Abstract
Antimicrobial resistance (AMR) plays an important role in the pathogenesis and spread of Clostridioides difficile infection (CDI), the leading healthcare-related gastrointestinal infection in the world. An association between AMR and CDI outbreaks is well documented, however, data is limited to a few ‘epidemic’ strains in specific geographical regions. Here, through detailed analysis of 10 330 publicly-available C. difficile genomes from strains isolated worldwide (spanning 270 multilocus sequence types (STs) across all known evolutionary clades), this study provides the first species-wide snapshot of AMR genomic epidemiology in C. difficile . Of the 10 330 C . difficile genomes, 4532 (43.9 %) in 89 STs across clades 1–5 carried at least one genotypic AMR determinant, with 901 genomes (8.7 %) carrying AMR determinants for three or more antimicrobial classes (multidrug-resistant, MDR). No AMR genotype was identified in any strains belonging to the cryptic clades. C. difficile from Australia/New Zealand had the lowest AMR prevalence compared to strains from Asia, Europe and North America (P<0.0001). Based on the phylogenetic clade, AMR prevalence was higher in clades 2 (84.3 %), 4 (81.5 %) and 5 (64.8 %) compared to other clades (collectively 26.9 %) (P<0.0001). MDR prevalence was highest in clade 4 (61.6 %) which was over three times higher than in clade 2, the clade with the second-highest MDR prevalence (18.3 %). There was a strong association between specific AMR determinants and three major epidemic C. difficile STs: ST1 (clade 2) with fluoroquinolone resistance (mainly T82I substitution in GyrA) (P<0.0001), ST11 (clade 5) with tetracycline resistance (various tet-family genes) (P<0.0001) and ST37 (clade 4) with macrolide-lincosamide-streptogramin B (MLSB) resistance (mainly ermB) (P<0.0001) and MDR (P<0.0001). A novel and previously overlooked tetM-positive transposon designated Tn6944 was identified, predominantly among clade 2 strains. This study provides a comprehensive review of AMR in the global C. difficile population which may aid in the early detection of drug-resistant C. difficile strains, and prevention of their dissemination worldwide.
Keywords: antimicrobial resistance, Clostridioides difficile, genomics
Data Summary
This study utilises publicly available raw sequence reads available at the NCBI Sequence Read Archive (SRA) as of January 2020. The details of all genomes are available in the Supplementary Data (10.6084 /m9 .figshare.14623533).
Impact Statement.
Utilising a publicly-available database of 10 330 sequence reads, this study provides the first species-wide evaluation of genotypic AMR in C. difficile . We report the most common AMR determinants and their genomic neighbourhood, associations between important AMR and MDR genotypes and specific clades or geographical regions, and rare AMR genotypes that may have been missed in earlier studies.
Introduction
Antimicrobial resistance (AMR) is one of the biggest threats to modern medicine. Without focused interventions and collaborations across all government sectors, AMR could be responsible for an estimated ten million deaths and the loss of up to US$210 trillion of annual global income by 2050 [1]. The US Centers for Disease Control and Prevention (CDC) reported on AMR health threats in 2013 [2], with an update in 2019 [3], highlighting organisms with the highest AMR burden and threat [3].
Clostridioides ( Clostridium ) difficile infection (CDI) causes major gastrointestinal illness worldwide [4], responsible for as many as 14 000 deaths annually in the US [2]. C. difficile has been classified by the CDC as an urgent threat, the highest threat level, in both the 2013 and 2019 CDC reports, responsible for the highest number of annual deaths among the pathogens listed [2, 3]. In contrast to other pathogens, AMR in C. difficile has some unique features. AMR usually leads to difficulties in treating infections [5], and although the treatment of CDI is also a challenge [6], such challenge is not due to AMR per se as resistance to antimicrobials predominantly used for the treatment of CDI (vancomycin, metronidazole and fidaxomicin) remains rare [7]. Instead, AMR plays a significant role in the pathogenesis and spread of CDI [8], as it allows C. difficile to survive antimicrobial exposure in the host, while selective pressure allows the emergence and spread of AMR strains. Several AMR strains have been associated with outbreaks; PCR ribotype (RT) 017 with clindamycin [9], RTs 017 and 027 with fluoroquinolones [10, 11], RT 027 with rifampicin [12] and RT 078 with tetracyclines [13].
Using multi-locus sequence typing (MLST), the population of C. difficile can be divided into five major clades (C1 – C5) and three smaller cryptic clades (C-I, C-II and C-III). The three cryptic clades are extremely divergent (Figs 1 and 2a) and likely represent independent species or subspecies based on the genomic data [14]. Three of the five major clades contain epidemic sequence types (STs); C2 contains ST 1 (corresponding to RT 027), C4 contains ST 37 (RT 017) and C5 contains ST 11 (several RTs, including RT 078) [14]. To date, studies have been conducted on the role of AMR in the emergence and spread of two epidemic STs, 1 and 11 [10, 12, 13]. A few studies have focused also on C. difficile ST 37 [9, 11], a third epidemic lineage [15], which shows a high prevalence of resistance to many antimicrobial classes [8]. Although these studies provided insights on how AMR impacts the spread of C. difficile , they are limited to a few strain types in specific geographical regions, and there has not been any study of AMR prevalence in the species-wide population of C. difficile . Here, through detailed analysis of 10 330 publicly-available genomes from C. difficile isolated worldwide, we provide the first species-wide snapshot of AMR genomic epidemiology in C. difficile .
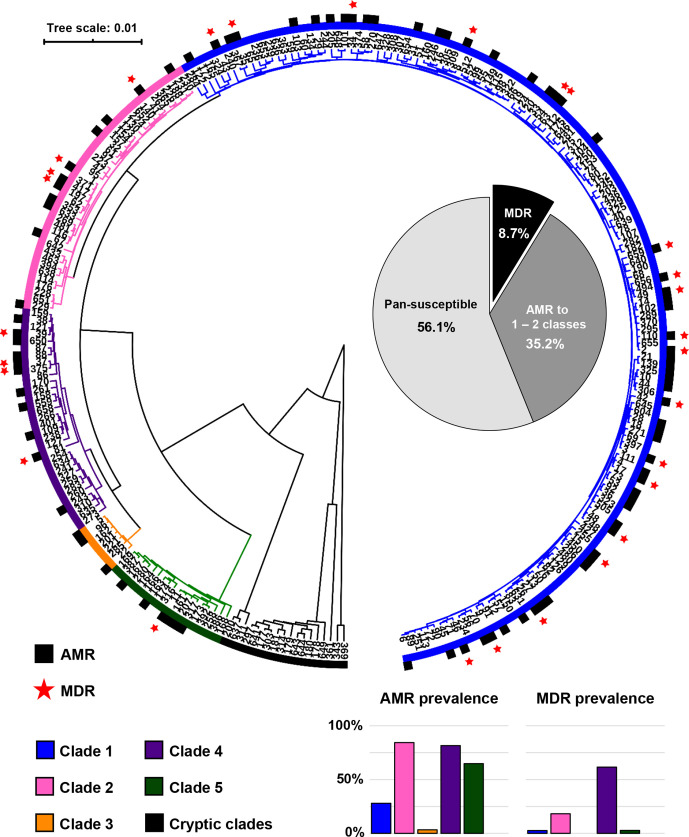
Fig. 1.
Distribution of resistant and multidrug-resistant (MDR) C. difficile . The UPGMA phylogenetic tree represents a total of 270 STs included in this study. The black sections indicate that at least one strain in the ST had acquired resistance (AMR) to at least one antimicrobial class. The red stars indicate that at least one strain in the ST was MDR (i.e. had acquired resistance to at least three antimicrobial classes). The pie chart in the middle shows the overall prevalence of MDR C. difficile (black), C. difficile resistance to 1–2 antimicrobial classes (dark grey) and pan-susceptible C. difficile (light grey) among 10 330 C . difficile strains. The bar charts below show the prevalence of resistant and MDR strains in each clade.
