Abstract
Background
The human protein transmembrane protease serine type 2 (TMPRSS2) plays a key role in SARS-CoV-2 infection, as it is required to activate the virus’ spike protein, facilitating entry into target cells. We hypothesized that naturally-occurring TMPRSS2 human genetic variants affecting the structure and function of the TMPRSS2 protein may modulate the severity of SARS-CoV-2 infection.
Methods
We focused on the only common TMPRSS2 non-synonymous variant predicted to be damaging (rs12329760 C>T, p.V160M), which has a minor allele frequency ranging from 0.14 in Ashkenazi Jewish to 0.38 in East Asians. We analysed the association between the rs12329760 and COVID-19 severity in 2,244 critically ill patients with COVID-19 from 208 UK intensive care units recruited as part of the GenOMICC (Genetics Of Mortality In Critical Care) study. Logistic regression analyses were adjusted for sex, age and deprivation index. For in vitro studies, HEK293 cells were co-transfected with ACE2 and either TMPRSS2 wild type or mutant (TMPRSS2V160M). A SARS-CoV-2 pseudovirus entry assay was used to investigate the ability of TMPRSS2V160M to promote viral entry.
Results
We show that the T allele of rs12329760 is associated with a reduced likelihood of developing severe COVID-19 (OR 0.87, 95%CI:0.79–0.97, p = 0.01). This association was stronger in homozygous individuals when compared to the general population (OR 0.65, 95%CI:0.50–0.84, p = 1.3 × 10−3). We demonstrate in vitro that this variant, which causes the amino acid substitution valine to methionine, affects the catalytic activity of TMPRSS2 and is less able to support SARS-CoV-2 spike-mediated entry into cells.
Conclusion
TMPRSS2 rs12329760 is a common variant associated with a significantly decreased risk of severe COVID-19. Further studies are needed to assess the expression of TMPRSS2 across different age groups. Moreover, our results identify TMPRSS2 as a promising drug target, with a potential role for camostat mesilate, a drug approved for the treatment of chronic pancreatitis and postoperative reflux esophagitis, in the treatment of COVID-19. Clinical trials are needed to confirm this.
Keywords: SARS-CoV-2, COVID-19, TMPRSS2, Targeting the host to prevent COVID19 severity
Introduction
The severe acute respiratory syndrome like coronavirus (SARS-CoV-2) has infected over 190 million individuals globally and has caused more than 4.2 million deaths as of August 2021 [1]. SARS-CoV-2 infection has a broad clinical spectrum, ranging from asymptomatic or mildly symptomatic, to a life-threatening presentation requiring admission to intensive care. Age, and to a much lesser extent male gender and various underlying clinical conditions, such as cardiovascular disease, obesity and diabetes, are known risk factors associated with an increased COVID-19 morbidity and mortality [2,3]. The role of an individual's genetic background has recently emerged as an additional, yet not clearly understood, risk factor for COVID-19 [4], [5], [6]. Rare genetic variants in genes involved in the regulation of type I interferon (IFN) immunity, including autosomal recessive IRF7 and IFNAR1 deficiencies, have been identified in patients with life-threatening COVID-19 [6]. Autoantibodies to type I IFNs also account for at least 10% of cases of critical COVID-19 pneumonia[7]. Genome-wide association studies (GWAS) have discovered genetic haplotypes spanning several genes that are associated with COVID-19 severity [3,4,8].
The transmembrane protease serine type 2 (TMPRSS2) protein plays a key role in coronavirus infections [9], [10], [11], including SARS-CoV-2, as it is required for priming the virus’ spike (S) glycoprotein through its cleavage, thus facilitating endosome-independent entry into target cells [12,13]. TMPRSS2, which is part of the type 2 transmembrane serine protease (TTSP) family, is characterized by androgen receptor elements located upstream to its transcription site [14]. As well as cleaving and activating viral glycoproteins of coronaviruses and influenza A and B viruses [15], TMPRSS2 is subject to autocleavage, which results in the liberation of its soluble catalytic domain [16]. The conditions under which autocleavage of TMPRSS2 and other members of the TTSPs family occurs are yet to be elucidated.
TMPRSS2 is expressed in lung and bronchial cells [17], but also in the colon, stomach, pancreas, salivary glands and numerous other tissues [18]. Moreover, it is co-expressed in bronchial and lung cells with the angiotensin-converting enzyme 2 (ACE2) [17], which is the best described SARS-CoV-2 cellular receptor [19]. In the olfactory epithelium of mice, the expression of TMPRSS2, but not ACE2, appears to be age-related and greater in older compared to younger animals [20]. Similarly, a recent study showed that expression of TMPRSS2 in mouse and human lung tissue is also age-related [21]. Studies in TMPRSS2 knock out (KO) mice reported reduced SARS-CoV and MERS-CoV replication in the lungs compared to wild-type mice, and a reduced proinflammatory viral response, especially cytokine and chemokine release via the Toll-like receptor 3 pathway [22,23]. We have recently shown that TMPRSS2 expression permits cell surface entry of SARS-CoV-2, allowing the virus to bypass potent endosomal restriction factors [24]. In vitro studies have shown that TMPRSS2 inhibitors prevent primary airway cell and organoid infection by SARS-CoV and SARS-CoV-2 [25,24,26]. In animal studies, mice infected with SARS-CoV and treated with the serine protease inhibitor camostat mesilate had a high survival rate [27]. Recently, camostat mesilate (which, in Japan, is already approved for patients with chronic pancreatitis and postoperative reflux esophagitis) was shown to block SARS-CoV-2 lung cell infection in vitro [12,24]. Furthermore, camostat mesylate and its metabolite GBPA have been shown to block SARS-CoV-2 spread in human lung tissue ex vivo [28]. Several clinical trials using camostate in COVID-19 patients are currently underway [29].
In view of the data from animal models and cell-based studies supporting a protective role of a knock out TMPRSS2 on coronavirus infection (including SARS and MERS), we hypothesized that naturally-occurring TMPRSS2 genetic variants affecting the structure and function of the TMPRSS2 protein may modulate the severity of SARS-CoV-2 infection.
Methods
TMPRSS2 three-dimensional structure and variant analysis
The recently released 3D structure of TMPRSS2 (PDB: 7meq) was used to assess the impact missense variants. The Phyre homology modelling algorithm [30] was used to resolve missing amino acid regions in the SRSC domain that were not experimentally solved (described in Supplementary material). The FASTA sequence of TMPRSS2 was obtained from the UniProt protein knowledge database [31] (UniProt Id O15393, corresponding to 492 amino acid transcript Ensembl ID ENST00000332149.10). The recently released AlphaFold model [32] was compared to the Phyre model. The impact of each missense variant on the TMPRSS2 protein structure was assessed by analysing the following 16 features, using our in-house algorithm Missense3D [33]: breakage of a disulfide bond, hydrogen bond or salt bridge, introduction of a buried proline, clash, introduction of hydrophilic residue, introduction of a buried charged residue, charge switch in a buried residue, alteration in secondary structure, replacement of a charged with uncharged buried residue, introduction of a disallowed phi/psi region, replacement of a buried glycine with any other residue, alteration in a cavity, replacement of cis proline, buried to exposed residue switch, replacement of a glycine located in a bend. In addition, we used the SIFT [34] and Polyphen2 [35] variant predictors, which mainly use evolutionary conservation to assess the effect of each variant. The effect of variant rs12329760 was further assess using: i. CONDEL [36], which reports a weighted average of the scores from fatHMM and MutationAssessor, and ii. FoldX5 force field [37], which calculates the stability of a protein based on the estimation of its free energy. A ΔΔG> 0.5 kcal/mol (calculated as: ΔΔG= ΔGwt - ΔGmut) was predicted to have a destabilizing effect.
Participants
Genetics Of Mortality In Critical Care (GenOMICC) and the International Severe Acute Respiratory Infection Consortium (ISARIC) Coronavirus Clinical Characterisation Consortium (4C) (ISARIC 4C)
Cases: this cohort was established between March 2020 and July 2020 (first COVID-19 wave) and comprises of 2244 critically ill, hospitalized COVID-19 positive patients from 208 UK intensive care units (ICUs): 2109, patients were recruited as part of the GenOMICC project, and an additional 135 cases as part of the International Severe Acute Respiratory Infection Consortium (ISARIC) Coronavirus Clinical Characterisation Consortium (4C) study. The clinical characteristics and comorbidities of these patients have been extensively reported in Pairo-Castineira et al. [8]. Only unrelated individuals (up to 3rd degree, based on kinship analysis (King 2.1)) were included. Samples were excluded if the genotype-based sex inference did not match the reported sex, or if a XXY karyotype was present. Moreover, patients of mixed genetic ancestry, and from ancestry groups with small numbers of cases (such as North American Indian, n = 13) defined using admixture supervised mode with 1000 genomes as reference, were excluded.
Controls: ancestry-matched controls (ratio 1 case to 5 controls) without a positive COVID-19 test were obtained from the UK BioBank population study. COVID-19 test results in BioBank are obtained from Public Health England, Public Health Scotland and SAIL for English, Scottish and Welsh data, respectively. The vast majority of results are from nose/throat swabs analysed by PCR. For patients admitted to hospital, results can also be from samples obtained from the lower respiratory tract. Only unrelated individuals (up to 3rd degree) were included. Individuals with sex mismatch were excluded. For validation, 45,875 unrelated individuals of European ancestry from the 100,000 Genomes Project were used as an alternative control group.
DNA extraction, genotyping and quality control have been described in detail previously[8]. Genetic ancestry was inferred using ADMIXTURE and reference individuals from the 1000 Genomes project. Imputation was performed using the TOPMed reference panel.
Cells, pseudovirus and plasmid
Human embryonic kidney 293T cells (293Ts; ATCC) were maintained in Dulbecco's modified Eagle's medium (DMEM), 10% foetal calf serum (FCS), 1% non-essential amino acids (NEAA), 1% penicillin-streptomycin (P/S). Human Caco-2 (ATCC HTB-37) and Calu-3 (ATCC HTB-55) were maintained in DMEM, 20% FCS, 1% NEAA, 1% P/S. All cell lines were maintained at 37 °C, 5% CO2.
Lentiviral pseudotype production was performed as previously described[24],[38]. Briefly, pseudovirus was generated by co-transfecting 293Ts with lentiviral packaging constructs pCSFLW (minimal HIV genome with firefly luciferase reporter), pCAGGs-GAGPOL (HIV packing proteins) and the relevant viral glycoprotein in pcDNA3.1 – either the G glycoprotein from Indiana vesiculovirus (VSV-G) or SARS-CoV-2 spike protein. Co-transfections were performed at a plasmid ratio of 1.5:1:1 for pCSFLW:GAGPOL:glycoprotein. Pseudovirus was harvested at 48 and 72 h post-transfection, pooled, filtered, then frozen down. ACE2 FLAG was used as previously described [24]. TMPRSS2 expression plasmid was a kind gift from Roger Reeves (Addgene plasmid #53,887; http://n2t.net/addgene:53887; RRID:Addgene_53,887) [39]. Non-cleavable ACE2-FLAG and TMPRSS2 mutants were generated by overlap extension PCR or site-directed mutagenesis.
Phenotypic assays
293Ts were co-transfected with FLAG-tagged, non-cleavable ACE2 and TMPRSS2, as previously described [24]. Briefly, confluent 10cm2 dishes of 293T cells were co-transfected with 1 µg each of TMPRSS2 and ACE2-FLAG. 24 h later, cells were resuspended in fresh media and either spun down for lysis and western blot or added to 96 well plates along with pseudovirus. 24 h later, media was refreshed and a further 24 h later, cells were lysed with reporter lysis buffer (Promega), and luminescence (measured as relative luminescence units, RLU) was read on a FLUOstar Omega plate reader (BMF Labtech) using the Luciferase Assay System (Promega).
Cell pellets for western blot were lysed in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM TRIS, pH 7.4) supplemented with an EDTA-free protease inhibitor cocktail tablet (Roche). Cell lysates were combined with 4x Laemmli buffer (Bio-Rad) with 10% β-mercaptoethanol and boiled for 5 min. Membranes were probed with mouse anti-tubulin (abcam; ab7291), rabbit anti-TMPRSS2 (abcam; ab92323) and/or mouse anti-FLAG (F1804, Sigma). Near infra-red (NIR) secondary antibodies, IRDye® 680RD Goat anti-mouse (abcam; ab216776) and IRDye® 800CW Goat anti-rabbit (abcam; ab216773) were subsequently used. Blots were imaged using the Odyssey Imaging System (LI-COR Biosciences). Densitometry was performed using ImageJ.
Reagent, cell lines and antibody validation
All reagents, cell lines and antibodies used in this study are commercially available and validation data are available on the manufacturers’ websites.
