Abstract
This study aimed to provide efficient recognition of bacterial strains on personal computers from MinION (Nanopore) long read data. Thanks to the fall in sequencing costs, the identification of bacteria can now proceed by whole genome sequencing. MinION is a fast, but highly error-prone sequencing device and it is a challenge to successfully identify the strain content of unknown simple or complex microbial samples. It is heavily constrained by memory management and fast access to the read and genome fragments. Our strategy involves three steps: indexing of known genomic sequences for a given or several bacterial species; a request process to assign a read to a strain by matching it to the closest reference genomes; and a final step looking for a minimum set of strains that best explains the observed reads. We have applied our method, called ORI, on 77 strains of Streptococcus thermophilus . We worked on several genomic distances and obtained a detailed classification of the strains, together with a criterion that allows merging of what we termed ‘sibling’ strains, only separated by a few mutations. Overall, isolated strains can be safely recognized from MinION data. For mixtures of several non-sibling strains, results depend on strain abundance.
Keywords: bacterial strain identification, long read, MinION, strain classification, Streptococcus thermophilus, bloom filters
Data Summary
The authors confirm all supporting data, code and protocols have been provided within the article or through supplementary data files:
The ORI code (Oxford Nanopore Reads Identification) is available at https://github.com/gsiekaniec/ORI.
All sequencing data used in our experiments (raw Nanopore fastq reads) can be downloaded on the Genouest server (https://data-access.cesgo.org/index.php/s/lApWiOf1BFYUpQV).
The interactive ITOL tree of S. thermophilus species is available at: https://itol.embl.de/tree/131254134671311597925585, and the complete list of associated ‘maximal biclusters’ (subset of S. thermophilus strains and subset of associated specific genes) is available on https://github.com/gsiekaniec/GeneTree.
Impact Statement.
This paper describes a new efficient method and the associated software for bacterial strain identification from few long read sequencing data without prior assembly. Very few identification software programs have been designed for long read data, a source that offers more information for the discrimination of strains at the cost of a higher error rate. Our work improves the state of the art by combining sensitivity and robustness, based on an efficient data index and exact optimization for parsimonious explanation of result strains. We have validated the entire process from sample sequencing to sample identification on a species whose strains are difficult to distinguish, Sreptococcus thermophilus. It is possible to address any set of bacterial species by changing the index, thanks to a compiler accepting any set of genomic sequences. Our tool can therefore be useful to all microbiologists wishing to control the presence or absence of certain strains possibly mixed within a sample. We chose to focus on data from the Oxford Nanopore MinION device since its low cost and real-time sequencing capacities are well suited for testing in any laboratory or even on site. As all programs can be freely downloaded from our sites, and the databases can be configured for the species of interest of each laboratory, we hope this tool will have considerable impact.
Introduction
For industrial, agri-food and clinically relevant bacteria, rapid identification at the fine level of strains is necessary and remains a great challenge. This is clear in the field of health (e.g. differentiating the non-pathogenic Escherichia coli strain MG1655 from enterohemorrhagic strain 278F2 [1]). It is also necessary in many other fields such as ecology, where adaptation to a given niche often leads to the emergence of new strains [2], or in food processes, such as fermentation [3]. Moreover, for newly isolated strains or species, looking for the closest known organism can yield important information for its functional characterization.
The development of next generation sequencing (NGS) technologies in the last two decades has revolutionized the characterization of microbial communities, without prior bacterial culturing. For instance, Illumina platforms that produce short reads (up to 300 bp) are now widely used for bacterial community taxonomic profiling [4–6] and de novo bacterial genome assembly [7, 8]. The overlapping reads produced are assembled into larger sequences that are mapped on sequence databases. However, read length makes these processes challenging, due to repetitive elements larger than reads, and this leads to fragmented assemblies. Third-generation sequencing technologies such as PacBio or Oxford Nanopore Technologies (ONT), producing read lengths reaching tens of kilobases or even more, can overcome this issue [9] but are error-prone. The best assemblers to date are hybrid, using both short and long reads [10].
Here we consider the issue of identification from long reads only, without prior assembly of these reads. Currently, identification from long reads hardly goes down to a level finer than genus or species [11]. Problems include the proximity of strains within a species, the prevalence of certain species in complex samples that can blur the recognition process, and databases thatare far from cover all microbial diversity. Therefore, there is still a gap between the promises of whole genome sequencing (WGS) for microbial identification, notably at the finer-grained taxonomic levels, and its large application in demanding operational contexts [12]. Among the possible obstacles to its diffusion is the very rapid advance in technology, which makes it difficult to produce stable identification software. While programs are becoming mature for work on short reads, the landscape is still very dynamic and uncertain for long reads.
Commonly used identification software can be broadly divided into two categories, the sequence alignment and the k-mers-based approaches. Sequence alignment, largely popularized by the program blast and its numerous variants [13], consists in optimally matching a read sequence with a fragment of a genomic sequence (representative software are MEGAN [14], MG-RAST [15], PanPhlAn [16], Centrifuge [17] and Kaiju [18]). To reduce their practical complexity, these algorithms are often limited to high-scoring alignments and loose sensitivity. Over the last few years, research effort has focused on simplified representations of genomes and reads, which has led to a significant improvement in comparison performance. The principle is to split genomic sequences into overlapping k-mers, short strings of fixed size (typically 30 nt). It is possible to store them compactly in an index for all known genomes and retrieve their presence very efficiently in sequenced reads in order to assign each read to its closest genomes. This powerful approach is followed by major current identification tools such as Kraken [19], Kraken 2 [20], CLARK [21], StrainSeeker [22] and Opal [23].
The majority of these tools use short reads because they have been available for a long time and with good accuracy. Unfortunately, they generally lack long-range genomic information to resolve differences at the strain level [24]. Paired-end reads, comprising short sequences at the end of a fragment separated with a variable gap, provide slightly more information since they can be extended to larger reads by trying to bridge the gap using other reads [25], but the process may introduce errors and these reads seem to have been used mainly for assembly and comparison purposes [26]. Relatively few identification software programs accept long reads. The most popular is Kraken and its more recent version Kraken 2 [20], since it is easy to instal, robust and fast. This software is based on the decomposition of genomes into k-mers and uses a very efficient storage structure. Some k-mers are specific to a unique species and some are linked to the lowest common ancestor (LCA) in terms of genus or family of all species where they occur. Thus, reads are classified at various levels of this hierarchy depending on their k-mer content. Kraken 2 rarely allows assigning a read at a level below the species because it uses a compression process (minimizers) that reduces the differences between genomes. Minimizers represent a sequence by a smaller included one, and serve to save space and time by retaining only part of the information, although they introduce a certain number of false positives during the identification process [27]. StrainSeeker [22] is a tool with a more restricted vocation, but which is of interest for strain identification from short reads. It uses a database of k-mer lists organized along a guide tree of bacteria (user-provided) where each leaf corresponds to a strain-specific set of k-mers. It is compatible with long reads, although we do not know of any application using it in this context. It was designed primarily for the identification of isolates but can handle mixtures of a few strains. Another tool, Centrifuge [17], follows a read alignment approach and is at the core of the species identification workflow in the platform EPI2ME (ONT). Unfortunately, its index size can become very large if it has to include strain-level genomes. Metamaps [28], specifically developed for the analysis of metagenomic datasets, also uses alignments to identify species/strains, is designed for long reads and, according to its authors [28], seems more precise than Centrifuge. However, the current version is not simple to instal and lacks robustness (users may suffer from ‘core dump’ errors).
