Abstract
Burkholderia pseudomallei is a Gram-negative soil-dwelling bacillus that causes melioidosis, a frequently fatal infectious disease, in tropical and subtropical regions. Previous studies have identified the overall genetic and evolutionary characteristics of B. pseudomallei on a global scale, including its origin and transmission routes. However, beyond its known hyperendemicity foci in northern Australia and Southeast Asia, the distribution and genetic characteristics of B. pseudomallei in most tropical regions remain poorly understood, including in southern China. Here, we sequenced the genomes of 122 B. pseudomallei strains collected from Hainan, an island in southern China, in 2002–2018, to investigate the population structure, relationships with global strains, local epidemiology, and virulence and antimicrobial-resistance factors. A phylogenetic analysis and hierarchical clustering divided the Hainan strains into nine phylogenic groups (PGs), 80 % of which were concentrated within five major groups (group 1: corresponding to minor sequence types [STs], 12.3 %; group 3: ST46 and ST50, 31.1 %; group 9: ST58, 13.1 %; group 11: ST55, 8.2 %; group 15: mainly ST658, 15.6%). A phylogenetic analysis that included global strains suggested that B. pseudomallei in Hainan originated from Southeast Asian countries, transmitted in multiple historical importation events. We also identified several mutual transmission events between Hainan and Southeast Asian countries in recent years, including three importation events from Thailand and Singapore to Hainan and three exportation events from Hainan to Singapore, Malaysia, and Taiwan island. A statistical analysis of the temporal distribution showed that the Hainan strains of groups 3, 9, and 15 have dominated the disease epidemic locally in the last 5 years. The spatial distribution of the Hainan strains demonstrated that some PGs are distributed in different cities on Hainan island, and by combining phylogenic and geographic distribution information, we detected 21 between-city transmission events, indicating its frequent local transmission. The detection of virulence factor genes showed that 56 % of the Hainan strains in group 1 encode a B. pseudomallei -specific adherence factor, boaB, confirming the specific pathogenic characteristics of the Hainan strains in group 1. An analysis of the antimicrobial-resistance potential of B. pseudomallei showed that various kinds of alterations were identified in clinically relevant antibiotic resistance factors, such as AmrR, PenA and PBP3, etc. Our results clarify the population structure, local epidemiology, and pathogenic characteristics of B. pseudomallei in Hainan, providing further insight into its regional and global transmission networks and improving our knowledge of its global phylogeography.
Keywords: Burkholderia pseudomallei, Hainan, whole genome sequencing, bacteria genetic diversity, transmission, genomic epidemiology
Data Summary
All sequence data for the 122 newly sequenced Hainan B. pseudomallei isolates have been uploaded to the National Center for Biotechnology Information (NCBI) database under BioProject number PRJNA669904. The isolate metadata and accession numbers are listed in supplementary data table S1 (available in the online version of this article).
Impact Statement.
Burkholderia pseudomallei is a deadly pathogen endemic to tropical and subtropical areas, especially northern Australia and Southeast Asia. However, our understanding of the large-scale genomics of B. pseudomallei strains in most other tropical regions outside these two hyperendemic foci is limited, including on Hainan island, China. In this study, we sequenced 122 B. pseudomallei isolates from Hainan and analysed them comparatively with 1532 B. pseudomallei genomes from the NCBI database. Our results reveal the population structure of the Hainan isolates, which constitute five major phylogenic groups, together with the minor populations of sporadic strains. We have extended our knowledge of the global transmission network and local epidemiology by identifying its mutual spread between Hainan island and Southeast Asian countries, and its frequent local transmission. We have also identified the virulence and antimicrobial-resistance genes of the Hainan B. pseudomallei isolates, which should clarify the pathogenic characteristics of this pathogen, which is highly significant for its clinical management and treatment.
Introduction
Burkholderia pseudomallei , the environmental Gram-negative bacillus that causes melioidosis, is commonly found in soil and water in tropical and subtropical regions, especially in northern Australia and Southeast Asia. The bacterium has been classified as a biothreat agent by the US Centers for Disease Control and Prevention because it has potential utility as a bioweapon [1]. B. pseudomallei causes an estimated 165 000 human cases of melioidosis per year worldwide, with a case fatality rate (CFR) of up to 40%, and ineffective antimicrobial treatment may cause the CFR to exceed 70 % [2–4]. Therefore, it is very important to clarify the evolutionary and pathogenic characteristics of B. pseudomallei to improve the surveillance and control of this pathogen.
Traditional molecular typing methods, such as pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), and multilocus variable-number tandem repeat analysis (MLVA), have greatly improved our understanding of the epidemiological characteristics of B. pseudomallei . PFGE was previously considered the gold standard method for detecting subtle differences in genetically related strains and characterizing the strains involved in outbreaks [5–8]. However, it has strict requirements for the experimental conditions used, and a single difference in these could result in different typing results, making communication among laboratories difficult. MLVA and MLST have been widely used to examine the epidemiological relatedness, population structures, and evolutionary characteristics of B. pseudomallei [9–12] because they can detect genetic variations in more detail than PFGE and the sequence data can be readily compared across laboratories worldwide [13]. However, scientists have noted that these typing methods are unable to determine fine-scale population structures because the genotyping resolution is low and the rates of recombination among local B. pseudomallei populations are high [14, 15]. Furthermore, a previous study showed that sequence type (ST) homoplasy can cause MLST to produce confounding results, albeit infrequently [16].
Whole-genome sequencing (WGS) provides the highest possible discriminatory power among bacterial isolates, and has been used in various aspects of B. pseudomallei research, including source tracking, the determination of population structures, and the inference of global transmission routes [4, 17]. Genome-wide single-nucleotide polymorphism (SNP) analyses pointed to Australia as the early reservoir of B. pseudomallei and subdivided the species into 19 phylogenic groups (PGs) with tree-independent hierarchical Bayesian clustering, based on the core genome mapping alignment of 469 global strains [14, 17]. Except for the Australasian group (group 1) which is located at the root of the phylogenetic tree, and an African-American group (group 19), which occurs on a separate branch within the Asian groups, the remaining 17 PGs are all Asian groups, mainly composed of strains from the Malay Peninsula and countries bordered by the Mekong River. A phylogeographic analysis based on WGS detected global and regional transmission routes, which extended from Australia to Southeast Asia, followed by its onward transmission to South Asia and East Asia [17]. African B. pseudomallei strains also originated from Asia and were then transmitted to the Americas between 1650 and 1850, driven by anthropogenic factors, such as human migration, trade, and slavery routes, and also by the introduction of pigs and via the flight paths of migratory birds [12, 17–21]. However, increasing numbers of melioidosis cases have been reported in other tropical regions, outside the hyperendemicity foci in northern Australia and Southeast Asia, but the corresponding distributions of B. pseudomallei remain unclear. Because the mortality rate of melioidosis is high, it is essential to investigate the distribution of this pathogen in other tropical regions, in order to extend disease surveillance and prevention.
B. pseudomallei outbreaks have been recognized in southern China (Hainan, Guangdong, Guangxi, Fujian, and Taiwan) for many years since the 1970s, especially in China’s southern island province, Hainan, which covers an area of 35 400 km2 and has a population of over 9.34 million native inhabitants. With the increasing number of tourists each year and the improvement in clinical diagnostic technology over the past two decades, there has been a substantial increase in the number of cases of infection diagnosed in Hainan [22] since the first case of human melioidosis was reported in 1989 [23]. Previous molecular typing studies with MLST divided the Hainan strains into a number of STs, some of which predominated (STs 55, 70, 46, 50, and 58) and some of which were novel (STs 1341, 1345, 1346, etc.) [11, 24, 25], indicating the high genetic diversity of the strains in Hainan. A global-scale phylogenetic analysis based on MLST suggested that China’s isolates were closely related to isolates from Southeast Asia, particularly from Thailand and Malaysia [11]. Although the relationships and genetic characteristics of the Australasian and Southeast Asian strains have been reasonably well characterized, and molecular typing methods have revealed some molecular characteristics of B. pseudomallei in Hainan, the origin, spread, and genetic characteristics of the Hainan strains, which are essential components of global genomic epidemiological studies of this pathogen, are still poorly understood. In this study, we used large-scale genomic epidemiological techniques to investigate the population structure, relationships with global strains, local epidemiology, virulence factors, and antimicrobial-resistance genes of B. pseudomallei in Hainan, to further characterize the global phylogeography and epidemiology of B. pseudomallei .