Statistical analysis
Sample size: Critically ill Covid-19 patients, n = 2,244; random controls matched by ancestry from UK Biobank, n = 11,220. The sample size was determined pragmatically by the number of cases recruited during the first wave of the outbreak in the UK (as described in [8]). No randomization was performed. Blinding was not used in this study because the exposure (genotype) and outcome (ICU admission) are objective. Confounding was controlled by the use of covariates: age, sex, deprivation score and genetic ancestry [8]. The association between the TMPRSS2 rs12329760 variant and COVID-19 severity was assessed using logistic regression. Genetic associations in the GenOMICC/ISARIC 4C cohort were analysed as previously described[8]. Briefly, logistic regression with additive and recessive models was performed in PLINKv1.9, adjusting for sex, age, mean-centred age-squared, top 10 principal components (principal component analysis [PCA] performed to adjust for population stratification) and deprivation index decile based on UK postcode. Each major ancestry group alternative in the 100,000 Genomes control group was performed with mixed model association tests in SAIGE (v0.39) [40], including age, sex, age-squared, age-sex interaction and the first 20 principal components as covariates. Trans-ethnic meta-analysis of GenOMICC data for different ancestries was performed by METAL using an inverse-variance weighted method and the P-value for heterogeneity was calculated with Cochran's Q-test for heterogeneity implemented in the same software [41].
Additional publicly available genetic data were obtained from the COVID-19 Host Genetics Initiative meta-analyses, release 6 (June 15, 2021) [42]. The COVID-19 Host genetics initiative classifies COVID-19 severity according to the use of invasive and non-invasive ventilation during hospital admission. Here we report the four different phenotype comparisons:
-
•
A2: 8,779 critically ill confirmed cases (inclusion criteria: hospitalized for COVID-19 and either death or on respiratory support including intubation, CPAP, BiPAP, continue external negative pressure, Optiflow/very high flow Positive End Expiratory Pressure Oxygen) versus 1,001,875 population controls,
-
•
B1: 14,480 hospitalised cases versus 73,191 non-hospitalised cases,
-
•
B2 24,274 hospitalised cases versus 2,061,529 population controls, and
-
•
C2: 112,443 COVID-19 cases of unspecified severity versus 2,473,889 population controls.
Analyses used all data with the exclusion of the 23&Me study, for which full data were not publicly available. Meta-analysis in all cases was performed using a fixed effect, inverse variance-weighted model, either as a trans-ethnic meta-analysis or subsetted by ancestry group.
Data are presented as mean ± standard deviation. Log-normality was assessed using the Shapiro-Wilk test and QQ plot. A two-tailed Student's t-test was used to compare the means of two groups. One-way ANOVA was used to compare the means of more than two groups.
Colocalisation analysis
Colocalisation analysis for genetic associations was performed by an Approximate Bayes Factor approach using the package coloc version 5, in R 4.1.0 [43]. Summary statistics (beta and variance) were from GWAS data [8] and from lung eQTL data from GTex v8 [44], in individuals of European ancestry. To reduce the likelihood of violation of the single causal variant assumption arising from multiple independent association signals, the analysis was restricted to a region extending to 5 kb upstream and downstream of the TMPRSS2 gene. With the assumption that exactly one measured SNP in the region was causative for each trait, SNP-level priors (p1 and p2) of 1/(n SNPs) were used for the probability of association with each individual trait, with an arbitrary prior of 0.1 x p1 for p12, the SNP-level prior probability of association with both traits. Sensitivity analysis was performed to assess the impact of prior selection, comparing the selected priors to the more stringent default priors (10−4 for p1 and p2, 10−5 for p12), and varying the p12 range from p1 to p1 x p2.
Ethics
Research ethics committees (Scotland 15/SS/0110, England, Wales and Northern Ireland: 19/WM/0247). Current and previous versions of the study protocol are available at genomicc.org/protocol. All participants gave informed consent.
Role of funders
The Wellcome Trust, UKRI, MRC/UKRI, Howard Hughes Medical Institute, Rockefeller University, St. Giles Foundation, Fisher centre for Alzheimer's Research Foundation, Meyer Foundation, Square Foundation, Grandir Fonds de solidarité pour l'enfance, SCOR Corporate Foundation for Science, Institut National de la Santé et de la Recherche Médicale (INSERM), University of Paris, National Institutes of Health, French Foundation for Medical Research, FRM and French National Research Agency (ANR) GENCOVID, Agence Nationale de la Recherche, Health Data Research UK and BBSRC provided funding to support the salaries of the authors but had no role in the design, data collection, analysis, interpretation of the results or writing of the report. The content of this publication is solely the responsibility of the authors.
Results
We extracted 377 TMPRSS2 genetic variants reported as loss of function (LoF), missense or inframe and indel in the database of population genetic variations GnomAD (v2.1.1). All variants passed quality filters in GnomAD. Nine variants flagged by GnomAD as carrying dubious annotation or quality were excluded. Forty variants were loss of function or indel and the remaining 328 were missense. All variants except for one (rs12329760 C>T) were very rare (MAF <0.001). We focused on missense variants and studied the evolutionary conservation of TMPRSS2 amino acids and the impact of amino acid substitution on TMPRSS2 protein structure (described in Methods). The first experimental structure of TMPRSS2 was released in the public domain in June 2021. Although the trypsin domain was well resolved, several unstructured loops were present in the SRCR domain. We, therefore, used homology modelling and the Phyre2 server to model the missing regions of SRCR (Fig. 1). For completeness, a comparison between the Phyre2 model and the recently released (August 2021) AlphaFold [32] model is presented in Figure S1 (Phyre2 versus AlphaFold model: root mean square deviation [RMSD] 0.5 Å). We identified the chemical and physical bonds that stabilize the TMPRSS2 structure (i.e. hydrogen bonds, cysteine and salt bridges, as detailed in the Methods) and are affected by amino acid substitutions naturally occurring in the human population. A total of 137 variants were predicted damaging to the structure and/or function of TMPRSS2. Of these, 136 variants are extremely rare in the human population, with an average minor allele frequency (MAF) of 9.67 × 10−6 (cumulative MAF of 7.3 × 10−4) and are, therefore, unlikely to be of use as a marker of COVID-19 infection severity in the general population. The remaining variant, rs12329760, (NC_000021.9:g.41480570 C>T [GRCh38.p13], p.Val160Met on Ensembl transcript ENST00000332149.5 and Val197Met on the Ensembl transcript ENST00000398585.3) is predicted damaging and causes the substitution of an evolutionary conserved valine to methionine (Figure S2). Overall, the minor allele frequency (MAF) of this variant is 0.25 in the human population, with 6.7% homozygous individuals (9,587 T/T homozygotes out of 141,456 individuals sequenced as part of the GnomAd project). Under Hardy-Weinberg equilibrium and a MAF of 0.25, it is expected that 37% of individuals will be heterozygous for this variant. The MAF of rs12329760 T varies according to ethnicities and ranges from 0.14 in Ashkenazi Jewish to 0.38 in East Asian populations (0.15 in Latino, 0.23 in non-Finnish Europeans, 0.25 in South Asians, 0.29 in African/African Americans and 0.37 in Finnish Europeans). This highly conserved valine occurs in the scavenger receptor cysteine-rich (SRCR) domain, whose function within TMPRSS2 is still not fully understood, although a role in ligand and/or protein interaction has been proposed [45]. Indeed, this domain is present in several proteins involved in host defence, such as CD5, CD6 and Complement factor I [46,47].
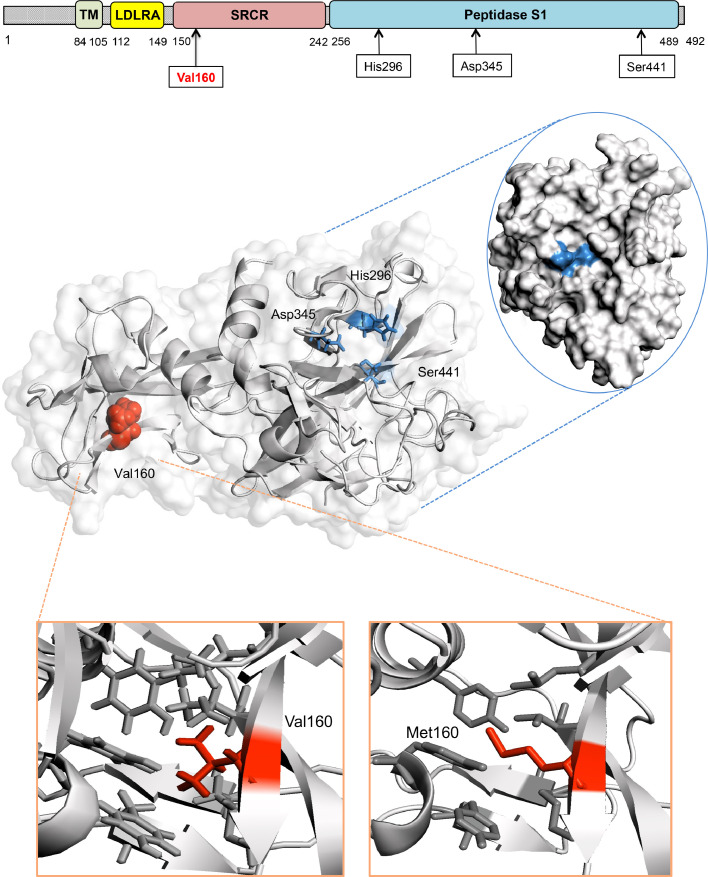
Fig. 1.
The TMPRSS2 protein and the p.Val160Met variant
The TMPRSS2 protein is composed of a cytoplasmic region (residues 1–84), a transmembrane region (TM, residues 85–105) and an extracellular region (residues 106–492). The latter is composed of three domains: the LDLR class A (residues 112–149), the scavenger receptor cysteine-rich domain (SRCR) (residues 150–242) and the Peptidase S1 (residues 256–489), which contains the protease active site: residues His296, Asp345 and Ser441. The three-dimensional structure of the extracellular region residues 145–491 corresponding to domains SRCR-2 (in green) and Peptidase S1 (in blue) is presented. Valine 160 (Val 160, depicted as a red sphere on the cartoon), which harbours variant p.Val160Met, occurs in the SRCR domain and spatially far from the TMPRSS2 catalytic site (mapped onto the surface of TMPRSS2).
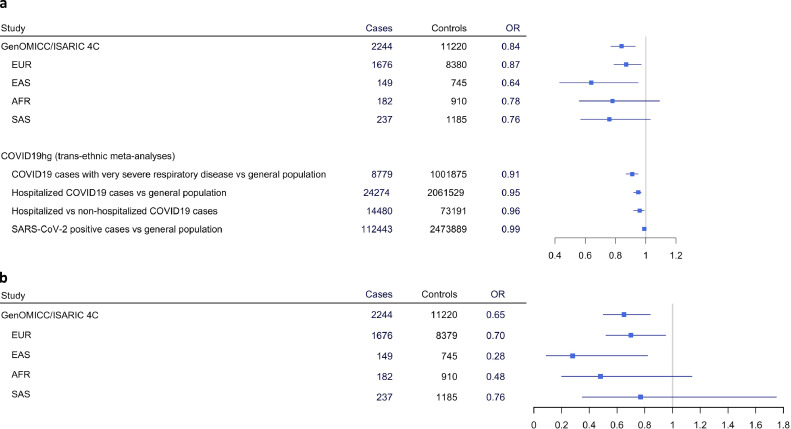
We first analysed the relation between TMPRSS2 rs12329760 and life-threatening SARS-CoV-2 infection in 2,244 critically ill, hospitalized, COVID-19 positive patients from 208 UK intensive care units (ICUs) (Table 1) recruited as part of the GenOMICC (genomicc.org) and ISARIC 4C (isaric4c.net) projects. These patients were representative of critically ill patients with COVID-19 in the UK population during the first Sars-Co-V2 outbreak of 2020[8]. Patients were treated in intensive care units (ICU/ITU) because of their propensity to critical respiratory failure due to COVID-19. Within the GenOMICC cohort (n = 2,109), mean age was 57.3 ± 12.1, 624 (30%) patients were females, and 396(19%) had comorbidities; 1,557 (74%) required invasive ventilation and 459 (22%) died within 60 days. Within the ISARIC 4C cohort (n = 135), mean age was 57.3 ± 2.9, 46 (34%) were females, and 40 (30%) had comorbidities; 25 (19%) required invasive ventilation and 22 (16%) died within 60 days, as described in[8]. 11,220 ancestry-matched individuals without a COVID-19-positive PCR test from the UK BioBank, acted as controls. Under an additive model, we found that the minor T allele of rs12329760 was significantly associated with a protective effect against severe COVID-19 in individuals of European ancestry (1,676 cases, 8,379 controls) with an OR of 0.87 (95%CI:0.79–0.97, p = 0.01). A protective effect was also observed in individuals of East Asian ancestry (149 cases, 745 controls; OR 0.64, 95%CI:0.43–0.95, p = 0.03). Similar effect sizes were observed in South Asians and Africans, but did not reach statistical significance, most likely as a result of the small sample size (Fig. 2). We further confirmed this protective effect on a trans-ethnic meta-analysis, using the entire cohort of 2,244 patients (OR 0.84, 95%CI:0.77–0.93, p = 5.8 × 10−4, Fig. 2, panel A). A heterogeneity analysis suggested that the T allele has a similar effect across different ethnicities (p = 0.47). To ascertain that this association was not an artefact due to population bias in the UK BioBank controls, the results from the European cohort were confirmed on an independent control population (45,875 unrelated individuals of European ancestry from 100 K Genomes [48]: OR 0.89, 95%CI:0.81–0.99, p = 0.02). Under a recessive model (i.e. individuals homozygous for the T allele), the trans-ethnic meta-analysis on 2,244 critically ill COVID-19 patients estimated an OR of 0.65 for TT homozygotes (95%CI:0.50–0.84, p = 1.3 × 10−3). In subset analyses, the OR was estimated at 0.70 (95%CI:0.52–0.95, p = 0.024) in Europeans, and 0.28 (95%CI:0.09–0.82, p = 0.019) in East Asians versus their corresponding ancestry-matched controls (Fig. 2, panel B).