This paper describes the use of long read sequencing technologies for the identification of bacteria at the strain level. We chose Streptococcus thermophilus as a model species given its importance in the food industry and the health sectors as a dairy starter and a probiotic [29] and because it encompasses closely related strains (see Text S1, available in the online version of this paper) with a low genomic diversity [30, 31]. We have developed a new strain identification method called ORI (Oxford Nanopore Reads Identification) and compared it to Kraken 2 and StrainSeeker. This approach dedicated to long reads has a technical background similar to Kraken, but differs in its goal to recognize at the strain level. The software for ORI can be found and downloaded from the following site: https://github.com/gsiekaniec/ORI. This work demonstrates that despite their significant error rate, the length of reads from MinION (ONT) allows fine-scale taxonomic assignment, even with mixtures and with closely related strains such as those from S. thermophilus . ORI can be easily adapted to other genera or species.
Methods
Bacterial strains, growth conditions and DNA extraction
A set of 46 genomes of S. thermophilus (Table S1) from public databases and 31 strains (Table S2) from the collection of the CIRM-BIA resource centre were used in this study. Streptococcus macedonicus PA (CIRM-BIA2314) and Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842 (CIRM-BIA658) from the CIRM-BIA collection were added as controls (different species/genus).
The 31 S . thermophilus strains and S. macedonicus PA were precultured anaerobically in LM17 medium (M17 medium supplemented with 2 %, w/v, lactose) [32] initially inoculated at 1 % (v/v), and incubated at 42 °C for 9 h. LM17 medium was further inoculated at 1 % (v/v) with the LM17 preculture and grown overnight at 42 °C. L. delbrueckii subsp. bulgaricus ATCC 11842 was grown in MRS medium [33] under the same conditions. To avoid excessive fragmentation, DNA extraction was performed using the Genomic-tip 100/G from Qiagen, according to the manufacturer's protocol with slight modifications. Briefly, lysozyme, proteinase K and RNAse A quantities were doubled and lysis incubation times were increased to 1.5 h. After DNA elution from the column, DNA was precipitated with isopropanol and spooled using a glass rod. It was immediately transferred in a clean microcentrifuge Eppendorf tube containing 200 μL EB buffer from Qiagen (10 mM Tris-Cl, pH 8.5) and allowed to dissolve overnight at 4 °C. Finally, DNA integrity and size (>10 kb) were assessed by electrophoresis on an agarose gel (0.7 %, w/v, TBE 0.5×, migration under 100 V). DNA purity was estimated by Nanodrop measurements (ND-1000 Spectrophotometer). DNA quantification was done using a 1× dsDNA high-sensitivity assay kit on a QUBIT 4 Fluorometer (Thermo Fisher Scientific).
MinION library preparation and sequencing
Libraries were prepared for MinION (ONT) sequencing according to the manufacturer’s protocol ‘Rapid Barcoding Sequencing (SQK-RBK004)’ (version RBK_9054_v2_revF_23 Jan2018), starting with 400 ng of DNA per S. thermophilus strain, using the 12 barcodes to multiplex 12 strains at each sequencing experiment. No additional purification step on AMPure beads XP was performed. Barcoded DNAs were pooled together. Finally, a flow cell (R9.4.1, FLO-MIN106D) was loaded with 200–500 ng of the library and run for 48 h, generating around 7–9 Gbp of sequencing data. The MinKNOW software (version release 19.05.0) was used to monitor the run and generate the fast5 files. Fastq files were obtained after basecalling with Guppy (version 4.4.1, default parameters) in high-accuracy mode.
DNA of the 31 CIRM-BIA strains has previously been sequenced with NGS Illumina technology. Illumina and Nanopore data allowed us to fully and correctly assemble their genomes, using the Unicycler hybrid assembler (version 0.4.7) [10]. Complete annotated genomes have been made available on the NCBI database (accession numbers listed in Table S2).
Filtering the raw reads
The fastq files generated by MinION sequencing were demultiplexed and adapter-trimmed using qcat (https://github.com/nanoporetech/qcat; version 1.1.0). MinION reads have a high error rate, in particular in homopolymers. In our sequencing data, basecalled with version 4.4.1 of Guppy in high-accuracy mode, the error rate was around 5 %, including around 3 % insertions/deletions. A selection step of best reads was performed in order to limit their global sequence error rate: each read must have an average quality of at least 9 and a length greater than 2000 bp (see details in Table 1). All data (raw Nanopore fastq reads and associated assembled genomes) can be downloaded on the Genouest server (see ‘Data summary’ section).
Table 1.
Average percentage error rate in the S. thermophilus sequences (calculated from an alignment of the reads against the reference genomes performed with Minimap2)
The filters retain only sequences with a quality greater than 9 and a size greater than 2000 bp.
|
Errors |
Mismatches |
Deletions |
Insertions |
Total |
|---|---|---|---|---|
|
All sequences |
1.50 % |
2.16 % |
1.40 % |
5.06 % |
|
With filters |
1.44 % |
2.08 % |
1.37 % |
4.89 % |
Creating a pan-genomic index for the sequence characterization of species
To compare reads and genomes efficiently, they were cut into small fragments of fixed size, called k-mers, which are the basic unit of comparison. However, given the high error rate of MinION sequences, we replaced k-mers with a more sensitive pattern, the spaced seeds [34], which introduce don’t care positions in k-mers that accept substitution errors [35]. Although genomes do not have this error rate problem, they have been indexed like the reads in order to make the two types of sequences comparable. We used a spaced seed pattern of size 15, 111111001111111, where 1 denotes a perfect nucleotide match and 0 a don’t care position. This means that the genomes and reads are cut into fragments (k-mers) of size 15, which are compared on the basis of the positions set to 1 in the pattern. This pattern was selected for optimal classification of the reads of JIM8332, using the iedera software [36, 37] on all the spaced seed patterns of size in the range [9 : 21] (see Fig. S1). The matching positions form a word called qgram. The index containing all these qgrams was built as follows: for each strain, its reference genome was cut into k-mers on which the spaced seed pattern was applied, which yielded qgrams. Each qgram was then inserted into a compact probabilistic data structure of fixed size (about 5×108 bits) based on Bloom filters [38, 39].
Data and index used for other methods
In our experiments, we adjusted the index choices for Kraken 2 and StrainSeeker in order to compare the results fairly. The genome databases were exactly the same for the three methods. StrainSeeker requires another input, a classification tree on the whole set of genomes guiding the identification process. This tree was generated on the MicroScope platform (see next section) in our experiments.
For Kraken 2 (version 2.0.9-beta), we used the default k-mer length of 35 and minimizer length 32. For the StrainSeeker (version 1.5) index, we used a k-mer length 16.
Comparing strains
Several measures were used to estimate the similarity between two genomes, the average identity between nucleotides (ANI) in their shared coding regions (at least 70 % of identity and 70 % coverage of the shorter gene), and two other distances taking the whole genome into account, Jaccard and Hamming distances. We used FastANI (ManyToMany mode) to estimate the ANI distance [40]. The Jaccard distance was computed on sets of qgrams (see Equation S1). The Hamming distance H was computed based on the proportion of positions that differ between the Bloom filters of the two genomes (i.e. a Bloom filter is a vector of 0/1 and the Hamming distance is the number of positions with two different values in the vector divided by the vector length) (see Equation S1).
The MicroScope platform [41] (https://mage.genoscope.cns.fr; access date May 2020, v3.14.0) was used to annotate the genome of the 77 S. thermophilus strains, to generate a strain classification tree and to compute the pangenome. Briefly, MicroScope uses the AMIGene software to predict protein-coding genes [42] and combines the results of several tools to assign molecular functions [41]. A strain classification tree is computed using the neighbour-joining algorithm with pairwise genome distances obtained from Mash software [43]. Three sets of genes corresponding to the core, variable and strain-specific genes were downloaded from the pangenome computed by MicroScope. It is based on MICFAM gene families, which are computed using a single linkage clustering algorithm of homologous genes sharing an amino acid alignment coverage and identity above 80 %. After standardizing the gene names of the pangenome families for all strains, we created a strain × gene matrix (see Fig. 1).