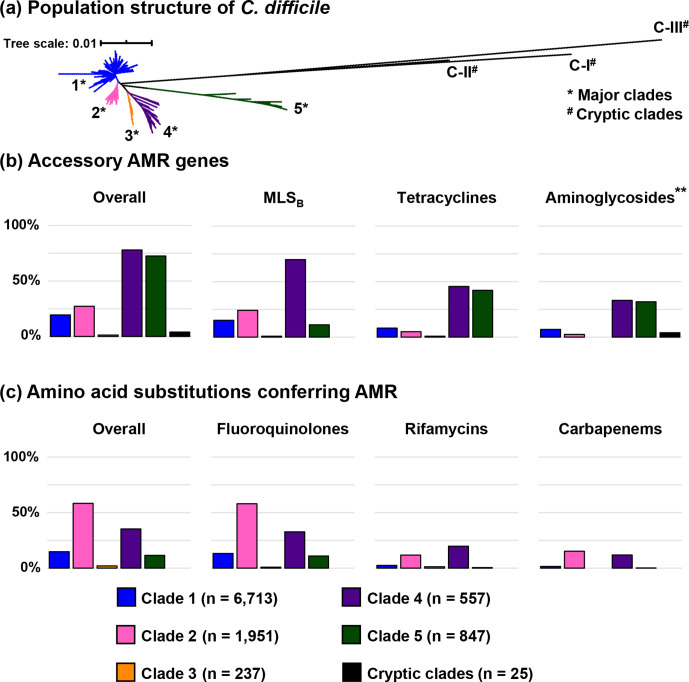
Fig. 2.
Summary of antimicrobial resistance genotype of C. difficile . (a) For evolutionary context, a neighbour-joining phylogeny based on MLST shows the global population structure of C. difficile . (b) The prevalence of C. difficile strains harbouring accessory AMR genes across different clades (leftmost) and the prevalence of resistance to important antimicrobial classes conferred mainly by accessory AMR genes. The presence of an aminoglycoside resistance gene (**) does not contribute to the definition of MDR C. difficile . (c) The prevalence of C. difficile strains having significant amino acid substitutions associated with AMR across different clades (leftmost) and the prevalence of resistance to important antimicrobial classes conferred mainly by amino acid substitution.
Methods
Genome collection and de-replication of clonal strains
The starting point for this analysis was an international collection of 12 098 C . difficile Illumina paired-end sequence reads sourced from the NCBI Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra/) in January 2020. All sequence reads were screened for contamination using Kraken2 v2.0.8-beta and only reads with >85 % of sequences classified as C. difficile were included. MLST was confirmed on these raw sequence reads by SRST2 v0.2.0 with the database available on PubMLST (https://pubmlst.org/organisms/clostridioides-difficile) as previously described [14, 16]. This dataset comprised a total of 270 STs spanning the eight currently described evolutionary clades with a relatively high number of reads from epidemic strains, particularly STs 1 (C2; n=2,532), 11 (C5; n=1,185) and 37 (C4; n=786), many of which were likely to be clonal. To adjust for this strain selection bias, pairwise average nucleotide identity (ANI) of reads from these three STs, as well as ST 2 (n=1153), the most common strain in C1, were compared using the Sketch algorithm included in BBtools (https://sourceforge.net/projects/bbmap/). Reads with an ANI of 99.98 % or higher were considered to be clonal and only one genome from each clonal complex was included in the final analysis. Based on a small dataset of 240 C . difficile reads (28 680 possible pairs, 531 of which were clonal pairs), this cut-off point had a sensitivity of 70.1 % and a specificity of 76.8 % for the detection of clonal strains as defined by Didelot et al. (data not shown) [17]. The 10 330 reads remaining in the dataset are summarised in Table 1.
Table 1.
C. difficile strains in the de-replicated NCBI database (January 2020)
|
C. C. difficile clade |
No. of genomes (%) |
Most prevalent STs |
|---|---|---|
|
C1 |
6713 (65.0 %) |
ST 2 (9.2 %)* |
|
|
ST 8 (6.0 %)* |
|
|
|
ST 3 (5.4 %)* |
|
|
|
ST 42 (4.1 %)* |
|
|
|
ST 6 (3.2 %)* |
|
|
|
ST 44 (2.5 %)* |
|
|
|
ST 14 (2.4 %)* |
|
|
C2 |
1951 (18.9 %) |
ST 1 (16.6 %)* |
|
|
ST 41 (0.8 %) |
|
|
C3 |
237 (2.3 %) |
ST 5 (2.0 %) |
|
|
ST 22 (0.2 %) |
|
|
C4 |
557 (5.4 %) |
ST 37 (4.3 %)* |
|
|
ST 39 (0.2 %) |
|
|
C5 |
847 (8.2 %) |
ST 11 (7.6 %)* |
|
|
ST 167 (0.1 %) |
|
|
Cryptic clades |
25 (0.2 %) |
ST 361 (<0.1 %) |
|
|
ST 177 (<0.1 %) |
|
|
Total |
10 330 |
– |
*Ten most prevalent sequence types (STs) in this dataset.
ST, sequence type.
Identification of multidrug-resistant C. difficile
Multidrug-resistant (MDR) C. difficile in this study refers to C. difficile strains with genotypic AMR determinants (both accessory genes and mutations in chromosomal genes) for at least three of the following antimicrobial classes: carbapenems, fluoroquinolones, glycopeptides (vancomycin), nitroimidazoles (metronidazole), oxazolidinones (linezolid), macrolide-lincosamide-streptogramin B (MLSB), phenicols, rifamycins, tetracyclines and sulfa-containing agents. Resistance determinants for aminoglycosides and cephalosporins were excluded from this definition as C. difficile is intrinsically resistant to these agents [18, 19].
Detection of accessory AMR genes and associated transposons
To detect the presence of accessory AMR genes, raw sequence reads were interrogated against ResFinder/ARGannot databases, with an addition of two newly-characterised AMR genes found in C. difficile , erm(52) and mefH, using SRST2 with default settings [16, 20–22]. These databases contain over 500 different genes conferring resistance to 15 different antimicrobial classes, covering all AMR genes known to be carried by the C. difficile population analysed so far [20, 21]. The spectrum of β-lactamase enzymes detected was confirmed against the CARD 2020 database [23]. To further characterise the genomic context of the most common accessory AMR genes, C. difficile strains with ermB, tetM and tet44 genes were interrogated using SRST2 against a database of C. difficile transposons carrying ermB (Tn5398 [GenBank accession AF109075.2], Tn6189 [MK895712.1], Tn6194 [HG475346.1], Tn6215 [KC166248.1] and Tn6218 [HG002387.1]), tetM (Tn916 [U09422.1], Tn5397 [AF333235.1] and Tn6190 [FN665653]) and tet44 (Tn6164 [FN665653]) [24, 25] with 80 % minimum coverage and 10 % maximum divergence [16], corresponding with 72 % minimum nucleotide identity (NI).
To detect the presence of a plasmid conferring metronidazole resistance (pCD-METRO) [26], a custom database was created consisting of all eight coding sequences (CDS) of pCD-METRO. SRST2 was used with default settings on all sequence reads against this customised database [16]. The 23 C . difficile genomes from the original study [26] were included in the analysis and used to evaluate the accuracy of the database.
Detection of amino acid substitutions conferring AMR
All genomes were screened for known point mutations in gyrA, gyrB, rpoB, pbp1 and pbp3 genes using customised databases in SRST2. The reference sequences for these genes were obtained from the PubMLST database (https://pubmlst.org/organisms/clostridioides-difficile/) as well as reference C. difficile genomes (CD630 [C1, GenBank accession AM180355], CD196 [C2, FN538970], M68 [C4, FN668375] and M120 [C5, FN665653]). C. difficile strains were categorized as resistant to an antimicrobial if they carried a gene allele with at least one significant point mutation listed in Table 2 [24, 27, 28].
Table 2.