Methods
Burkholderia pseudomallei isolates and genomic sequencing
A total of 122 Hainan B. pseudomallei strains were collected and characterized in this study, 119 of which were obtained from patients and the remaining three from well water samples. The clinical cases were treated in three hospitals in 2002–2018, and included patients from other parts of China or other countries who had travelled to Hainan. DNA libraries of the 122 Hainan isolates were prepared with the Illumina protocol and sequenced on the Illumina NovaSeq 6000 system, with paired-end runs and a read length of 150 bp. On average 1.7 Gb of clean data was generated for each strain.
A total of 1532 publicly available B. pseudomallei genome sequences were downloaded from the National Center for Biotechnology Information (NCBI) database (up to March 2019), which had been collected for various purposes and were isolated from various sources in 1942–2018. The sources of the published genomes included 22 countries or regions, especially the known hyperendemicity foci in northern Australia and Southeast Asia. The details of the genomes of all the strains analysed in this study are listed in supplementary data table S1.
Genome assembly, variant calling, and MLST
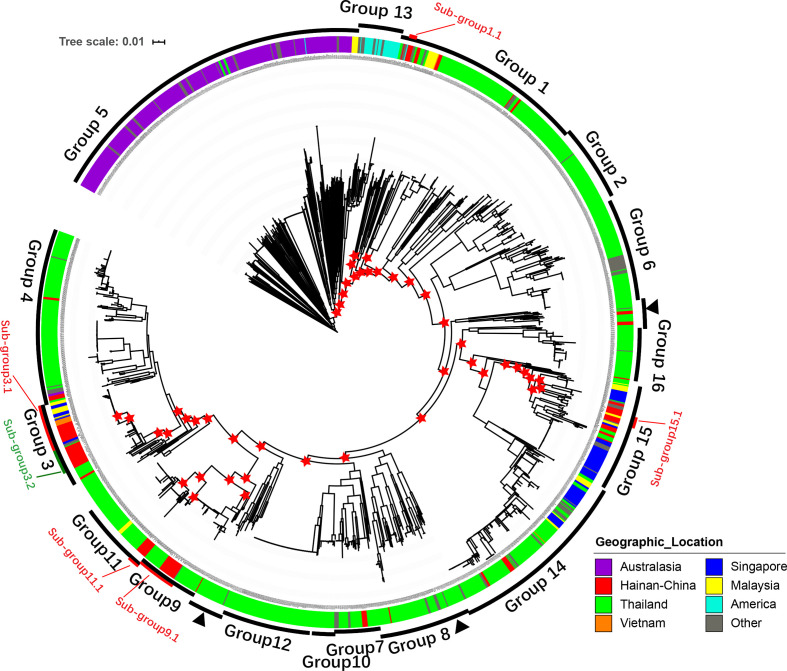
We assembled the genomes de novo with SOAPdenovo v2.04 [26], as previously described [27], and then identified the SNPs with a previously described method [26, 28]. First, the assemblies were aligned against the reference genome, using MUMmer v3.1 [29], to generate a whole-genome alignment and identify the SNPs in the core genome (regions present in 99 % of isolates). Second, the clean sequencing reads of the Hainan isolates were mapped to the assemblies to evaluate the SNP accuracy with SOAPaligner [30]. Only high-quality SNPs (supported by >10 reads, quality value >30) were retained in the analysis. The repetitive regions in the reference genome were identified with TRF v4 [31] and a blastn search against itself. The SNPs in the repetitive regions were excluded and only biallelic SNPs were used in the subsequent analysis. B. pseudomallei K96243 (NC_006350.1, NC_006351.1) was used as the reference sequence in the analysis of 1654 genomes. A total of 325 036 SNPs (5.8 Mb core genome) were identified in the 1654 isolates, which were then used to construct a maximum likelihood (ML) tree with FastTree V2 [32], using the default settings. The phylogenetic tree was visualized with iToL [33] (Fig. 1). We also inferred the multilocus STs of the 122 newly sequenced Hainan strains using the software mlst (Seemann T., https://github.com/tseemann/mlst), which incorporates components of the PubMLST database (https://pubmlst.org/).
Fig. 1.
Phylogeny and population structure of B. pseudomallei . A core SNP-based maximum likelihood phylogeny of 1654 genomes, with geographic origins highlighted. Differences in the levels of bacterial diversity across different geographic origins: Australasia (purple), Hainan, China (red), Thailand (green), Vietnam (orange), Singapore (blue), Malaysia (yellow), America (cyan), and other (grey). Outer ring represents population clusters based on Bayesian Analysis of Population Structure (BAPS) hierarchical clustering (groups 1–16) and six subgroups of the five major groups based on a threshold of 5000 pairwise SNP distances (PSDs; see Methods). Except for group 8, which is paraphyletic and marked by three black arrowheads, each group forms a monophyletic branch. One hundred iterations of bootstrap were performed and red asterisks indicate bootstrap values of major nodes that are greater than 80 in the five major groups.
Hierarchical Bayesian clustering
Tree-independent hierarchical Bayesian clustering with hierarchical Bayesian Analysis of Population Structure (hierBAPS) [34, 35] was used to determine the population structure generated from the core genome mapping alignment. hierBAPS was run on the 325 036 SNPs of the 1654 isolates with a maximum number of populations (K) of 100, 200, or 300. The 1654 isolates were clustered into 14 (K=100), 13 (K=200), or 12 (K=300) lineages. To separate the strains into lineages that shared much closer genetic backgrounds, we first took K=100 and clustered the strains into 14 groups (Fig. S1). The results clustered the Australian strains into three groups, including two clonal groups of strains with very small numbers of SNPs, which seemed to have originated from two outbreaks. The Hainan strains in group 7, which was paraphyletic, clustered into multiple clades. To simplify the analytical process without affecting the accuracy of the results, we focused on the Hainan strains in this study, and tried to divide them into monophyletic groups that shared much closer genetic backgrounds. Therefore, we combined all the Australian strains into one group (group 5) and continued the hierarchical clustering of group 7 into secondary clusters. This resulted in 16 groups (supplementary data table S1), which were used for the subsequent lineage-specific analyses.
Analysis of individual lineages
To explore the phylogenetic relationships between the Hainan and global B. pseudomallei strains with greater resolution, we calculated the pairwise SNP distances (PSDs) among all the strains in the five major PGs (which contained 80 % of all the Hainan strains, as described above), and subdivided them into several subgroups (group 1, 56 subgroups; group 3, six subgroups; group 9, four subgroups; group 11, nine subgroups; group 15, 19 subgroups) based on a threshold of 5000 PSDs, which ensured that the population of strains was subdivided into groups with closely related genetic backgrounds and allowed the recombination detection tool Gubbins [36] to operate within its best performance range [17, 37]. Here, we explored the phylogenetic relationships within the six subgroups that contained at least five Hainan strains (subgroup 1.1, subgroup 3.1, subgroup 3.2, subgroup 9.1, subgroup 11.1, and subgroup 15.1; Fig. 1).