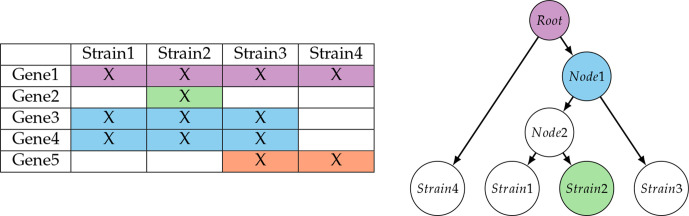
Fig. 1.
Biclusters in a strain × gene matrix and associated labelling of nodes in a classification tree.
Clustering strains by gene content
In addition to the strain tree produced by MicroScope, we produced a biclustering of strains and genes, using formal concept analysis [44]. Once biclusters are determined, they are added to nodes of the strain classification tree, showing how close the strains are with respect to their gene content. The root of the tree contains the core genome genes and the leaves contain the genes specific to this leaf (strain). Note that some nodes may stay unlabelled because they cannot be characterized in terms of a subset of genes. In our example, this is the case for Node2 since any characterization of it would also cover Strain3 (Fig. 1). Conversely, {Strain3, Strain4} is not present in the tree although it can be uniquely characterized by the presence of Gene5. The tree can be displayed using an online tool for the presentation of annotated phylogenetic trees called iTOL [45] and biclusters can be recovered through popups associated with each node.
Experimentation design
To test our method and compare it to Kraken 2 [20] and StrainSeeker [22], 180 strain identification experiments were performed (see Table S3). They were based on the construction of various sets of reads extracted from the MinION sequencing data by random draws performed uniformly across the filtered sequenced reads. The distribution of read lengths in the sets had the following characteristics: median 6436 bp, mean 8842 bp, standard deviation 7861 bp, for a value in the range [2000–189 000]. Each identification result for a fixed set of parameter values is an average that has been calculated on five different sets of strains randomly selected from the whole set of possible strains. The parameters were the number of reads (1000, 4000 or 16 000 reads), the number of strains to be identified (four or six strains), the proximity of the strains (distant, moderately close or close strains) and finally the distribution of strain abundances in the experiments (uniform distribution or distribution with dominant and subdominant strains). The set of experiments has been split into two parts to present the results, 90 with a uniform distribution and 90 with low-abundance strains. For close proximity experiments each strain was required to be very close to at least one other strain but could be distant from the others (see Table S4).
The percentage of subdominant strains was 12.5 % of the reads for each of the two strains in mixtures of four strains and respectively 6.25, 3.12 and 3.12 % for three strains in mixtures of six strains. For the validation of identification methods, we used the sum of Hamming distances between each predicted strain and the closest real, target one in the sample. We also calculated the Matthews correlation coefficient (MCC) to measure the precision/sensitivity trade-off of the method (see Equation S2).
Similar to Kraken 2, StrainSeeker does not always identify a single target but proposes a group of strains as an identification result. To take this behaviour into account, we counted in this calculation one true positive for each correct strain and one false positive for each incorrect strain. As a correlation coefficient, MCC ranges from 1 for a perfect prediction to −1 for complete disagreement between the observation and reality and 0 indicates no relationship. It is balanced with what we called an ambiguity ratio, which was calculated as the ratio of the number of predicted to number of actual strains.
Results
We first describe the new method we propose for the identification of strains, then the identification results themselves, which are compared to those obtained by two other methods, Kraken 2 and StrainSeeker.
ORI, a new method for the identification of bacterial strains from long reads
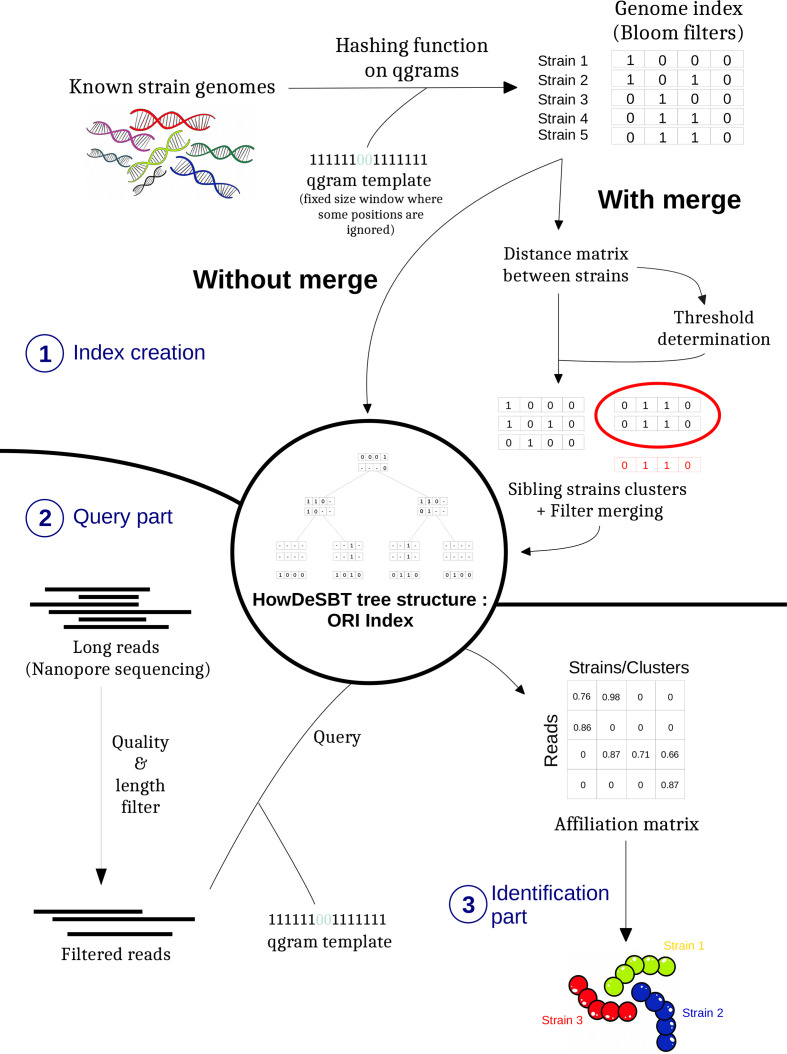
We propose a new method and software, ORI, based on indexing fragments of a set of known bacterial genomes in a compact data structure. ORI is therefore in line with the principles of a method such as Kraken, although it differs in many ways. The first step (index creation in Fig. 2) takes as input the sequences of known genomes and builds a data index based on a hierarchical structure of Bloom filters [38, 39], each one being of fixed size (5×108 bits). They code for the presence in each genome of small fragments of fixed size, called qgrams, which are similar to k-mers but allow for mismatched characters at some positions. There is an optional but recommended operation during this step, the merging of very closely related strains (distance below a given threshold) in a single pangenome (see sections below). The second step (Query part in Fig. 2) takes as input the sequenced reads of an unknown sample and consists of filtering (for length and quality), qgram extraction, and finally querying against the index. It results in a large score matrix crossing known genomes and reads, estimating for each read its probability of originating from a given genome and thus for the genome to be present in the sample. The last step is the identification process itself, which takes as input the genome × read matrix and looks for a minimal set of strains that best explain the reads in the observed sample (see below for more details).
Fig. 2.
Overview of the ORI method in three steps: (1) genome indexing, (2) query the index from filtered reads, and (3) identification of strains.
Compact pan-genomic indexes of bacterial sequences
In all our experiments, two indexes were produced. The first index has been built with all S. thermophilus genomes (77 strains) plus two strains from other species/order, S. macedonicus and L. delbrueckii subsp. bulgaricus , for a total size of 23.4 Mb. Note that we have excluded plasmids in the current version since plasmids are considered part of the mobilome (horizontal transfer) and not specific of a strain. The second index was designed for experiments where very closely related isolates (such as mutant strains or variants that are indistinguishable) were merged under a single strain identifier (we call such strains ‘sibling’ strains, see section below). This has been achieved by merging all Bloom filters from closely related strains. This index had a similar size of 22.7 Mb. Both index sizes were comparable to the index of Kraken 2 (18.1 Mb for exactly the same strains), and much smaller than StrainSeeker’s index (2.1 Gb).