Table 1.
Characteristics of 2,244 GenOMICC/ISARIC patients and 11,220 BioBank controls included in the study.
| Patient characteristics | Cases (n = 2,244) | Controls (n = 11,220) | |
|---|---|---|---|
| missing data | |||
| Females, n. (%) | 670[30] | 6,075[54] | |
| Age (years), mean ± SD | 57.3 ± 11.6 | 66.1 ± 8.0 | |
| Invasive ventilation, n. (%) | 1,582 (70.50) | 66 (2.94) | n.a |
| Death, n. (%) | 481 (21.44) | 368 (16.40) | n.a |
| Ancestry | |||
| European, n. (MAF) | 1,676 (0.20) | 8,380 (0.23) | |
| South Asian, n. (MAF) | 237 (0.21) | 1,185 (0.24) | |
| African, n. (MAF) | 182 (0.26) | 910 (0.29) | |
| East Asian, n. (MAF) | 149 (0.28) | 745 (0.38) | |
MAF, minor allele frequency; n.a, not available.
Fig. 2.
Association of TMPRSS2 rs12329760 to COVID-19 severity
Results are presented for the additive (a) and the recessive (b) model using different COVID-19-positive patient cohorts. The results from large GWAS meta-analyses performed as part of the COVID-19hg initiative (1) are also shown.
OR, odds ratio; EUR, European; EAS, East Asian; AFR, African; SAS, South Asian.
To assess whether the rs12329760 could be a proxy for an association with a nearby expression quantitative trait locus (eQTL), colocalisation analysis was performed to compare the GWAS signal at the locus to eQTL associations for TMPRSS2 and neighbouring gene MX1 in GTex version 8 [44] (Figure S3 A-C), using an Approximate Bayes Factor approach [43]. Under an assumption of a single causal variant within the locus for each trait, the posterior probability of a common causal variant was 1.1% for TMPRSS2 expression and 2.0% for MX1 expression, compared to posterior probabilities of 67% and 42% respectively for independent associations. Sensitivity analysis showed that the analysis was robust to choice of prior probabilities: more stringent software-default single-trait priors increased the posterior probabilities of null or single-trait-only association hypotheses, but had little impact on the colocalisation probability (1.1% for TMPRSS2 and 0.2% for MX1); varying prior probability for colocalisation (Figure S3 D-F) had an impact only when approaching the prior for single-trait associations, and did not result in posterior probabilities for colocalisation exceeding those for separate associations. Although independent contributions from multiple variants towards the genetic association cannot be excluded, this indicates that any genetic association between rs12327960 and severe COVID-19 is unlikely to be attributable to linkage disequilibrium with an eQTL and, thus, modification of protein function is more likely.
For additional corroboration of the genetic signal, we investigated the results of large GWAS meta-analyses performed in the context of the COVID-19 Host Genetics Initiative (COVID-19hg, available at https://www.COVID-19hg.org/, release 6, June 2021) [42]. Compared with the general population, in a trans-ethnic meta-analysis, the minor T allele of rs12329760 was associated with a significantly protective effect against severe COVID-19 (patients requiring hospitalization for COVID-19): 24,274 cases versus 2,061,529 controls; OR 0.95, 95%CI:0.92–0.97, p = 4.72 × 10−6. In ancestry-specific sub-group analyses, this effect was significant for a European population (OR 0.94, 95%CI:0.91–0.96, p = 5.66 × 10−6), but not for individuals of African, Hispanic-American, or admixed African/Hispanic/American ancestry; however, lower sample sizes for these groups limited study power, and subgroup analyses were not available for Asian populations. The protective effect was particularly evident in confirmed, critically ill cases (8,779 cases versus 1,001,875 population controls; OR 0.91, 95%CI:0.87–0.95, p = 8.18 × 10−6). Furthermore, the rs12329760 T allele was associated with reduced risk of hospitalisation after confirmed infection (14,480 hospitalised versus 73,191 cases not requiring hospitalization within 21 days after the test): OR 0.96, 95%CI:0.92–0.99, p = 0.012. Finally, there was no significant difference (p = 0.056) in the prevalence of the T allele between the general population (n = 2,473,889) and pooled individuals with a laboratory-confirmed SARS-CoV-2 infection (including hospitalized and life-threatening COVID-19 cases from the metanalyses previously described) or with a self-reported or physician-confirmed COVID diagnosis (total n = 112,443 cases, Fig. 2, panel A).
Although these meta-analyses include UK Biobank data and all except the hospitalised versus non-hospitalised comparison include data from the GenOMICC/ISARIC 4C cohort, and thus do not provide completely independent replication, these cohorts only comprise less than 25% of the total cases, limiting the impact of this single study on the overall results. These data therefore provide further support for our hypothesis that the TMPRSS2 rs12329760 variant has a protective effect against severe and/or life-threatening COVID-19. However, studies examining the prevalence of this variant in SARS-CoV-2 infected asymptomatic or pauci-symptomatic individuals are needed to ascertain its protective effect against mild viral infection.
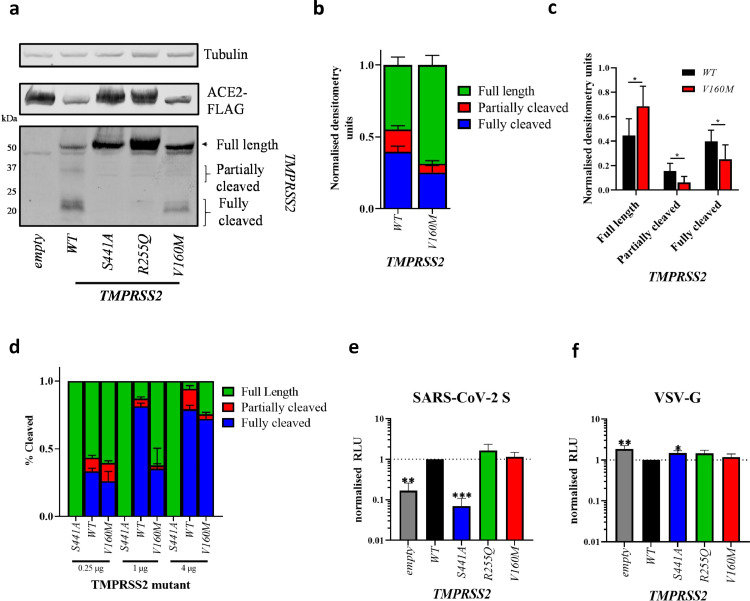
To investigate the phenotypic effect of the TMPRSS2 V160M variant, we co-transfected 293Ts cells, which we have previously confirmed that they do not endogenously express ACE2 or TMPRSS2 [24], with ACE2 and either TMPRSS2 wild type (TMPRSS2WT) or V160M (TMPRSS2V160M), as previously described [24]. We and others previously observed that co-expression of TMPRSS2 and ACE2 results in rapid cleavage of ACE2. We, therefore, used a mutant ACE2 that is more poorly degraded by TMPRSS2[49]. Two additional TMPRSS2 variants were included as controls: the catalytically inactive S441A (TMPRSS2S441A) and the catalytically active R255Q (TMPRSS2R255Q), that is unable to autocleave[16]. First, we investigated the autocleavage pattern of the different TMPRSS2 variants. The N-terminal membrane-bound part of TMPRSS2 can exist as different cleaved intermediates: a full-length uncleaved form of approximately 55 kDa, a partially cleaved form, and a fully cleaved form of 20 kDa. The latter is the product of TMPRSS2 autocleavage at arginine 255, which results in the liberation of the catalytically active protease domain in the extracellular space, leaving a small transmembrane N-terminal domain [16]. Wild type TMPRSS2 is expressed as roughly equal amounts of full-length and fully cleaved forms, with a small amount of partially cleaved product. As expected, the catalytically inactive TMPRSS2S441A and the non-autocleavable TMPRSS2R255Q resulted in only the full-length TMPRSS2 being expressed. However, TMPRSS2V160M resulted in a significantly higher proportion of full-length (55 kDa), and a significantly lower proportion of fully cleaved protein (20 kDa) (p < 0.05, Student's t-test). This difference was clear across a range of TMPRSS2 concentrations, with TMPRSS2 showing a concentration-dependant autocleavage phenotype: the higher the concentration of TMPRSS2, the higher the amount of autocleavage. Overall, these data suggest the V160M substitution exerts a partial inhibitory effect on the proteolytic autocleavage of TMPRSS2 (see Fig. 3A-D, Supplementary Figure S4).
Fig. 3.
Phenotypic impact of the TMPRSS2 V160M variant on autocleavage and SARS-CoV-2 spike-mediated entry; (A) Western blot analysis of TMPRSS2 autocleavage after expression in HEK 293Ts. (B,C, D) densitometry was determined in ImageJ and shows mean±standard deviation from N = 6 (B,C) or N = 3 (D) independent repeats. Statistics determined by two-tailed Student's t-test.
Entry of lentiviral pseudotypes expressing (E) SARS-CoV-2 spike glycoprotein or (F) Vesicular stomatitis virus glycoprotein (VSV-G) into HEK 293Ts co-expressing ACE2-FLAG and either empty vector or TMPRSS2 variants. Data shows mean±standard deviation of 3 independent repeats from different weeks, normalised to WT TMPRSS2. (E,F) Statistics determined by one-way ANOVA with multiple comparisons against WT on Log-transformed data (after determining log normality by the Shapiro-Wilk test and QQ plot). Values in µg indicate the amount of TMPRSS2 plasmid added to each condition. RLU, relative luminescence units.
*, 0.05 ≥ P > 0.01; **, 0.01 ≥ P > 0.001, ***, 0.001 ≥ P.
Subsequently, we investigated the effect of TMPRSS2V160M on promoting viral entry, using a previously described SARS-CoV-2 pseudovirus entry assay [24]. Pseudovirus expressing the glycoprotein from the vesicular stomatitis virus (VSV-G) was used as a control, as this virus enters cells in a TMPRSS2-independent manner [24]. Briefly, cells co-transfected with ACE2 and TMPRSS2 wild type or variants were incubated with the pseudovirus (as described in [24,38]) and, after 48 h, luminescence was measured. TMPRSS2WT enhanced viral entry by ∼5-fold compared to empty vector, while the catalytically dead TMPRSS2S441A showed no enhancement (Fig. 3E). The non-autocleavable mutant TMPRSS2R255Q showed similar enhancement, suggesting that autocleavage is dispensable for optimal TMPRSS2-mediated enhancement. TMPRSS2V160M showed no significant difference in viral entry compared to the TMPRSS2WT. Overall, the expression of catalytically active TMPRSS2 proteins only slightly inhibited VSV-G mediated entry (Fig. 3F).