Summary of known non-synonymous chromosomal point mutations conferring AMR
|
Protein |
Substitution |
Clade distribution* |
Comment |
|||||
|---|---|---|---|---|---|---|---|---|
|
C1 |
C2 |
C3 |
C4 |
C5 |
Cryptic |
|||
|
Fluoroquinolone resistance | ||||||||
|
GyrA |
Val43Asp |
○ |
○ |
○ |
○ |
○ |
○ |
Absent in this dataset |
|
|
Asp71Val |
● |
○ |
○ |
● |
○ |
○ |
Found in <10 strains in this dataset |
|
|
Asp81Asn |
● |
○ |
○ |
○ |
○ |
○ |
Found in <10 strains in this dataset |
|
|
Thr82Ile |
● |
● |
● |
● |
● |
○ |
Most common substitution |
|
|
Thr82Val |
● |
○ |
○ |
○ |
● |
○ |
Found in <10 strains in this dataset |
|
|
Ala118Thr |
● |
○ |
○ |
○ |
● |
○ |
Found in <10 strains in this dataset |
|
|
Ala384Asp |
● |
○ |
○ |
○ |
○ |
○ |
Found in <10 strains in this dataset |
|
GyrB |
Arg377Gly |
○ |
○ |
○ |
○ |
○ |
○ |
Absent in this dataset |
|
|
Asp426Asn |
● |
● |
○ |
● |
○ |
○ |
Most common substitution |
|
|
Asp426Val |
● |
○ |
○ |
● |
○ |
○ |
|
|
|
Arg447Lys |
● |
● |
○ |
○ |
● |
○ |
|
|
|
Glu466Val |
○ |
○ |
○ |
○ |
● |
○ |
Found in <10 strains in this dataset |
|
Rifamycin resistance | ||||||||
|
RpoB |
Asp492Asn |
○ |
○ |
○ |
○ |
○ |
○ |
Absent in this dataset |
|
|
Asp492Val |
○ |
○ |
○ |
○ |
○ |
○ |
Absent in this dataset |
|
|
His502Asn |
● |
● |
○ |
● |
○ |
○ |
|
|
|
His502Arg |
○ |
○ |
○ |
○ |
○ |
○ |
Absent in this dataset |
|
|
His502Leu |
○ |
○ |
○ |
○ |
○ |
○ |
Absent in this dataset |
|
|
His502Tyr |
● |
○ |
○ |
○ |
○ |
○ |
Found in <10 strains in this dataset |
|
|
Arg505Lys |
● |
● |
● |
● |
● |
○ |
Most common substitution |
|
|
Ser550Phe |
● |
● |
○ |
○ |
● |
○ |
Found in <10 strains in this dataset |
|
|
Ser550Tyr |
● |
● |
○ |
● |
○ |
○ |
Found in <10 strains in this dataset |
|
Fidaxomicin resistance | ||||||||
|
RpoB |
Gln1073Arg |
○ |
○ |
○ |
○ |
○ |
○ |
Absent in this dataset |
|
Carbapenem resistance | ||||||||
|
Pbp1 |
Leu543His |
● |
○ |
○ |
○ |
○ |
○ |
|
|
|
Ala555Thr |
● |
● |
○ |
● |
● |
○ |
Most common substitution |
|
Pbp3 |
Tyr721Ser |
● |
○ |
○ |
● |
○ |
○ |
|
*Based on significant findings in this study. Solid circles refer to the presence of the substitution in the clade.
Assessment of AMR prevalence in different geographical areas
Data on geographical regions of isolation was available for 6227 (60.3 %) C . difficile strains: Asia (n=355), Europe (n=3548), North America (n=2212) and Australia/New Zealand (n=112). The clade distribution was notably different in these regions (Table 3). Thus, multiple logistic regression analyses were performed using R to assess the clade-adjusted AMR prevalence for major antimicrobial classes (MLSB, tetracyclines, fluoroquinolones and rifamycins), as well as MDR prevalence. From the initial analysis, the overall AMR prevalence was lowest in strains from Australia/New Zealand. Thus, they were used as the reference group in this analysis.
Table 3.
Clade distribution in four major geographical regions
|
Region |
Clade |
|||||
|---|---|---|---|---|---|---|
|
C1 |
C2 |
C3 |
C4 |
C5 |
Cryptic |
|
|
Asia |
76.6 % |
3.4 % |
3.9 % |
15.2 % |
0.6 % |
0.3 % |
|
Europe |
74.4 % |
11.0 % |
3.4 % |
2.7 % |
8.3 % |
0.2 % |
|
North America |
68.9 % |
24.7 % |
0.1 % |
2.1 % |
4.0 % |
0.2 % |
|
Australia/New Zealand |
39.3 % |
26.8 % |
1.8 % |
3.6 % |
28.6 % |
0.0 % |
Results
Summary of AMR and MDR prevalence
Of the 10 330 C . difficile genomes evaluated, 4532 (43.9 %) contained acquired resistance genes for at least one antimicrobial class, with 89 STs across five major clades having at least one resistant strain (Fig. 1). A total of 901 strains (8.7 %) across 28 STs harboured resistance determinants for three or more antimicrobial classes and were therefore classified as MDR. Based on resistance prevalence, C. difficile could be divided into clades with an overall resistance prevalence of ≥50 %, which included C2, C4 and C5, each of which contained an epidemic ST (ST 1 in C2, ST 37 in C4 and ST 11 in C5), and clades with an overall resistance prevalence of <50 %, which included C1 and C3, as well as all three cryptic clades. The prevalence of MDR C. difficile was highest in C4 C. difficile (61.6 % [343/557] compared to an overall 5.7 % [558/9,773] in other clades), over three times higher than in C2 which had the second-highest prevalence of MDR strains (356/1951; 18.3 %). The overall resistance prevalence of important antimicrobial classes is shown in Fig. 2.
AMR prevalence in different geographical regions
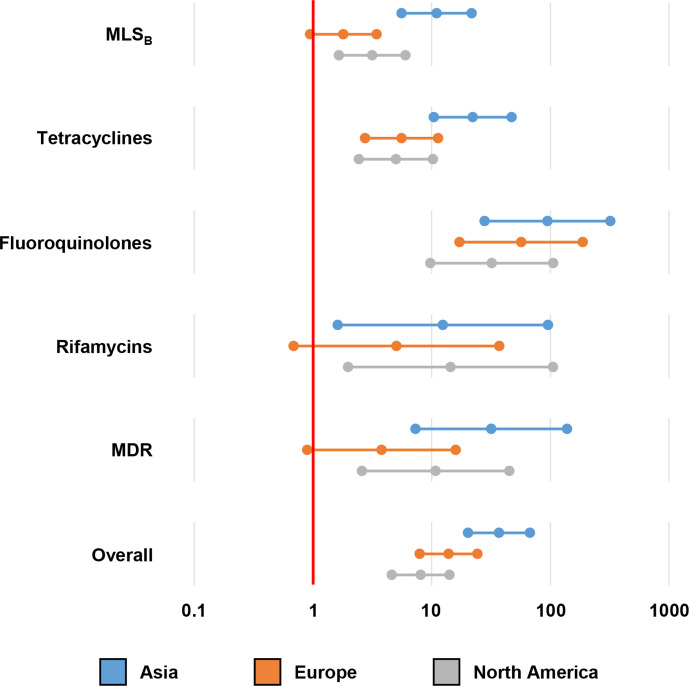
Fig. 3 shows the results of logistic regression analyses of the clade-adjusted AMR and MDR prevalence compared to strains from Australia/New Zealand as the reference. Overall, strains from Asia, Europe and North America all had higher AMR prevalence (P<0.0001). The difference in AMR prevalence was most pronounced for fluoroquinolones, where the prevalence of substitution associated with fluoroquinolone resistance (FQR) in the three continents (collectively 1491/6115; 24.4 %) was estimated to be at least nine times higher than in Australia/New Zealand (3/112; 2.7 %). In Asia, Europe and North America, AMR prevalence was not significantly different, with AMR prevalence in Asia (99/355; 27.9 %) marginally higher than in Europe (814/3548; 22.9 %) and North America (578/2212; 26.1 %).
Fig. 3.