The core regions of the different genome sets varied, which affected both SNP calling and phylogenetic reconstruction. To achieve high phylogenetic resolution, we reselected reference genomes for the subgroups to increase the sensitivity of variant calling. Four strains within the five major PGs with complete chromosomal contigs were chosen as the reference genomes for subgroup 1.1 (GCA_003606245.1), subgroup 3.1 (GCA_003546995.1), subgroup 3.2 (GCA_003546995.1), subgroup 9.1 (GCA_000954175.1), and subgroup 11.1 (GCA_001277975.1). However, no relevant reference genome was available as a complete genome for group 15, so strain TOMS (GCA_002900605.1) in group 15 was selected, assembled, and ordered relative to its closest reference (GCA_003547015.1) using ABACAS v1.3.1 [38] and ACT [39], and then manually curated. We then re-identified the core genome and SNPs of each subgroup with the method described above. Recombination fragments were called and removed from the core genome alignments using Gubbins [36]. Subgroup-specific phylogenies were constructed with the remaining variants.
Phylogeographic analysis
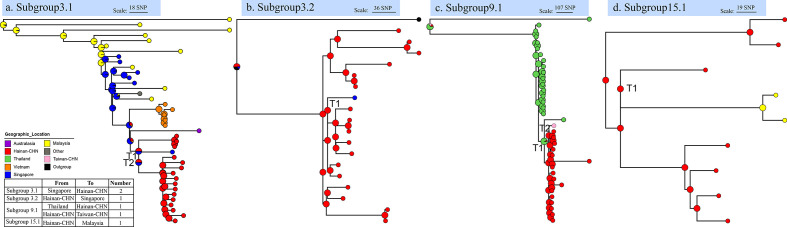
To examine the relationships between the Hainan and global B. pseudomallei isolates, we constructed ML trees separately for subgroup 3.1, subgroup 3.2, subgroup 9.1, and subgroup 15.1, which each contained global isolates and more than five Hainan isolates, based on the new SNP sets without recombination, using RAxML-NG v.0.9 [40]. As in previous studies [41, 42], the transmission of B. pseudomallei between geographic regions was assessed separately for each subgroup tree using an implementation of stochastic mapping on phylogenies (SIMMAP) with the phytools v0.5 package in R [43, 44]. The region of origin was treated as a discrete trait and mapped to each tree with the ARD model (which allows each region-to-region transfer rate to vary independently), with 100 replicates. The results reported (Fig. 2) are the median values for the number of mutual transitions among Hainan and other countries or regions, summarized from 100 replicate mappings for each tree. To explore the local transmission of B. pseudomallei within Hainan island, we constructed ML trees of only Hainan strains separately for subgroup 1.1, subgroup 3.1, subgroup 3.2, subgroup 9.1, subgroup 11.1, and subgroup 15.1, which contained more than five Hainan isolates and their exact sampling locations, based on the re-called SNP sets without recombination. We then estimated the average numbers of between-city transmission events (Fig. 3) with phytools v0.5, as described above. Evolutionary time was calculated based on the average pairwise SNP distance (aPSD) of isolates in corresponding transmissions, the mutation rate of 6.8 (95 % HPD, 4.6 to 8.9) SNPs per year generated from 1.17×10−6 (95 % HPD, 7.91×10−7 to 1.53×10−6) substitutions per site per year reported by Chewapreecha et al. in 2017 [17], and 5.4 Mb core genome in this study.
Fig. 2.
Relationships between global and Hainan B. pseudomallei isolates. Phylogenetic trees of four subgroups that contain global isolates and more than five Hainan isolates, and estimates of the average numbers of between-country transmission events, determined with phytools (see Methods). Pie charts in the ancestral nodes of the phylogenetic trees represent the likelihood proportion of geographical origins. Nodes with ‘T’ represent transmission events, and the table summarizes the transmission events, with directions and numbers.
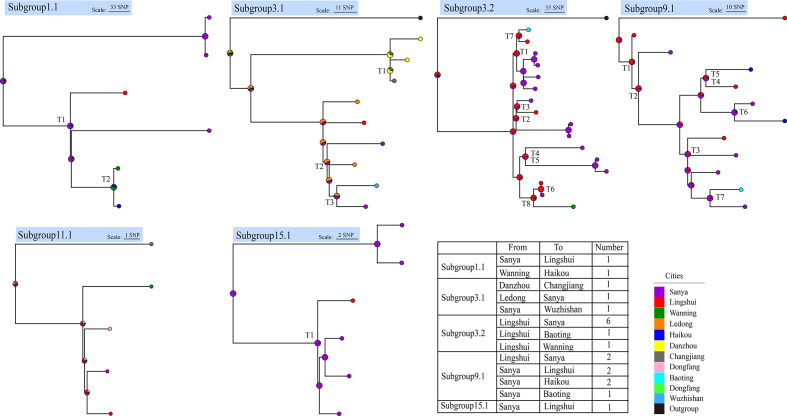
Fig. 3.
Local transmission events within Hainan island. Phylogenetic trees of Hainan strains with exact sampling locations in the six subgroups of the five major groups, and estimates of the average numbers of between-city transmission events, determined with phytools (see Methods). Pie charts in the ancestral nodes of the phylogenetic trees represent the likelihood proportion of geographical origins. Nodes with ‘T’ represent transmission events, and the table summarizes the transmission events, with directions and numbers.
Identification of virulence factors and antimicrobial-resistance genes
We compared the assembled genomes of all 1654 strains with the core dataset of the Virulence Factors Database (VFDB) [45] and the Comprehensive Antibiotic Resistance Database (CARD, https://card.mcmaster.ca/) to detect possible virulence factors and antimicrobial-resistance genes, using blastn and Resistance Gene Identifier (RGI) v5.1.1 [46], respectively. We compared all the assembled genomes with the four regulators (AmrR [BPSL1805], BpeR [BPSL0812], BpeT [BPSS0290] and BpeS [BPSL0731]) of three efflux pumps (AmrAB-OprA, BpeAB-OprB and BpeEF-OprC) in B. pseudomallei . We also compared the assembled genomes with the β-lactamase PenA [BPSS0946] and the penicillin-binding protein 3 (PBP3, [BPSS1219]), to identify the mutations through blastn. Sequence alignment was performed using mega v7.0.14 [47]. All the software settings used were the default parameters, unless otherwise stated. A gene/segment was considered present if the overall hit coverage and identity were both ≥70 % [48].
Results
Population structure and genetic diversity of B. pseudomallei in Hainan
We collected and sequenced 122 B. pseudomallei isolates from patients with melioidosis or from the environment, distributed across 12 cities on Hainan island, between 2002 and 2018. After assembling, the number of contigs and average sizes of the assemblies were 173 (88–1305, >500 bp) and 7.1 Mb (7.0–7.6 Mb), with an average depth of 254-fold (126–439-fold) for B. pseudomallei genome. The average G+C content and N50 were 68.16 % (67.68–68.35 %) and 150 813 bp (15 385–261 885 bp), respectively. To determine the relationships between the Hainan and global B. pseudomallei strains, we compared the newly sequenced genomes with 1532 published B. pseudomallei genomes, which were mainly from strains collected in the hyperendemicity foci in northern Australia and Southeast Asia. The total dataset consisted of 1654 strains (see supplementary data table S1 for details of the isolates).
We established the population relationships across all 1654 genomes using both ML trees and a hierBAPS analysis [35] based on genome-wide SNPs (Fig. 1). Sixteen PGs (groups 1–16) were detected, 12 of which were defined (supplementary data table S3) in a previous study [17]. Except for group 8 and a bin cluster (43 isolates), groups 1–7 and 9–16 each formed a monophyletic group in the phylogeny (Fig. 1). All the Hainan strains were distributed within nine PGs, and 80 % of them were concentrated in five major PGs (>5 %; group 1, 12.3 %; group 3, 31.1 %; group 9, 13.1 %; group 11, 8.2 %; group 15, 15.6 %). The remaining strains were scattered sporadically throughout the other four PGs (Fig. 1). With MLST, the 122 isolates were resolved into 38 STs, six of which (>5 strains, STs 46, 50, 55, 58, 70, 658) were predominant (supplementary data table S2). We found that the Hainan strains in group 1 corresponded to multiple minor STs; the Hainan strains in group 3 corresponded to ST46 and ST50; and the Hainan strains in groups 9, 11, and 15 corresponded to ST58, ST55, and mainly ST658, respectively (supplementary data table S3). A phylogenetic analysis and hierarchical clustering supported the division of the major population of B. pseudomallei in Hainan into five PGs, with minor populations of sporadically distributed strains, suggesting the remarkably high genetic diversity of the local endemic strains.