Affiliation of MinION reads to strain genomes by index queries
Once an index has been created, our method identifies the bacteria present in a sample by querying the index with the sample reads. As with genomes, each MinION read is cut into k-mers on which a spaced seed is applied, which provides the qgrams of the read. The index is then requested using all these qgrams. A minimum number of qgrams from a read (half of them; parameter --threshold=0.5 in ORI) must be found in a strain genome before it can be retained for identification purposes. This threshold allows efficient filtering of reads that concern the species of interest. It takes full advantage of the length of the reads and allows in particular to remove contaminants, which would be difficult on short reads [46]. The output of a request is a list for each read of possible strains, weighted by their number of qgrams, summarized in a read × strain matrix.
Identification of one or several strains in samples
Starting from a set of reference bacterial strains and a sequenced sample, the goal of identification is to find a subset of that best explains the observed reads. We say that a read is explained by a strain if this strain is a possible answer to the read query in the index, i.e. it matches with a number of qgrams in the read greater than the threshold (50 %).
From a theoretical point of view, this issue may be seen as a set cover optimization problem: find a minimal subset to fully explain the reads, a difficult problem that can be solved if the number of operational taxonomic units in the sample is of moderate size (see Text S2).
We followed a declarative problem solving approach called Answer Set Programming (ASP) for this [47]. A quick preprocessing step was applied beforehand to filter reads and strains that provided little information: (1) the reads found in too many strains were not taken into account during the identification (by default, if a read can be affiliated to 18 strains or more, they are assumed to be part of the core genome of the species) and (2) only the best strains (12 by default, parameter --nbchoices) were taken into account for each remaining read, according to the proportion of qgrams matching them in the read.
Measuring the proximity between strains
To evaluate the difficulty of identification and assess its accuracy, it is necessary to measure the proximity of genomes. This has been achieved at the coarse level of genes and at the finer level of genomic content.
The gene level
We have computed the complete list of maximal biclusters (see section Comparing and clustering strains in the Methods) comprising a subset of S. thermophilus strains and a subset of associated specific genes (available on https://github.com/gsiekaniec/GeneTree). Genes specific to the set of strains under each node of the strain classification tree produced by MicroScope are available in an interactive version of this tree via clickable links for each node (see https://itol.embl.de/tree/131254134671311597925585). All the maximal biclusters cannot be represented by a node of the tree because they follow a different topology. It is already possible to perceive in this tree that some strains are very close if not identical and differ only by few unknown genes.
The genomic fragment level
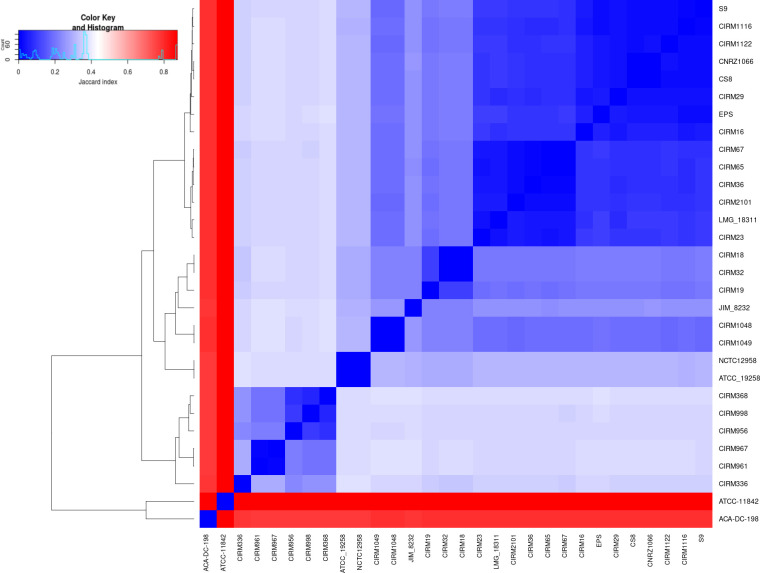
The proximity between genomes is usually measured with the ANI distance. Our genome index offers other ways to evaluate this proximity. We have produced a slightly more precise Jaccard distance matrix using spaced seeds. Fig. 3 shows an extract of this matrix for 28 strains in the form of a heatmap (eight strains from public genome database and 20 strains from the CIRM-BIA collection). The complete heatmap, containing the 77 S . thermophilus strains, can be found in Fig. S2. Two genomes, one from S. macedonicus and the other from L. delbrueckii subsp. bulgaricus , have been added as an external group. As expected, strains of different species or order are easily distinguished. The group of six CIRM-BIA strains at the bottom left of the heatmap that is distant from other strains clearly forms a new subgroup comprising strains that were mostly isolated from traditional Italian dairy products.
Fig. 3.
Heatmap of the Jaccard distance for 28 S. thermophilus strains + S. macedonicus ACA-DC 198 + L. delbrueckii subsp. bulgaricus ATCC 11842.
We measured the coherence of Jaccard and ANI distance with respect to the Mash distance used by MicroScope on our strain set. The observed Pearson correlation is very good (r=0.967, P=1e-04 for ANI, and r=0.987, P=1e-04 for Jaccard distance). As expected, Jaccard is slightly better than ANI distance (ANI only compares orthologous genes).
In our work, it is more efficient to compute the Hamming distance between the Bloom filters rather than the Jaccard distance (see sections Comparing and Clustering strains in the Methods). We checked that this distance corresponded to the Jaccard distance for strain comparison. There was indeed a very good correlation between the two distances (r=0.99, P=1e-04). Using quadratic regression, we obtained the equation H=3.81e −03 (J 2+J)between Hamming H and Jaccard J distance values (see Fig. S3). For small values (J0.15), a linear relationship fitted the values very well: H=4.26e −03 J.
Sibling strains: a threshold to establish two isolates as indistinguishable
To improve detection accuracy, we grouped closely related strains into groups called ‘sibling strains’. Identification results depend on their definition, which involves some threshold for the maximum distance between the genome of sibling strains, much lower than that between species. Fortunately, we have shown that the choice of the distance used seems not to be crucial for this task.
In the remainder, we will compare results without or with clustering. The first results are obtained with a Hamming distance threshold of 0 ( = ), and the second results are those that regroup all isolates based on a Hamming distance less than = (0.05 for Jaccard). This default threshold has been determined empirically for S. thermophilus by trying different threshold values and looking for a minimum value for which the identification error rate remains low on isolated strains. Below this distance, the sequencer error level prevents reliable affiliation of reads. ORI produces a graph that shows the distribution of distances between strains and helps to select a reasonable maximum distance threshold, which leads to grouping of a limited number of strains with close genomes. Clusters of sibling strains were formed as follows: (1) a graph is created whose vertices are the strains and edges connect those less than away from each other; (2) maximum cliques of this graph are computed and intersecting cliques are merged; and (3) Bloom filters of the genomes belonging to these cliques are merged, corresponding to the pangenome of the sibling strains (see index creation in Fig. 2 and Table S5, Fig. S2).
Identification results
We present in this section the results of ORI in various contexts and compare them on the same database with the leading method Kraken 2 [20] and the program dedicated to strain identification, StrainSeeker [22]. As an indication, we have provided tables showing the precision of identification of S. thermophilus strains in all 180 experiments (see Table S6) and the precision of identification of the subdominant S. thermophilus strains (see Table S7).
Identification of isolated strains: ORI is the most robust method
The first test was to identify a single strain, S. thermophilus JIM 8232, from a moderate quantity of MinION reads (4000), randomly selected among all reads sequenced for this strain (around 200 000 reads). This strain is quite distant from the others (see Fig. 3): it is not closely related to any other known strain. Almost all reads were retained by the three tested methods. For Kraken 2, 50.10 % of reads were classified as JIM 8232, 26.56 % of reads were classified as S. thermophilus (it stops at the species level) and the rest had an incorrect strain classification. For ORI, 99.7 % of reads were classified as JIM 8232, the rest being unclassified. For StrainSeeker, JIM 8232 was recognized as the sole strain of the sample (100 % of JIM 8232). Although StrainSeeker was developed specifically for Illumina reads, it provided excellent results with ONT reads on this test.
