Abstract
Invasive non-typhoidal Salmonella (iNTS) disease continues to be a significant public health problem in sub-Saharan Africa. Common clinical misdiagnosis, antimicrobial resistance, high case fatality and lack of a vaccine make iNTS a priority for global health research. Using whole genome sequence analysis of 164 invasive Salmonella isolates obtained through population-based surveillance between 2008 and 2016, we conducted genomic analysis of the serovars causing invasive Salmonella diseases in rural Gambia. The incidence of iNTS varied over time. The proportion of atypical serovars causing disease increased over time from 40 to 65 % compared to the typical serovars Enteritidis and Typhimurium that decreased from 30 to 12 %. Overall iNTS case fatality was 10%, but case fatality associated with atypical iNTS alone was 10 %. Genetic virulence factors were identified in 14/70 (20 %) typical serovars and 45/68 (66 %) of the atypical serovars and were associated with: invasion, proliferation and/or translocation (Clade A); and host colonization and immune modulation (Clade G). Among Enteritidis isolates, 33/40 were resistant to four or more of the antimicrobials tested, except ciprofloxacin, to which all isolates were susceptible. Resistance was low in Typhimurium isolates, but all 16 isolates were resistant to gentamicin. The increase in incidence and proportion of iNTS disease caused by atypical serovars is concerning. The increased proportion of atypical serovars and the high associated case fatality may be related to acquisition of specific genetic virulence factors. These factors may provide a selective advantage to the atypical serovars. Investigations should be conducted elsewhere in Africa to identify potential changes in the distribution of iNTS serovars and the extent of these virulence elements.
Keywords: atypical serovar, cytolethal distending toxin gene, invasive non-typhoidal Salmonella , whole genome sequencing
Data Summary
Accession numbers for all sequences used in this study can be found under BioProject PRJEB39996: SAMEA8226912 to SMEA8227075. The authors confirm all supporting data, code and protocols have been provided within the article or through supplementary data files.
Impact Statement.
In sub-Saharan Africa, in addition to causing gastroenteritis, non-typhoidal Salmonella cause life-threatening infections including septicaemia, pneumonia and meningitis. As such, insight into the genomic characteristics of serovars causing invasive non-typhoidal Salmonella (iNTS) disease is of importance to public health. Using genomics and epidemiological data through population-based surveillance, the study showed an increased in the proportion of Salmonella disease caused by atypical serovars over time. The increased proportion of atypical serovars and the high associated case fatality may be related to acquisition of specific genetic virulence factors. The shift in serovar prevalence could have implications for vaccine development and thus represent a public health concern. Therefore, investigations should be made to identify potential changes in the distribution of iNTS serovars elsewhere in Africa and the prevalence of these virulence elements.
Introduction
Salmonella enterica is a phenotypically diverse Gram-negative bacterial species, consisting of more than 2600 serovars. Some serovars are implicated in life-threatening systemic infections and are host-restricted to humans [1]. These include S. enterica serovar Typhi and S. enterica serovar Paratyphi (S. Paratyphi A–C). In contrast, non-typhoidal Salmonella species infect both humans and animals [2]; S. enterica serovar Typhimurium and S. enterica serovar Enteritidis are the most commonly reported in association with Salmonella gastroenteritis. Globally [3], these serovars are responsible for about 75 million cases and 27 000 deaths annually [3].
In sub-Saharan Africa, in addition to causing gastroenteritis, non-typhoidal Salmonella (NTS) cause life-threatening infections including septicaemia, pneumonia and meningitis [4]. About 3.4 million cases of invasive Salmonella [i.e. infection of Salmonella outside the gut (e.g. in the blood, meninges or lungs)] caused by NTS (iNTS) are reported annually, with Typhimurium and Enteritidis being responsible for 80–90 % of these cases [5]. The majority of these infections affect children, and are often associated with human immunodeficiency virus (HIV) infection, prior malarial infection, severe anaemia or malnutrition, and case fatality of up to 25% [6–9]. In adults, HIV infection is associated with iNTS disease, and case fatality up to 50 % has been reported [7–9]. In some parts of Africa, the burden of iNTS disease is higher than that of pneumococcus, infecting tens of thousands of people [7–9]. In The Gambia, iNTS disease in children ranks third after Streptococcus pneumoniae and Staphylococcus aureus as a cause of invasive bacterial disease [10]. Despite the burden of this disease in our setting, the genomic epidemiology of NTS is still poorly understood.
Susceptibility to invasive Salmonella disease could be attributed to host genetic background and immunological status [4]. However, some serovars are known to cause bacteraemia more frequently than others, signifying the importance of pathogen characteristics. For example, a high burden of invasive disease caused by a specific genotype of S. Typhimurium has been associated with host adaptation as a result of extensive genomic degradation and acquisition of resistance genes [11]. In addition, the virulence factor cytolethal distending toxin gene (CdtB) is known to contribute to variation in disease severity in some NTS serovars [12]. The CdtB gene, which was thought to be unique to Salmonella Typhi, has been associated with increased host colonization, tumorigenesis, neoplastic lesions [13] and DNA damage similar to that caused by serovar Typhi [13]. The presence of the gene in Typhi is associated with host immune modulation as well as persistence of the pathogen in vivo [12]. Recently, the presence of CdtB has also been documented in NTS serovars and is believed to be clade-associated [12]. Thus, the presence of this virulence gene in NTS serovars could influence the virulence of these strains.
During population-based invasive bacterial disease surveillance in rural Gambia between 2008 and 2016, we observed changes in the incidence, case fatality and distribution of iNTS serovars. Surveillance in the same location from 2000 to 2004 documented Enteritidis and Typhimurium as the dominant iNTS serovars [14]. Although shifts in Salmonella serovar prevalence and dominance have been documented in The Gambia and elsewhere in the world [14 15, 16], the genomic characteristics and epidemiological factors responsible for this shift are unclear. We used whole genome sequencing and bioinformatic analyses to investigate changes in pathogen characteristics between 2008 and 2016.
Methods
Disease surveillance
The surveillance methodology has been previously described [17]. We conducted population-based surveillance for invasive bacterial disease in individuals aged 2 months and older resident in the Basse Health and Demographic Surveillance System (BHDSS) in the Upper River Region, The Gambia [17]. Similarly, surveillance criteria included infants aged 0–60 days if there was suspected meningitis, although these infants were not included in calculations of sepsis or pneumonia incidence. We used standardized criteria to identify and investigate patients presenting with suspected pneumonia, septicaemia or meningitis to all health facilities in the study area between 12 May 2008 and 31 December 2016. Blood, cerebrospinal fluid (CSF), and lung aspirates (LA) were collected according to standardized criteria, and we used conventional microbiological methods to culture and identify bacterial pathogens. Gram-negative isolates were identified as Salmonella biochemically using a commercial kit (Analytic Profile Index 20E) and antimicrobial susceptibility testing was done using the disc diffusion method and following CLSI reference thresholds [18].