Relationships between Hainan and global B. pseudomallei isolates
A phylogenic tree (Fig. 1) showed that all the Hainan isolates, distributed in nine distinct lineages, were surrounded by isolates from the Malay Peninsula or countries bordered by the Mekong River, suggesting close genetic relationships and transmission events in the history of the Hainan and Southeast Asian isolates, confirming previous hypotheses [11, 24, 25] at the WGS level. The minor Hainan isolates, scattered in the minor lineages (groups 4, 7, 8, 14) on the phylogenetic tree, with long branches and more than 5000 PSDs, suggested the historical transmission of B. pseudomallei from Southeast Asian countries to Hainan, especially from Thailand (Fig. 1). This indicates that the introduction of B. pseudomallei into Hainan was not recent and that melioidosis has been endemic to Hainan island for a long time [22]. However, to study the transmission relationships of the Hainan isolates, we focused on the recently emerged clonal groups containing Hainan isolates. We analysed the five major groups and divided the Hainan isolates into six subgroups (subgroups 1.1, 3.1, 3.2, 9.1, 11.1, and 15.1) based on a threshold of 5000 PSDs (see Methods).
To infer the origins of the B. pseudomallei isolates on Hainan island and the direction of dissemination between the global and Hainan isolates, we re-called the SNPs and constructed ML trees of four recently emerged subgroups (subgroups 3.1, 3.2, 9.1, and 15.1; Fig. 1), which each contained global and ≥5 Hainan strains with sampling location information. We then estimated the average numbers of between-country transmission events with phytools [44]. We detected a total of six transmission events with the stochastic mapping approach (SIMMAP) [43, 44] in all four subgroups (Fig. 2, supplementary data table S4). We detected two transmission events from Singapore to Hainan in subgroup 3.1 (Fig. 2a) and one transmission event from Hainan to Singapore in subgroup 3.2 (Fig. 2b). In subgroup 9.1 and subgroup 15.1, we detected one transmission event from Thailand to Hainan, one from Hainan to Taiwan, and one from Hainan to Malaysia (Fig. 2c, d). Our results indicate that the two original sources of Hainan strains in recent years were Thailand and Singapore, and also show that there were not only import events from Southeast Asian countries (Thailand and Singapore) to Hainan, but also export events from Hainan to Southeast Asian countries or other regions (Singapore, Malaysia, and Taiwan-CHN). These data demonstrate the mutual transmission of isolates from Hainan and those from geographically close countries or regions. We also calculated the evolutionary time of these transmissions ranging from 10.1 to 39.1 years (supplementary data table S8) based on the average pairwise SNP distance and the mutation rate of 6.8 SNPs per year.
Local epidemiology of B. pseudomallei in Hainan
The sampling times of 81 Hainan isolates, which accounted for 66.4 % of all isolates, were documented. The temporal distribution of these strains (Fig. S2a) showed that those of group 1 (containing the minor STs) have rarely appeared in the past two decades, and were not sampled after 2014. The strains of group 11 (ST55) were only detected in four years (2008, 2009, 2012, and 2013) in the past two decades and were also not sampled after 2014. However, the strains of group 3 (ST46, ST50, etc.), group 9 (ST58), and group 15 (ST366, ST658, etc.) accounted for a major proportion of the strains detected since 2008, especially those of group 3, which have been locally epidemic since 2002. Our sample set included only 15 strains collected after 2013. To avoid the effects of any sampling bias, we also investigated previous molecular epidemiological studies that included the STs of B. pseudomallei in Hainan from 2014 to 2016 [25, 49]. These studies also showed a predominance of STs in groups 3, 9, and 15 (Fig. S2a1), suggesting that most attention should be paid to the strains of these three major PGs in the future surveillance and prevention of B. pseudomallei infections.
The geographic sampling sites or the patients’ home addresses were documented for 85 Hainan isolates (69.7 % of all isolates), and involved 12 cities, the majority of which were in coastal areas. We analysed the spatial distribution of these recorded strains statistically on a map of Hainan island (Fig. S2b). The strains of groups 1 and 3 were distributed widely in northern, central, and southern coastal areas, whereas the distributions of the strains in groups 9, 11, and 15 were uneven. To avoid any inaccuracies arising from sampling bias (see details in the Discussion), we then examined previous studies. We found that although B. pseudomallei has been detected in coastal areas all around the island (northern, central, and southern), its distribution strongly suggests that southern cities (Sanya and Lingshui) are hotspots on the island. Therefore, we inferred that multiple B. pseudomallei populations have colonized Hainan island and spread widely within it.
To confirm local transmission events and infer the directions of transmission, we re-called the SNPs and constructed ML trees of only the Hainan isolates with sampling location information in the six recently emerged subgroups, clustered based on a threshold of 5000 PSDs, as described above. Using phytools [44], we detected a total of 21 between-city transmission events (Fig. 3, supplementary data table S5), 18 of which originated in the two southern hotspot cities, Sanya and Lingshui, and spread to the northern, western, and eastern areas of the island. Besides, the calculated evolutionary time showed that all of the local transmissions occurred in recent two decades, except two transmissions from Sanya to Lingshui, which occurred 73.1 and 40.7 years ago (supplementary data table S8). This movement may have been driven by anthropogenic factors. For instance, Sanya is a famous tourist city and attracts millions of tourists each year. Thus, our results show that in addition to multiple import events from other Southeast Asian countries, there have been frequent local transmission events within Hainan island.
Virulence factors and antimicrobial resistance
To assess the virulence potential of B. pseudomallei in Hainan compared with that of global strains, we mapped the assembled genomes of all 1654 strains against the core dataset of the VFDB. We detected a total of 160 virulence factor genes (supplementary data table S6), including those encoding capsular polysaccharide I (wcb gene cluster), a diverse complement of autotransporters (bimA, boaA etc.), multiple specialized secretion systems (T3SS, T4SS, and T6SS), a flagellum-related gene cluster, etc. When the identified virulence factors were annotated based on the Clusters of Orthologous Genes (COG) database, the frequencies of the following terms were elevated: ‘cell motility’, ‘intracellular trafficking, secretion and vesicular transport’, ‘cell wall, membrane, envelope biogenesis’, and ‘signal transduction mechanism’ (Fig. S3).
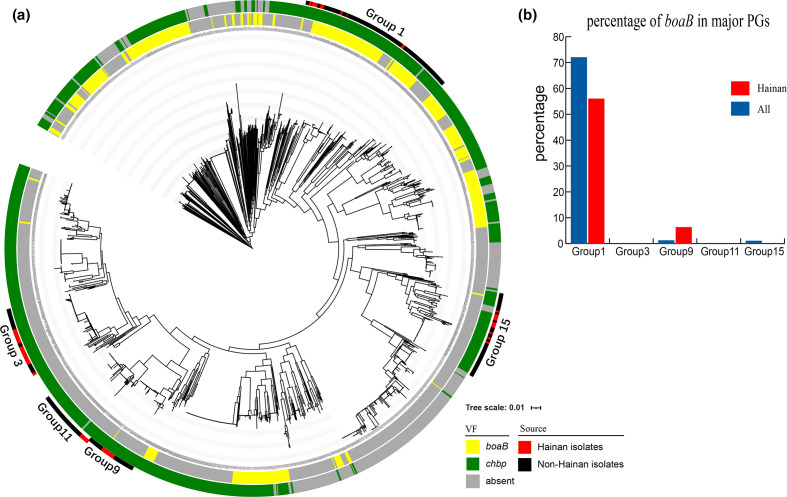
We identified 152 virulence factors within >95 % of the total strains and 100 % of the Hainan strains (supplementary data table S6), suggesting the impressive virulence arsenal of B. pseudomallei . Six virulence factors were present in only a few global strains (<5 %; supplementary data table S6), and not in any Hainan strain, and three (ybtT, chuS, gspC) of the six minority virulence factors were concentrated in a single strain (strain 110, SAMN05715376). The remaining two virulence factor genes (boaB, chbp) were variably present among the global and Hainan strains (Fig. 4a). The gene boaB was present in ~33 % of the global strains and ~12 % of the Hainan strains, and most of the Hainan strains that contained boaB (56 %) were in major group 1 (Fig. 4b). The gene boaB (BPSL1705) appears to be a B. pseudomallei -specific virulence factor gene, encoding a trimeric autotransporter adhesin that mediates the bacterium’s adherence to the epithelial cells of the human respiratory tract [50], and also may play a role in its invasion and survival in macrophages [51]. We inferred that boaB may enhance the strain virulence of Hainan strains in group 1, and promote the adhesin to host cells, which could be verified in combination with clinical phenotypes. Another variably occurring virulence factor gene, chbp, which encodes a secretion effector of the type III secretion system (T3SS), was identified in ~72 % of the global strains and ~89 % of the Hainan strains, with a scattered distribution on the phylogenic tree (Fig. 4).