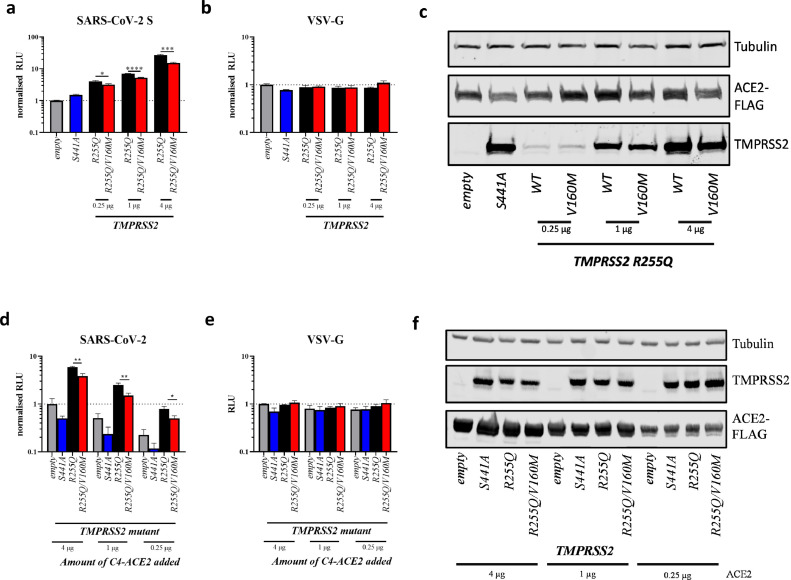
The partial inhibitory effect exerted by the V160M variant on the proteolytic autocleavage of TMPRSS2 resulted in a far greater proportion of uncleaved, surface-expressed TMPRSS2V160M compared to TMPRSS2WT. We compared this autocleavage seen in transfected 293T cells, to that seen in several epithelial cell lines that naturally express endogenous ACE2 and TMPRSS2 [24]: the human lung cell line, Calu-3, and the human colorectal adenocarcinoma cell line, Caco-2, both of which are extensively used for SARS-CoV-2 research. Interestingly, no fully cleaved TMPRSS2 could be detected as opposed to 293T cells, while both cell lines expressed mostly full length or partially cleaved TMPRSS2. This again suggests that the high levels of autocleavage seen in 293T cells may be, in part, an artefact of overexpression (Supplementary Figure S5). Therefore, we re-assessed whether TMPRSS2V160M affects SARS-CoV-2 S-expressing pseudovirus entry by using the double mutant TMPRSS2R255Q/V160M (which does not autocleave and is, therefore, more similar to endogenous TMPRSS2 in Calu-3 and Caco-2 cells), to control for protein cell-surface expression. Under these conditions, and across a range of plasmid titrations of both TMPRSS2 mutants and ACE2, TMPRSS2R255Q/V160M showed a significantly reduced ability to promote SARS-CoV-2 S-expressing pseudovirus compared to TMPRSS2R255Q alone, despite equal protein expression (Fig. 4A,C,D,F). Again, TMPRSS2R255Q/V160M had no effect on VSV-G-mediated entry (Fig. 4B,E).
Fig. 4.
Phenotypic impact of the TMPRSS2 non-autocleavable version of the V160M variant on SARS-CoV-2 spike-mediated entry
Entry of lentiviral pseudotypes expressing (A,D) SARS-CoV-2 spike glycoprotein or (B,E) vesicular stomatitis virus glycoprotein (VSV-G) into HEK 293Ts co-expressing ACE2-FLAG and either empty vector or TMPRSS2 variants. Data shows mean±standard deviation of 3 independent repeats from different weeks, normalised to empty vector. A-C shows titrations of mutant TMPRSS2 with constant ACE2 expression, while D-E show titrations of ACE2 with constant levels of TMPRSS2 expressed.
(A,B,D,E) statistics determined by two-tailed Student's t-test. (C,F) Western blot analysis of TMPRSS2 autocleavage mutant (R255Q) titration with or without the V160M substitution. µg values indicate the amount of TMPRSS2 or ACE2 plasmid added to each condition. RLU, relative luminescence units.
*, 0.05 ≥ P > 0.01; **, 0.01 ≥ P > 0.001, ***, 0.001 ≥ P.
Discussion
Overall, our results suggest that the rs12329760 C>T variant results in a moderately less catalytically active TMPRSS2, which is less able to autocleave and prime the SARS-CoV-2 spike protein. This may explain the protective effect against life-threatening COVID-19 observed in our cohort of patients admitted to ICU, compared to the general population. Such an effect was more prominent in homozygotes (recessive model) for the rs12329760 C>T in whom a 30% (OR 0.70) risk reduction was observed. Unfortunately, we did not have samples from asymptomatic/pauci symptomatic patients, but data from COVID-19hg meta-analyses appear to suggest that the rs12329760 variant has no protective effect against SARS-Co-V2 infection per se.
The allele frequency of TMPRSS2 rs12329760 (data from GnomAD population database) varies across different populations and is higher in East Asian and Finnish individuals (MAF 0.38 and 0.37, respectively) compared to south Asians (MAF 0.25) and non-Finnish Europeans (MAF 0.23). The lowest frequency of the T allele is reported in Latino and Jewish-Ashkenazi individuals (MAF 0.15). However, in our study, the small sample size of populations of non-European ancestry does not allow conclusions on the effect size of TMPRSS2 rs12329760 in different ethnicities. Genotyping of the TMPRSS2 rs12329760 variant on large COVID19 cohorts of patients of non-European genetic ancestry is, therefore, needed to assess its role in determining the differences in the severity of COVID-19 across various populations (e.g. between East Asia and Europe [50]). Indeed, a recent study showed a lower T allele frequency in a small cohort of Chinese patients with life-threatening COVID-19 compared to the population frequency [51]. Although the differences in the proportion of SARS-CoV-2 patients who develop severe COVID-19 across different populations [50] are more likely to be explained by social behaviour, public health measures to curb outbreaks, exposure to other viruses and immunological factors, human genetic variation across different populations may also contribute to the observed differences.
The pharmacological inhibition of TMPRSS2 using serine protease inhibitors, such as camostat and nafamostat, has been proposed as a pharmacological treatment of COVID-19 patients. In vitro [12] and animal studies have demonstrated that camostat can block viral entry (reviewed in [52]), and initial reports on the repurposing of camostat in COVID-19 patients have provided promising results [53]. However, a recently completed clinical trial using camostat in patients hospitalized for severe COVID-19, did not demonstrate a significant reduction in time-to-clinical improvement compared to placebo [54]. As the authors suggest, these patients were likely to have passed the most active stage of viral replication at the time of treatment and were in the hyper-inflammatory stage of the COVID-19, thus possibly explaining the lack of camostat efficacy. Several additional clinical trials of camostat in COVID-19 are currently underway [29]. Recently, the placebo-controlled phase III trial conducted in Japan on pauci symptomatic COVID-19 patients administering camostat mesilate 600 mg 4 times a day did not meet its primary end point of time to negative Sars-CoV-2 test [55], however data on secondary end points, such as progression to severe or life-threatening COVID-19, are still not publicly available.
Very little is known on TMPRSS2 and further extensive in vitro and in vivo studies on its pathophysiology are necessary. Since the beginning of the COVID-19 pandemic, the interest in TMPRSS2 has focused only on its role as a serine protease involved in the activation of the SARS-CoV-2 spike protein. However, as a soluble protease, TMPRSS2 may have additional substrates, and in vitro studies have demonstrated that PAR2 is one of these substrates [56,57]. PAR2 is expressed in several tissues, including lung, vascular endothelial and vascular smooth muscle cells [58,59] and its protease-mediated activation promotes inflammation by inducing prostaglandin synthesis and cytokine production in the lungs and other organs [60], [61], [62], [63], [64]. An intriguing hypothesis is that, similar to other soluble serine proteases, such as the human airway trypsin-like protease HAT (also known as TMPRSS11D), the soluble wild type TMPRSS2 protease may also have a role in promoting inflammation in the lungs and other tissues.
Since May 2020, when we reported the TMPRSS2 variant rs12329760 as possibly damaging to protein structure/function and raised the possibility that it could partly explain host susceptibility to COVID-19 severity [65], several other studies have also supported this hypothesis [66], [67], [68], [69], [70], [71] . In this study we have confirmed our initial hypothesis and provided a mechanistic effect to explain how this variant may contribute to the host susceptibility to severe COVID-19.
As previously discussed, one limitation of our study was the lack of access to a cohort of asymptomatic/pauci symptomatic COVID-19 patients. In the absence of such a cohort, we considered the general population as a good proxy and used this for comparison with COVID-19 severe cases. Indeed, a recent systematic review and metanalysis shows that one third of COVID-19 positive cases do not develop symptoms [72]. When well-characterized cohorts of asymptomatic/pauci symptomatic COVID-19 patients become available, it will be possible to further investigate the role of TMPRSS2 variant rs12329760 on Sars-Co-V2 infection. Another limitation of this study is that we did not directly validate our results in endogenously expressing cell lines, such as like Calu-3, as this would require gene editing the endogenous TMPRSS2. Calu-3 cells are extremely slow growing and highly resistant to single cell cloning, thus making this cell line not particularly suitable for gene editing.
In conclusion, the T allele of the common TMPRSS2 variant rs12329760 confers a reduced risk of severe COVID-19. Similar to what observed in the TMPRSS2 KO mouse, the Val160Met substitution, which exerts a partial inhibitory effect on the proteolytic autocleavage of TMPRSS2 and the priming of the SARS-CoV-2 spike protein, is associated with a milder COVID-19 infection compared to the wild type. Differences in population frequency of this genetic variant may contribute to the reported variability in COVID-19 severity across various ethnicities and studies on large COVID-19 cohorts of patients of non-European genetic ancestry are needed to clarify this. Further studies are needed to assess the expression of TMPRSS2 across different age groups; indeed a reduced TMPRSS2 expression in younger compared to older individuals, as observed in mice and in preliminary human studies, could help explain age-related differences in COVID-19 morbidity. Moreover, TMPRSS2 could be a viable drug target in COVID-19 patients, and camostat mesilate, or other novel TMPRSS2 inhibitors, may have a role in the treatment of COVID-19. Clinical trials are needed to confirm this.
Funding
Wellcome Trust, BBSRC, UKRI Future Leader's Fellowship, Health Data Research UK
AD and NP were supported by the Wellcome Trust (grants 104,955/Z/14/Z, 218,242/Z/19/Z and 211,496/Z/18/Z) and TK by the BBSRC (grants BB/P011705/1 and BB/P023959/1), VSS is supported by UKRI Future Leader's Fellowship (MR/S032304/1), J-LC is supported by Howard Hughes Medical Institute, Rockefeller University, St. Giles Foundation, Fisher centre for Alzheimer's Research Foundation, Meyer Foundation, Square Foundation, Grandir - Fonds de solidarité pour l'enfance, SCOR Corporate Foundation for Science, Institut National de la Santé et de la Recherche Médicale (INSERM), University of Paris, National Institutes of Health (R01AI088364), French Foundation for Medical Research (EQU201903007798), FRM and French National Research Agency (ANR) GENCOVID project (ANR-20-COVI-0003); LA is supported by the Agence Nationale de la Recherche (ANR-10-IAHU-01, ANR-10-LABX-62-IBEID), TPP and WSB are supported by BBSRC grants BB/R013071/1 and BBSRC and the G2P-UK National Virology consortium (funded by MRC/UKRI, grant ref: MR/W005611/1), AT was supported by Roslin Institute Strategic Programme Grants from the BBSRC (BBS/E/D/10,002,070 and BBS/E/D/30,002,275) and Health Data Research UK (references HDR-9004 and HDR-9003).
Author contributions
NP, EP-C, AT and JKB contributed to population data analysis. AD, TK and MJES contributed to 3D modelling and structural analysis. TPP and WSB contributed to laboratory work. NP, EP-C, TPP and AD contributed to data analysis. NP, TPP, WSB, AD contributed to study design. NP, TPP, EP-C, AC, VS-S, J-LC, LA, WSB, JKB, MJES and AD contributed to interpretation of findings and manuscript preparation. AD conceived the study, contributed to study coordination and wrote the first draft of the manuscript. All authors approved the final version of the manuscript.
Data availability
Full summary-level data in support of the findings of this study are available for download from https://genomicc.org/data. Individual level data can be analysed by qualified researchers in the ISARIC 4C/GenOMICC data analysis platform by application at https://genomicc.org/data. BioBank data and Genomics England data are available to registered researchers at https://www.ukbiobank.ac.uk/ and https://www.genomicsengland.co.uk/. The COVID-19 Host Genetics Initiative2 (COVID-19hg) summary statistics are available at https://www.COVID-19hg.org/.
Supplementary Information is available for this paper.
Declaration of Competing Interest
Dr. David reports grants from Wellcome Trust during the conduct of the study; Dr. Parkinson reports grants from Wellcome Trust during the conduct of the study; Dr. Peacock reports grants from MRC/UKRI, grants from BBSRC during the conduct of the study; Dr. Pairo-Castineira has nothing to disclose. Dr. Khanna reports grants from BBSRC during the conduct of the study; Dr. Cobat has nothing to disclose. Dr. Tenesa reports grants from BBSRC, grants from Health Data Research UK during the conduct of the study. Dr. Sancho-Shimizu reports grants from UKRI Future Leader's Fellowship during the conduct of the study; Dr. Casanova reports other from Howard Hughes Medical Institute, other from Rockefeller University, other from St. Giles Foundation, other from Fisher centre for Alzheimer's Research Foundation, other from Meyer Foundation, other from Square Foundation, other from Grandir - Fonds de solidarité pour l'enfance, other from SCOR Corporate Foundation for Science, other from Institut National de la Santé et de la Recherche Médicale (INSERM), other from University of Paris, other from National Institutes of Health (NIH), other from French Foundation for Medical Research (FRM), other from FRM and French National Research Agency (ANR) GENCOVID project during the conduct of the study; Dr. Abel reports other from Agence Nationale de la Recherche during the conduct of the study; Dr. Barclay reports grants from BBSRC during the conduct of the study; Dr. Baillie has nothing to disclose. Dr. Sternberg reports grants from Wellcome Trust, grants from BBSRC, during the conduct of the study.