Difference in antimicrobial resistance (AMR) prevalence in different geographical regions. Multiple logistic regression analyses were performed to compare the clade-adjusted AMR prevalence in four regions (Asia, Europe, North America and Australia/New Zealand). The Forest plot represents the estimated AMR prevalence in each continent compared to Australia/New Zealand.
Fluoroquinolone resistance
Overall, 2959 C . difficile strains (28.6 %) carried known DNA gyrase substitutions associated with FQR. The prevalence of FQR was highest in clade C2 (1606/1951; 82.3 %), followed by C4 (296/557; 53.1 %). Most resistance was conferred by point substitutions solely within the GyrA subunit of the enzyme (2771/2959; 93.7 %), followed by point substitutions solely within the GyrB subunit (104/2959; 3.5 %). Only 2.8 % (84/2959) had substitutions on both gyrase subunits. The prevalence of GyrB subunit substitution (both alone and in addition to GyrA substitution) was highest in C4 (59/557; 10.6 %). The most common GyrA substitution was Thr82Ile (2843/2855; 99.6 % of strains with GyrA substitution) and the most common GyrB substitution was Asp426Asn (131/188; 69.7 % of strains with GyrB substitution), followed by Asp426Val (44/188; 23.4 %), the latter was almost exclusive to C4 (40/44; 90.9 % of strains with Asp426Val substitution belonged to C4). Interestingly, a Ser416Ala substitution, a polymorphism that does not confer resistance, was found in a majority of C5 (825/847; 94.9 %) and cryptic clades (20/25; 80.0 %), but in only one clade C1 strain and none of the other major clades.
MLSB resistance
Table 4 summarises the major genotypic determinants for MLSB antimicrobials detected in our survey. The most common determinants were ermB (1775 strains, 17.2 %) followed by erm(52) (145 strains, 1.4 %) and ermG (25 strains, 0.2 %). The erm class genes, which methylate 23S rRNA and prevent the binding of MLSB antimicrobials, are associated with high-level resistance to all MLSB antimicrobials, as shown by high-level resistance to both clindamycin and erythromycin [29]. The most common non-erm genes were mefH (156 strains, 1.5 %), mefA (24 strains, 0.2 %), msrD (21 strains, 0.2 %) and lnuC (17 strains, 0.2 %). In total, 1979 C . difficile strains (19.2 %) across 65 STs (23.9%) in five major clades carried acquired MLSB resistance determinants.
Table 4.
Summary of resistance determinants for MLSB antimicrobials
|
Gene |
Clade distribution [N (%)] |
Overall |
|||||
|---|---|---|---|---|---|---|---|
|
C1 |
C2 |
C3 |
C4 |
C5 |
Cryptic |
||
|
ermB |
953 (14.2%) |
421 (21.6%) |
0 (0.0%) |
328 (58.9%) |
73 (8.6%) |
0 (0.0%) |
1776 (17.2%) |
|
Tn5398 |
168 (2.5%) |
1 (0.1%) |
0 (0.0%) |
0 (0.0%) |
1 (0.1%) |
0 (0.0%) |
170 (1.6%) |
|
Tn6189 |
259 (3.9%) |
104 (5.3%) |
0 (0.0%) |
44 (7.9%) |
17 (2.0%) |
0 (0.0%) |
424 (4.1%) |
|
Tn6194 |
204 (3.0%) |
270 (13.8%) |
0 (0.0%) |
268 (48.1%) |
46 (5.4%) |
0 (0.0%) |
788 (7.6%) |
|
Tn6215 |
106 (1.6%) |
0 (0.0%) |
0 (0.0%) |
0 (0.0%) |
2 (0.2%) |
0 (0.0%) |
108 (1.0%) |
|
Tn6218 |
200 (3.0%) |
4 (0.2%) |
0 (0.0%) |
10 (1.8%) |
2 (0.2%) |
0 (0.0%) |
216 (2.1%) |
|
Unknown |
16 (0.2%) |
42 (2.2%) |
0 (0.0%) |
6 (1.1%) |
5 (0.6%) |
0 (0.0%) |
69 (0.7%) |
|
Other erm genes |
86 (1.3%) |
17 (0.9%) |
1 (0.4%) |
66 (11.8%) |
4 (0.5%) |
0 (0.0%) |
175 (1.7%) |
|
Non-erm genes |
76 (1.1%) |
104 (5.3%) |
1 (0.4%) |
22 (3.9%) |
18 (2.1%) |
0 (0.0%) |
222 (2.1%) |
Among ermB-positive strains, known ermB-carrying transposons were identified in 1706 strains (96.5 %) (range, 77.6–100.0 % NI). Transposon diversity was highest in C1 (Table 4). The most common ermB-positive transposon was Tn6194 (788/1775; 44.4 %; 81.9–100.0 % NI), followed by Tn6189 (424/1775; 23.9 %; 77.6–99.9 % NI) and Tn6218 (216/1775; 12.2 %; 85.3–100.0 % NI). Tn5398, which contains two copies of the ermB gene, was found in 170 strains (9.6 %; 81.2–100.0 % NI), most of which belonged to clade C1 (168/170; 98.8 %).
Tetracycline resistance
Table 5 summarises the genotypic determinants found for tetracyclines. The most common tetracycline resistance determinant was tetM (1447 strains, 14.0 %), followed by tet40 (214 strains, 2.1 %) and tet44 (125 strains, 1.2 %). These three genes encode ribosomal protection proteins which prevent the binding of tetracyclines to 16S rRNA. In total, 1645 C . difficile strains (15.9 %) across 68 STs (25.0 %) in five major clades carried at least one tet gene, with 333 strains (3.2 %) carrying more than one gene, 81.4 % of which (271/333) belonged to clade C5. Five ST11 C. difficile strains (C5) carried four different tet genes, the highest number of tet genes per genome in this dataset. Interestingly, tet40 and tet44 were almost exclusively found in clade C5 C. difficile (94.9 and 98.4 % of tet40- and tet44-positive C. difficile belonged to C5, respectively).
Table 5.