A second test concerned S. thermophilus CIRM-BIA67, which is much more difficult to identify than JIM 8232 because of its proximity to other strains (see Fig. 3). In particular, CIRM-BIA67 and CIRM-BIA65 are sibling strains. The differences stem mainly from a contraction in CIRM-BIA67 of two tandem repeats from a 13.5 kb fraction of the genome. Table 2 shows detailed results for an increasing number of reads.
Table 2.
Identification of reads from S. thermophilus strain CIRM-BIA67 from various numbers of reads
|
Method |
100 reads |
1000 reads |
10 000 reads |
20 000 reads |
|---|---|---|---|---|
|
Kraken 2 |
C67 2 % |
C67 1.70 % |
C67 1.31 % |
C67 1.23 % |
|
StrainSeeker |
One group of many indistinguishable strains (C67 and 66 other strains) 100% |
C67 100 % |
C67 13.13 % +4 groups (other strains) |
C67 2.2 % +C65 1.86% +22 other strains +2 groups |
|
ORI |
C67 100 % |
C67 100 % |
C65 100 % |
C65 100 % |
C67, CIRM-BIA67; C65, CIRM-BIA65.
Kraken 2 assigned most of the reads to the species S. thermophilus and less than 2 % to strain CIRM-BIA67. StrainSeeker recognized the species level (a large group of strains that cannot be distinguished) for 100 reads, found a perfect result for 1000 reads, and then a decreasing proportion of CIRM-BIA67 for more reads. It is thus very sensitive to the number of reads. ORI identification is almost perfect in each case, with identification of the correct CIRM-BIA67 strain up to 1000 reads and the sibling strain CIRM-BIA65 with more reads. For all methods, the error level of reads introduces limitations in the identification of closely related strains. For ORI, this can be corrected by merging them (see Identification of strain mixtures after merging).
Identification of strain mixtures: ORI needs at least 500 reads for each strain
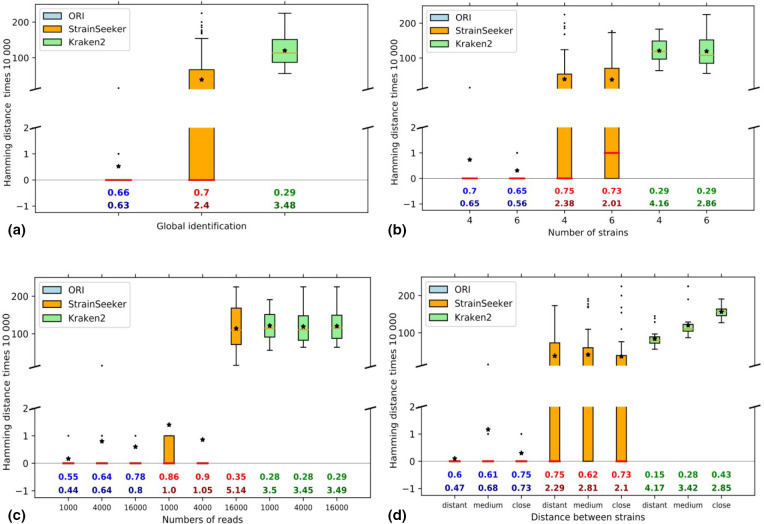
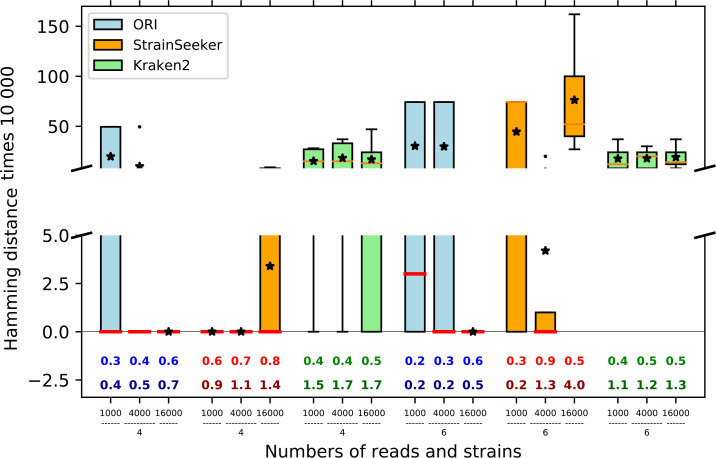
We ran the 90 experiments containing a uniform number of reads for each strain (see Experimental design in Methods). The diagrams in Fig. 4 display boxplots for the sum of the Hamming distances showing how close the predicted strains are from real ones. Below each boxplot, two numbers are given, MCC and the Ambiguity ratio. The MCC, which measures the adequacy of the binary decision on the presence/absence of strains, shows the balance between specificity and sensitivity. The ambiguity ratio is necessary because Kraken 2 and StrainSeeker propose ambiguous answers with more strains than actually exist, and it tends to artificially increase the MCC, whereas ORI tries to minimize the number of predicted strains.
Fig. 4.
Identification results on a balanced mix of S. thermophilus strains. The Hamming distance between observed and expected strains, on the y-axis, has been multiplied by 10 000 (in blue for ORI, orange for StrainSeeker and green for Kraken 2). Stars represent mean values. Matthews correlation coefficient (MCC) values are given on the first line just above the x-axis at the bottom of the diagrams, followed by the ambiguity ratio (number of strains identified/number of strains present).
We computed the Hamming distance, the MCC and the ambiguity measures over all experiments (global identification, Fig. 4a), then detailed the results along three parameters: the number of strains, the number of reads and the proximity of the strains (Fig. 4b–d). Overall, identification results are very good for ORI (Fig. 4a), from the point of view of both mean and standard deviation, showing the robustness of the method. StrainSeeker also gives good results and has the best MCC but is penalized by the production of multiple solutions and wider variations than ORI. Kraken 2 is clearly less well adapted to the recognition of strains.
Fig. 4b shows the effect of increasing the number of strains in the mixture. Results are very similar for four or six strains.
For Fig. 4c, different amounts of read data were used. No difference was observed for Kraken 2, using 1000, 4000 or 16 000 reads. By contrast, ORI seems to be sensitive to read number: the more data, the better strain identification with regard to MCC and ambiguity ratio. For StrainSeeker, using a large number of reads clearly leads to a drop in accuracy and results are optimal with 4000 reads.
Finally, the resolution power of methods was measured for mixtures containing increasingly close target strains (increasing the difficulty of identification). Fig. 4(d) shows that Kraken 2 is sensitive to this parameter and provides poor results for a mixture of closely related strains. With ORI and StrainSeeker, which are more sensitive, the effect is not marked but ORI shows a slight degradation of MCC and the ambiguity ratio. This behaviour, due to the existence of S. thermophilus genomes with high proximity, motivated the introduction of a merging procedure prior to strain identification (see next section).
We end with the most difficult context, the identification of subdominant strains in a mixture of four or six strains (Fig. 5). The mixture of four strains contains two dominant strains (representing respectively 50 and 25 % of the total reads each) and two subdominant strains (12.5 % of the reads each), while the mixture of six strains contains three dominant strains (50/25/12.5 % respectively) and three subdominant strains (6.25 %/3.125/3.125 % respectively). Of note is that Kraken 2 behaves better in this situation, reducing the difference compared to ORI and especially StrainSeeker. ORI continues to perform well for strains with at least 500 reads but its results are degraded below this. For StrainSeeker, the number of reads used for identification is crucial: it works well with 4000 reads (even better than ORI) but works surprisingly poorly with 16 000 reads.
Fig. 5.