Fig. 4.
Variably-presented virulence factors of 1654 B. pseudomallei isolates. (a) Phylogenetic tree with two rings shows the distribution of two variably occurring virulence factors (VFs): boaB (yellow) and chbp (green). The outer ring represents the five major groups, which contain global strains (black) and Hainan isolates (red). (b) Histogram shows the percentage of Hainan B. pseudomallei isolates in the five major groups that contain the virulence factor gene boaB; boaB is present in 56 % of the Hainan strains in group 1, but is almost absent from the other major groups.
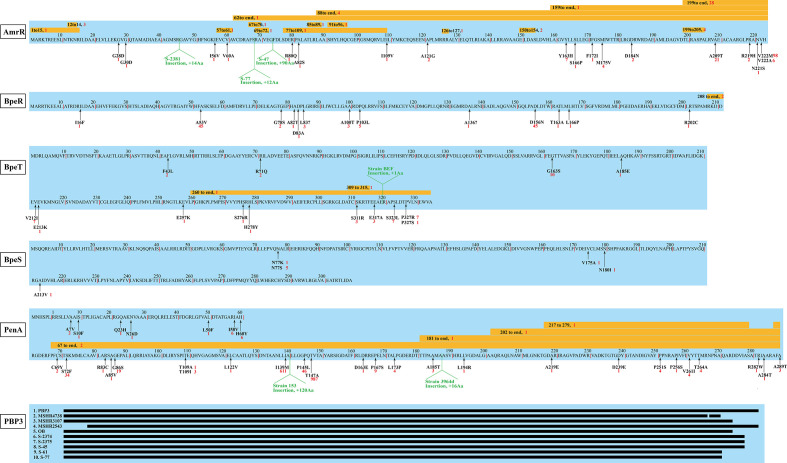
To clarify the antimicrobial-resistance (AMR) characteristics of the Hainan B. pseudomallei strains, we mapped the assembled genomes of all 1654 strains to the CARD using RGI. We identified a total of eight antimicrobial-resistance genes among all 1654 B. pseudomallei genomes (supplementary data table S7). AmrAB-OprA multidrug efflux pump is ubiquitous to B. pseudomallei [52, 53] and mediated resistance to aminoglycoside and macrolide antibiotics. The identified genes amrA and amrB are the two essential components of the AmrAB-OprA multidrug efflux pump. The three efflux pumps in B. pseudomallei (AmrAB-OprA, BpeAB-OprB and BpeEF-OprC) confer clinically relevant resistance to multiple antibiotics through alterations in their respective regulators: AmrR, BpeR, and BpeT and BpeS. We found a complete absence of amrR in a Thailand genome 708 a, which was caused by a large deletion on chromosome 1 reported by Schweizer et al. in 2009 [54]. Moreover, we found a duplicate copy of amrR in an Australian genome BEC, 17 point mutations on amrR in 148 genomes, three insertions in three Hainan genomes, and 15 fragment deletions in 51 genomes (Fig. 5). A total of 13 Hainan strains were involved in the identified mutations, including two point mutations (AmrRV222M: S-2365, S-301–17, S-57, S-110, S-59; AmrRA121G: S-69), three insertions (Insertion45: S-2381, Insertion68: S-77, Insertion73: S-47) and two deletions (Deletion159-end: S-36, Deletion199-205: S-25, S-2371, S-2372). We infer that all the mutations found in amrR may relate with varied level of resistance to macrolides or meropenem in B. pseudomallei except for AmrRV222M which has been proved not to cause AMR [55]. This assumption needs to be experimentally verified in the future. Compared with the regulator AmrR of AmrAB-OprA, the other three regulators, BpeR, BpeT, and BpeS, were less well studied and the mutations in them identified in current study have been summarized in Fig. 5. The identified gene omp38 is a porin present in every B. pseudomallei strain and has no relevance to clinically relevant antimicrobial resistance [52]. The gene oxa that is ubiquitous to B. pseudomallei strains encodes the class D β-lactamases (oxacillinases named OXA-42, –43 and −57) [56]. However, no oxacillinase has been shown to cause resistance to ceftazidime in clinical B. pseudomallei isolates [52] and the mechanism of clinically relevant resistance to ceftazidime is caused by the mutations within the class A β-lactamase PenA [52, 57–59] and the deletion of PBP3 [60]. We identified a total of 32 point mutations and seven indels in PenA (Fig. 5), among which, four point mutations (I139M, P145L, T147A and T264A) were present in but not limited to 64 Hainan genomes (supplementary data table S9). Previously reported mutations (C69Y, P167S and S72F, etc.) [52, 61] that have been experimentally verified were not identified in newly-sequenced Hainan genomes except for PenAT147A, which was caused by a nucleotide transition from A to G at position 457. PenAT147A was identified in 987 genomes (59.7 %) in current study, including 64 (52 %) newly sequenced Hainan genomes (supplementary data table S9). However, multiple studies [53, 55] have proved that PenAT147A was a natural variant present in >50 % B . pseudomallei genomes and did not confer clinically relevant AMR. The sequence analysis of PBP3 of all the 1654 genomes showed that there was a duplicate copy of PBP3 residing in two contigs of an Australian genome MSHR2138, and the result also revealed nine deletions to varying degrees in nine genomes, including five newly sequenced Hainan genomes (Fig. S5, S-2374, S-2375, S-45, S-61, S-77). However, the relationship between the AMR and the alteration in PenA and PBP3 requires further exploration through phenotype experiments. The AMR analysis in our study provided helpful suggestions in antibiotic selection during clinical therapy.
Fig. 5.
Mutations on clinically relevant antibiotic resistance genes of 1654 B. pseudomallei isolates. The black arrows point to the locations of point mutations, the yellow boxes indicate the positions of deletions, and the green markers indicate the positions of insertions, the red numbers refer to the number of genomes involved in the specific mutations. The positions of amino acid substitutions are numbered according to the Ambler scheme [75].
