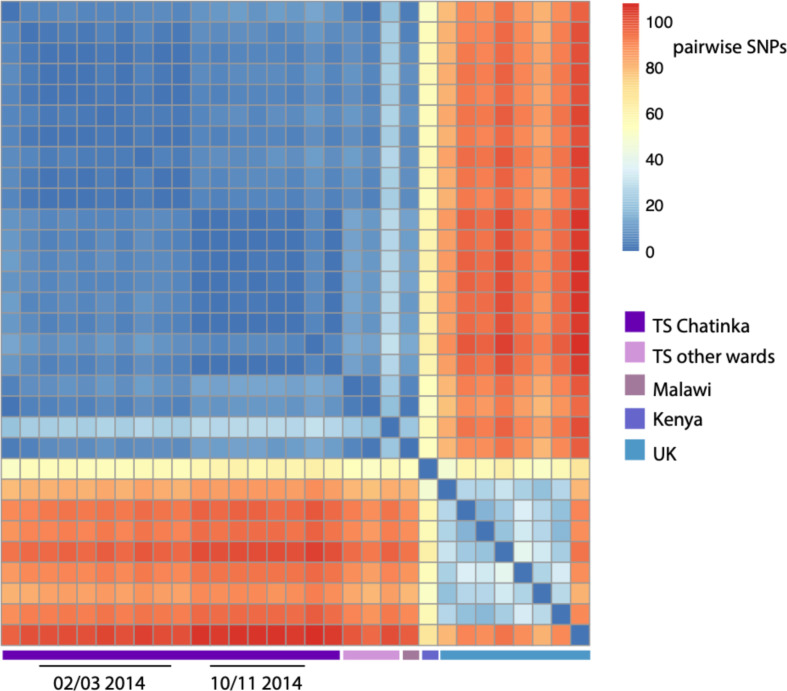
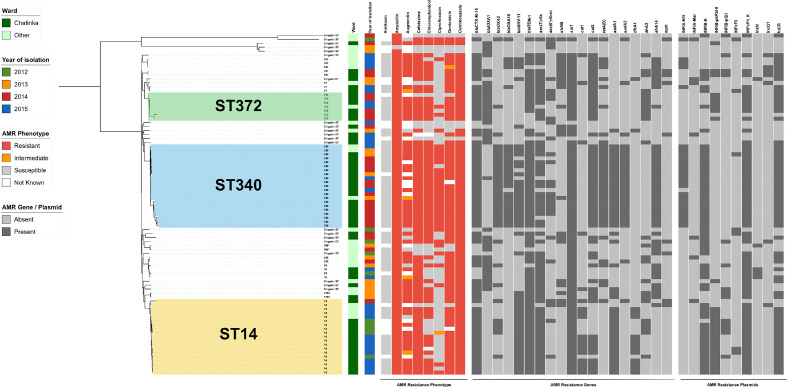
ST340, an ST within CC258 that has been associated with MDR hospital infections worldwide [17, 18], accounted for almost a third (30%, n=17/56) of all K. pneumoniae isolated from Chatinkha during the study period. All ST340 isolates were capsular type 15 and serotype O4 and differed from one another by ≤30 SNPs (Fig. 3), indicating that a single ST340 lineage was circulating in the ward over the entire study period. Over the suspected outbreak period, ST340 accounted for more than half of the K. pneumoniae BSIs from Chatinkha (58%, n=14/24), strongly suggesting that dissemination of this clonal strain within Chatinkha was responsible for the peak in BSIs observed from February to November 2014. The ST340 isolates were resistant to augmentin, ampicillin, ceftriaxone, chloramphenicol, ciprofloxacin, cotrimoxazole and gentamicin and susceptible only to amikacin, an antibiotic not locally available. Consistent with this, all ST340 isolates harboured multiple antibiotic resistance genes, conserved between all ST340 isolates, namely bla CTX-M-15, bla OXA-1, bla OXA-2, bla OXA-16, bla SHV-11, bla TEM-1 (beta-lactams), aac(6')-Ib-cr (quinolone), strAB (streptomycin), sul1 (sulphonamides), catA1, catA2 (phenicols), tetA(D) (tetracycline), aadA1, aadA2, aac(3’)-IIa (aminoglycosides), dfrA1, dfrA2 (trimethoprim) and mph (macrolides), and a number of plasmids, including IncR and IncF types (see Fig. 4). Of the ST340 BSI cases reported in neonates on Chatinkha, outcome data were available for 13, four of whom died, bringing the case fatality rate to 31%, which is equivalent to other reports of MDR K. pneumoniae [2].