Identification of subdominant strains in a mixture of S. thermophilus strains using various numbers of reads. The Hamming distance between observed and expected strains, on the y-axis, has been multiplied by 10 000 (in blue for ORI, orange for StrainSeeker and green for Kraken 2). Matthews correlation coefficient (MCC) values are given on the first line just above the x-axis at the bottom of the diagrams, followed by the ambiguity ratio (number of strains identified/number of strains present).
Identification of strain mixtures after merging highly similar strains: best results for ORI
As previously observed in Fig. 5, the main issues for ORI come from very closet related isolates. We have tested the impact of preprocessing the data by merging them as one strain type. The results obtained with this new merged index were compared to the previous results (Tables 3 and 4).
Table 3.
S. thermophilus strain identification by ORI, with and without merge index, in a balanced mixture of four or six strains more or less genetically close, by using 1000, 4000 or 16 000 sequencing reads
Best results are in bold type. Values of Hamming distance (0=perfect identification); MCC: Matthews correlation coefficient (1=perfect correlation); Ambiguity: number of strains identified/number of strains present.
|
(a) Global identification results (mean over all 90 experiments): |
||||||
|---|---|---|---|---|---|---|
|
Method |
ORI |
ORI_merge |
||||
|
Distance |
0.52 |
0.41 |
||||
|
(MCC/Ambiguity) |
0.66/0.63 |
0.92/0.91 |
||||
|
(b) Heterogeneity, mean results (variable number of strains mixed): |
||||||
|
Method |
ORI |
ORI_merge |
||||
|
Number of strains |
4 |
6 |
4 |
6 |
||
|
Distance |
0.73 |
0.31 |
0.53 |
0.29 |
||
|
(MCC/Ambiguity) |
0.70/0.65 |
0.65/0.56 |
0.94/0.93 |
0.96/0.96 |
||
|
(c) Data quantity, mean results (variable number of .fastq reads): |
||||||
|
Method |
ORI |
ORI_merge |
||||
|
Number of reads |
1000 |
4000 |
16 000 |
1000 |
4000 |
16 000 |
|
Distance |
0.17 |
0.8 |
0.6 |
0 |
0.43 |
0.8 |
|
(MCC/Ambiguity) |
0.55/0.44 |
0.64/0.64 |
0.78/0.80 |
0.86/0.77 |
0.93/0.92 |
0.98/1.05 |
|
(d) Resolution power, mean results (variable proximity between strains within the mixture): |
||||||
|
Method |
ORI |
ORI_merge |
||||
|
Proximity |
Distant |
Medium |
Close |
Distant |
Medium |
Close |
|
Distance |
0.10 |
1.17 |
0.30 |
0 |
0.90 |
0.33 |
|
(MCC/Ambiguity) |
0.75/0.73 |
0.61/0.68 |
0.6/0.47 |
0.93/0.89 |
0.87/0.85 |
0.97/1 |
Table 4.
Subdominant S. thermophilus strain identification by ORI, without/with merge, in a mixture of four or six strains, by using 1000, 4000 or 16 000 Nanopore sequencing reads
Best results are in bold type. Values of Hamming distance: in all experiments, minimum value is 0 (perfect identification); MCC: Matthews correlation coefficient (1=perfect correlation); Ambiguity ratio: number of strains identified/number of strains present; sd: standard deviation
|
No. of strains |
4 (ORI/ORI_merge) |
6 (ORI/ORI_merge) |
||||
|---|---|---|---|---|---|---|
|
Number of reads |
1000 |
4000 |
16 000 |
1000 |
4000 |
16 000 |
|
Distance |
19.8/19.8 |
9.9/0 |
0/0 |
30.3/15.4 |
26.7/0 |
0/0 |
|
MCC |
0.28/0.38 |
0.42/0.78 |
0.57/0.9 |
0.22/0.38 |
0.34/0.65 |
0.63/0.8 |
|
Ambiguity |
0.4/0.4 |
0.5/0.8 |
0.7/1 |
0.2/0.33 |
0.2/0.47 |
0.53/0.67 |
Overall, merging leads to almost perfect identification results, with a Hamming distance that decreases with respect to the version of ORI without merging and it is mostly null, except for experiments with an insufficient number of reads. The MCC increases with the number of available reads, and is close to 1 on average, showing that ORI with merge combines high accuracy with high sensitivity. Note that it is possible to get a null average distance and a MCC less than 1; this points to perfect accuracy but loss of sensitivity: strains are identified but some are lacking.
The method is more robust in the sense that the variations in parameter values have less influence on the results.
The identification of subdominant strains is shown in Table 4 (full results can be found in Tables S8 and S9). The results are clearly better with merging for samples of 4000 reads; in our experiments, merging sibling strains allowed us to achieve strain identification from as few as 250 reads. For experiments with 1000 reads, which need a recognition from 125 reads or fewer, the method is limited and the gain is smaller.
Discussion
In this work, we have compared three methods that have a common methodological basis. They split genomes and reads into small fragments that are inserted in an index, a very compact and efficient data structure for making comparisons. However, ORI seems to be better than Kraken 2 in terms of identifying bacterial strains. StrainSeeker behaves correctly with parameters carefully tuned. It has been designed for short reads of high quality and it will return poor results either if there are too few or too many reads. Moreover, StrainSeeker does not scale to a large number of genomes, and its index requires 100 times more memory than the other methods. ORI features three major choices adapted to strain-level identification from long read noisy data. A first one is to replace k-mers by qgrams, which tolerate a few substitution errors, and is particularly useful given the higher error level of ONT sequencing. Note that it is effective for relatively short k-mers since indel errors are not taken into account. Another important difference relates to the choice of the tradeoff to be made between the sensitivity and the computational cost of the method. Kraken 2 is an excellent choice for large-scale species recognition, based on the use of minimizers. This choice leads to a relative loss of sensitivity which does not allow it to recognize closely related strains. ORI uses a first identification step for each read, utilizing the fact that all the k-mers in a long read must belong to a single species. A last difference is that ORI uses an exact optimization step in order to select a minimum number of species/strains that explain most of the reads. This is a unique point of ORI that increases its robustness (very few false positive) and also reduces the list of identified strains, which can be very useful for subsequent analysis of the results.
There is a need to achieve ID resolution of a group of highly similar isolates [48], rather than the exact isolate itself. Clinically it is useful for identifying outbreak strains, which is a major use of MinION sequencing (error-prone Nanopore reads). In a recent paper [49] the authors worked on almost 2000 samples of Streptococcus agalactiae , characterized by different degrees of virulence and preferred host. However, even if the microbiologists are demanding regarding the distinction of strains, they only distinguish a few dozen different types at most. Stopping strain distinction at the individual isolate level is of little practical value. Another recent application of clustering close strains is in the inference of antibiotic resistance and susceptibility [50]. We have presented two versions of ORI, one including a preprocessing step merging in a single pangenome the sequences of known isolates that appear to be very similar. Our results showed that the version that merges seems the most appropriate with regard to the high error rate of current ONT sequencing technology. The identification results are robust and the biologist may conduct additional investigations to discriminate the strain among a limited number of possibilities. Considering these sibling strains in the iTOL tree, they are annotated mostly by specific genes of unknown function, contrary to other strains (e.g. S. thermophilus JIM 8232). Two unknown genes labelled differently could nevertheless be related and sibling strains, and therefore could usefully point to groups with a compact pangenome, analysable in fine detail, valuable knowledge for strain selection. An interesting perspective would be to check that merging allows us to distinguish strains for important phenotypic traits such as antimicrobial resistance profiles [51].
Another important parameter is the number of reads to be used for identification. We proposed a set of 4000 reads to be sufficient for a mixture and 500 reads for an isolate, something that helps in fast identification of the content of a sample. In fact, it seems that a good information/noise ratio is needed in practice: either the number of reads is insufficient to detect the accessory genome, or it is too high and noisy reads tend to introduce ghost recognitions. We still need to work on this aspect so that ORI does not decrease its identification quality when the number of reads increases. As identification requires a good information/noise ratio, one way to improve it is to pre-filter the reads and strains in order to obtain the fewest erroneous and most strain-specific reads.