Acknowledgement
This research was conducted using the UK BioBank Resource under project 788
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.retram.2022.103333.
Appendix 1: GenOMICC Consortium
GenOMICC Co-Investigators
J. Kenneth Baillie1,2, Colin Begg3, Sara Clohisey1, Charles Hinds4, Peter Horby5, Julian Knight6, Lowell Ling7, David Maslove8, Danny McAuley9,10, Johnny Millar1, Hugh Montgomery11, Alistair Nichol12, Peter J.M. Openshaw13,14, Alexandre C Pereira15, Chris P Ponting16, Kathy Rowan17, Malcolm G Semple18,19, Manu Shankar-Hari20, Charlotte Summers21, Timothy Walsh2.
Management, Laboratory and Data team
Latha Aravindan22, Ruth Armstrong1, J. Kenneth Baillie1,2, Heather Biggs23, Ceilia Boz1, Adam Brown1, Richard Clark24, Sara Clohisey1, Audrey Coutts24, Judy Coyle1, Louise Cullum1, Sukamal Das22, Nicky Day1, Lorna Donnelly24, Esther Duncan1, Angie Fawkes24, Paul Finernan1, Max Head Fourman1, Anita Furlong23, James Furniss1, Bernadette Gallagher1, Tammy Gilchrist24, Ailsa Golightly1, Fiona Griffiths1, Katarzyna Hafezi24, Debbie Hamilton1, Ross Hendry1, Andy Law1, Dawn Law1, Rachel Law1, Sarah Law1, Rebecca Lidstone-Scott1, Louise Macgillivray24, Alan Maclean24, Hanning Mal1, Sarah McCafferty24, Ellie Mcmaster1, Jen Meikle1, Shona C Moore18, Kirstie Morrice24, Lee Murphy24, Sheena Murphy22, Mybaya Hellen1, Wilna Oosthuyzen1, Chenqing Zheng25, Jiantao Chen25, Nick Parkinson1, Trevor Paterson1, Katherine Schon23, Andrew Stenhouse1, Mihaela Das22, Maaike Swets1,26, Helen Szoor-McElhinney1, Filip Taneski1, Lance Turtle18, Tony Wackett1, Mairi Ward1, Jane Weaver1, Nicola Wrobel24, Marie Zechner1, Mybaya Hellen1.
Guys and St Thomas’ Hospital, London, UK
Gill Arbane27, Aneta Bociek27, Sara Campos27, Neus Grau27, Tim Owen Jones27, Rosario Lim27, Martina Marotti27, Marlies Ostermann27, Manu Shankar-Hari27, Christopher Whitton27.
Barts Health NHS Trust, London, UK
Zoe Alldis28, Raine Astin-Chamberlain28, Fatima Bibi28, Jack Biddle28, Sarah Blow28, Matthew Bolton28, Catherine Borra28, Ruth Bowles28, Maudrian Burton28, Yasmin Choudhury28, David Collier28, Amber Cox28, Amy Easthope28, Patrizia Ebano28, Stavros Fotiadis28, Jana Gurasashvili28, Rosslyn Halls28, Pippa Hartridge28, Delordson Kallon28, Jamila Kassam28, Ivone Lancoma-Malcolm28, Maninderpal Matharu28, Peter May28, Oliver Mitchelmore28, Tabitha Newman28, Mital Patel28, Jane Pheby28, Irene Pinzuti28, Zoe Prime28, Oleksandra Prysyazhna28, Julian Shiel28, Melanie Taylor28, Carey Tierney28, Suzanne Wood28, Anne Zak28, Olivier Zongo28.
James Cook University Hospital, Middlesbrough, UK
Stephen Bonner29, Keith Hugill29, Jessica Jones29, Steven Liggett29, Evie Headlam29.
Royal Stoke University Hospital, Staffordshire, UK
Nageswar Bandla30, Minnie Gellamucho30, Michelle Davies30, Christopher Thompson30.
North Middlesex University Hospital NHS trust, London, UK
Marwa Abdelrazik31, Dhanalakshmi Bakthavatsalam31, Munzir Elhassan31, Arunkumar Ganesan31, Anne Haldeos31, Jeronimo Moreno-Cuesta31, Dharam Purohit31, Rachel Vincent31, Kugan Xavier31, kumar Rohit32, Frater Alasdair31, Malik Saleem31, Carter David31, Jenkins Samuel31, Zoe Lamond31, Wall Alanna31.
The Royal Liverpool University Hospital, Liverpool, UK
Jaime Fernandez-Roman33, David O. Hamilton33, Emily Johnson33, Brian Johnston33, Maria Lopez Martinez33, Suleman Mulla33, David Shaw33, Alicia A.C. Waite33, Victoria Waugh33, Ingeborg D. Welters33, Karen Williams33.
King's College Hospital, London, UK
Anna Cavazza34, Maeve Cockrell34, Eleanor Corcoran34, Maria Depante34, Clare Finney34, Ellen Jerome34, Mark McPhail34, Monalisa Nayak34, Harriet Noble34, Kevin O'Reilly34, Evita Pappa34, Rohit Saha34, Sian Saha34, John Smith34, Abigail Knighton34.
Charing Cross Hospital, St Mary's Hospital and Hammersmith Hospital, London, UK
David Antcliffe35, Dorota Banach35, Stephen Brett35, Phoebe Coghlan35, Ziortza Fernandez35, Anthony Gordon35, Roceld Rojo35, Sonia Sousa Arias35, Maie Templeton35.
Nottingham University Hospital, Nottingham, UK
Megan Meredith36, Lucy Morris36, Lucy Ryan36, Amy Clark36, Julia Sampson36, Cecilia Peters36, Martin Dent36, Margaret Langley36, Saima Ashraf36, Shuying Wei36, Angela Andrew36.
John Radcliffe Hospital, Oxford, UK
Archana Bashyal37, Neil Davidson37, Paula Hutton37, Stuart McKechnie37, Jean Wilson37.
Kingston Hospital, Surrey, UK
David Baptista38, Rebecca Crowe38, Rita Fernandes38, Rosaleen Herdman-Grant38, Anna Joseph38, Denise O'Connor39, Meryem Allen38, Adam Loveridge38, India McKenley38, Eriko Morino38, Andres Naranjo38, Richard Simms38, Kathryn Sollesta38, Andrew Swain38, Harish Venkatesh38, Jacyntha Khera38, Jonathan Fox38.
Royal Infirmary of Edinburgh, Edinburgh, UK
Gillian Andrew40, J. Kenneth Baillie40, Lucy Barclay40, Marie Callaghan40, Rachael Campbell40, Sarah Clark40, Dave Hope40, Lucy Marshall40, Corrienne McCulloch40, Kate Briton40, Jo Singleton40, Sohphie Birch 40.
Queen Alexandra Hospital, Portsmouth, UK
Lutece Brimfield41, Zoe Daly41, David Pogson41, Steve Rose41.
Morriston Hospital, Swansea, UK
Ceri Battle42, Elaine Brinkworth42, Rachel Harford42, Carl Murphy42, Luke Newey42, Tabitha Rees42, Marie Williams42, Sophie Arnold42.
Addenbrooke's Hospital, Cambridge, UK
Petra Polgarova43, Katerina Stroud43, Charlotte Summers43, Eoghan Meaney43, Megan Jones43, Anthony Ng43, Shruti Agrawal43, Nazima Pathan43, Deborah White43, Esther Daubney43, Kay Elston43.
BHRUT (Barking Havering) - Queens Hospital and King George Hospital, Essex, UK
Lina Grauslyte44, Musarat Hussain44, Mandeep Phull44, Tatiana Pogreban44, Lace Rosaroso44, Erika Salciute44, George Franke44, Joanna Wong44, Aparna George44.
Royal Sussex County Hospital, Brighton, UK
Laura Ortiz-Ruiz de Gordoa45, Emily Peasgood45, Claire Phillips45, Laura Ortiz-Ruiz de Gordoa45, Emily Peasgood45, Claire Phillips45.
Queen Elizabeth Hospital, Birmingham, UK
Michelle Bates46, Jo Dasgin46, Jaspret Gill46, Annette Nilsson46, James Scriven46.
St George's Hospital, London, UK
Carlos Castro Delgado47, Deborah Dawson47, Lijun Ding47, Georgia Durrant47, Obiageri Ezeobu47, Sarah Farnell-Ward47, Abiola Harrison47, Rebecca Kanu47, Susannah Leaver47, elena Maccacari47, Soumendu Manna47, Romina Pepermans Saluzzio47, Joana Queiroz47, Tinashe Samakomva47, Christine Sicat47, Joana Texeira47, Edna Fernandes Da Gloria47, Ana Lisboa47, John Rawlins47, Jisha Mathew47, Ashley Kinch47, William James Hurt47, Nirav Shah47, Victoria Clark47, Maria Thanasi47, Nikki Yun47, Kamal Patel47.
Stepping Hill Hospital, Stockport, UK
Sara Bennett48, Emma Goodwin48, Matthew Jackson48, Alissa Kent48, Clare Tibke48, Wiesia Woodyatt48, Ahmed Zaki48.
Countess of Chester Hospital, Chester, UK
Azmerelda Abraheem49, Peter Bamford49, Kathryn Cawley49, Charlie Dunmore49, Maria Faulkner49, Rumanah Girach49, Helen Jeffrey49, Rhianna Jones49, Emily London49, Imrun Nagra49, Farah Nasir49, Hannah Sainsbury49, Clare Smedley49.
Royal Blackburn Teaching Hospital, Blackburn, UK
Tahera Patel50, Matthew Smith50, Srikanth Chukkambotla50, Aayesha Kazi50, Janice Hartley50, Joseph Dykes50, Muhammad Hijazi50, Sarah Keith50, Meherunnisa Khan50, Janet Ryan-Smith50, Philippa Springle50, Jacqueline Thomas50, Nick Truman50, Samuel Saad50, Dabheoc Coleman50, Christopher Fine50, Roseanna Matt50, Bethan Gay50, Jack Dalziel50, Syamlan Ali50, Drew Goodchild50, Rhiannan Harling50, Ravi Bhatterjee50, Wendy Goddard50, Chloe Davison50, Stephen Duberly50, Jeanette Hargreaves50, Rachel Bolton50.
The Tunbridge Wells Hospital and Maidstone Hospital, Kent, UK
Miriam Davey51, David Golden51, Rebecca Seaman51.
Royal Gwent Hospital, Newport, UK
Shiney Cherian52, Sean Cutler52, Anne Emma Heron52, Anna Roynon-Reed52, Tamas Szakmany52, Gemma Williams52, Owen Richards52, Yusuf Cheema52.
Pinderfields General Hospital, Wakefield, UK
Hollie Brooke53, Sarah Buckley53, Jose Cebrian Suarez53, Ruth Charlesworth53, Karen Hansson53, John Norris53, Alice Poole53, Alastair Rose53, Rajdeep Sandhu53, Brendan Sloan53, Elizabeth Smithson53, Muthu Thirumaran53, Veronica Wagstaff53, Alexandra Metcalfe53.
Royal Berkshire NHS Foundation Trust, Berkshire, UK
Mark Brunton54, Jess Caterson54, Holly Coles54, Matthew Frise54, Sabi Gurung Rai54, Nicola Jacques54, Liza Keating54, Emma Tilney54, Shauna Bartley54, Parminder Bhuie54.
Broomfield Hospital, Chelmsford, UK
Sian Gibson55, Amanda Lyle55, Fiona McNeela55, Jayachandran Radhakrishnan55, Alistair Hughes55.
Northumbria Healthcare NHS Foundation Trust, North Shields, UK
Bryan Yates56, Jessica Reynolds56, Helen Campbell56, Maria Thompsom56, Steve Dodds56, Stacey Duffy56.
Whiston Hospital, Prescot, UK
Sandra Greer57, Karen Shuker57, Ascanio Tridente57.
Croydon University Hospital, Croydon, UK
Reena Khade58, Ashok Sundar 58, George Tsinaslanidis58.