Discussion
In this study, newly sequenced B. pseudomallei samples from Hainan included 119 clinical strains from melioidosis patients and three environmental strains from well water. There has only been a single report of a handful of suspected cases of human-to-human transmission of B. pseudomallei [62], and skin inoculation, the oral ingestion of B. pseudomallei-contaminated water [4, 63, 64], and the inhalation of B. pseudomallei during extreme weather events [65, 66] are considered the main routes of infection. Therefore, we inferred that the sampling locations of these clinical isolates represented the distribution of B. pseudomallei in the environment of Hainan. All the samples analysed in this study were collected widely across 12 districts, ten of which were coastal regions, and notably, most of the isolates were collected in the southern region of Hainan. The uneven geographic distribution of B. pseudomallei was consistent with previous soil sampling in 360 rice paddy fields all around the Hainan island in 2016, which also revealed an uneven distribution of B. pseudomallei [67]. The organism was frequently isolated from coastal regions, especially southern cities, which may be attributable to multiple environmental and anthropogenic factors. First, a previous study demonstrated that the presence of B. pseudomallei is strongly associated with various environmental factors, including high rainfall, high temperatures, anthrosol and acrisol soil types, high salinity, high proportion of gravel, and soil pH [4, 68, 69]. The higher salinity and the soil type in the coastal regions of Hainan may be more suitable for this pathogen than those in the central mountainous regions. Second, Hainan island enjoys a tropical monsoonal climate in its southern part, and experiences typhoons (mean annual frequency, eight), with preferential landing sites in the east and south of the island, which consequently receive high rainfall and high wind speeds [67]. Furthermore, data from clinical and climate studies have highlighted a potential link between typhoons or high rainfall and melioidosis [70, 71]. Finally, Sanya, the southernmost city of Hainan island, has the most severe B. pseudomallei endemicity. It is an international tourist city, attracting millions of international tourists every year, which increases the possible introduction of B. pseudomallei from other endemic areas around the world.
When studying the transmission relationship between Hainan and global B. pseudomallei genomes, we identified several mutual transmission events between Hainan island and neighbouring countries or regions, such as Malaysia, Singapore and Taiwan island, etc. This may be driven by anthropogenic factors, including cross-border trade and travel. Since the Han Dynasty about 2200 years ago, Hainan island was an important supply station on the trade route between Southeast Asian countries and the mainland China. Hainan island is also an important hub in the network of ‘One Belt One Road’ that was proposed in 2013 and formally implemented since 2015. Frequent commercial contacts have increased the risk of B. pseudomallei dissemination and promoted gene transfer among the local strains on Hainan island. Moreover, as a popular tourist attraction, Hainan island attracts millions of domestic and foreign tourists each year, which also potentially increased spread risk of B. pseudomallei . Globalization has greatly promoted the communication between different countries and regions, which has also increased the risk of the spread of B. pseudomallei . The occurrence of cross-border transmission between Hainan island and Southeast Asian countries has deepened our understanding of the genomic epidemiological characteristics of B. pseudomallei and will facilitate our prevention and control efforts.
We suppose that more researches are needed to study the historical introduction events of B. pseudomallei from Australia to Asia. Previous studies [12, 17, 21] have used WGS analyses to identify single introduction event of B. pseudomallei from Australia to Asia. The oceanic biogeographic barrier between Asian and Australasian B. pseudomallei populations, the Wallace line, has limited the dissemination of B. pseudomallei , encouraged its divergent evolution, and ensured the paucity of genotypic or phylogenetic overlap between the two known hyperendemicity foci [72, 73]. However, in the phylogeny presented in this study, five B. pseudomallei strains (SAMN04208629, SAMN04208630, SAMN04208605, SAMN04208593, and SAMN02443741) from Thailand clustered into two distinct clades inside the Australian group (group 5), suggesting a close genetic background and several historical transmission events from Australia to Thailand. Furthermore, although the sampling site of one strain was unclear, the other four strains were collected from the environment, supporting the notion that these strains were more likely to have been transmitted to Thailand via environment-related routes, and not by Thai melioidosis patients who had ever travelled to Australia and brought B. pseudomallei back to Thailand. Therefore, multiple historical introduction events from Australia to Southeast Asia might be detected if larger sample sets were considered.
When assessing the spatial distribution of B. pseudomallei on Hainan island, we found an uneven distribution of the strains in groups 9, 11, and 15 (Fig. S2b), unlike the strains in groups 1 and 3, which were widely distributed in the northern, central, and southern coastal areas. The strains of group 9 only occurred in the northern and southern coastal areas; the strains of group 11 were only distributed in the central and southern coastal areas; and the strains of group 15 only appeared in three southern cities. To avoid inaccurate results caused by sampling bias, we also perused previous studies, which showed that strains of ST58 (probably group 9) appeared repeatedly in central coastal cities (Danzhou and Qionghai) in 2010 and 2014 [11, 25], strains of ST55 (probably group 11) only appeared, and infrequently, in the northern city of Haikou in 2012 [24], and strains of ST658 and ST1105 (probably group 15) appeared in several central and northern coastal cities (Haikou, Wenchang, Dingan, and Danzhou) [11, 24, 25, 49]. Thus, the spatial distribution of B. pseudomallei in Hainan suggests that it has no preferential geographic distribution in the coastal areas on the island.
Previous studies [24, 25] based on MLST revealed the close relationship between isolates of Hainan island and Southeast Asian countries, especially Thailand, but there have never been direct evidences proving the mutual transmissions, or even the directions of transmissions. However, based on the whole genome sequences in our study, we not only clarified the population structure and predominant phylogenetic groups of B. pseudomallei in Hainan island, but also proved frequent between-country and between-city transmissions with exact directions. We also used large-scale comparative genomics to identify the virulence factors and antimicrobial-resistance genes of B. pseudomallei in Hainan, especially those experimentally verified AMR genes that confer clinically relevant resistance (amrR, penA, PBP3, etc.). Although a variety of antibiotic resistance testing on Hainan isolates have proved resistance to varies kinds of common antibiotics [74], but researches about the mutations on AMR genes were limited. Our study provided a feasible method for future research on resistance mechanisms of Hainan isolates and the AMR analysis in current study provided helpful suggestions in antibiotic selection during clinical therapy. These data provide further insight into the pathogenic and epidemiological characteristics of the B. pseudomallei strains in Hainan, which are extremely important for the surveillance, prevention, and clinical treatment of melioidosis. Demonstration on the relationships between global and Hainan B. pseudomallei isolates have filled some research gaps in tropical or subtropical regions outside the known hyperendemicity foci of northern Australia and Southeast Asia. We thus provide further insight into the regional and global transmission network of B. pseudomallei , improving our understanding of its global phylogeography.
Supplementary Data
Funding information
This work was supported by the National Key Program for Infectious Diseases of China (no. 2018ZX10101003 and 2018ZX10714-002) and the Hainan Natural Science Foundation (no. 817398).
Author contributions
Y.C. and R.Y., conceived and designed the project. H.Z., analysed the data and wrote the initial manuscript. Y.C. and Y.T., coordinated and managed this research. Y.T., H.C., H.H., Y.L., S.L., H.K., H.Z., D.S. and K.S., collected and isolated the strains for whole genome sequencing. T.Z., Y.S., J.Q., X.Z, C.Y. and Y.W., provided invaluable feedback and insights into the analysis. All authors approved the final version of the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AMR, antimicrobial resistance; CARD, comprehensive antibiotic resistance database; CFR, case fatality rate; MLST, multilocus sequence typing; MLVA, multilocus variable-number tandem repeat analysis; PFGE, pulsed-field gel electrophoresis; PG, phylogenic group; PSD, pairwise SNP distance; SNP, single nucleotide polymorphism; ST, sequence type; VFDB, virulence factors database; WGS, whole genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Nine supplementary data files and three supplementary figures are available with the online version of this article.