Domestic animal ownership
Given that NTS also infect domestic animals, they can represent an important route of transmission. Data from the Global Enteric Multicentre Study [19] collected in the study area between 2007 and 2012 were used to compare changes in the prevalence of domestic animal ownership and invasive Salmonella over time.
Sample population
We analysed 164 Salmonella genomes from isolates obtained from blood, CSF or LA samples collected during the surveillance. Genomic DNA was extracted in-house from all isolates and sent to the Wellcome Sanger Institute, UK, for whole genome sequencing.
Quality control, assembly and resistance genes
Extracted DNA was sequenced using the lllumina Hiseq 2500 platform, to produce sequencing reads of 125 bp in FASTQ format [20], with a minimum target depth of coverage of 50×. The reads and genomes were quality checked using FASTQC (v0.11.5) and an in-house pipeline, with manual review. The reads were of high quality with an average Phred score of 30 and thus did not require any trimming. Spades (v3.13.1) was used to perform de novo assembly with default settings [21] to produce draft assemblies in FASTA format. Quast (v5.0.2) [22] was used to assess the quality of assemblies. Contigs shorter than 300 bp were removed from the assemblies as per Page et al. [23]. Four genomes were significantly larger (6 Mb) than the rest of the genomes, indicating contamination, and were therefore removed from the analysis.
We used Abricate (v0.9.8) to identify antimicrobial resistance genes, plasmids and virulence genes for each assembly using the comprehensive antimicrobial resistance database (CARD) [24] (downloaded 24 October 2019), Resfinder [25] (downloaded 10 September 2019), PlasmidFinder [26] (downloaded 10 September 2019) and the virulence factor database (VFDB) [27] (downloaded 18September 2019). A minimum nucleotide identity and coverage of 98 % was used for all databases. Virulence factors universally present in Salmonella were excluded. The multilocus sequence type (MLST) of each draft genome was predicted using mlst (v2.8) with default settings against the S. enterica MLST scheme in the PubMLST database [28].
Phylogenetic analysis
Sequencing reads were mapped to the S. enterica serovar Typhimurium LT2 reference genome (accession number GCF_000006945.2) using Snippy (v4.0.7) with default settings. SNPs from the core genome alignment were used to reconstruct a maximum-likelihood phylogenetic tree using the general time-reversible model with IQTREE (v1.3.11.1) [29] and 1000 bootstraps for branch length. Interactive Tree of Life (ITOL) (v5) [30] was used to visualize and annotate the phylogenetic tree. Where serovars appeared to have developed into an outbreak they were analysed phylogenetically with other isolates from outside our study. In addition, when genotypes (or STs) were identified that were known to be restricted elsewhere in the world, phylogenetic comparisons were made to determine whether they were related.
Pan and accessory genome analysis
We used Prokka (v1.13.3) [31] to annotate and predict coding genes from the assembled genomes using S. Typhimurium LT2 protein sequences from GenBank to provide high-quality species-specific gene name annotation. The resulting GFF3 files were used as input to Roary (v3.13.2) [32] to generate a pan-genome, producing an analysis of the core and accessory genome.
Statistical analysis
Summary statistics were prepared using proportions for categorical and mean/median/range for continuous variables including demographic and baseline characteristics. We used Fisher’s exact test for associations between categorical variables. All data management and statistical analyses were performed using the R statistical package.
Results
Demographic data
Between 2008 and 2016, 22 305 patients were enrolled in the surveillance study with 20 199 microbiological cultures, an average of 2244 per year (range: 1047–2370) (Table 1). In total, 164 Salmonella isolates were obtained from 157 patients. Patient age ranged from 3 days to 42 years with children aged <5 years representing more than 90 % (n=145) of the cases. By sample type, 157 isolates were from blood, six from CSF and one from LA. Six patients had isolates detected from more than one clinical sample type. The baseline patient characteristics are summarized in Table 2.
Table 1.
Numbers of patients enrolled, blood cultures collected and Salmonella isolates detected each year
|
Year |
Total enrolled |
Total blood cultures taken |
Enteritidis |
Typhimurium |
Typhi |
Atypical |
Paratyphi |
Total |
|---|---|---|---|---|---|---|---|---|
|
2008 |
1212 |
1047 |
0 |
0 |
3 |
3 |
1 |
7 |
|
2009 |
2099 |
1898 |
1 |
2 |
1 |
4 |
0 |
8 |
|
2010 |
1869 |
1605 |
6 |
3 |
1 |
2 |
0 |
12 |
|
2011 |
2688 |
2385 |
23 |
2 |
4 |
4 |
0 |
33 |
|
2012 |
2899 |
2592 |
7 |
3 |
7 |
20 |
0 |
37 |
|
2013 |
2580 |
2200 |
2 |
3 |
5 |
17 |
0 |
27 |
|
2014 |
2707 |
2536 |
7 |
3 |
3 |
9 |
0 |
22 |
|
2015 |
3742 |
3566 |
3 |
2 |
2 |
5 |
0 |
12 |
|
2016 |
2509 |
2370 |
0 |
1 |
1 |
4 |
0 |
6 |
|
Total |
22 305 |
20 199 |
49 |
19 |
27 |
68 |
1 |
164 |
Table 2.
Summary of baseline patient characteristics
|
Variable |
Characteristic |
N (%) |
|---|---|---|
|
Sex |
Male |
84 (53.5) |
|
|
Female |
73 (46.5) |
|
Diagnosis |
Pneumonia |
93 (59.2) |
|
|
Meningitis |
11 (7.0) |
|
|
Septicaemia |
46 (29.3) |
|
|
Other focal sepsis |
6 (3.8) |
|
|
Other |
1 (0.6) |
|
Disease outcome |
Dead |
16 (10.2) |
|
|
Discharged and/or recovered |
111 (70.7) |
|
|
Not admitted |
22 (14.0) |
|
|
Absconded |
1 (0.6) |
|
|
Transferred |
6 (3.8) |
|
|
Missing |
1 (0.6) |
|
Age range |
0–5 years |
144 (91.7) |
|
|
6–15 years |
7 (4.5) |
|
|
>15 years |
6 (3.8) |
|
Nutritional status |
Acute malnutrition |
51 (32.5) |
|
|
Moderate acute malnutrition |
32 (20.4) |
|
|
Well nourished |
64 (40.8) |
|
|
Missing |
10 (6.4) |
|
Reside within the surveillance area |
Yes |
136 (86.6) |
|
|
No |
21 (13.4) |
|
Sample type |
Blood |
157 (95.7) |
|
|
Cerebospinal fluid |
6 (3.7) |
|
|
Lung aspirate |
1 (0.6) |
|
Infection rate by serotype |
Enteritidis |
47 (29.9) |
|
|
Typhimurium |
18 (11.5) |
|
|
Typhi |
27 (17.2) |
|
|
Paratyphi C |
1 (0.6) |
|
|
Atypical |
64 (40.8) |
Genomic analysis
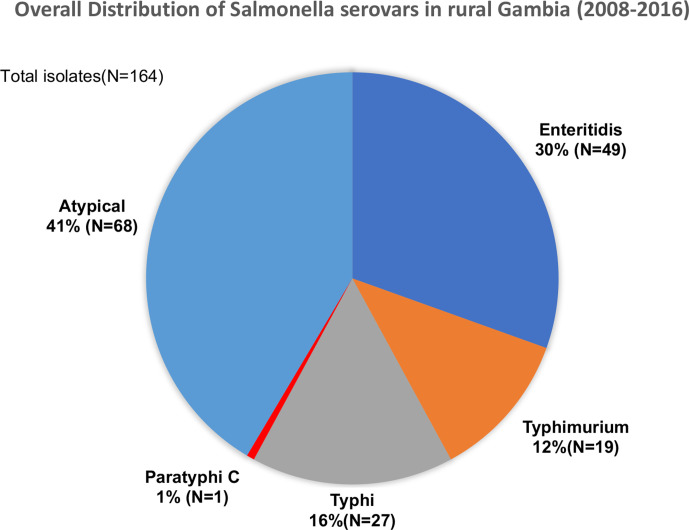
MLST analysis revealed 31 distinct serovars and 45 sequence types (STs). We detected 27 serovars that were not Enteritidis, Typhimurium, Typhi or Paratyphi. We grouped these isolates and called them atypical serovars. A considerable proportion, 41 % (n=68), of isolates were atypical. The atypical serovars most often isolated were Dublin (n=14), Virchow (n=7) and Poona (n=5). Enteritidis, Typhimurium and Typhi constituted 30 % (n=49), 12 % (n=19) and 16 % (n=27) of the isolates, respectively. Only one isolate was S . enterica serovar Paratyphi C of ST3039 (Fig. 1).