York Hospital, York, UK
Isobel Birkinshaw59, Joseph Carter59, Kate Howard59, Joanne Ingham59, Rosie Joy59, Harriet Pearson59, Samantha Roche59, Zoe Scott59.
Heartlands Hospital, Birmingham, UK
Hollie Bancroft60, Mary Bellamy60, Margaret Carmody60, Jacqueline Daglish60, Faye Moore60, Joanne Rhodes60, Mirriam Sangombe60, Salma Kadiri60, James Scriven60.
Ashford and St Peter's Hospital, Surrey, UK
Maria Croft61, Ian White61, Victoria Frost61, Maia Aquino61.
Barnet Hospital, London, UK
Rajeev Jha62, Vinodh Krishnamurthy62, Lai Lim62, Rajeev Jha62, Vinodh Krishnamurthy62, Li Lim62.
East Surrey Hospital, Redhill, UK
Edward Combes63, Teishel Joefield63, Sonja Monnery63, Valerie Beech63, Sallyanne Trotman63.
Ninewells Hospital, Dundee, UK
Christine Almaden-Boyle64, Pauline Austin64, Louise Cabrelli64, Stephen Cole64, Matt Casey64, Susan Chapman64, Stephen Cole64, Clare Whyte64.
Worthing Hospital, Worthing, UK and St Richard's Hospital, Chichester, UK
Yolanda Baird65, Aaron Butler65, Indra Chadbourn65, Linda Folkes65, Heather Fox65, Amy Gardner65, Raquel Gomez65, Gillian Hobden65, Luke Hodgson65, Kirsten King65, Michael Margarson65, Tim Martindale65, Emma Meadows65, Dana Raynard65, Yvette Thirlwall65, David Helm65, Jordi Margalef65.
Southampton General Hospital, Southampton, UK
Kristine Criste66, Rebecca Cusack66, Kim Golder66, Hannah Golding66, Oliver Jones66, Samantha Leggett66, Michelle Male66, Martyna Marani66, Kirsty Prager66, Toran Williams66, Belinda Roberts66, Karen Salmon66.
The Alexandra Hospital, Redditch and Worcester Royal Hospital, Worcester, UK
Peter Anderson67, Katie Archer67, Karen Austin67, caroline Davis67, Alison Durie67, Olivia Kelsall67, Jessica Thrush67, Charlie Vigurs67, Laura Wild67, Hannah-Louise Wood67, Helen Tranter67, Alison Harrison67, Nicholas Cowley67, Michael McAlindon67, Andrew Burtenshaw67, Stephen Digby67, Emma Low67, Aled Morgan67, Naiara Cother67, Tobias Rankin67, Sarah Clayton67, Alex McCurdy67.
Sandwell General Hospital and City Hospital, Birmingham, UK
Cecilia Ahmed68, Balvinder Baines68, Sarah Clamp68, Julie Colley68, Risna Haq68, Anne Hayes68, Jonathan Hulme68, Samia Hussain68, Sibet Joseph68, Rita Kumar68, Zahira Maqsood68, Manjit Purewal68.
Blackpool Victoria Hospital, Blackpool, UK
Leonie Benham69, Zena Bradshaw69, Joanna Brown69, Melanie Caswell69, Jason Cupitt69, Sarah Melling69, Stephen Preston69, Nicola Slawson69, Emma Stoddard69, Scott Warden69.
Royal Glamorgan Hospital, Pontyclun, UK
Bethan Deacon70, Ceri Lynch70, Carla Pothecary70, Lisa Roche70, Gwenllian Sera Howe70, Jayaprakash Singh70, Keri Turner70, Hannah Ellis70, Natalie Stroud70.
The Royal Oldham Hospital, Manchester, UK
Jodie Hunt71, Joy Dearden71, Emma Dobson71, Andy Drummond71, Michelle Mulcahy71, Sheila Munt71, Grainne O'Connor71, Jennifer Philbin71, Chloe Rishton71, Redmond Tully71, Sarah Winnard71.
Glasgow Royal Infirmary, Glasgow, UK
Susanne Cathcart72, Katharine Duffy72, Alex Puxty72, Kathryn Puxty72, Lynne Turner72, Jane Ireland72, Gary Semple72.
St James's University Hospital and Leeds General Infirmary, Leeds, UK
Kate Long73, Simon Whiteley73, Elizabeth Wilby73, Bethan Ogg73.
University Hospital North Durham, Darlington, UK and Darlington Memorial Hospital, Darlington, UK
Amanda Cowton74, Andrea Kay74, Melanie Kent74, Kathryn Potts74, Ami Wilkinson74, Suzanne Campbell74, Ellen Brown 74.
Fairfield General Hospital, Bury, UK
Julie Melville75, Jay Naisbitt75, Rosane Joseph75, Maria Lazo75, Olivia Walton75, Alan Neal75.
Wythenshawe Hospital, Manchester, UK
Peter Alexander76, Schvearn Allen76, Joanne Bradley-Potts76, Craig Brantwood76, Jasmine Egan76, Timothy Felton76, Grace Padden76, Luke Ward76, Stuart Moss76, Susannah Glasgow 76.
Royal Alexandra Hospital, Paisley, UK
Lynn Abel77, Michael Brett77, Brian Digby77, Lisa Gemmell77, James Hornsby77, Patrick MacGoey77, Pauline O'Neil77, Richard Price77, Natalie Rodden77, Kevin Rooney77, Radha Sundaram77, Nicola Thomson77.
Good Hope Hospital, Birmingham, UK
Bridget Hopkins78, James Scriven78, Laura Thrasyvoulou78, Heather Willis78.
Tameside General Hospital, Ashton Under Lyne, UK
Martyn Clark79, Martina Coulding79, Edward Jude79, Jacqueline McCormick79, Oliver Mercer79, Darsh Potla79, Hafiz Rehman79, Heather Savill79, Victoria Turner79.
Royal Derby Hospital, Derby, UK
Charlotte Downes80, Kathleen Holding80, Katie Riches80, Mary Hilton80, Mel Hayman80, Deepak Subramanian80, Priya Daniel80.
Medway Maritime Hospital, Gillingham, UK
Oluronke Adanini81, Nikhil Bhatia81, Maines Msiska81, Rebecca Collins81.
Royal Victoria Infirmary, Newcastle Upon Tyne, UK
Ian Clement82, Bijal Patel 82, A Gulati82, Carole Hays82, K Webster82, Anne Hudson82, Andrea Webster82, Elaine Stephenson82, Louise McCormack82, Victoria Slater82, Rachel Nixon82, Helen Hanson82, Maggie fearby82, Sinead Kelly82, Victoria Bridgett82, Philip Robinson82.
Poole Hospital, Poole, UK
Julie Camsooksai83, Charlotte Humphrey83, Sarah Jenkins83, Henrik Reschreiter83, Beverley Wadams83, Yasmin Death83.
Bedford Hospital, Bedford, UK
Victoria Bastion84, Daphene Clarke84, Beena David84, Harriet Kent84, Rachel Lorusso84, Gamu Lubimbi84, Sophie Murdoch84, Melchizedek Penacerrada84, Alastair Thomas84, Jennifer Valentine84, Ana Vochin84, Retno Wulandari84, Brice Djeugam84.
Queens Hospital Burton, Burton-On-Trent, UK
Gillian Bell85, Katy English85, Amro Katary85, Louise Wilcox85.
North Manchester General Hospital, Manchester, UK
Michelle Bruce86, Karen Connolly86, Tracy Duncan86, Helen T-Michael86, Gabriella Lindergard86, Samuel Hey 86, Claire Fox86, Jordan Alfonso86, Laura Jayne Durrans86, Jacinta Guerin86, Bethan Blackledge86, Jade Harris86, Martin Hruska86, Ayaa Eltayeb86, Thomas Lamb86, Tracey Hodgkiss86, Lisa Cooper86, Joanne Rothwell86.
Aberdeen Royal Infirmary, Aberdeen, UK
Angela Allan87, Felicity Anderson87, Callum Kaye87, Jade Liew87, Jasmine Medhora87, Teresa Scott87, Erin Trumper87, Adriana Botello87.
Derriford Hospital, Plymouth, UK
Liana Lankester88, Nikitas Nikitas88, Colin Wells88, Bethan Stowe88, Kayleigh Spencer88.
Manchester Royal Infirmary, Manchester, UK
Craig Brandwood89, Lara Smith89, Richard Clark89, Katie Birchall89, Laurel Kolakaluri89, Deborah Baines 89, Anila Sukumaran89.
Salford Royal Hospital, Manchester, UK
Elena Apetri90, Cathrine Basikolo90, Bethan Blackledge90, Laura Catlow90, Bethan Charles90, Paul Dark90, Reece Doonan90, Jade Harris90, Alice Harvey90, Daniel Horner90, Karen Knowles90, Stephanie Lee90, Diane Lomas90, Chloe Lyons90, Tracy Marsden90, Danielle McLaughlan90, Liam McMorrow90, Jessica Pendlebury90, Jane Perez90, Maria Poulaka90, Nicola Proudfoot90, Melanie Slaughter90, Kathryn Slevin90, Melanie Taylor90, Vicky Thomas90, Danielle Walker90, Angiy Michael 90, Matthew Collis90.
William Harvey Hospital, Ashford, UK
Tracey Cosier91, Gemma Millen91, Neil Richardson91, Natasha Schumacher91, Heather Weston91, James Rand91.
Queen Elizabeth University Hospital, Glasgow, UK
Nicola Baxter92, Steven Henderson92, Sophie Kennedy-Hay92, Christopher McParland92, Laura Rooney92, Malcolm Sim92, Gordan McCreath92.
Bradford Royal Infirmary, Bradford, UK
Louise Akeroyd93, Shereen Bano93, Matt Bromley93, Lucy Gurr93, Tom Lawton93, James Morgan93, Kirsten Sellick93, Deborah Warren93, Brian Wilkinson93, Janet McGowan93, Camilla Ledgard93, Amelia Stacey93, Kate Pye93, Ruth Bellwood93, Michael Bentley93.
Bristol Royal Infirmary, Bristol, UK
Jeremy Bewley94, Zoe Garland94, Lisa Grimmer94, Bethany Gumbrill94, Rebekah Johnson94, Katie Sweet94, Denise Webster94, Georgia Efford94.
Norfolk and Norwich University hospital (NNUH), Norwich, UK
Karen Convery95, Deirdre Fottrell-Gould95, Lisa Hudig95, Jocelyn Keshet-Price95, Georgina Randell95, Katie Stammers95.
Queen Elizabeth Hospital Gateshead, Gateshead, UK
Maria Bokhari96, Vanessa Linnett96, Rachael Lucas96, Wendy McCormick96, Jenny Ritzema96, Amanda Sanderson96, Helen Wild96.
Sunderland Royal Hospital, Sunderland, UK
Anthony Rostron97, Alistair Roy97, Lindsey Woods97, Sarah Cornell97, Fiona Wakinshaw97, Kimberley Rogerson97, Jordan Jarmain97.
Aintree University Hospital, Liverpool, UK
Robert Parker98, Amie Reddy98, Ian Turner-Bone98, Laura Wilding98, Peter Harding98.
Hull Royal Infirmary, Hull, UK
Caroline Abernathy99, Louise Foster99, Andrew Gratrix99, Vicky Martinson99, Priyai Parkinson99, Elizabeth Stones99, Llucia Carbral-Ortega100.
University College Hospital, London, UK
Georgia Bercades101, David Brealey101, Ingrid Hass101, Niall MacCallum101, Gladys Martir101, Eamon Raith101, Anna Reyes101, Deborah Smyth101.
Royal Devon and Exeter Hospital, Exeter, UK
Letizia Zitter102, Sarah Benyon102, Suzie Marriott102, Linda Park102, Samantha Keenan102, Elizabeth Gordon102, Helen Quinn102, Kizzy Baines102.
The Royal Papworth Hospital, Cambridge, UK
Lenka Cagova103, Adama Fofano103, Lucie Garner103, Helen Holcombe103, Sue Mepham103, Alice Michael Mitchell103, Lucy Mwaura103, Krithivasan Praman103, Alain Vuylsteke103, Julie Zamikula103.
Ipswich Hospital, Ipswich, UK
Bally Purewal104, Vanessa Rivers104, Stephanie Bell104.
Southmead Hospital, Bristol, UK
Hayley Blakemore105, Borislava Borislavova105, Beverley Faulkner105, Emma Gendall105, Elizabeth Goff105, Kati Hayes105, Matt Thomas105, Ruth Worner105, Kerry Smith105, Deanna Stephens105.
Milton Keynes University Hospital, Milton Keynes, UK
Louise Mew106, Esther Mwaura106, Richard Stewart106, Felicity Williams106, Lynn Wren106, Sara-Beth Sutherland 106.