Fig. 3.
Heatmap showing the number of SNP differences between the K. pneumoniae ST340 isolates sequenced as part of the outbreak investigation. To bring the relatedness of our samples into context with other ST340 isolates, the analysis includes a community-acquired Malawian ST340 sequenced as part of a previous study at QECH [6], a single ST340 isolate from a study in Kenya [20] and eight ST340 genomes from the UK [21].
Fig. 4.
The distribution of antimicrobial resistance elements within the K. pneumoniae isolates from QECH. The phylogeny is based on a core genome SNP alignment of the Malawian isolates (n=86), and the branches are labelled with the ST. The panel adjoining the phylogeny shows the antimicrobial resistance phenotype and the absence/presence of key antimicrobial resistance genes and plasmids amongst the isolates. Beta-lactamase genes: bla CTXM, bla TEM, bla OXA, bla SHV. Quinolone resistance genes: aac3, aac6. Streptomycin resistance gene: strAB. Sulphonamide resistance gene: sul1. Chloramphenicol resistance genes: cat1, cat2. Tetracycline resistance gene: tetA. Aminoglycoside resistance genes: aadA, aad1. Trimethoprim resistance genes: dfrA1, dfrA2, dfrA14. Macrolide resistance gene: mph.
Prior to the outbreak, ST340 BSIs had only been isolated from Chatinkha on two previous occasions (May and November 2013) (Fig. 1). These isolates differed from the first two ST340 cases reported in February 2014 by fewer than five SNPs (Fig. 3), confirming that closely related strains were circulating in Chatinkha at least 6 months prior to the outbreak. Previous to this, ST340 was isolated on a single occasion from two other paedeatric wards, the earliest in February 2013. These two ST340s isolates differed from the presumed outbreak ST340 precursor isolate by fewer than seven SNPs. Whilst we cannot confirm the exact date at which ST340 was seeded into QECH, this indicates that ST340 was circulating in the hospital at least a year prior to the outbreak. Following the outbreak, ST340 was identified twice more on Chatinkha: a single case in April 2015 and again a single case in October 2015. October 2015 saw six cases of K. pneumoniae BSIs identified on Chatinkha, hinting at the start of another outbreak, but these six cases were caused by five different STs. The fact that ST340 continued to circulate in the ward after November 2014 but did not to contribute to a further peak in cases suggests that the success of this clone during the outbreak period was not driven by genomic factors alone.
Interestingly, a recent study characterizing MDR Enterobacteriaceae , using PFGE and PCR, across northern and central Malawi, identified seven isolates of ST340 K. pneumoniae in 2016–2017 [19]. This indicates that although MDR ST340 strains appeared to be circulating across the whole country during this time period, these strains were probably not identifical clones of the outbreak in QECH because PCR confirmed that the nothern isolates carried different resistance genes (eg bla KPC-2).
ST14 did not contribute to the outbreak
Despite being the second most commonly isolated ST from Chatinkha (27%, n=15/56) during the study period, only a single ST14 case was reported during the outbreak period, in March 2014. ST14 showed a greater level of variation in their resistance profiles relative to ST340. These isolates were predominantly susceptible to locally available antibiotics (chloramphenicol and ciprofloxacin) in addition to amikacin and, thus, the ST14 infections would have been treatable (Fig. 4).
ST372 related to a peak in cases
ST372 (capsular type: 43, serotype: O2V1) also appeared to contribute to the peak in K. pneumoniae BSIs over the outbreak period as ST372 was responsible for 20.8 % (n=5/24) of cases. ST372 was first observed in February 2014 in Chatinkha and caused four BSI in 1 month and the remaining five K. pneumoniae BSIs reported during the outbreak period belonged to five different STs. ST372 isolates displayed relatively disparate resistance and plasmid profiles and may not be clonal (Fig. 4).