We end this section with a few perspectives that could help extend this work.
Rapid identification at low cost
In our experiments, DNA has been extracted by a kit specific for long reads, using the ONT flow cell R9.4 for sequencing. Such DNA extraction requires long passage times through the column by gravity (6–8 h, depending on the strain) and is relatively expensive, including all costs. In practice, it is crucial to decrease costs and time as much as possible while maintaining ORI’s good performance. Using a DNA extraction kit designed for short reads would lead to fragments three times shorter but save time and money (4 h extraction, 5 € per sample). A good compromise could be a rapid extraction with magnetic beads (e.g. MagAttract HMW from Qiagen). For the library sequencing kit, we used a rapid barcoding sequencing kit, which reduces size by a factor of 3 and time by a factor of 20 (15 min) compared to the classic SQK-LSK109 kit. Once loaded with 12 samples, the flow cell produced about 3 million reads (9 Gb of sequences) after 48 h of run. In fact, less than 10 min of sequencing is sufficient to produce the 4000 reads necessary for strain identification. Furthermore, ONT suggest the low-cost, low-throughput Flongle flow cell that could be an interesting alternative for producing the required data in about 30 min. Overall, from sample collection (e.g. a fermented food product) to sequence files, for one sample containing four strains, requires less than 6 h (for a total cost of maximum 200 €, using Flongle). Then, depending on the processor used, identification could be achieved within 1 h.
Quantification of strains in a mixture
We plan to estimate strain abundances in ORI by using an expectation-maximization (EM) algorithm similar to the one used in Centrifuge [17]. Users already have access to indicative abundance values in conjunction with strain classification to help interpret the identification results. For instance, consider a sample that contains 16 000 reads of six S . thermophilus strains equitably distributed and making two clusters of sibling strains: A={CIRM-BIA18, CIRM-BIA32} and B={CIRM-BIA2101, CIRM-BIA23, CIRM-BIA65 and CIRM-BIA67}. Thus, the sample comprises 1/3 of A and 2/3 of B. For this sample ORI found 25 % of A, 72 % of B and 3 % of a third cluster C={CIRM-BIA1116, CIRM-BIA1122, CIRM-BIA16, CIRM-BIA29, CNRZ1066, CS8, EPS, S9} (MCC=0.807). Since the abundance of C is quite low and it is quite close to cluster B (see Fig. 3), it is reasonable to assume that the reads originated in fact from B. By increasing the merging threshold τ, B and C would indeed be merged, leading to perfect identification.
Towards a generalization to other species and genera?
The index can be tailored to any set of bacterial genomes by modifying the genome index, as explained on the website. We tested our method on Streptococcus pyogenes (around 200 genomes in NCBI; some closely related and mostly different by their phage content). A known S. pyogenes emm75 strain of our collection was correctly identified by ORI. We also used 1600 complete genomes to create a specific E. coli index. Reads were from a piglet gut microbiota. This gave promising results in about 5 h: all the reference strains identified were also from a piglet gut microbiota. The next challenge is to test ORI on a larger number of species in order to fit users’ study species. As a first step, we plan to increase our database to the whole order Lactobacillales . We aim at keeping good accuracy and sensitivity for identification at the strain level while staying efficient with respect to memory and computation time. This will require some modifications in the means of requesting the index which, currently, remains the most time-consuming part.
Supplementary Data
Funding information
This research was funded by a PhD grant from an INRAE-INRIA consortium agreement (G. Siekaniec) and an INRIA delegation position (E. Roux). The France Génomique and French Bioinformatics Institute national infrastructures are funded as part of the Investissement d’Avenir programme managed by Agence Nationale pour la Recherche, contracts ANR-10-INBS-09 and ANR-11-INBS-0013.
Acknowledgements
All data were processed on the Genouest bioinformatics platform (https://www.genouest.org/). LABGeM (CEA/Genoscope and CNRS UMR8030), the France Génomique and French Bioinformatics Institute national infrastructures are acknowledged for support within the MicroScope annotation platform [41]. We thank in particular David Vallenet, scientific manager of the MicroScope platform, who provided us with text to describe its functioning. We also thank Genoscope (Corinne Cruaud, Stefan Engelen and Jean-Marc Aury) and CNRS I2BC (Delphine Naquin, Erwin Van Dijk and Yan Jaszczyszyn) for their generous support for training in the use of the MinION and associated bioinformatics tools. Finally, the reviewers greatly helped to improve the first manuscript.
Author contributions
Conceptualization, J.N.; methodology, G.S., J.N. and T.L.; software, G.S. and T.L.; validation, G.S.; resources, E.R.; data curation, G.S. and E.R.; writing–original draft preparation, J.N., G.S. and E.R.; writing–review and editing, J.N., E.R., G.S. and E.G.; supervision, J.N., E.R. and E.G.; project administration, J.N.; funding acquisition, J.N. and E.G. All authors have read and agreed to the published version of the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ANI, average nucleotide identity; ASP, answer set programming; HMW, high molecular weight; LCA, lowest common ancestor; MCC, matthews correlation coefficient; MICFAM, microScope gene families; NGS, next generation sequencing; ONT, Oxford Nanopore Technologies; ORI, Oxford Nanopore reads identification; SD, standard deviation; WGS, whole genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Nine supplementary tables, two supplementary text and three supplementary figures are available with the online version of this article.
References
- 1.Stromberg ZR, Van Goor A, Redweik GAJ, Wymore Brand MJ, Wannemuehler MJ, et al. Pathogenic and non-pathogenic Escherichia coli colonization and host inflammatory response in a defined microbiota mouse model. Dis Model Mech. 2018;11:11. doi: 10.1242/dmm.035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siezen RJ, Starrenburg MJC, Boekhorst J, Renckens B, Molenaar D, et al. Genome-scale genotype-phenotype matching of two Lactococcus lactis isolates from plants identifies mechanisms of adaptation to the plant niche. Appl Environ Microbiol. 2008;74:424–436. doi: 10.1128/AEM.01850-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, Liu M, Xu J, Qi Y, Zhao N, et al. First insight into the probiotic properties of ten Streptococcus thermophilus strains based on in vitro conditions. Curr Microbiol. 2020;77:343–352. doi: 10.1007/s00284-019-01840-3. [DOI] [PubMed] [Google Scholar]
- 4.Meola M, Rifa E, Shani N, Delbès C, Berthoud H, et al. DAIRYdb: a manually curated reference database for improved taxonomy annotation of 16S rRNA gene sequences from dairy products. BMC Genomics. 2019;20:560. doi: 10.1186/s12864-019-5914-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lesker TR, Durairaj AC, Gálvez EJC, Lagkouvardos I, Baines JF, et al. An integrated metagenome catalog reveals new insights into the murine gut microbiome. Cell Rep. 2020;30:2909–2922. doi: 10.1016/j.celrep.2020.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma B, France MT, Crabtree J, Holm JB, Humphrys MS, et al. A comprehensive non-redundant gene catalog reveals extensive within-community intraspecies diversity in the human vagina. Nat Commun. 2020;11:940. doi: 10.1038/s41467-020-14677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pérez-Cobas AE, Gomez-Valero L, Buchrieser C. Metagenomic approaches in microbial ecology: an update on whole-genome and marker gene sequencing analyses. Microb Genom. 2020;6 doi: 10.1099/mgen.0.000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quince C, Walker AW, Simpson JT, Loman NJ, Segata N. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017;35:833–844. doi: 10.1038/nbt.3935. [DOI] [PubMed] [Google Scholar]