Summary of resistance determinants for tetracyclines
|
Gene |
Clade distribution [N (%)] |
Overall |
|||||
|---|---|---|---|---|---|---|---|
|
C1 |
C2 |
C3 |
C4 |
C5 |
Cryptic |
||
|
tetM |
457 (6.8%) |
128 (6.6%) |
0 (0.0%) |
402 (72.2%) |
460 (54.3%) |
0 (0.0%) |
1447 (14.0%) |
|
Tn916 |
146 (2.2%) |
25 (1.3%) |
0 (0.0%) |
95 (17.1%) |
298 (35.2%) |
0 (0.0%) |
564 (5.5%) |
|
Tn5397 |
215 (3.2%) |
1 (0.1%) |
0 (0.0%) |
1 (0.2%) |
8 (0.9%) |
0 (0.0%) |
225 (2.2%) |
|
Tn6190 |
7 (0.1%) |
2 (0.1%) |
0 (0.0%) |
297 (53.3%) |
150 (17.7%) |
0 (0.0%) |
456 (4.4%) |
|
Tn6944 |
52 (0.8%) |
97 (5.0%) |
0 (0.0%) |
6 (1.1%) |
1 (0.1%) |
0 (0.0%) |
156 (1.5%) |
|
Unknown |
37 (0.6%) |
3 (0.2%) |
0 (0.0%) |
3 (0.5%) |
3 (0.4%) |
0 (0.0%) |
46 (0.4%) |
|
tet44 |
2 (<0.1 %) |
0 (0.0%) |
0 (0.0%) |
0 (0.0%) |
123 (14.5%) |
0 (0.0%) |
125 (1.2%) |
|
Tn6164 |
2 (<0.1 %) |
0 (0.0%) |
0 (0.0%) |
0 (0.0%) |
123 (14.5%) |
0 (0.0%) |
125 (1.2%) |
|
Other tet genes |
129 (1.9%) |
12 (0.6%) |
2 (0.8%) |
14 (2.5%) |
336 (39.7%) |
0 (0.0%) |
493 (4.8%) |
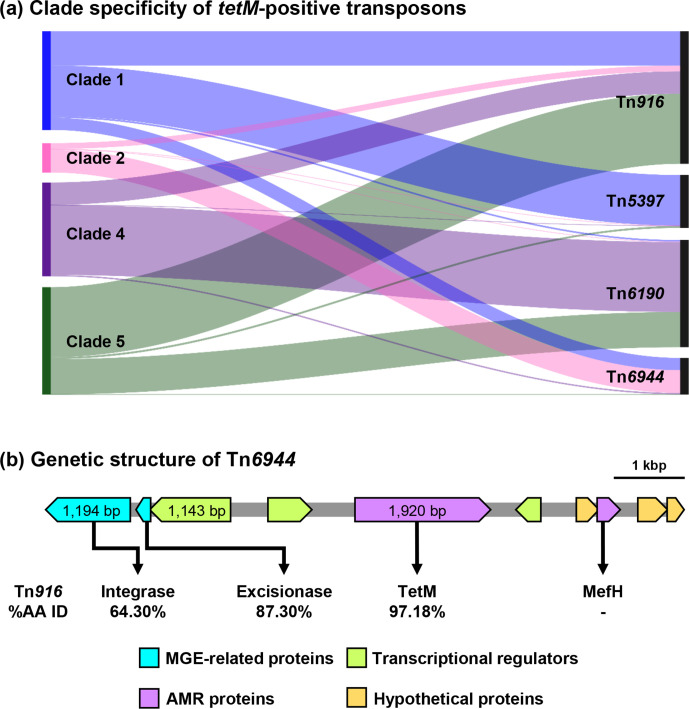
Known tetM-positive transposons and their variants were detected in 1245 (86.0 %) tetM-positive C. difficile (78.0–100.0 % NI). Transposon diversity was highest in clade C1 (Table 5). The most common transposons were Tn916 (564/1447; 39.0 %; 83.3–100.0 % NI) and Tn6190 (456/1447; 31.5 %; 81.5–100.0 % NI). In contrast to the prevalence of ermB-positive transposons above, the distribution of tetM-positive transposons was different in clades C2, C4 and C5 (Fig. 4a). Known tetM-positive transposons could not be identified in 78.1 % of tetM-positive clade C2 C. difficile (100/128). Analysis of the assembled genome of ST1 strain C00008355, a clinical isolate from the UK [SRA accession ERR347593], showed that the tetM gene was located on a 9013 bp element with an overall 37.1 % GC which did not match any transposons in the NCBI database or published literature (Fig. 4b). The annotated sequence of this novel Tn, designated Tn6944 by the Liverpool transposon repository [30], was submitted to GenBank and is available in the DDBJ/ENA/GenBank databases under the accession number BK013348. Besides tetM, Tn6944 also carries mefH which encodes a macrolide efflux protein [22]. Tn6944 was identified in an additional 156 C . difficile strains (78.0–100.0 % NI), 97 of which belonged to clade C2 (Table 5). All tet44-positive C. difficile harboured Tn6164 (80.3–100.0 % NI), a 100 kbp genomic island containing tet44 and ant [6]-Ib, a streptomycin resistance determinant [31].
Fig. 4.
Clade specificity of tetM-positive transposons in C. difficile . (a) Sankey diagram shows the prevalence of four tetM-positive transposons commonly found in C. difficile . The left and right axes represent C. difficile clades and the transposons, respectively. The height of the left axis corresponds to the number of tetM-positive C. difficile strains in each clade, excluding strains with unknown transposons (clade 1, n=419; clade 2, n=208; clade 4, n=711; clade 4, n=688). (b) The genetic structure of the novel tetM-positive Tn, Tn6944 [BK013348]. The amino acid sequences of the key elements in this transposon were compared to the elements found in Tn916 [U09422.1].
Vancomycin resistance
A complete vanB operon (vanRB , vanSB , vanYB , vanW, vanHB , vanB and vanXB genes) was identified in one C. difficile strain, belonging to ST 11 (clade C5). This vanB operon was previously described to be phenotypically silent due to a ~2.1 kbp disruption of the vanRB gene which is a response regulator and part of a key two-component system [32, 33]. This strain was thus considered susceptible to vancomycin.
Metronidazole resistance
SRST2 with the customised pCD-METRO plasmid database correctly identified the plasmid in 14 C . difficile genomes from the Boekhoud et al. study [26] (nine belonged to ST 15 and five belonged to ST 2). Apart from these strains, the pCD-METRO plasmid was found in only one C. difficile strain belonging to ST15 (clade C1, RT 010, non-toxigenic), the same RT reported in the Boekhoud et al. study [26]. In total, only ten of 223 C. difficile ST 15 strains (4.5 %) contained the pCD-METRO plasmid.
Rifamycin resistance
Points mutation in rpoB were found in 688 C. difficile strains (6.7 %), with the highest prevalence in clade C4 (179/557; 32.1 %), followed by C2 (327/1951; 16.8 %). The most common substitution was Arg505Lys found in 68.0 % of resistant strains (468/688), followed by His502Asn (340/688; 49.4 %), with 44.5 % of resistant strains (306/688) having both substitutions. Besides rifamycins, a Gln1073Arg substitution in RpoB was also reported to be associated with reduced susceptibility to fidaxomicin [28]. This substitution was not detected in this dataset.
Carbapenem resistance
A total of 643 C. difficile strains (6.2 %) had substitutions in either Pbp1 or Pbp3 conferring imipenem resistance, with the prevalence slightly higher in clades C2 and C4 (21.6 and 19.4 %, respectively, P=0.2786) than the other clades (collectively 1.4 %, P<0.0001); 504 C. difficile strains had a substitution in Pbp1 (492 having A555T and 12 having L543H), 125 strains had a Y721S substitution in Pbp3 and 12 strains from ST 37 (C4) had substitutions on both Pbp1 (all A555T) and Pbp3.
In addition to the detection of point substitutions, carbapenemase-encoding genes were identified in two C. difficile strains; an unnamed strain [accession ERR2703875; ST 2, C1] carried SHV-1 and CD72 [accession SRR5367248; ST 81, C4] carried PER-1. By NCBI blast approach, the SHV-1 encoding gene was found on an element resembling a Klebsiella pneumoniae plasmid tig00001208_pilon [CP036443.1, 99.7 % sequence identity, 35 % coverage] and the PER-1 encoding gene was found on an element resembling Acinetobacter haemolyticus plasmid pAHTJR1 [CP038010.1, 99.8 % sequence identity, 5 % coverage].
Other resistance types
Genotypic resistance determinants for five other antimicrobials were also identified. First, 124 C. difficile strains (1.2 %) were positive for the cfrB gene which confers linezolid resistance [34]. Resistance determinants for trimethoprim were identified in 147 (1.4 %) C. difficile strains, six of which also harboured sulphonamide resistance determinants. Ninety-eight C. difficile strains (1.0 %) carried chloramphenicol resistance determinants. The most common determinant was catP (92/124; 93.9 %).
In addition to the C. difficile class D β-lactamases which confer intrinsic cephalosporin resistance in C. difficile [18], a few C. difficile strains also had other classes of β-lactamases. Forty-three C. difficile strains carried genes encoding extended-spectrum β-lactamases (ESBL), the most common type belonging to the TEM family (36 strains), and five strains carried AmpC β-lactamase genes.
Finally, 1250 C. difficile strains (12.1 %) carried various aminoglycoside-resistance determinants. The most common determinants were aac6-aph2 (666 strains, 6.5 %), aph-III (279 strains, 2.7 %) and sat4 (271 strains, 2.6 %) genes. Notably, 270 strains carried a locus containing aph-III and sat4 genes adjacent to one another, 68.2 % of which (184/270) belonged to clade C5 (183 ST 11 strains and one ST 163 strain). This locus had 99.91 % nucleic acid identity to a gene cluster found in Erysipelothrix rhusiopathiae , as described in a previous study [35].