Fig. 1.
Breakdown of invasive Salmonella serovars isolated between 2008 and 2016 from patients in rural Gambia.
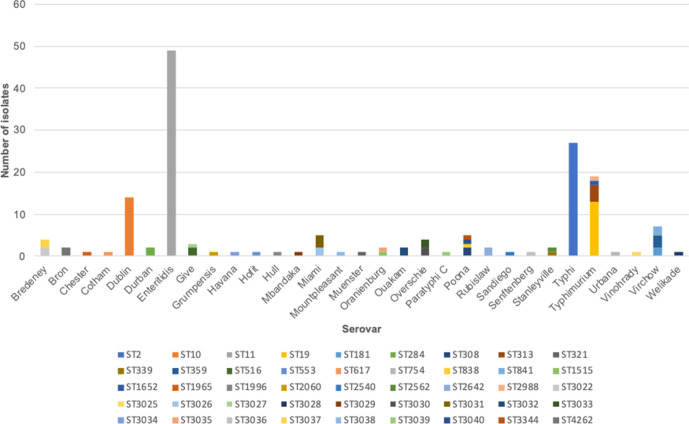
Of all the STs, ST11 was dominant, representing 30 % (n=49) of the isolates, followed by ST2, which accounted for 16 % (n=27). ST10 and ST19 represented 9 % (n=14) and 8 % (n=13) of the isolates, respectively. Other STs included ST313 (n=4), ST3031 (n=3) and ST359 (n=3). Isolates of Typhimurium were represented by four STs: ST19, ST313, ST2988 and ST165. Serovars Virchow and Poona were represented by three and four STs, respectively. Some atypical serovars, including Bredeney, Give, Miami, Oranienburg, Overschie, Poona, Stanleyville and Virchow, were represented by two or more STs each. In contrast, serovars Enteritidis, Typhi and Dublin were represented by only one ST each: ST11, ST2 and S10, respectively (Fig. 2).
Fig. 2.
Representation of STs amongst invasive Salmonella serovars isolated between 2008 and 2016 from patients in rural Gambia.
Distribution of Salmonella serovars over time
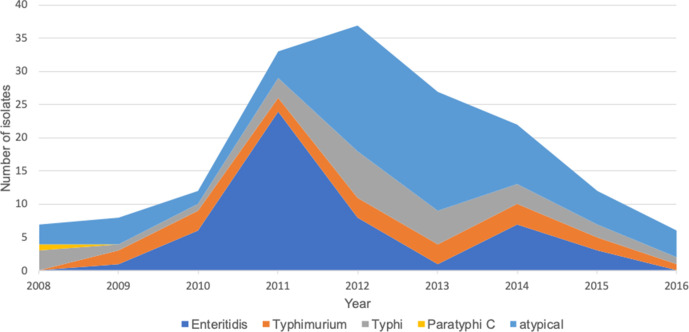
During 2000–2004 serovars Enteritidis (81%) and Typhimurium (8%) were the dominant iNTS serovars [14]. Over the study period, we observed an increase in the proportion of atypical serovars (Fig. 3). In 2008 and 2009, invasive Salmonella infection caused by atypical serovars accounted for most cases compared with infection caused by Enteritidis and Typhimurium. However, this trend changed in 2011 when Enteritidis became predominant and accounted for about 80 % of all Salmonella cases. A high proportion of atypical serovars was then observed between 2012 and 2014. Overall, from 2012 to 2014, atypical serovars were responsible for almost 50 % of Salmonella infections. The major serovars within this group included Dublin, Bredeney, Miami and Overchie. From 2015 to 2016, we observed a further decline in the proportion of Enteritidis and Typhimurium serovars in the population, while atypical serovars were associated with over 50 % of cases.
Fig. 3.
Case counts of each type of invasive Salmonella serovar in Basse, rural Gambia, between 2008 and 2016.
Incidence and case fatality rate
Amongst all cases of invasive Salmonella disease, the case fatality rate was 10 % (16/157). Case fatality for atypical serovars was 10 % (7/68) and 12 % (6/49) for Enteritidis. Typhi, Typhimurium and Paratyphi C were associated with only one death each. Amongst hospitalized patients, Enteritidis and atypical serovars accounted for 42 % (32/77) and 31 % (24/77) of cases while Typhi and Typhimurium accounted for 16 % (12/77) and 13 % (10/77) of cases, respectively. Amongst atypical serovars, those with the CdtB gene were responsible for 10 % (3/31) of all deaths while atypical serovars without the toxin gene accounted for 11 % (4/37) of all deaths.
The majority of the patients (59%) had suspected pneumonia or septicaemia (29%). Of the 46 patients with septicaemia, 26 (56 %) were infected with atypical serovars; Dublin, Overchie, Bredeney and Poona accounted for most of these cases. Overall, we did not find a statistically significant association between malnutrition and any specific serovar, though this should be interpreted with caution due to the small numbers. However, comparing typical vs. atypical serovars, the proportion of children with severe acute malnutrition appeared to be higher in the atypical group 19/32 (59 %) compared to Enteritidis 6/32 (18 %), Typhimurium 3/32 (9 %) or Typhi 4/32 (12 %) groups (P=0.05).