The outbreak strains in a global context
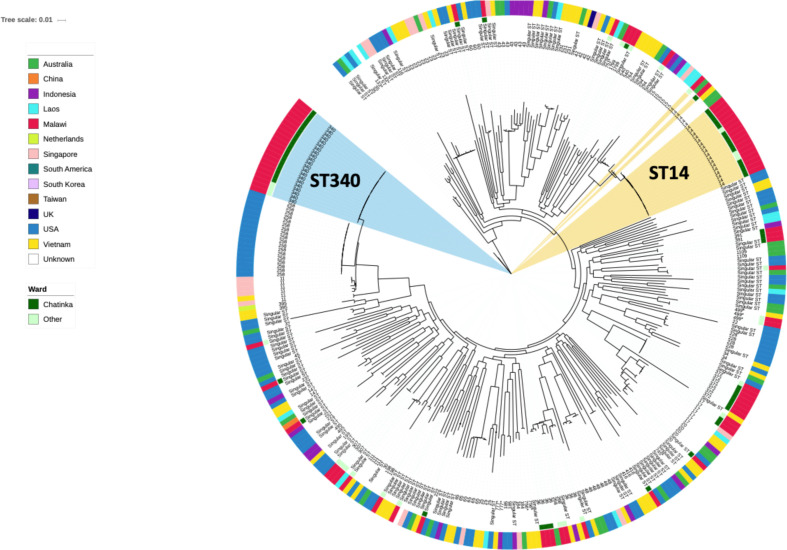
A core phylogeny was constructed to place isolates into a global context using a sequencing dataset of diverse K. pneumoniae isolates [2] (Fig. 5). Although no ST340 isolates were present in this dataset, the Malawian ST340 isolates were most closely related to ST258 isolates from the USA. Interestingly, ST14 strains from Malawi were clonally related (<100 SNPs) to international isolates with >99.9 % identity to isolates from Australia and the Netherlands, indicating a global spread. All ST372 isolates assayed in this study were from Malawi and we were unable to place them in a global context as no comparative international strains were available in this dataset.
Fig. 5.
Population structure of K. pneumoniae . A core genome phylogeny of the Malawian KP-I isolates (n=81) in the context of a previously published global dataset. Branch labels are annotated with ST. The inner coloured circle indicates if the Malawian isolates were recovered from the Chatinkha neonatal care unit, while the outer ring indicates the country of isolation.
Conclusion
We studied K. pneumoniae BSIs isolated from neonates admitted to a neonatal unit over a 4 year period, encompassing a suspected outbreak in 2014, as well as a representative subset of K. pneumoniae BSI reported from other paediatric wards within the same hospital.
We show that ST340, and to a lesser extent ST372, caused an increase in BSIs reported on the neonatal ward in 2014. ST340 was observed in other wards prior to the outbreak, suggesting that it was circulating in the hospital prior to the outbreak. ST372 was not identified in other wards prior to the outbreak, but this may be due to the limitations of our sampling. In addition to the outbreak lineages, we observed a large cluster of ST14 isolates, which intermittently contributed to no more than two BSI cases per month on Chatinkha. All other STs identified in Chatinkha over the sampling period were only observed in sporadic single isolate clusters. From the sequencing analysis alone, it is unclear what exact factors allowed the specific lineages to persist or successfully transmit within Chatinkha beyond antibiotic selection and this is under further investigation by performing genomics of environmental samples and staff swabs. Furthermore, continual monitoring of the hospital environment, especially for the re-emergence of MDR ST340, is urgently needed in order to implement measures to prevent the persistence and/or spread of these locally untreatable lineages.
Funding information
Funding for this study was from an LSTM research development grant, RDF160301AC12 (A.K.C., J.C. and N.F.), and from the Wellcome Trust through the MLW Core Grant Training Scheme 206 545 (A.K.C., C.P.). A.K.C. was supported partially by an ARC DECRA Fellowship, DE180100929.
Acknowledgements
The authors would like to thank Professor Julian Parkhill for assistance facilitating sequencing at Wellcome Sanger Institute and Professor Christopher Tang for assistance facilitating the sequencing done at the Oxford Genomics Centre. We also acknowledge the expert informatics support from the Pathogen Informatics team at the Wellcome Sanger Institute. The MLW bacteraemia archive arises from a routine diagnostic microbiology service provided to QECH by MLW, and we would like to acknowledge the work of the doctors, clinical officers and blood culture nurses in collecting these samples, without whom this work would not be possible. We would also like to acknowledge the support of guardians, especially the mothers of the babies admitted to the Chatinkha neonatal unit, for this work.