Discussion
The success of several epidemic C. difficile strains is thought to be associated with an AMR phenotype which provides a survival advantage for these C. difficile strains in the presence of antimicrobials while imposing little fitness cost [36–38]. Resistance to several antimicrobial classes has been associated with specific C. difficile lineages: fluoroquinolone and rifamycin resistance and C. difficile ST 1 (C2) [10, 12], tetracycline resistance and C. difficile ST 11 (C5) [13], as well as resistance to various antimicrobial classes and MDR and C. difficile ST 37 (C4) [8]. This study provides genotypic evidence to support these associations, demonstrated by the higher resistance prevalence and, especially in the case of tetracycline resistance in C. difficile ST 11, a higher diversity of resistance determinants in the associated clades.
Although the metadata was not complete (only 60.3 % of strains had information on geographical origin and there was inadequate information on host species), some interesting findings can be seen in this genome subset. Fig. 3 demonstrates the difference in AMR prevalence in different continents which may reflect the use of antimicrobials in these regions. The most prominent example is fluoroquinolones which are strictly regulated in Australia and New Zealand but widely used elsewhere [39]. Consequently, there was a stark difference in the prevalence of FQR between Australia and the other three regions. Besides fluoroquinolones, the high prevalence of MLSB and tetracycline resistance, especially in Asia, is suggestive of the overuse of these antimicrobials in the region [40]. We compared the prevalence of AMR genotypes in Australia/New Zealand with a surveillance study from the same region and found that the prevalence in this study correlates with the phenotypic data (P>0.05 for clindamycin [high-level resistance], moxifloxacin and rifaximin resistance) [41]. A similar correlation was seen when comparing the AMR prevalence in Asia and North America with studies from Thailand [22] and the United States [42], respectively. On the contrary, this study underestimated the AMR prevalence in Europe [43, 44]. It should be noted that there was a difference in the number of sequenced strains from various regions. For instance, there were 3548 strains from Europe, many of which were from non-clinical sources, and only 112 strains from Australia/New Zealand in this dataset. As next-generation sequencing (NGS) becomes more accessible [45] and the collection of metadata becomes more systematic, a future study should represent a more complete picture of AMR in the global C. difficile population.
Based on a large sample size, which should give an accurate representation of the C. difficile population, this study provides a global atlas of genotypic AMR determinants in C. difficile . In general, one resistance determinant appeared to dominate in most antimicrobial classes. For example, ermB and tetM genes were found in almost 90 % of C. difficile strains with genotypic resistance to MLSB and tetracycline, respectively. Fluoroquinolone and rifamycin resistance was also mainly determined by a single substitution in GyrA (Thr82Ile) and RpoB (Arg505Lys), respectively. This is similar to other Gram-positive bacteria, such as Staphylococcus aureus [46], where one genotypic determinant is responsible for a resistance phenotype in a majority of the bacterial population and is in contrast to many Gram-negative bacteria, such as several members in the Enterobacteriaceae [47], where resistance to an antimicrobial class can be conferred by several genotypic determinants. The dominance of a single genotypic determinant accommodates the development of genotype-based rapid detection kits for drug-resistant C. difficile , similar to real-time PCR assays for methicillin-resistant S. aureus [48]. Such tools can be beneficial for surveillance for C. difficile outbreaks in the future.
Another benefit of large sample size and NGS is the power to detect rare genotypic determinants. The most notable finding was the detection of carbapenemase-encoding genes in two C. difficile strains, STs 2 and 81, comprising approximately 0.02 % of the population. Previously, carbapenem resistance in C. difficile has been mainly associated with point substitutions on Pbp1 and Pbp3 which cannot be transferred horizontally and only confer imipenem resistance [27]. On the contrary, many carbapenemases provide resistance to a wide range of carbapenem antimicrobials and are capable of horizontal transfer [49]. The detection of carbapenemase-encoding genes is concerning, as C. difficile mainly resides in the colon, the same habitat as many pathogenic Enterobacteriaceae , and transfer of these genes could give rise to carbapenem-resistant Enterobacteriaceae (CRE), another urgent threat in AMR [3]. Conversely, C. difficile can also serve as a reservoir of these resistance genes. Indeed, the gene encoding SHV-1, one of the carbapenemases found in this study, was found on an element similar to a K. pneumoniae plasmid (tig00001208, GenBank accession CP036443.1; 99.7 % NI), suggesting a possible inter-phylum transfer event between these two organisms, although this plasmid was classified as an IncF plasmid according to PlasmidFinder [50]. Generally, the host range for IncF plasmids is limited to only within the Family Enterobacteriaceae [51]. Further study is thus needed to confirm that this horizontal transfer is possible.
Recently, two novel resistance determinants for MLSB antimicrobials were found in Asian C. difficile isolates; erm(52) and mefH [22]. In a larger population of C. difficile , these two genes were found in 1.4–1.5 % of C. difficile strains, approximately six times more prevalent than ermG, a gene previously believed to be the second most prevalent resistance determinant in C. difficile [8]. Failing to detect these two determinants could partially explain the discrepancy between resistance genotype and phenotype in earlier studies [24]. Indeed, the inclusion of erm(52) improved the concordance between clindamycin resistance genotype and high-level clindamycin resistance phenotype to 100 % and mefH provided concordant genotype to C. difficile strains with isolated erythromycin resistance [22]. Further characterisation of mefH revealed that the gene was located adjacent to tetM on a newly defined transposon Tn6944 (Fig. 4b). This transposon has also escaped detection and characterisation despite being present mainly in ST 1 (clade C2), a strain that has been extensively studied [10, 52]. Interestingly, even though tetracycline resistance was a key factor in the evolution of the epidemic C. difficile ST 11 due to its use in agricultural practices [13], this antimicrobial was not included in the antimicrobial susceptibility panel in a pan-European study [43, 44]. Tetracycline resistance was also never mentioned in studies involving C. difficile ST 1, perhaps because the prevalence in this lineage was much lower than that of FQR mutations (7.1 vs 82.3 %, respectively).
A recent study explored the genomic architectures of several accessory AMR genes in 2190 publicly-available C. difficile assemblies and suggested that horizontal gene transfer played a crucial role in the spread of AMR both within C. difficile and among intestinal bacteria in general [53]. This study provides more supporting evidence, as there was a high diversity of ermB-positive transposons throughout the four major clades, suggesting a constant exchange of genes among the population. Evidence of gene transfer could also be seen among tetM-positive transposons. For instance, Tn6190 was shared between C4 and C5, despite their divergence over a million years ago [14].
We also identified key antimicrobials, resistance to which can potentially lead to outbreaks of CDI; fluoroquinolones, MLSB, rifamycins and tetracyclines, as well as the specific C. difficile clades associated with such resistance. This provides an opportunity to develop a focused antimicrobial stewardship policy, targeting specific antimicrobial classes based on the prevalent C. difficile strains in the region. A real-world example can be seen in the US, where the reduction of fluoroquinolone use led to a significant reduction in the number of CDI cases and the associated cost [3].
As an obligate anaerobe, C. difficile is intrinsically resistant to aminoglycosides. Additional resistance determinants to these antimicrobials are not beneficial to the bacterium and are unlikely to be conserved in the genome. Thus, the presence of aminoglycoside resistance determinants should reflect recent, and likely continuous, inter-species gene transfer with taxa in diverse environments such as the animal gut and soils. The most common aminoglycoside resistance determinant was aac6-aph2, a bifunctional gene found in Staphylococcus spp. and Enterococcus spp. [54], commensal species commonly found in the human and animal gut. Interestingly, many ST 11 (C5) strains also carried an aph-III and sat4 cluster, a gene cluster found in E. rhusiopathiae which inhabits the porcine gut [55], supporting the animal origin and One Health importance of this lineage [35]. Indeed, aminoglycosides have been heavily used in both agricultural and veterinary practices [56]. The presence of aminoglycoside resistance determinants in C. difficile highlights another aspect of AMR in C. difficile ; the role of C. difficile as a reservoir of AMR genes. Aminoglycosides remain a key treatment option for serious staphylococcal and enterococcal infections, such as infective endocarditis, in conjunction with β-lactams antimicrobials [57]. Resistance to aminoglycosides in these pathogens complicates treatment of these infections which may result in adverse clinical outcomes. Thus, colonisation with C. difficile carrying these resistance determinants may pose an additional risk of treatment failure in these patients.