Domestic animal ownership and prevalence of NTS over time
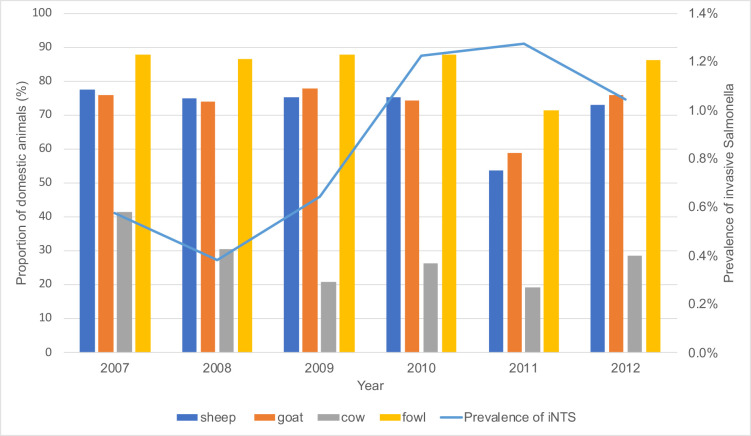
The prevalence of invasive Salmonella increased from 2007 to 2010 while domestic animal ownership by households remained constant throughout this period (Fig. 4).
Fig. 4.
Relationship between invasive Salmonella disease incidence (blue line) and the proportion of different species of domestic animals reared in rural Gambia between 2007 and 2012.
Phylogenetic analysis
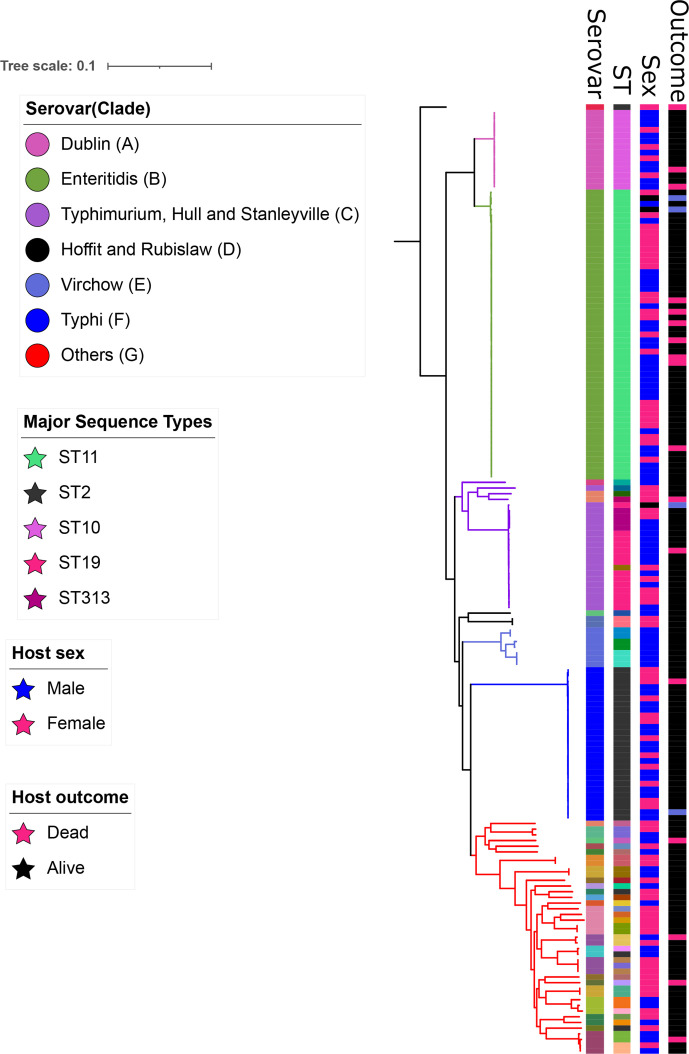
We constructed a pan- Salmonella phylogenetic tree using SNPs generated from 3331 sites in the core genome, excluding repeated regions and transposable elements. The tree resolved seven distinct clades, A–G. Clades A and B comprised Dublin and Enteritidis serovars, respectively. Typhimurium clustered with Hull and Stanleyville in clade C. Clade D included serovars Hofit and Rubislaw while clade E comprised only Virchow isolates. All the Typhi isolates formed a distinct clade, clade F, and the remaining serovars formed a separate clade, clade G (Fig. 5).
Fig. 5.
Maximum-likelihood phylogenetic tree of 164 Salmonella genomes isolated from patients in rural Gambia between 2008 and 2016. Seven distinct clades were resolved from the tree and denoted by different colours (see key). Metadata are shown alongside the phylogenetic tree and include host sex and disease status. The serovars and most prevalent STs are annotated on the tree and denoted using different colours. The tree was rooted on the Salmonella Paratyphi C isolate.
Genomic analysis of Enteritidis isolates
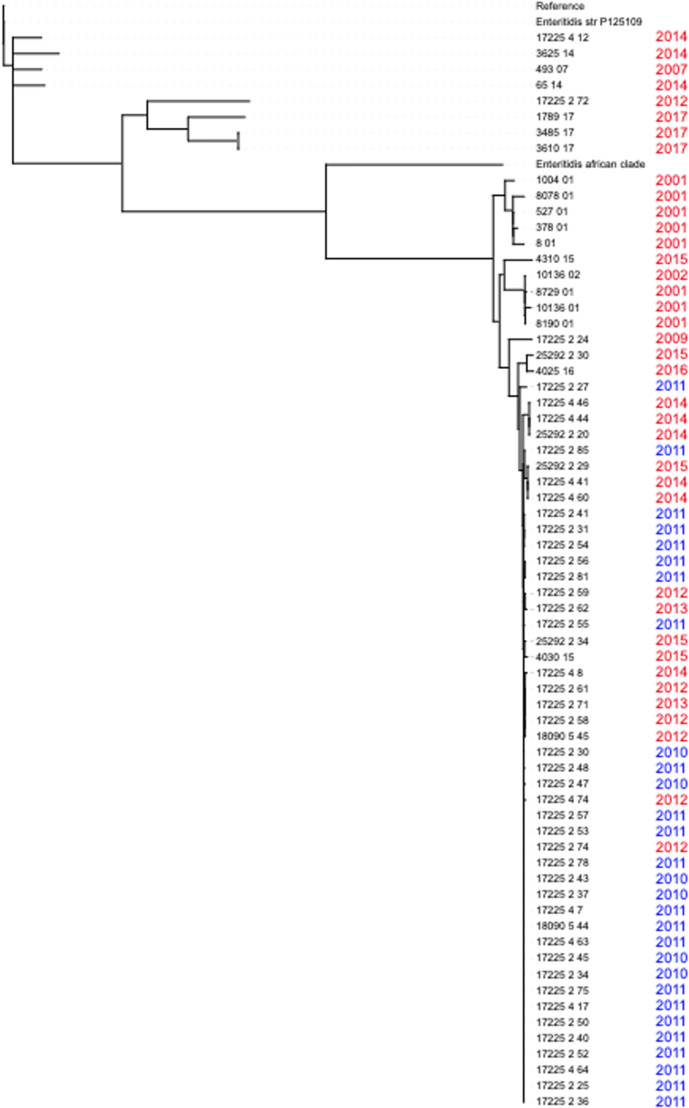
To understand the reason for the high proportion of Enteritidis between 2010 and 2011, we used phylogenetic analysis to compare the 2010 and 2011 Enteritidis genomes in our dataset with Enteritidis genomes collected in The Gambia before and after 2010. This analysis indicated a potential outbreak (Fig. 6) with more than 70 % (21/29) of the Enteritidis isolates collected during the surveillance in 2010 and 2011 clustered closely on the tree with short branch lengths, suggesting closely related strains circulating during this time frame. All the surveillance isolates except two (17225 4 12 and 17225 2 72) clustered with the West African reference clade (here called Enteritidis African Clade).