References
- 1.Wiersinga W, van der Poll T, White N, Day N, Peacock S. Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei . Nat Rev Microbiol. 2006;4:272–282. doi: 10.1038/nrmicro1385. [DOI] [PubMed] [Google Scholar]
- 2.White NJ, Chaowagul W, Wuthiekanun V, Dance DAB, Wattanagoon Y. Halving of mortality of severe melioidosis by ceftazidime. The Lancet. 1989;2:697–701. doi: 10.1016/S0140-6736(89)90768-X. [DOI] [PubMed] [Google Scholar]
- 3.Lipsitz R, Garges S, Aurigemma R, Baccam P, Blaney DD, et al. Workshop on treatment of and postexposure prophylaxis for Burkholderia pseudomallei and B. mallei infection, 2010. Emerging Infectious Diseases, Article. 2012;18 doi: 10.3201/eid1812.120638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Limmathurotsakul D, Golding N, Dance DAB, Messina JP, Pigott DM. Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat Microbiol. 2016;1:15008. doi: 10.1038/nmicrobiol.2015.8. [DOI] [PubMed] [Google Scholar]
- 5.Talon D, Cailleaux V, Thouverez M, Michel-Briand Y. Discriminatory power and usefulness of pulsed-field gel electrophoresis in epidemiological studies of Pseudomonas aeruginosa . J Hosp Infect. 1996;32:135–145. doi: 10.1016/s0195-6701(96)90055-9. [DOI] [PubMed] [Google Scholar]
- 6.Inglis TJ, Garrow SC, Adams C, Henderson M, Mayo M. Acute melioidosis outbreak in Western Australia. Epidemiol Infect. 1999;123:437–443. doi: 10.1017/s0950268899002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chua KH, See KH, Thong KL, Puthucheary SD. DNA fingerprinting of human isolates of Burkholderia pseudomallei from different geographical regions of Malaysia. Trop Biomed. 2010;27:517–524. [PubMed] [Google Scholar]
- 8.Azura MN, Norazah A, Kamel AG, Zorin SA. DNA fingerprinting of septicemic and localized Burkholderia pseudomallei isolates from Malaysian patients. Southeast Asian J Trop Med Public Health. 2011;42:114–121. [PubMed] [Google Scholar]
- 9.Currie BJ, Haslem A, Pearson T, Hornstra H, Leadem B. Identification of Melioidosis outbreak by multilocus variable number tandem repeat analysis. Emerging Infect Dis. 2009;15:169–174. doi: 10.3201/eid1502.081036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao C, Hu Z, Chen J, Tang M, Li Q. Molecular epidemiology and antibiotic resistance of Burkholderia pseudomallei isolates from Hainan, China: A STROBE compliant observational study. Medicine (Baltimore) 2019;98:e14461. doi: 10.1097/MD.0000000000014461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu X, Chen H, Li S, Wang LC, Liu ZG. Molecular characteristics of Burkholderia pseudomallei collected from humans in Hainan, China. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarovich DS, Garin B, De Smet B, Kaestli M, Mayo M. Phylogenomic analysis reveals an asian origin for African Burkholderia pseudomallei and further supports melioidosis endemicity in Africa. mSphere. 2016;1:e00089-15. doi: 10.1128/mSphere.00089-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maiden MCJ, Bygraves JA, Feil E, Morelli G, Russell JE. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearson T, Giffard P, Beckstrom-Sternberg S, Auerbach R, Hornstra H. Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biol. 2009;7:78. doi: 10.1186/1741-7007-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelthaler DM, Bowers J, Schupp JA, Pearson T, Ginther J, et al. Molecular investigations of a locally acquired case of melioidosis in Southern AZ, USA. PLoS Negl Trop Dis. 2011;5:e1347. doi: 10.1371/journal.pntd.0001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Smet B, Sarovich DS, Price EP, Mayo M, Theobald V, et al. Whole-genome sequencing confirms that Burkholderia pseudomallei multilocus sequence types common to both Cambodia and Australia are due to homoplasy. J Clin Microbiol. 2015;53:323–326. doi: 10.1128/JCM.02574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chewapreecha C, Holden MT, Vehkala M, Valimaki N, Yang Z. Global and regional dissemination and evolution of Burkholderia pseudomallei . Nat Microbiol. 2017;2:16263. doi: 10.1038/nmicrobiol.2016.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanagida T, Carod J-F, Sako Y, Nakao M, Hoberg EP, et al. Genetics of the pig tapeworm in Madagascar reveal a history of human dispersal and colonization. PLoS One. 2014;9:e109002. doi: 10.1371/journal.pone.0109002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choy JL, Mayo M, Janmaat A, Currie BJ. Animal melioidosis in Australia. 2000;74:153–158. doi: 10.1016/s0001-706x(99)00065-0. [DOI] [PubMed] [Google Scholar]
- 20.Hampton V. Melioidosis in birds and Burkholderia pseudomallei dispersal, Australia. Emerging Infect Dis. 2011;17:1310–1312. doi: 10.3201/eid1707.100707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price EP, Currie BJ, Sarovich DS. Genomic insights into the melioidosis pathogen, Burkholderia pseudomallei . Curr Trop Med Rep. 2017;4:95–102. doi: 10.1007/s40475-017-0111-9. [DOI] [Google Scholar]
- 22.Zheng X, Xia Q, Xia L, Li W. Endemic melioidosis in Southern China: Past and Present. Trop Med Infect Dis. 2019;4:E39. doi: 10.3390/tropicalmed4010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Lu Z, Han O. Epidemiology of melioidosis in China. Zhonghua Liu Xing Bing Xue Za Zhi. 1994;15:292–295. [PubMed] [Google Scholar]
- 24.Fang Y, Zhu P, Li Q, Chen H, Li Y. Multilocus sequence typing of 102 Burkholderia pseudomallei strains isolated from China. Epidemiol Infect. 2016;144:1917–1923. doi: 10.1017/S0950268815003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang Y, Hu Z, Chen H, Gu J, Hu H, et al. Multilocus sequencing-based evolutionary analysis of 52 strains of Burkholderia pseudomallei in Hainan, China. Epidemiol Infect. 2018:1–6. doi: 10.1017/S0950268818002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo R, Liu B, Xie Y, Li Z, Huang W. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui Y, Yang X, Didelot X, Guo C, Li D. Epidemic Clones, Oceanic Gene Pools, and Eco-LD in the Free Living Marine Pathogen Vibrio parahaemolyticus . Mol Biol Evol. 2015;32:1396–1410. doi: 10.1093/molbev/msv009. [DOI] [PubMed] [Google Scholar]
- 28.Yang C, Zhang X, Fan H, Li Y, Hu Q. Genetic diversity, virulence factors and farm-to-table spread pattern of Vibrio parahaemolyticus food-associated isolates. Food Microbiol. 2019;84:103270. doi: 10.1016/j.fm.2019.103270. [DOI] [PubMed] [Google Scholar]
- 29.Delcher AL, Salzberg SL, Phillippy AM. Using MUMmer to identify similar regions in large sequence sets. Current Protocs in Bioinformatics. 2003;10:10. doi: 10.1002/0471250953.bi1003s00. [DOI] [PubMed] [Google Scholar]
- 30.Li R, Yu C, Li Y, Lam TW, Yiu SM. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 31.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corander J, Marttinen P, Sirén J, Tang J. Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics. 2008;9:539. doi: 10.1186/1471-2105-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng L, Connor TR, Siren J, Aanensen DM, Corander J. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol. 2013;30:1224–1228. doi: 10.1093/molbev/mst028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nuclc Acids Research. 2015:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Croucher NJ, Finkelstein JA, Pelton SI, Mitchell PK, Lee GM. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat Genet. 2013;45:656. doi: 10.1038/ng.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Assefa S, Keane TM, Otto TD, Newbold C, Berriman M. ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics. 2009;25:1968–1969. doi: 10.1093/bioinformatics/btp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG. ACT: the Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 40.Kozlov AM, Diego D, Tomáš F, Benoit M, Alexandros S. RAxML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;(21) doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moura A, Criscuolo A, Pouseele H, Maury MM, Leclercq A. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat Microbiol. 2016;2:16185. doi: 10.1038/nmicrobiol.2016.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holt KE, McAdam P, Thai PVK, Thuong NTT, DTM H. Frequent transmission of the Mycobacterium tuberculosis Beijing lineage and positive selection for the EsxW Beijing variant in Vietnam. Nat Genet. 