This study utilised the direct analysis of raw sequence reads without the need for genome assembly which enabled the characterisation of a large dataset within a relatively short time (approximately 5 min of CPU time [16 cores] per strain as opposed to more than 30 min of CPU time per strain for a de novo assembly pipeline). SRST2 provides rapid MLST and AMR genotyping [16]. SRST2-based AMR genotyping can be performed using three types of databases: well-characterised databases of accessory AMR genes [20, 21, 23], species-specific gene allele databases (e.g. the PubMLST database), as well as customised databases. The latter was used in a previous study on a smaller dataset, the results of which were similar to a standard approach using blast on annotated draft genomes [58].
Besides the lack of complete metadata, another limitation of this study was the lack of comparative phenotypic data, as the study was performed on a publicly-available genome dataset. However, many key AMR genotypes were reported to have a high correlation with phenotypic characteristics [24, 58]. Thus, the prevalence values reported in this study should reflect the resistance prevalence in C. difficile population. Also, this study only reports the presence or absence of genotypic AMR determinants and does not take into account the different alleles of the genes, as the alleles were not included in the databases used in the analyses [20, 21]. Further analyses on the allelic distribution across C. difficile population may provide additional information on the spread of AMR genes.
In conclusion, almost half of C. difficile strains studied carried at least one genotypic resistant determinant. The resistance prevalence was higher among clades C2, C4 and C5 which have been associated with epidemic C. difficile STs 1, 37 and 11, respectively. Though resistance to antimicrobials for treatment of CDI is rare, this study provides evidence to support the role of AMR in the spread of C. difficile , as well as the role of C. difficile as a reservoir of accessory AMR genes, most notably aminoglycoside resistance determinants and carbapenemase-encoding genes.
Supplementary Data
Funding information
This work was supported, in part, by funding from The Raine Medical Research Foundation (RPG002-19) and a Fellowship from the National Health and Medical Research Council (APP1138257) awarded to D.R.K. K.I. is a recipient of the Mahidol Scholarship from Mahidol University, Thailand. This research used the facilities and services of the Pawsey Supercomputing Centre [Perth, Western Australia].
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AMR, antimicrobial resistance; CARD, comprehensive antibiotic resistance database; CDC, centers for disease control and prevention; CDI, clostridioides difficile infection; CDS, coding sequence; FQR, fluoroquinolone resistance; MDR, multidrug resistance; MLST, multilocus sequence typing; NI, nucleotide identity; RT, PCR ribotyping; ST, sequence type.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Supplementary material is available with the online version of this article.
References
- 1.Dadgostar P. Antimicrobial resistance: implications and costs. Infect Drug Resist. 2019;12:3903–3910. doi: 10.2147/IDR.S234610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention (CDC) Antibiotic Resistance Threats in the United States, 2013. Atlanta, GA: US: Department of Health and Human Services, CDC; 2013. [Google Scholar]
- 3.Centers for Disease Control and Prevention (CDC) Antibiotic Resistance Threats in the United States. Atlanta, GA: US: Department of Health and Human Services, CDC; 2019. [Google Scholar]
- 4.Leffler DA, Lamont JT. Clostridium difficile infection. N Engl J Med. 2015;372:1539–1548. doi: 10.1056/NEJMra1403772. [DOI] [PubMed] [Google Scholar]
- 5.Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109:309–318. doi: 10.1179/2047773215Y.0000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Beurden YH, Nieuwdorp M, van de Berg P, Mulder CJJ, Goorhuis A. Current challenges in the treatment of severe Clostridium difficile infection: early treatment potential of fecal microbiota transplantation. Therap Adv Gastroenterol. 2017;10:373–381. doi: 10.1177/1756283X17690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banawas SS. Clostridium difficile infections: a global overview of drug sensitivity and resistance mechanisms. Biomed Res Int. 2018;2018:8414257. doi: 10.1155/2018/8414257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imwattana K, Knight DR, Kullin B, Collins DA, Putsathit P, et al. Antimicrobial resistance in Clostridium difficile ribotype 017. Expert Rev Anti Infect Ther. 2020;18:17–25. doi: 10.1080/14787210.2020.1701436. [DOI] [PubMed] [Google Scholar]
- 9.Kuijper EJ, de Weerdt J, Kato H, Kato N, van Dam AP, et al. Nosocomial outbreak of Clostridium difficile-associated diarrhoea due to a clindamycin-resistant enterotoxin A-negative strain. Eur J Clin Microbiol Infect Dis. 2001;20:528–534. doi: 10.1007/s100960100550. [DOI] [PubMed] [Google Scholar]
- 10.He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile . Nat Genet. 2013;45:109–113. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drudy D, Harnedy N, Fanning S, Hannan M, Kyne L. Emergence and control of fluoroquinolone-resistant, toxin A-negative, toxin B-positive Clostridium difficile . Infect Control Hosp Epidemiol. 2007;28:932–940. doi: 10.1086/519181. [DOI] [PubMed] [Google Scholar]
- 12.Curry SR, Marsh JW, Shutt KA, Muto CA, O’Leary MM, et al. High frequency of rifampin resistance identified in an epidemic Clostridium difficile clone from a large teaching hospital. Clin Infect Dis. 2009;48:425–429. doi: 10.1086/596315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dingle KE, Didelot X, Quan TP, Eyre DW, Stoesser N, et al. A role for tetracycline selection in recent evolution of agriculture-associated Clostridium difficile PCR ribotype 078. MBio. 2019;10:e02790-18. doi: 10.1128/mBio.02790-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knight DR, Imwattana K, Kullin B, Guerrero-Araya E, Paredes-Sabja D, et al. Major genetic discontinuity and novel toxigenic species in Clostridioides difficile taxonomy. eLife. 2021;10:e64325. doi: 10.7554/eLife.64325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imwattana K, Knight DR, Kullin B, Collins DA, Putsathit P, et al. Clostridium difficile ribotype 017 - characterization, evolution and epidemiology of the dominant strain in Asia. Emerg Microbes Infect. 2019;8:796–807. doi: 10.1080/22221751.2019.1621670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Didelot X, Eyre DW, Cule M, Ip CLC, Ansari MA, et al. Microevolutionary analysis of Clostridium difficile genomes to investigate transmission. Genome Biol. 2012;13:R118. doi: 10.1186/gb-2012-13-12-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toth M, Stewart NK, Smith C, Vakulenko SB. Intrinsic class D beta-lactamases of Clostridium difficile . mBio. 2018;9:e01803-18. doi: 10.1128/mBio.01803-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khanafer N, Daneman N, Greene T, Simor A, Vanhems P, et al. Susceptibilities of clinical Clostridium difficile isolates to antimicrobials: a systematic review and meta-analysis of studies since 1970. Clin Microbiol Infect. 2018;24:110–117. doi: 10.1016/j.cmi.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 20.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imwattana K, Putsathit P, Knight DR, Kiratisin P, Riley TV. Molecular characterization of, and antimicrobial resistance in, Clostridioides difficile from Thailand, 2017-2018. [Epub ahead of print] Microb Drug Resist. 2021 doi: 10.1089/mdr.2020.0603. [DOI] [PubMed] [Google Scholar]