Fig. 6.
Phylogenetic tree of 49 Salmonella Enteritidis isolates collected during the surveillance period and 16 other isolates collected from The Gambia (both within the surveillance area and outside) at different time points. Isolates collected in the present study between 2010 and 2011 are coloured blue and those collected before or after the surveillance period are coloured red. The tree is rooted on the Salmonella Typhimurium LT2 reference genome.
Genomic analysis of S. Typhimurium ST313 isolates
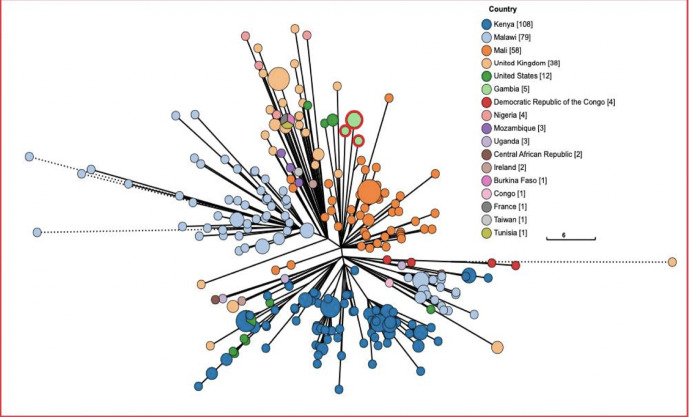
We found that five isolates had the ST313 genotype, which has been implicated as the causative agent of invasive Salmonella disease in Kenya and Malawi. For this reason, we used phylogenetic analysis to compare the ST313 isolates in our study with other global strains in Enterobase [33]. We found that the isolates circulating in The Gambia are of the lineage 1 type and different from the type circulating in Kenya and Malawi, which are of lineage 2 (Fig. 7).
Fig. 7.
Phylogenetic tree of five Salmonella Typhimurium ST313 isolates from our study and all ST313 isolates from other countries (as indicated in the key).
Isolates from our study are highlighted in green with a red ring and are clustered away from the Kenyan (dark blue) and Malawian (sky blue) ST313 strains. The size of the node indicates the number of genomes. Bigger circles represent more genomes.
Distribution of virulence, resistance and plasmid genes
A total of 124 virulence genes within and outside the Salmonella pathogenicity islands (SPIs) were detected. The distribution of virulence genes detected and how they grouped based on the loci present can be found in Table S1 (available in the online version of this article). Some virulence genes were conserved in the Salmonella isolates evaluated while others were only present in some serovars. For example, SPI-7, which encodes vex and tvi genes, was found in Typhi serovars only while SPI-11, which encodes the CdtB gene, was found in several serovars within the atypical group.
Some genes found outside the SPIs, including fimbriae- and adhesin-encoding genes as well as the type 1 fimbriae, were conserved in all isolates. Most of the genes that were variable in their distribution were found residing outside the pathogenicity islands. These genes included Gifsy-1 found in Typhimurium and Paratyphi C serovars only, and Gifsy-2 effector genes found only in Bron, Dublin, Enteritidis, Paratyphi C and Typhimurium isolates. Interestingly, we found 42 % (31/68) of serovars in the atypical group had the virulence gene cdtB and that this gene was present in all our Typhi isolates.
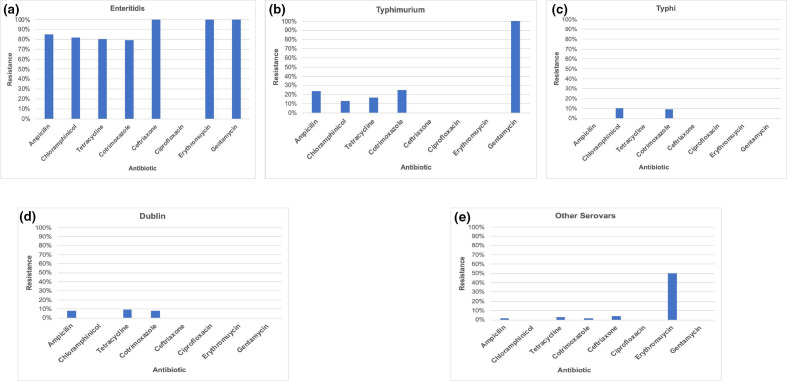
Genomic analysis indicated more antimicrobial resistance genes in Enteritidis than any other serovar. Analysis of phenotypic data showed a similar pattern, with 80–100 % (n=40) of Enteriditis isolates being resistant to all the antimicrobials tested except ciprofloxacin. We observed 100 % (n=40) sensitivity in all Enteriditis isolates tested against ciprofloxacin (Fig. 8a). Some of the resistance genes present in Enteritidis were also found in Typhimurium ST313 isolates but were present in only few of the atypical serovars. All Typhimurium isolates (n=16) tested were resistant to gentamycin. We found only few plasmid genes in our dataset. This was more pronounced in some serovars such as Dublin, Enteritidis and Typhimurium. In fact, none of the Typhi strains had a plasmid gene and only a few of the atypical serovars had one or two plasmids. We found that some plasmids were specific to serovars. For example, IncX1 was found only in Dublin isolates. IncFIIB was common in Typhimurium isolates while Incl1 and IncQ were found in all Enteritidis isolates (see Table 3 for a full summary).
Fig. 8.
Antibiotic resistance patterns in invasive Salmonella serovars isolated in rural Gambia between 2008 and 2016.
Table 3.