Author contributions
Original conceptualization: A.K.C., J.C., N.F.; patient observations and data collection: A.B., N.K., E.S., P.I.T.; patient sample collection and storage: S.B., A.B., N.K., E.S., P.I.T.; sample handling and DNA extraction: C.P., A.K.C.; genomic analysis and interpretation: J.C., E.H., P.M., A.K.C., N.F.; writing – original draft preparation: J.C., A.K.C., E.H., N.F.; writing – review and editing: all authors.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Ethical approval for the study was awarded by the University of Malawi College of Medicine Research Ethics Committee (P.08/14/1614 and P.018/17/2255).
Footnotes
Abbreviations: BSI, bloodstream infection; CC, clonal complex; ESBL, extended spectrum beta-lactamase; MDR, multi-drug resistance; MLST, multi-locus sequence type; QECH, Queen Elizabeth Central Hospital; ST, sequence type.
All supporting data, code and protocols have been provided within the article or through supplementary data files.
References
- 1.Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41:252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 2.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112:E3574–81. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stapleton PJM, Murphy M, McCallion N, Brennan M, Cunney R. Outbreaks of extended spectrum beta-lactamase-producing Enterobacteriaceae in neonatal intensive care units: a systematic review. Arch Dis Child Fetal Neonatal Ed. 2016;101:F72–8. doi: 10.1136/archdischild-2015-308707. [DOI] [PubMed] [Google Scholar]
- 4.Zheng B, Dai Y, Liu Y, Shi W, Dai E. Molecular epidemiology and risk factors of carbapenem-resistant Klebsiella pneumoniae infections in Eastern China. Front Microbiol. 2017;8:1–11. doi: 10.3389/fmicb.2017.01061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wyres KL, Gorrie C, Edwards DJ, Wertheim HFL, Hsu LY. Extensive capsule locus variation and large-scale genomic recombination within the Klebsiella pneumoniae clonal group 258. Genome Biol Evol. 2015;7:1267–1279. doi: 10.1093/gbe/evv062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Musicha P, Cornick JE, Bar-Zeev N, French N, Masesa C. Trends in antimicrobial resistance in bloodstream infection isolates at a large urban hospital in Malawi (1998-2016): a surveillance study. Lancet Infect Dis. 2017;17:1042–1052. doi: 10.1016/S1473-3099(17)30394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis MPA, van Dongen S, Abreu-Goodger C, Bartonicek N, Enright AJ. Kraken: A set of tools for quality control and analysis of high-throughput sequence data. Methods. 2013;63:41–49. doi: 10.1016/j.ymeth.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:11. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 11.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58 doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom. 2016;2:e000102. doi: 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wick RR, Heinz E, Holt KE, Wyres KL. Kaptive web: User-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J Clin Microbiol. 2018;56:e00197. doi: 10.1128/JCM.00197-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12:4188. doi: 10.1038/s41467-021-24448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tolentino FM, Bueno MFC, Franscisco GR, Barcelos DD de P, Lobo SM, et al. Endemicity of the high-risk clone Klebsiella pneumoniae ST340 coproducing QnrB, CTX-M-15, and KPC-2 in a Brazilian hospital. Microbial Drug Resistance. 2019:528–537. doi: 10.1089/mdr.2018.0006. [DOI] [PubMed] [Google Scholar]
- 18.Netikul T, Kiratisin P. Genetic characterization of carbapenem-resistant Enterobacteriaceae and the spread of carbapenem-resistant Klebsiella pneumonia ST340 at a university hospital in thailand. PLoS One. 2015;10:e0139116. doi: 10.1371/journal.pone.0139116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumwenda GP, Sugawara Y, Abe R, Akeda Y, Kasambara W, et al. First Identification and genomic characterization of multidrug-resistant carbapenemase-producing Enterobacteriaceae clinical isolates in Malawi, Africa. J Med Microbiol. 2019;68:1707–1715. doi: 10.1099/jmm.0.001087. [DOI] [PubMed] [Google Scholar]
- 20.Henson SP, Boinett CJ, Ellington MJ, Kagia N, Mwarumba S. Molecular epidemiology of Klebsiella pneumoniae invasive infections over a decade at Kilifi County Hospital in Kenya. Int J Med Microbiol. 2017;307:422–429. doi: 10.1016/j.ijmm.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moradigaravand D, Martin V, Peacock SJ, Parkhill J. Evolution and epidemiology of multidrug-resistant Klebsiella pneumoniae in the United Kingdom and Ireland. mBio. 2017;8:e01976. doi: 10.1128/mBio.01976-16. [DOI] [PMC free article] [PubMed] [Google Scholar]