- 23.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48:D25. :D517. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spigaglia P. Recent advances in the understanding of antibiotic resistance in Clostridium difficile infection. Ther Adv Infect Dis. 2016;3:23–42. doi: 10.1177/2049936115622891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, et al. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci U S A. 2010;107:7527–7532. doi: 10.1073/pnas.0914322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boekhoud IM, Hornung BVH, Sevilla E, Harmanus C, Bos-Sanders I, et al. Plasmid-mediated metronidazole resistance in Clostridioides difficile . Nat Commun. 2020;11:598. doi: 10.1038/s41467-020-14382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isidro J, Santos A, Nunes A, Borges V, Silva C, et al. Imipenem resistance in Clostridium difficile ribotype 017, Portugal. Emerg Infect Dis. 2018;24:741–745. doi: 10.3201/eid2404.170095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leeds JA, Sachdeva M, Mullin S, Barnes SW, Ruzin A. In vitro selection, via serial passage, of Clostridium difficile mutants with reduced susceptibility to fidaxomicin or vancomycin. J Antimicrob Chemother. 2014;69:41–44. doi: 10.1093/jac/dkt302. [DOI] [PubMed] [Google Scholar]
- 29.Solomon K, Fanning S, McDermott S, Murray S, Scott L, et al. PCR ribotype prevalence and molecular basis of macrolide-lincosamide-streptogramin B (MLSB) and fluoroquinolone resistance in Irish clinical Clostridium difficile isolates. J Antimicrob Chemother. 2011;66:1976–1982. doi: 10.1093/jac/dkr275. [DOI] [PubMed] [Google Scholar]
- 30.Tansirichaiya S, Rahman MA, Roberts AP. The Transposon Registry. Mobile DNA-UK. 2019;10 doi: 10.1186/s13100-019-0182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corver J, Bakker D, Brouwer MSM, Harmanus C, Hensgens MP, et al. Analysis of a Clostridium difficile PCR ribotype 078 100 kilobase island reveals the presence of a novel transposon, Tn6164. Bmc Microbiol. 2012;12:130. doi: 10.1186/1471-2180-12-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knight DR, Androga GO, Ballard SA, Howden BP, Riley TV. A phenotypically silent vanB2 operon carried on a Tn1549-like element in Clostridium difficile . mSphere. 2016;1:e00177-16. doi: 10.1128/mSphere.00177-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Courvalin P. Vancomycin resistance in gram-positive cocci. Clin Infect Dis. 2006;42 Suppl 1:34. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- 34.Marin M, Martin A, Alcala L, Cercenado E, Iglesias C, et al. Clostridium difficile isolates with high linezolid MICs harbor the multiresistance gene cfr. Antimicrob Agents Chemother. 2015;59:586–589. doi: 10.1128/AAC.04082-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knight DR, Kullin B, Androga GO, Barbut F, Eckert C, et al. Evolutionary and genomic insights into Clostridioides difficile sequence type 11: a diverse zoonotic and antimicrobial-resistant lineage of global one health importance. mBio. 2019;10:e00446-19. doi: 10.1128/mBio.00446-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dang UT, Zamora I, Hevener KE, Adhikari S, XQ W, et al. Rifamycin resistance in Clostridium difficile is generally associated with a low fitness burden. Antimicrob Agents Chemother. 2016;60:5604–5607. doi: 10.1128/AAC.01137-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wasels F, Kuehne SA, Cartman ST, Spigaglia P, Barbanti F, et al. Fluoroquinolone resistance does not impose a cost on the fitness of Clostridium difficile in vitro . Antimicrob Agents Chemother. 2015;59:1794–1796. doi: 10.1128/AAC.04503-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wasels F, Spigaglia P, Barbanti F, Mastrantonio P. Clostridium difficile erm(B)-containing elements and the burden on the in vitro fitness. J Med Microbiol. 2013;62:1461–1467. doi: 10.1099/jmm.0.057117-0. [DOI] [PubMed] [Google Scholar]
- 39.Collins DA, Putsathit P, Elliott B, Riley TV. Laboratory-based surveillance of Clostridium difficile strains circulating in the Australian healthcare setting in 2012. Pathology. 2017;49:309–313. doi: 10.1016/j.pathol.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 40.Li GH, Hou DJ, Fu GH, Guo JY, Guo XB, et al. A review of prophylactic antibiotics use in plastic surgery in China and a systematic review. Int J Surg. 2014;12:1300–1305. doi: 10.1016/j.ijsu.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 41.Putsathit P, Hong S, George N, Hemphill C, Huntington PG, et al. Antimicrobial resistance surveillance of Clostridioides difficile in Australia, 2015-18. J Antimicrob Chemother. 2021;76:1815–1821. doi: 10.1093/jac/dkab099. [DOI] [PubMed] [Google Scholar]
- 42.Tickler IA, Obradovich AE, Goering RV, Fang FC, Tenover FC, et al. Changes in molecular epidemiology and antimicrobial resistance profiles of Clostridioides (Clostridium) difficile strains in the United States between 2011 and 2017. Anaerobe. 2019;60:S1075-9964(19)30100-3. doi: 10.1016/j.anaerobe.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 43.Freeman J, Vernon J, Morris K, Nicholson S, Todhunter S, et al. Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin Microbiol Infect. 2015;21:248–e16. doi: 10.1016/j.cmi.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 44.Freeman J, Vernon J, Pilling S, Morris K, Nicholson S, et al. The ClosER study: results from a three-year pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes, 2011-2014. Clin Microbiol Infect. 2018;24:724–731.:S1198-743X(17)30570-0. doi: 10.1016/j.cmi.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 45.Imwattana K, Knight DR, Riley TV. Can sequencing improve the diagnosis and management of Clostridioides difficile infection. Expert Rev Mol Diagn. 2021;21:429–431. doi: 10.1080/14737159.2021.1915774. [DOI] [PubMed] [Google Scholar]
- 46.Foster TJ. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. Fems Microbiol Rev. 2017;41:430–449. doi: 10.1093/femsre/fux007. [DOI] [PubMed] [Google Scholar]
- 47.Wilson H, Torok ME. Extended-spectrum beta-lactamase-producing and carbapenemase-producing Enterobacteriaceae. Microb Genom. 2018;4:000197. doi: 10.1099/mgen.0.000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galia L, Ligozzi M, Bertoncelli A, Mazzariol A. Real-time PCR assay for detection of Staphylococcus aureus, Panton-Valentine leucocidin and methicillin resistance directly from clinical samples. AIMS Microbiol. 2019;5:138–146. doi: 10.3934/microbiol.2019.2.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Queenan AM, Bush K. Carbapenemases: The versatile beta-lactamases. Clin Microbiol Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, et al. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae . J Antimicrob Chemother. 2018;73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 52.Valiente E, Cairns MD, Wren BW. The Clostridium difficile PCR ribotype 027 lineage: a pathogen on the move. Clin Microbiol Infect. 2014;20:396–404. doi: 10.1111/1469-0691.12619. [DOI] [PubMed] [Google Scholar]
- 53.Kartalidis P, Skoulakis A, Tsilipounidaki K, Florou Z, Petinaki E, et al. Clostridioides difficile as a dynamic vehicle for the dissemination of antimicrobial-resistance determinants: review and in silico analysis. Microorganisms. 2021;9:1383. doi: 10.3390/microorganisms9071383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Daigle DM, Hughes DW, Wright GD. Prodigious substrate specificity of AAC(6’)-APH(2"), an aminoglycoside antibiotic resistance determinant in enterococci and staphylococci. Chem Biol. 1999;6:99–110. doi: 10.1016/S1074-5521(99)80006-4. [DOI] [PubMed] [Google Scholar]
- 55.Zhang B, Ku X, Yu X, Sun Q, Wu H, et al. Prevalence and antimicrobial susceptibilities of bacterial pathogens in Chinese pig farms from 2013 to 2017. Sci Rep. 2019;9:9908. doi: 10.1038/s41598-019-45482-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Economou V, Gousia P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect Drug Resist. 2015;8:49–61. doi: 10.2147/IDR.S55778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baddour LM, Wilson WR, Bayer AS, Fowler VG, Tleyjeh IM, et al. Infective endocarditis in adults: Diagnosis, antimicrobial therapy, and management of complications: A scientific statement for healthcare professionals from the American Heart Association. Circulation. 2015;132:1435–1486. doi: 10.1161/CIR.0000000000000296. [DOI] [PubMed] [Google Scholar]
- 58.Imwattana K, Kiratisin P, Riley TV, Knight DR. Genomic basis of antimicrobial resistance in non-toxigenic Clostridium difficile in Southeast Asia. Anaerobe. 2020;66:S1075-9964(20)30146-3. doi: 10.1016/j.anaerobe.2020.102290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.