Summary of resistance and plasmid genes in each serovar
|
Clade |
Serovar |
Gene name |
Total (%) |
Plasmid genes |
Total (%) |
|---|---|---|---|---|---|
|
A |
Dublin |
fosA7_1 |
1/14 (7.1) |
IncFII(S)_1 |
14/14 (100) |
|
|
|
|
IncI1_1_Alpha |
1/14 (7.1) |
|
|
|
|
|
IncX1_1 |
14/14 (100) |
|
|
B |
Enteritidis |
aph(3'')-Ib_5 |
45/49 (91.8) |
ColpVC_1 |
1/49 (2.1) |
|
|
|
aph(6)6-Id_1 |
45/49 (91.8) |
IncFIB(S)_1 |
2/49 (4.1) |
|
|
|
blaTEM-1B_1 |
49/49 (100) |
IncFII(S)_1 |
2/49 (4.1) |
|
|
|
catA1_1 |
46/49 (93.8) |
IncI1_1_Alpha |
47/49 (95.9) |
|
|
|
dfrA7_5 |
46/49 (93.8) |
IncQ1_1 |
45/49 (91.8) |
|
|
|
sul1_5 |
46/49 (93.8) |
rep21_9_rep(pKH12) |
2/49 (4.1) |
|
|
|
sul2_6 |
45/49 (91.8) |
|
|
|
|
|
tet(B)_2 |
46/49 (93.8) |
|
|
|
C |
Typhimurium |
aph(3'')-Ib_5 |
3/19 (15.8) |
IncFIB(S)_1 |
18/19 (94.7) |
|
|
|
aph(6)6-Id_1 |
3/19 (15.8) |
IncFII(S)_1 |
18/19 (94.7) |
|
|
|
blaTEM-1B_1 |
3/19 (15.8) |
IncQ1_1 |
1/19 (5.3) |
|
|
|
catA1_1 |
3/19 (15.8) |
|
|
|
|
|
dfrA7_5 |
3/19 (15.8) |
|
|
|
|
|
sul1_5 |
3/19 (15.8) |
|
|
|
|
|
sul1_3 |
3/19 (15.8) |
|
|
|
|
|
fosA7_1 |
1/19 (5.3) |
|
|
|
|
Stanleyvelle |
fosA7_1 |
2/2 (100) |
|
|
|
D |
Hofit |
fosA7_1 |
1/1 (100) |
IncFIB(S)_1 |
1/1 (100) |
|
|
|
|
IncFII(S)_1 |
1/1 (100) |
|
|
E |
Virchow |
No gene |
pSL483_1 |
1/7 (14.3) |
|
|
F |
Typhi |
catA1_1 |
2/27 (7.4) |
No plasmid |
|
|
|
|
dfrA7_5 |
2/27 (7.4) |
|
|
|
|
|
sul1_5 |
2/27 (7.4) |
|
|
|
G |
Others: |
|
|
||
|
|
Mountpleasant |
fosA7_1 |
1/41 (100) |
IncFII(S)_1 |
2/41 (4.8) |
|
|
Senftenberg |
fosA7_1 |
1/41 (100) |
IncFII(pCoo)_1_pCoo |
1/41 (2.4) |
|
|
Grumpensis |
fosA7_1 |
1/41 (100) |
|
|
|
|
Paratyphi C |
fosA7_1 |
1/1 (100) |
IncFIB(S)_1 |
1/1 (100) |
|
|
|
|
IncFII(S)_1 |
1/1 (100) |
Discussion
In The Gambia, NTS is an important cause of invasive bacterial infections, especially in children [14, 34–37]. Using population-based epidemiological data and whole genome sequencing, we found an increase in the proportion of atypical NTS serovars causing invasive disease in rural Gambia between 2008 and 2016. We also observed changes in the incidence of disease over time. We identified sets of virulence genes in atypical serovar isolates that may be responsible for the increased prevalence of these serovars.
Few studies have described the distribution of NTS serovars in The Gambia [14, 37]. Between 2000 and 2004, Ikumapayi et al., reported Enteritidis as the major cause of invasive disease in rural Gambia while Typhimurium and other serovars accounted for only a few cases [14]. Interestingly, the present study showed a significant reduction in the proportion of invasive Salmonella disease caused by Enteritidis. To identify serovars, Ikumapayi et al., used conventional antisera agglutination methods while PCR methods were used for MLST [14]. This could underestimate the proportion of some serovars as antisera-based methods are limited in their ability to distinguish between closely related and polyphyletic serovars [38]. By exploiting the advantages of whole genome sequencing, we identified 31 different serovars and thus a greater diversity of Salmonella serovars causing invasive disease. Between 2005 and 2015, Kwambana-Adams et al., reported Typhimurium to be the predominant invasive serovar in the coastal parts of The Gambia [37], with 25 % of isolates being serovars other than Typhi, Typhimurium or Enteritidis. In comparison, our data show temporal and/or regional differences in the prevalence of Salmonella which could be attributed to many factors including host and pathogen genetic characteristics.
Globally, Typhimurium and Enteritidis are the two major serovars associated with invasive Salmonella disease [39, 40]. However, this trend was different in rural Gambia where atypical serovars including Dublin, Virchow and Poona are increasing in prevalence. Studies have shown that genetic factors and immune status predispose individuals to invasive Salmonella disease [4]. For example, malnutrition and HIV have been associated with increased susceptibility to invasive Salmonella disease [41]. However, in The Gambia, the prevalence of malnutrition and HIV has not changed over the years, suggesting that increased incidence of invasive Salmonella disease may be attributable to other environmental factors or the genetic characteristics of the pathogen. We observed an increase in atypical serovars with most cases occurring between 2012 and 2014. However, genomic analysis revealed various virulence factors implicated in invasion, proliferation and or translocation by Type III secretion systems in all Dublin isolates. Between 2012 and 2014, Dublin was the most common serovar isolated within the atypical group. Studies have reported that Dublin is associated with more severe disease and more frequently the cause of invasive disease than other types of non-Typhi Salmonella [42, 43]. The present study reported two deaths associated with the Dublin serovar, ranking second in mortality after Enteritidis.
Moreover, this study identified the cytolethal distending toxin gene (CdtB) in a high percentage of atypical serovars (Clade G). This gene encodes cytolethal distending toxin (CDT) which activates host DNA damage and thus leads to G2/M phase arrest [12]. Analysis of all Salmonella genome assemblies in RefSeq (accessed 26 March 2020) showed overall prevalence of cdtB to be 35 % (3832/10882), and when Typhi is excluded, this falls to 14 % (1628/8678). This shows an uncommonly high level of CdtB in our atypical serovars. Experimental studies show that populations of HeLa cells infected with cytolethal distending toxin-positive NTS serovars have a significantly larger proportion of cells with DNA damage response protein (53BP1) and γH2AX foci than CDT-negative serotypes [12]. More importantly, in vivo analysis showed increased colonization of the host by CDT-producing pathogens that was associated with tumorigenesis and neoplastic lesions that led to chronic infections [12]. Thus, we speculate that increased prevalence of cdtB genes in our study may provide these serovars with a fitness advantage over Enteritidis and Typhimurium, potentially contributing to the shift we observed.
In contrast, we observed a high proportion of Enteritidis between 2010 and 2011. This period coincided with heavy rains resulting in severe flooding in the Upper River Region. Subsequent high rates of malaria infection may have influenced the population’s susceptibility to iNTS disease. Phylogenetic analysis of the Enteritidis isolates suggests a potential outbreak. All Enteritidis isolates recovered during this period were isolated within the Basse area with similar virulence and antimicrobial resistance patterns. A study in Mali highlighted that, in contrast to Salmonella Typhimurium, iNTS disease caused by Salmonella Enteritidis started to increase from 2008 with the highest peak seen in 2010 and 2011 [16]. The finding in Mali corresponds with our observed increase in Enteritidis in 2010 and 2011, suggesting the potential combination of a regional increase in Enteritidis exacerbated by the impact of the flood in our setting.