Royal Hampshire County Hospital, Hampshire, UK
Emily Bevan107, Jane Martin107, Dawn Trodd107, Geoff Watson107, Caroline Wrey Brown107.
Queen Elizabeth Hospital, Woolwich, London, UK
Amy Collins108, Waqas Khaliq108, Estefania Treus Gude108.
Great Ormond St Hospital and UCL Great Ormond St Institute of Child Health NIHR Biomedical Research Centre, London, UK
Olugbenga Akinkugbe109, Alasdair Bamford109, Emily Beech109, Holly Belfield109, Michael Bell109, Charlene Davies109, Gareth A. L. Jones109, Tara McHugh109, Hamza Meghari109, Lauran O'Neill109, Mark J. Peters109, Samiran Ray109, Ana Luisa Tomas109.
Stoke Mandeville Hospital, Buckinghamshire, UK
Iona Burn110, Geraldine Hambrook110, Katarina Manso110, Ruth Penn110, Pradeep Shanmugasundaram110, Julie Tebbutt110, Danielle Thornton110.
University Hospital of Wales, Cardiff, UK
Jade Cole111, Michelle Davies111, Rhys Davies111, Donna Duffin111, Helen Hill111, Ben Player111, Emma Thomas111, Angharad Williams111.
Basingstoke and North Hampshire Hospital, Basingstoke, UK
Denise Griffin112, Nycola Muchenje112, Mcdonald Mupudzi112, Richard Partridge112, Jo-Anna Conyngham112, Rachel Thomas112, Mary Wright112, Maria Alvarez Corral112.
Arrowe Park Hospital, Wirral, UK
Reni Jacob113, Cathy Jones113, Craig Denmade113.
Chesterfield Royal Hospital Foundation Trust, Chesterfield, UK
Sarah Beavis114, Katie Dale114, Rachel Gascoyne114, Joanne Hawes114, Kelly Pritchard114, Lesley Stevenson114, Amanda Whileman114.
Musgrove Park Hospital, Taunton, UK
Patricia Doble115, Joanne Hutter115, corinne Pawley115, Charmaine Shovelton115, Marius Vaida115.
Peterborough City Hospital, Peterborough, UK and Hinchingbrooke Hospital, Huntingdon, UK
Deborah Butcher116, Susie O'Sullivan116, Nicola Butterworth-Cowin116.
Royal Hallamshire Hospital and Northern General Hospital, Sheffield, UK
Norfaizan Ahmad117, Joann Barker117, Kris Bauchmuller117, Sarah Bird117, Kay Cawthron117, Kate Harrington117, Yvonne Jackson117, Faith Kibutu117, Becky Lenagh117, Shamiso Masuko117, Gary H Mills117, Ajay Raithatha117, Matthew Wiles117, Jayne Willson117, Helen Newell117, Alison Lye117, Lorenza Nwafor117, Claire Jarman117, Sarah Rowland-Jones117, David Foote117, Joby Cole117, Roger Thompson117, James Watson117, Lisa Hesseldon117, Irene Macharia117, Luke Chetam 117, Jacqui Smith117, Amber Ford117, Samantha Anderson117, Kathryn Birchall117, Kay Housley117, Sara Walker117, Leanne Milner117, Helena Hanratty117, Helen Trower117, Patrick Phillips117, Simon Oxspring117, Ben Donne117.
Dumfries and Galloway Royal Infirmary, Dumfries, UK
Catherine Jardine118, Dewi Williams118, Alasdair Hay118.
Royal Bolton Hospital, Bolton, UK
Rebecca Flanagan119, Gareth Hughes119, scott Latham119, Emma McKenna119, Jennifer Anderson119, Robert Hull119, Kat Rhead119.
Lister Hospital, Stevenage, UK
Carina Cruz120, Natalie Pattison120.
Craigavon Area Hospital, County Armagh, NI
Rob Charnock121, Denise McFarland121, Denise Cosgrove121.
Southport and Formby District General Hospital, Ormskirk, UK
Ashar Ahmed122, Anna Morris122, Srinivas Jakkula122.
Calderdale Royal Hospital, Halifax, UK and Huddersfield Royal Infirmary, Huddersfield, UK
Asifa Ali123.
Calderdale Royal Hospital, Halifax, UK
Megan Brady123, Sam Dale123, Annalisa Dance123, Lisa Gledhill123, Jill Greig123, Kathryn Hanson123, Kelly Holdroyd123, Marie Home123, Diane Kelly123, Ross Kitson123, Lear Matapure123, Deborah Melia123, Samantha Mellor123, Tonicha Nortcliffe123, Jez Pinnell123, Matthew Robinson123, Lisa Shaw123, Ryan Shaw123, Lesley Thomis123, Alison Wilson123, Tracy Wood123, Lee-Ann Bayo123, Ekta Merwaha123, Tahira Ishaq123, Sarah Hanley123.
Prince Charles Hospital, Merthyr Tydfil, UK
Bethan Deacon124, Meg Hibbert124, Carla Pothecary124, Dariusz Tetla124, Chrsitopher Woodford124, Latha Durga124, Gareth Kennard-Holden 124.
Royal Bournemouth Hospital, Bournemouth, UK
Debbie Branney125, Jordan Frankham125, Sally Pitts125, Nigel White125.
Royal Preston Hospital, Preston, UK
Shondipon Laha126, Mark Verlander126, Alexandra Williams126.
Whittington Hospital, London, UK
Abdelhakim Altabaibeh127, Ana Alvaro127, Kayleigh Gilbert127, Louise Ma127, Loreta Mostoles127, Chetan Parmar127, Kathryn Simpson127, Champa Jetha127, Lauren Booker127, Anezka Pratley127.
Princess Royal Hospital, Telford and Royal Shrewsbury Hospital, Shrewsbury, UK
Colene Adams128, Anita Agasou128, Tracie Arden128, Amy Bowes128, Pauline Boyle128, Mandy Beekes128, Heather Button128, Nigel Capps128, Mandy Carnahan128, Anne Carter128, Danielle Childs128, Denise Donaldson128, Kelly Hard128, Fran Hurford128, Yasmin Hussain128, Ayesha Javaid128, James Jones128, Sanal Jose128, Michael Leigh128, Terry Martin128, Helen Millward128, Nichola Motherwell128, Rachel Rikunenko128, Jo Stickley128, Julie Summers128, Louise Ting128, Helen Tivenan128, Louise Tonks128, Rebecca Wilcox128.
Macclesfield District General Hospital, Macclesfield, UK
Maureen Holland129, Natalie Keenan129, Marc Lyons129, Helen Wassall129, Chris Marsh129, Mervin Mahenthran129, Emma Carter129, Thomas Kong129.
Royal Surrey County Hospital, Guildford, UK
Helen Blackman130, Ben Creagh-Brown130, Sinead Donlon130, Natalia Michalak-Glinska130, Sheila Mtuwa130, Veronika Pristopan130, Armorel Salberg130, Eleanor Smith130, Sarah Stone130, Charles Piercy130, Jerik Verula130, Dorota Burda130, Rugia Montaser130, Lesley Harden130, Irving Mayangao130, Cheryl Marriott130, Paul Bradley130, Celia Harris130.
Hereford County Hospital, Hereford, UK
Susan Anderson131, Eleanor Andrews131, Janine Birch131, Emma Collins131, Kate Hammerton131, Ryan O'Leary131.
University Hospital of North Tees, Stockton on Tees, UK
Michele Clark132, Sarah Purvis132.
Lincoln County Hospital, Lincoln, UK
Russell Barber133, Claire Hewitt133, Annette Hilldrith133, Karen Jackson-Lawrence133, Sarah Shepardson133, Maryanne Wills133, Susan Butler 133, Silvia Tavares133, Amy Cunningham 133, Julia Hindale 133, Sarwat Arif 133.
Royal Cornwall Hospital, Truro, UK
Sarah Bean134, Karen Burt134, Michael Spivey134.
Royal United Hospital, Bath, UK
Carrie Demetriou135, Charlotte Eckbad135, Sarah Hierons135, Lucy Howie135, Sarah Mitchard135, Lidia Ramos135, Alfredo Serrano-Ruiz135, Katie White135, Fiona Kelly135.
Royal Brompton Hospital, London, UK
Daniele Cristiano136, Natalie Dormand136, Zohreh Farzad136, Mahitha Gummadi136, Kamal Liyanage136, Brijesh Patel136, Sara Salmi136, Geraldine Sloane136, Vicky Thwaites136, Mathew Varghese136, Anelise C Zborowski136.
University Hospital Crosshouse, Kilmarnock, UK
John Allan137, Tim Geary137, Gordon Houston137, Alistair Meikle137, Peter O'Brien137.
Basildon Hospital, Basildon, UK
Miranda Forsey138, Agilan Kaliappan138, Anne Nicholson138, Joanne Riches138, Mark Vertue138, Miranda Forsey138, Agilan Kaliappan138, Anne Nicholson138, Joanne Riches138, Mark Vertue138.
Glan Clwyd Hospital, Bodelwyddan, UK
Elizabeth Allan139, Kate Darlington139, Ffyon Davies139, Jack Easton139, Sumit Kumar139, Richard Lean139, Daniel Menzies139, Richard Pugh139, Xinyi Qiu139, Llinos Davies139, Hannah Williams 139, Jeremy Scanlon139, Gwyneth Davies 139, Callum Mackay139, Joannne Lewis139, Stephanie Rees139.
West Middlesex Hospital, Isleworth, UK
Metod Oblak140, Monica Popescu140, Mini Thankachen140.
Royal Lancaster Infirmary, Lancaster, UK
Andrew Higham141, Kerry Simpson141, Jayne Craig141.
Western General Hospital, Edinburgh, UK
Rosie Baruah142, Sheila Morris142, Susie Ferguson142, Amy Shepherd142.
Chelsea & Westminster NHS Foundation Trust, London, UK
Luke Stephen Prockter Moore143, Marcela Paola Vizcaychipi143, Laura Gomes de Almeida Martins143, Jaime Carungcong143.
The Queen Elizabeth Hospital, King's Lynn, UK
Inthakab Ali Mohamed Ali144, Karen Beaumont144, Mark Blunt144, Zoe Coton144, Hollie Curgenven144, Mohamed Elsaadany144, Kay Fernandes144, Sameena Mohamed Ally144, Harini Rangarajan144, Varun Sarathy144, Sivarupan Selvanayagam144, Dave Vedage144, Matthew White144.
King's Mill Hospital, Nottingham, UK
Mandy Gill145, Paul Paul145, Valli Ratnam145, Sarah Shelton145, Inez Wynter145.
Watford General Hospital, Watford, UK
Siobhain Carmody146, Valerie Joan Page146.
University Hospital Wishaw, Wishaw, UK
Claire Marie Beith147, Karen Black147, Suzanne Clements147, Alan Morrison147, Dominic Strachan147, Margaret Taylor147, Michelle Clarkson147, Stuart D'Sylva147, Kathryn Norman147.
Forth Valley Royal Hospital, Falkirk, UK
Fiona Auld148, Joanne Donnachie148, Ian Edmond148, Lynn Prentice148, Nikole Runciman148, Dario Salutous148, Lesley Symon148, Anne Todd148, Patricia Turner148, Abigail Short148, Laura Sweeney148, Euan Murdoch148, Dhaneesha Senaratne148.
George Eliot Hospital NHS Trust, Nuneaton, UK
Michaela Hill149, Thogulava Kannan149, Wild Laura149.
Barnsley Hospital, Barnsley, UK
Rikki Crawley150, Abigail Crew150, Mishell Cunningham150, Allison Daniels150, Laura Harrison150, Susan Hope150, Ken Inweregbu150, Sian Jones150, Nicola Lancaster150, Jamie Matthews150, Alice Nicholson150, Gemma Wray150.
The Great Western Hospital, Swindon, UK
Helen Langton151, Rachel Prout151, Malcolm Watters151, Catherine Novis151.
Harefield Hospital, London, UK
Anthony Barron152, Ciara Collins152, Sundeep Kaul152, Heather Passmore152, Claire Prendergast152, Anna Reed152, Paula Rogers152, Rajvinder Shokkar152, Meriel Woodruff152, Hayley Middleton 152, Oliver Polgar152, Claire Nolan 152, Vicky Thwaites152, Kanta Mahay152.
Rotherham General Hospital, Rotherham, UK
Dawn Collier153, Anil Hormis153, Victoria Maynard153, Cheryl Graham153, Rachel Walker153, Victoria Maynard153.
Ysbyty Gwynedd, Bangor, UK
Ellen Knights154, Alicia Price154, Alice Thomas154, Chris Thorpe154.
Diana Princess of Wales Hospital, Grimsby, UK
Teresa Behan155, Caroline Burnett155, Jonathan Hatton155, Elaine Heeney155, Atideb Mitra155, Maria Newton155, Rachel Pollard155, Rachael Stead155.