- 9.Latorre-Pérez A, Villalba-Bermell P, Pascual J, Vilanova C. Assembly methods for nanopore-based metagenomic sequencing: a comparative study. Sci Rep. 2020;10:13588. doi: 10.1038/s41598-020-70491-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol [Internet] 2017;13:1–22. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anyansi C, Straub TJ, Manson AL, Earl AM, Abeel T. Computational methods for strain-level microbial detection in colony and metagenome sequencing data. Front Microbiol. 2020;11:1925. doi: 10.3389/fmicb.2020.01925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balloux F, Brynildsrud OB, van DL, Shaw LP, Chen H, et al. From Theory to Practice: Translating Whole-Genome Sequencing (WGS) into the Clinic. Trends in Microbiology. 2018;26:1035–1048. doi: 10.1016/j.tim.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh GB. Fundamentals of Bioinformatics and Computational Biology. Cham: Springer International Publishing; 2015. Alignment tools; pp. 159–170. [Google Scholar]
- 14.Daniel HH, Alexander FA, Ji Q, Stephan CS. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilke A, Bischof J, Gerlach W, Glass E, Harrison T, et al. The mg-rast metagenomics database and portal in 2015. Nucleic Acids Res. 2015;44:4. doi: 10.1093/nar/gkv1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scholz M, Ward DV, Pasolli E, Tolio T, Zolfo M, et al. Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat Methods. 2016;13:435–438. doi: 10.1038/nmeth.3802. [DOI] [PubMed] [Google Scholar]
- 17.Kim D, Song L, Breitwieser FP, Salzberg SL. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26:1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menzel P, Ng KL, Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun. 2016;7:1–9. doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ounit R, Wanamaker S, Close TJ, Lonardi S. CLARK: Fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers. BMC Genomics. 2015;16:236. doi: 10.1186/s12864-015-1419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roosaare M, Vaher M, Kaplinski L, Möls M, Andreson R, et al. Strainseeker: Fast identification of bacterial strains from raw sequencing reads using user-provided guide trees. PeerJ. 2017;5:e3353. doi: 10.7717/peerj.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer F, Bremges A, Belmann P, Janssen S, McHardy AC, et al. Assessing taxonomic metagenome profilers with OPAL. Genome Biol. 2019;20:51. doi: 10.1186/s13059-019-1646-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pollard MO, Gurdasani D, Mentzer AJ, Porter T, Sandhu MS. Long reads: their purpose and place. Hum Mol Genet. 2018;27:R234–41. doi: 10.1093/hmg/ddy177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vandervalk BP, Yang C, Xue Z, Raghavan K, Chu J, et al. Konnector v2.0: Pseudo-long reads from paired-end sequencing data. BMC Med Genomics. 2015;8:S1. doi: 10.1186/1755-8794-8-S3-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olm MR, Crits-Christoph A, Bouma-Gregson K, Firek BA, Morowitz MJ, et al. inStrain profiles population microdiversity from metagenomic data and sensitively detects shared microbial strains. Nat Biotechnol. 2021;39:727–736. doi: 10.1038/s41587-020-00797-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts M, Hayes W, Hunt BR, Mount SM, Yorke JA. Reducing storage requirements for biological sequence comparison. Bioinformatics. 2004;20:3363–3369. doi: 10.1093/bioinformatics/bth408. [DOI] [PubMed] [Google Scholar]
- 28.Dilthey AT, Jain C, Koren S, Phillippy AM. Strain-level metagenomic assignment and compositional estimation for long reads with MetaMaps. Nat Commun. 2019;10:3066. doi: 10.1038/s41467-019-10934-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinović A, Cocuzzi R, Arioli S, Mora D. Streptococcus thermophilus: To survive, or not to survive the gastrointestinal tract, that is the question! Nutrients. 2020;12:E2175. doi: 10.3390/nu12082175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexandraki V, Kazou M, Blom J, Pot B, Papadimitriou K, et al. Comparative genomics of Streptococcus thermophilus support important traits concerning the evolution, biology and technological properties of the species. Front Microbiol. 2019;10:2916. doi: 10.3389/fmicb.2019.02916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Junjua M, Kechaou N, Chain F, Awussi AA, Roussel Y, et al. A large scale in vitro screening of Streptococcus thermophilus strains revealed strains with a high anti-inflammatory potential. LWT. 2016;70:78–87. doi: 10.1016/j.lwt.2016.02.006. [DOI] [Google Scholar]
- 32.Terzaghi BE, Sandine WE. Improved medium for lactic Streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Man JC, Rogosa M, Sharpe ME. A medium for the cultivation of Lactobacilli . J Appl Bacteriol. 1960;23:130–135. doi: 10.1111/j.1365-2672.1960.tb00188.x. [DOI] [Google Scholar]
- 34.Leimeister C-A, Boden M, Horwege S, Lindner S, Morgenstern B. Fast alignment-free sequence comparison using spaced-word frequencies. Bioinformatics. 2014;30:1991–1999. doi: 10.1093/bioinformatics/btu177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Břinda K, Sykulski M, Kucherov G. Spaced seeds improve k-mer-based metagenomic classification. Bioinformatics. 2015;31:3584–3592. doi: 10.1093/bioinformatics/btv419. [DOI] [PubMed] [Google Scholar]
- 36.Kucherov G, Noé L, Roytberg M. A unifying framework for seed sensitivity and its application to subset seeds. J Bioinform Comput Biol. 2004:21. doi: 10.1142/s0219720006001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noé L. Best hits of 11110110111: Model-free selection and parameter-free sensitivity calculation of spaced seeds. Algorithms Mol Biol. 2017;12:1. doi: 10.1186/s13015-017-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crainiceanu A, Lemire D. Bloofi: Multidimensional Bloom filters. Information Systems. 2015;54:311–324. doi: 10.1016/j.is.2015.01.002. [DOI] [Google Scholar]
- 39.Harris RS, Medvedev P. Improved representation of sequence bloom trees. Bioinformatics. 2020;36:721–727. doi: 10.1093/bioinformatics/btz662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vallenet D, Calteau A, Dubois M, Amours P, Bazin A, et al. Microscope: An integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res. 2019;48:D579–89. doi: 10.1093/nar/gkz926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bocs S, Cruveiller S, Vallenet D, Nuel G, Médigue C. AMIGene: Annotation of MIcrobial Genes. Nucleic Acids Research. 2003;31:3723–3726. doi: 10.1093/nar/gkg590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ignatov DI. Russian Summer School in Information Retrieval. Springer; 2014. Introduction to formal concept analysis and its applications in information retrieval and related fields; pp. 42–141. [Google Scholar]
- 45.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–9. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marcelino VR, Holmes EC, Sorrell TC. The use of taxon-specific reference databases compromises metagenomic classification. BMC Genomics. 2020;21:1–5. doi: 10.1186/s12864-020-6592-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gebser M, Kaminski R, Kaufmann B, Schaub T. Answer set solving in practice. Synth Lect Artif Intell Mach Learn. 2012;6:1–238. [Google Scholar]
- 48.Van Rossum T, Ferretti P, Maistrenko OM, Bork P. Diversity within species: interpreting strains in microbiomes. Nat Rev Microbiol. 2020;18:491–506. doi: 10.1038/s41579-020-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gori A, Harrison OB, Mlia E, Nishihara Y, Chan JM, et al. Pan-gwas of streptococcus agalactiae highlights lineage-specific genes associated with virulence and niche adaptation. mBio. 2020;11:e00728-20. doi: 10.1128/mBio.00728-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Břinda K, Callendrello A, Ma KC, MacFadden DR, Charalampous T, et al. Rapid inference of antibiotic resistance and susceptibility by genomic neighbour typing. Nat Microbiol. 2020;5:455–464. doi: 10.1038/s41564-019-0656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greig DR, Dallman TJ, Hopkins KL, Jenkins C. Minion Nanopore sequencing identifies the position and structure of bacterial antibiotic resistance determinants in a multidrug-resistant strain of enteroaggregative Escherichia coli . Microb Genom. 2018;4:e000213. doi: 10.1099/mgen.0.000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.