Antibiotic resistance in some Salmonella serotypes has been reported in many parts of Africa including The Gambia [14, 44]. Our Enteritidis serovars had more resistance genes than other serovars. Similar findings were also reported in previous studies done in The Gambia which showed high percentages of multidrug resistance among Salmonella Enteritidis isolates [14]. However, five of our Typhimurium isolates of the ST313 genotype had resistance genes like those found in Enteritidis. In Kenya and Malawi, a distinct genotype of Typhimurium ST313 was reported to have a multidrug resistance gene located on a virulence plasmid [11]. Genomic analysis of all ST313 isolates in our study and those found in Enterobase suggests that this unique Typhimurium ST313 is restricted to eastern Africa. Nonetheless, continued monitoring of these genotypes in other parts of Africa is vital. It is, however, reassuring that many of the atypical serovars did not acquire resistance genes, although continued monitoring is essential as antimicrobial resistance is increasing, and has a high global health burden.
Limitations
The annual number of blood cultures performed was not constant and varied due to variable numbers of patients presenting to health facilities each year. Thus, annual counts of events were subjected to sampling bias with outbreaks, most notably meningococcus group W135 in 2012 and Respiratory syncytial virus (RSV)-related lower respiratory infections in 2015.
Plasmids were screened using short reads, which are difficult to reconstruct due to many repeats and are sometimes found in the chromosomal DNA [45]. Thus, long read sequencing such as Oxford Nanopore or PacBio is most efficient as repetitive regions can be spanned by long reads generated by these technologies [46].
Conclusion
Overall, this study has shown a wide distribution of invasive Salmonella serovars circulating in The Gambia. More importantly, an increase over time in atypical serovars with high case fatality rates was also documented. The study highlighted the potential effect of some virulence genes in contributing to the shift we observed. However, experimental and functional studies could shed more light on the role of such virulence genes and the evolutionary pressures on these serovars. The shift in serovar prevalence could have implications for vaccine development and thus represent a public health concern. Therefore, investigations need to identify potential changes in the distribution of iNTS serovars elsewhere in Africa and the prevalence of these virulence elements.
Supplementary Data
Funding information
The surveillance study was sponsored by GAVI’s Pneumococcal vaccines Accelerated Development and Introduction Plan (PneumoADIP), the Bill and Melinda Gates Foundation, and the UK Medical Research Council. A.J.P. and N.F.A. gratefully acknowledge the support of the Biotechnology and Biological Sciences Research Council (BBSRC); this research was funded by the BBSRC Institute Strategic Programme Grant Microbes in the Food Chain BB/R012504/1 and its constituent project(s) BBS/E/F/000PR10348 and BBS/E/F/000PR10352. A.K. was partially supported by a BBSRC Impact Acceleration Account award.
Acknowledgements
We thank Gordon Dougan and team for sequencing the isolates, Thanh Le Viet for bioinformatics support, Abdul Khalid Muhammad and Nuredin Mohammed for statistics support and Jahangir Hussain for providing GEMS domestic data. A.K. also wishes to acknowledge the management of MRCG at LSHTM for partly funding his internship at Quadram while working on this project.
Author contributions
A.K. and G.M. conceived the research idea and A.K. wrote the first draft of the manuscript. A.K., A.P. and N.F.A. did the bioinformatics analysis. U.N.I., R.S. and J.M. did the microbiology. G.D. and team did the sequencing. A.K.S. supervised A.K. and reviewed the manuscript. All authors have read and approved the final version of the manuscript.
Conflicts of interest
The author(s) declare that there are no conflicts of interest.
Ethical statement
The parent project consented participants (numbers 1087 and 1411) before enrolling them in the study. This study does not require any ethical approval.
Footnotes
Abbreviations: CDT, cytolethal distending toxin; CSF, cerebrospinal fluid; HIV, human immunodeficiency virus; iNTS, invasive non-typhoidal Salmonella; LA, lung aspirates; MLST, multilocus sequence typing; NTS, non-typhoidal Salmonella; SPI, Salmonella pathogenicity island; ST, sequence type.
All supporting data, code and protocols have been provided within the article or through supplementary data files. One supplementary table is available with the online version of this article.
References
- 1.Kingsley RA, Bäumler AJ. Host adaptation and the emergence of infectious disease: The Salmonella paradigm. Mol Microbiol. 2000;36:1006–1014. doi: 10.1046/j.1365-2958.2000.01907.x. [DOI] [PubMed] [Google Scholar]
- 2.Popoff MY, Bockemühl J, Gheesling LL. Supplement 2002 (no.46) to the Kauffmann-White scheme. Res Microbiol. 2004;155:568–570. doi: 10.1016/j.resmic.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, et al. The global burden of nontyphoidal salmonella gastroenteritis. Clin Infect Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 4.Gordon M. Europe PMC funders group invasive non-typhoidal salmonella disease – epidemiology, pathogenesis and diagnosis. Curr Opin Infect Dis. 2012;24:484–489. doi: 10.1097/QCO.0b013e32834a9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uche IV, MacLennan CA, Saul A. A systematic review of the incidence, risk factors and case fatality rates of invasive nontyphoidal salmonella (INTS) disease in Africa (1966 to 2014) PLoS Negl Trop Dis. 2017;11:1–28. doi: 10.1371/journal.pntd.0005118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon MA, Graham SM, Walsh AL, Wilson L, Phiri A, et al. Epidemics of invasive Salmonella enterica serovar enteritidis and S. enterica Serovar typhimurium infection associated with multidrug resistance among adults and children in Malawi. Clin Infect Dis. 2008;46:963–969. doi: 10.1086/529146. [DOI] [PubMed] [Google Scholar]
- 7.Berkley JA, Lowe BS, Mwangi I, Williams T, Bauni E, et al. Bacteremia among children admitted to a rural hospital in Kenya. N Engl J Med. 2005;352:39–47. doi: 10.1056/NEJMoa040275. [DOI] [PubMed] [Google Scholar]
- 8.Kiehlbauch JA, Vugia DJ, Yeboue K, N’Gbichi JM, Lacina D, et al. Pathogens and Predictors of Fatal Septicemia Associated with Human Immunodeficiency Virus Infection in Ivory Coast, West Africa. J Infect Dis. 1993;168:564–570. doi: 10.1093/infdis/168.3.564. [DOI] [PubMed] [Google Scholar]