2018;50:849–856. doi: 10.1038/s41588-018-0117-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bollback JP. SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics. 2006;7:88. doi: 10.1186/1471-2105-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3:217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- 45.Liu B, Zheng D, Jin Q, Chen L, Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47:D687–D692. doi: 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H, Yang C, Sun Z, Zheng W, Zhang W, et al. Genomic epidemiology of Vibrio cholerae reveals the regional and global spread of two epidemic non-toxigenic lineages. PLoS Negl Trop Dis. 2020;14:e0008046. doi: 10.1371/journal.pntd.0008046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang XM, Zheng X, Wu H, Zhou XJ, Kuang HH. Multilocus sequence typing of clinical isolates of Burkholderia pseudomallei collected in Hainan, a tropical Island of Southern China. Am J Trop Med Hyg. 2016;95:760–764. doi: 10.4269/ajtmh.16-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balder R, Lipski S, Lazarus JJ, Grose W, Wooten RM, et al. Identification of Burkholderia mallei and Burkholderia pseudomallei adhesins for human respiratory epithelial cells. 2010;10:250. doi: 10.1186/1471-2180-10-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andrea J, Dowling A. genome-wide analysis reveals loci encoding anti-macrophage factors in the human pathogen Burkholderia pseudomallei K96243. PLoS One. 2010 doi: 10.1371/journal.pone.0015693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schweizer HP. Mechanisms of antibiotic resistance in Burkholderia pseudomallei: implications for treatment of melioidosis. Future Microbiol. 2012;7:1389–1399. doi: 10.2217/fmb.12.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sarovich DS, Webb JR, Pitman MC, Viberg LT, Mayo M. Raising the stakes: loss of efflux pump regulation decreases meropenem susceptibility in Burkholderia pseudomallei . Clin Infect Dis. 2018;67:243–250. doi: 10.1093/cid/ciy069. [DOI] [PubMed] [Google Scholar]
- 54.Trunck LA, Propst KL, Wuthiekanun V, Tuanyok A, Beckstrom-Sternberg SM. Molecular basis of rare aminoglycoside susceptibility and pathogenesis of Burkholderia pseudomallei clinical isolates from Thailand. PLoS Negl Trop Dis. 2009;3:e519. doi: 10.1371/journal.pntd.0000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Madden DE, Webb JR, Steinig EJ, Currie BJ, Price EP. Taking the next-gen step: Comprehensive antimicrobial resistance detection from Burkholderia pseudomallei. EBioMedicine. 2021;63:103152. doi: 10.1016/j.ebiom.2020.103152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keith KE, Oyston PC, Crossett B, Fairweather NF, Brown KA. Functional Characterization of OXA-57, a Class D β-Lactamase from Burkholderia pseudomallei . Antimicrob Agents Chemother. 2005;49:1639–1641. doi: 10.1128/AAC.49.4.1639-1641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sarovich DS, Price EP, Von Schulze AT, Cook JM, Mayo M, et al. Characterization of ceftazidime resistance mechanisms in clinical isolates of Burkholderia pseudomallei from Australia. PLoS One. 2012;7:e30789. doi: 10.1371/journal.pone.0030789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sarovich DS, Price EP, Limmathurotsakul D, Cook JM, Von Schulze AT. Development of ceftazidime resistance in an acute Burkholderia pseudomallei infection. Infect Drug Resist. 2012;5:129–132. doi: 10.2147/IDR.S35529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rholl DA, Papp-Wallace KM, Tomaras AP, Vasil ML, Bonomo RA. Molecular investigations of PenA-mediated β-lactam resistance in Burkholderia pseudomallei . Front Microbiol. 2011;2:139. doi: 10.3389/fmicb.2011.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chantratita N, Rholl DA, Sim B, Wuthiekanun V, Limmathurotsakul D. Antimicrobial resistance to ceftazidime involving loss of penicillin-binding protein 3 in Burkholderia pseudomallei . Proc Natl Acad Sci U S A. 2011;108:17165–17170. doi: 10.1073/pnas.1111020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bugrysheva JV, Sue D, Gee JE, Elrod MG, Hoffmaster AR. Antibiotic resistance markers in Burkholderia pseudomallei strain Bp1651 identified by genome sequence analysis. Antimicrob Agents Chemother. 2017;61:e00010-17. doi: 10.1128/AAC.00010-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aziz A, Currie BJ, Mayo M, Sarovich DS, Price EP. Comparative genomics confirms a rare melioidosis human-to-human transmission event and reveals incorrect phylogenomic reconstruction due to polyclonality. Microb Genom. 2020;6 doi: 10.1099/mgen.0.000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Limmathurotsakul D, Kanoksil M, Wuthiekanun V, Kitphati R, deStavola B, et al. Activities of daily living associated with acquisition of melioidosis in northeast Thailand: a matched case-control study. PLoS Negl Trop Dis. 2013;7:e2072. doi: 10.1371/journal.pntd.0002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Limmathurotsakul D, Wongsuvan G, Aanensen D, Ngamwilai S, Saiprom N. Melioidosis caused by Burkholderia pseudomallei in drinking water, Thailand, 2012. Emerg Infect Dis. 2014;20:265–268. doi: 10.3201/eid2002.121891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen PS, Chen YS, Lin HH, Liu PJ, Ni WF. Airborne transmission of melioidosis to humans from environmental aerosols contaminated with B. PLoS Negl Trop Dis. 2015;9:e0003834. doi: 10.1371/journal.pntd.0003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng AC, Jacups SP, Gal D, Mayo M, Currie BJ. Extreme weather events and environmental contamination are associated with case-clusters of melioidosis in the Northern Territory of Australia. Int J Epidemiol. 2006;35:323–329. doi: 10.1093/ije/dyi271. [DOI] [PubMed] [Google Scholar]
- 67.Dong S, Wu L, Long F, Wu Q, Liu X. The prevalence and distribution of Burkholderia pseudomallei in rice paddy within Hainan, China. Acta Trop. 2018;187:165–168. doi: 10.1016/j.actatropica.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 68.Pumirat P, Cuccui J, Stabler RA, Stevens JM, Muangsombut V. Global transcriptional profiling of Burkholderia pseudomallei under salt stress reveals differential effects on the Bsa type III secretion system. BMC Microbiol. 2010;10:171. doi: 10.1186/1471-2180-10-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stopnisek N, Bodenhausen N, Frey B, Fierer N, Eberl L. Genus-wide acid tolerance accounts for the biogeographical distribution of soil Burkholderia populations. Environ Microbiol. 2014;16:1503–1512. doi: 10.1111/1462-2920.12211. [DOI] [PubMed] [Google Scholar]
- 70.Chen YL, Yen YC, Yang CY, Lee MS, CK H. The concentrations of ambient Burkholderia pseudomallei during typhoon season in endemic area of melioidosis in Taiwan. PLoS Negl Trop Dis. 2014;8:e2877. doi: 10.1371/journal.pntd.0002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu X, Pang L, Sim SH, Goh KT, Ravikumar S. Association of melioidosis incidence with rainfall and humidity, Singapore, 2003-2012. Emerg Infect Dis. 2015;21:159–162. doi: 10.3201/eid2101.140042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baker A, Mayo M, Owens L, Burgess G, Norton R, et al. Biogeography of Burkholderia pseudomallei in the torres strait Islands of Northern Australia. J Clin Microbiol. 2013;51:2520–2525. doi: 10.1128/JCM.00418-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baker AL, Pearson T, Sahl JW, Hepp C, Price EP, et al. Burkholderia pseudomallei distribution in Australasia is linked to paleogeographic and anthropogenic history. PLoS ONE. 2018;13:11. doi: 10.1371/journal.pone.0206845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rao C, Hu Z, Chen J, Tang M, Chen H. Molecular epidemiology and antibiotic resistance of Burkholderia pseudomallei isolates from Hainan, China: A STROBE compliant observational study. Medicine (Baltimore) 2019;98:e14461. doi: 10.1097/MD.0000000000014461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ambler RP, Coulson AF, Frère JM, Ghuysen JM, Joris B. A standard numbering scheme for the class A beta-lactamases. Biochem J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.