- 9.Gordon MA. Salmonella infections in immunocompromised adults. J Infect. 2008;56:413–422. doi: 10.1016/j.jinf.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 10.Darboe S, Okomo U, Muhammad AK, Ceesay B, Jallow M, et al. Community-acquired invasive bacterial disease in urban Gambia, 2005-2015: A hospital-based surveillance. Clin Infect Dis. 2019;69:13. doi: 10.1093/cid/ciz463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kingsley RA, Msefula CL, Thomson NR, Kariuki S, Holt KE, et al. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 2009;19:2279–2287. doi: 10.1101/gr.091017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller RA, Wiedmann M. The cytolethal distending toxin produced by nontyphoidal Salmonella serotypes javiana, montevideo, oranienburg, and mississippi induces DNA damage in a manner similar to that of serotype Typhi. MBio. 2016;7:1–11. doi: 10.1128/mBio.02109-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge Z, Feng Y, Whary MT, Nambiar PR, Xu S, et al. Cytolethal distending toxin is essential for Helicobacter hepaticus colonization in outbred Swiss Webster mice. Infect Immun. 2005;73:3559–3567. doi: 10.1128/IAI.73.6.3559-3567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikumapayi UN, Antonio M, Sonne-Hansen J, Biney E, Enwere G, et al. Molecular epidemiology of community-acquired invasive non-typhoidal Salmonella among children aged 2-29 months in rural Gambia and discovery of a new serovar. J Med Microbiol. 2007;56:1479–1484. doi: 10.1099/jmm.0.47416-0. [DOI] [PubMed] [Google Scholar]
- 15.Le Bacq F, Louwagie B, Verhaegen J. Salmonella typhimurium and Salmonella enteritidis: Changing epidemiology from 1973 until 1992. Eur J Epidemiol. 1994;10:367–371. doi: 10.1007/BF01719658. [DOI] [PubMed] [Google Scholar]
- 16.Bornstein K, Tennant SM, Hazen TH, Sorkin JD, Tapia MD, et al. Genetic changes associated with the temporal shift in invasive non-typhoidal salmonella serovars in Bamako Mali. PLoS Negl Trop Dis. 2019;13:1–15. doi: 10.1371/journal.pntd.0007297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackenzie GA, Plumb ID, Sambou S, Saha D, Uchendu U, et al. Monitoring the introduction of pneumococcal conjugate vaccines into West Africa: Design and implementation of a population-based surveillance system. PLoS Med. 2012;9:e1001161. doi: 10.1371/journal.pmed.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolinsky AL. M100 Performance Standards for Antimicrobial Susceptibility Testing. Journal of Services Marketing. 2017;Vol. 8:27–39. [Google Scholar]
- 19.Kotloff KL, Nasrin D, Blackwelder WC, Wu Y, Farag T, et al. The incidence, aetiology, and adverse clinical consequences of less severe diarrhoeal episodes among infants and children residing in low-income and middle-income countries: a 12-month case-control study as a follow-on to the Global Enteric Multicenter St. Lancet Glob Heal. 2019;7:84. doi: 10.1016/S2214-109X(19)30076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cock PJA, Fields CJ, Goto N, Heuer ML, Rice PM. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2009;38:1767–1771. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Page AJ, De Silva N, Hunt M, Quail MA, Parkhill J, et al. Robust high-throughput prokaryote de novo assembly and improvement pipeline for illumina data. Microb genomics. 2016;2:e000083. doi: 10.1099/mgen.0.000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:73. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carattoli A, Zankari E, Garciá-Fernández A, Larsen MV, Lund O, et al. In Silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu B, Zheng D, Jin Q, Chen L, Yang J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47:92. doi: 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jolley KA, Maiden MCJ. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letunic I, Bork P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019;47:256–259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 32.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alikhan NF, Zhou Z, Sergeant MJ, Achtman M. A genomic overview of the population structure of Salmonella. PLoS Genet. 2018;14:1–13. doi: 10.1371/journal.pgen.1007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enwere G, Biney E, Cheung Y, Zaman SMA, Okoko B, et al. Epidemiologic and clinical characteristics of community-acquired invasive bacterial infections in children aged 2-29 months in The Gambia. Pediatr Infect Dis J. 2006;25:700–705. doi: 10.1097/01.inf.0000226839.30925.a5. [DOI] [PubMed] [Google Scholar]
- 35.Okomo UA, Garba D, Fombah AE, Secka O, Ikumapayi UNA, et al. Bacterial Isolates and Antibiotic Sensitivity among Gambian Children with Severe Acute Malnutrition. Int J Pediatr. 2011;2011:1–8. doi: 10.1155/2011/825123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hill PC, Onyeama CO, Ikumapayi UNA, Secka O, Ameyaw S, et al. Bacteraemia in patients admitted to an urban hospital in West Africa. BMC Infect Dis. 2007;7:1–8. doi: 10.1186/1471-2334-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwambana-Adams B, Darboe S, Nabwera H, Foster-Nyarko E, Ikumapayi UN, et al. Salmonella infections in The Gambia, 2005-2015. Clin Infect Dis. 2015;61:62. doi: 10.1093/cid/civ781. [DOI] [PubMed] [Google Scholar]
- 38.Shi C, Singh P, Ranieri ML, Wiedmann M, Moreno Switt AI. Molecular methods for serovar determination of Salmonella. Crit Rev Microbiol. 2015;41:309–325. doi: 10.3109/1040841X.2013.837862. [DOI] [PubMed] [Google Scholar]
- 39.Galanis E, Lo Fo Wong DMA, Patrick ME, Binsztein N, Cieslik A, et al. Web-based surveillance and global Salmonella distribution, 2000-2002. Emerg Infect Dis. 2006;12:381–388. doi: 10.3201/eid1205.050854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stanaway JD, Parisi A, Sarkar K, Blacker BF, Reiner RC, et al. The global burden of non-typhoidal salmonella invasive disease: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis. 2019;19:1312–1324. doi: 10.1016/S1473-3099(19)30418-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bronzan RN, Taylor TE, Mwenechanya J, Tembo M, Kayira K, et al. Bacteremia in Malawian children with severe malaria: Prevalence, etiology, HIV coinfection, and outcome. J Infect Dis. 2007;195:895–904. doi: 10.1086/511437. [DOI] [PubMed] [Google Scholar]
- 42.Suez J, Porwollik S, Dagan A, Marzel A, Schorr YI, et al. Virulence gene profiling and pathogenicity characterization of non-typhoidal salmonella accounted for invasive disease in humans. PLoS One. 2013;8:e58449. doi: 10.1371/journal.pone.0058449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feldman MJB, Salmonella Bacteremia RA. Reports to the Centers for Disease Control, 1968-1979 author (s): Martin J. Blaser and Roger A. Feldman published by: Oxford University Press Stable. J Infect Dis. 1981;143:743–746. doi: 10.1093/infdis/143.5.743. [DOI] [PubMed] [Google Scholar]
- 44.Feasey NA, Hadfield J, Keddy KH, Dallman TJ, Jacobs J, et al. Distinct Salmonella Enteritidis lineages associated with enterocolitis in high-income settings and invasive disease in low-income settings. Nat Genet. 2016;48:1211–1217. doi: 10.1038/ng.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arredondo-alonso S, Willems RJ, Van SW, Schürch AC. On the (im) possibility of reconstructing plasmids from whole- genome short-read sequencing data. Microb Genom. 2017;3:e000128. doi: 10.1099/mgen.0.000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karlsson E, Lärkeryd A, Sjödin A, Forsman M, Stenberg P. Scaffolding of a bacterial genome using minion nanopore sequencing. Nat Publ Gr. 2015:1–8. doi: 10.1038/srep11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.