Russell's Hall Hospital, Dudley, UK
Vishal Amin156, Elena Anastasescu156, Vikram Anumakonda156, Komala Karthik156, Rizwana Kausar156, Karen Reid156, Jacqueline Smith156, Janet Imeson-Wood156.
Princess Royal Hospital
Denise Skinner157, Jane Gaylard157, Dee Mullan157, Julie Newman157.
Princess Royal Hospital, Haywards Heath, UK
Denise Skinner157, Jane Gaylard157, Dee Mullan157, Julie Newman157.
St Mary's Hospital, Newport, UK
Alison Brown158, Vikki Crickmore158, Gabor Debreceni158, Joy Wilkins158, Liz Nicol158.
University Hospital Lewisham, London, UK
Waqas Khaliq159, Rosie Reece-Anthony159, Mark Birt159.
Colchester General Hospital, Colchester, UK
Alison Ghosh160, Emma Williams160.
Queen Elizabeth the Queen Mother Hospital, Margate, UK
Louise Allen161, Eva Beranova161, Nikki Crisp161, Joanne Deery161, Tracy Hazelton161, Alicia Knight161, Carly Price161, Sorrell Tilbey161, Salah Turki161, Sharon Turney161.
Royal Albert Edward Infirmary, Wigan, UK
Joshua Cooper162, Cheryl Finch162, Sarah Liderth162, Alison Quinn162, Natalia Waddington162.
Victoria Hospital, Kirkcaldy, UK
Tina Coventry163, Susan Fowler163, Michael MacMahon163, Amanda McGregor163.
Eastbourne District General Hospital, East Sussex, UK and Conquest Hospital, East Sussex, UK
Anne Cowley164, Judith Highgate164, Anne Cowley164, Judith Highgate164.
Cumberland Infirmary, Carlisle, UK
Alison Brown165, Jane Gregory165, Susan O'Connell165, Tim Smith165, Luigi Barberis165.
New Cross Hospital, Wolverhampton, UK
Shameer Gopal166, Nichola Harris166, Victoria Lake166, Stella Metherell166, Elizabeth Radford166.
The Princess Alexandra Hospital, Harlow, UK
Amelia Daniel167, Joanne Finn167, Rajnish Saha167, Nikki White167, Amy Easthope167.
Salisbury District Hospital, Salisbury, UK
Phil Donnison168, Fiona Trim168, Beena Eapen168.
Dorset County Hospital, Dorchester, UK
Jenny Birch169, Laura Bough169, Josie Goodsell169, Rebecca Tutton169, Patricia Williams169, Sarah Williams169, Barbara Winter-Goodwin169.
University College Dublin, St Vincent's University Hospital, Dublin, Ireland
Ailstair Nichol170, Kathy Brickell170, Michelle Smyth 170, Lorna Murphy170.
Glangwili General Hospital, Camarthen, UK
Samantha Coetzee171, Alistair Gales171, Igor Otahal171, Meena Raj171, Craig Sell171.
Gloucestershire Royal Hospital, Gloucester, UK
Paula Hilltout172, Jayne Evitts172, Amanda Tyler172, Joanne Waldron172.
Yeovil Hospital, Yeovil, UK
Kate Beesley173, Sarah Board173, Agnieszka Kubisz-Pudelko173, Alison Lewis173, Jess Perry173, Lucy Pippard173, Di Wood173, Clare Buckley173.
Leicester Royal Infirmary, Leicester, UK
Peter Barry174, Neil Flint174, Patel Rekha 174, Dawn Hales174.
Royal Manchester Children's Hospital, Manchester, UK
Lara Bunni175, Claire Jennings175, Monica Latif175, Rebecca Marshall175, Gayathri Subramanian175.
Royal Victoria Hospital, Belfast, NI
Peter J McGuigan176, Christopher Wasson176, Stephanie Finn176, Jackie Green176, Erin Collins176, Bernadette King176.
Wrexham Maelor Hospital, Wrexham, Wales
Andy Campbell177, Sara Smuts177, Joseph Duffield177, Oliver Smith177, Lewis Mallon177, Watkins Claire177.
Walsall Manor Hospital, Walsall, UK
Liam Botfield178, Joanna Butler178, Catherine Dexter178, Jo Fletcher178, Atul Garg178, Aditya Kuravi178, Poonam Ranga178, Emma Virgilio178.
Darent Valley Hospital, Dartford, UK
Zakaula Belagodu179, Bridget Fuller179, Anca Gherman179, Olumide Olufuwa179, Remi Paramsothy179, Carmel Stuart179, Naomi Oakley179, Charlotte Kamundi179, David Tyl179, Katy Collins179, Pedro Silva179, June Taylor179, Laura King179, Charlotte Coates179, Maria Crowley179, Phillipa Wakefield179, Jane Beadle 179, Laura Johnson179, Janet Sargeant179, Madeleine Anderson179.
Warrington General Hospital, Warrington, UK
Ailbhe Brady180, Rebekah Chan180, Jeff Little180, Shane McIvor180, Helena Prady180, Helen Whittle180, Bijoy Mathew180.
Warwick Hospital, Warwick, UK
Ben Attwood181, Penny Parsons181.
University Hospitals Coventry & Warwickshire NHS Trust, Coventry, UK
Geraldine Ward182, Pamela Bremmer182.
University Hospital Monklands, Airdrie, UK
West Joe183, Baird Tracy183, Ruddy Jim183.
Princess of Wales Hospital, Llantrisant, UK
Ellie Davies184, Lisa Roche184, Sonia Sathe184.
Northwick Park Hospital, London, UK
Catherine Dennis185, Alastair McGregor185, Victoria Parris185, Sinduya Srikaran185, Anisha Sukha185.
Raigmore Hospital, Inverness, UK
Rachael Campbell186, Noreen Clarke186, Jonathan Whiteside186, Mairi Mascarenhas186, Avril Donaldson186, Joanna Matheson186, Fiona Barrett186, Marianne O'Hara186, Laura Okeefe186, Clare Bradley186.
Royal Free Hospital, London, UK
Christine Eastgate-Jackson187, Helder Filipe187, Daniel Martin187, Amitaa Maharajh187, Sara Mingo Garcia187, Glykeria Pakou187, Mark De Neef187.
Scunthorpe General Hospital, Scunthorpe, UK
Kathy Dent188, Elizabeth Horsley188, Muhmmad Nauman Akhtar188, Sandra Pearson188, Dorota Potoczna188, Sue Spencer188.
West Cumberland Hospital, Whitehaven, UK
Melanie Clapham189, Rosemary Harper189, Una Poultney189, Polly Rice189, Tim Smith189, Rachel Mutch189, Luigi Barberis189.
Airedale General Hospital, Keighley, UK
Lisa Armstrong190, Hayley Bates190, Emma Dooks190, Fiona Farquhar190, Brigid Hairsine190, Chantal McParland190, Sophie Packham190.
Birmingham Children's Hospital, Birmingham, UK
Rehana Bi191, Barney Scholefield191, Lydia Ashton191.
Liverpool Heart and Chest Hospital, Liverpool, UK
Linsha George192, Sophie Twiss192, David Wright192.
Pilgrim Hospital, Lincoln, UK
Manish Chablani193, Amy Kirkby193, Kimberley Netherton193.
Prince Philip Hospital, Lianelli, UK
Kim Davies194, Linda O'Brien194, Zohra Omar194, Igor Otahal194, Emma Perkins194, Tracy Lewis194, Isobel Sutherland194.
Furness General Hospital, Barrow-in-Furness, UK
Karen Burns195, Andrew Higham195.
Scarborough General Hospital, Scarborough, UK
Dr Ben Chandler196, Kerry Elliott196, Janine Mallinson196, Alison Turnbull196.
Southend University Hospital, Westcliff-on-Sea, UK
Prisca Gondo197, Bernard Hadebe197, Abdul Kayani197, Bridgett Masunda197.
Alder Hey Children's Hospital, Liverpool, UK
Taya Anderson198, Dan Hawcutt198, Laura O'Malley198, Laura Rad198, Naomi Rogers198, Paula Saunderson198, Kathryn Sian Allison198, Deborah Afolabi198, jennifer whitbread198, Dawn jones 198, Rachael Dore 198.
Torbay Hospital, Torquay, UK
Matthew Halkes199, Pauline Mercer199, Lorraine Thornton199.
Borders General Hospital, Melrose, UK
Joy Dawson200, Sweyn Garrioch200, Melanie Tolson200, Jonathan Aldridge200.
Kent & Canterbury Hospital, Canterbury, UK
Ritoo Kapoor201, David Loader201, Karen Castle201.
West Suffolk Hospital, Bury St Edmunds, UK
Sally humphreys202, Ruth Tampsett202.
James Paget University Hospital NHS Trust, Great Yarmouth, UK
Katherine Mackintosh203, Amanda Ayers203, Wendy Harrison203, Julie North203.
The Christie NHS Foundation Trust, Manchester, UK
Suzanne Allibone204, Roman Genetu204, Vidya Kasipandian204, Amit Patel204, Ainhi Mac204, Anthony Murphy204, Parisa Mahjoob204, Roonak Nazari204, Lucy Worsley204, Andrew Fagan204.
The Royal Marsden Hospital, London, UK
Thomas Bemand205, Ethel Black205, Arnold Dela Rosa205, Ryan Howle205, Shaman Jhanji205, Ravishankar Rao Baikady205, Kate Colette Tatham205, Benjamin Thomas205.
University Hospital Hairmyres, East Kilbride, UK
Dina Bell206, Rosalind Boyle206, Katie Douglas206, Lynn Glass206, Emma Lee206, Liz Lennon206, Austin Rattray206.
Withybush General Hospital, Pembrokeshire, Wales
Abigail Taylor207, Rachel Anne Hughes207, Helen Thomas207, Alun Rees207, Michaela Duskova207, Janet Phipps207, Suzanne Brooks207, Michelle Edwards207.
Ealing Hospital, Southall, UK
Victoria Parris208, Sheena Quaid208, Ekaterina Watson208.
North Devon District Hospital, Barnstaple, UK
Adam Brayne209, Emma Fisher209, Jane Hunt209, Peter Jackson209, Duncan Kaye209, Nicholas Love209, Juliet Parkin209, Victoria Tuckey209, Lynne Van Koutrik209, Sasha Carter209, Benedict Andrew209, Louise Findlay209, Katie Adams209.
St John's Hospital Livingston, Livingston, UK
Jen Service210, Alison Williams210, Claire Cheyne210, Anne Saunderson210, Sam Moultrie210, Miranda Odam210.
Northampton General Hospital NHS Trust, Northampton, UK
Kathryn Hall211, Isheunesu Mapfunde 211, Charlotte Willis211, Alex Lyon211.
Harrogate and District NHS Foundation Trust, Harrogate, UK
Chunda Sri-Chandana212, Joslan Scherewode212, Lorraine Stephenson 212, Sarah Marsh 212.
National Hospital for Neurology and Neurosurgery, London, UK
David Brealey213, John Hardy213, Henry Houlden213, Eleanor Moncur213, Eamon Raith213, Ambreen Tariq213, Arianna Tucci213.
Bronglais General Hospital, Aberystwyth, UK
Maria Hobrok214, Ronda Loosley214, Heather McGuinness214, Helen Tench214, Rebecca Wolf-Roberts214.
Golden Jubilee National Hospital, Clydebank, UK
Val Irvine215, Benjamin Shelley215.
Homerton University Hospital Foundation NHS Trust, London UK
Amy Easthope216, Claire Gorman216, Abhinav Gupta216, Elizabeth Timlick216, Rebecca Brady216.
Royal Hospital for Children, Glasgow, UK
Colin Begg3, Barry Milligan3.
Sheffield Children's Hospital, Sheffield, UK
Arianna Bellini217, Jade Bryant217, Anton Mayer217, Amy Pickard217, Nicholas Roe217, Jason Sowter217, Alex Howlett 217.
The Royal Alexandra Children's Hospital, Brighton, UK
Katy Fidler218, Emma Tagliavini218, Kevin Donnelly218.
1Roslin Institute, University of Edinburgh, Easter Bush, Edinburgh, EH25 9RG, UK
2Intensive Care Unit, Royal Infirmary of Edinburgh, 54 Little France Drive, Edinburgh, EH16 5SA, UK
3Royal Hospital for Children, Glasgow, UK
4William Harvey Research Institute, Barts and the London School of Medicine and Dentistry, Queen Mary University of London, London EC1M 6BQ, UK
5Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, University of Oxford, Old Road Campus, Roosevelt Drive, Oxford, OX3 7FZ, UK
6Wellcome Centre for Human Genetics, University of Oxford, Oxford, UK
7Department of Anaesthesia and Intensive Care, The Chinese University of Hong Kong, Prince of Wales Hospital, Hong Kong, China
8Department of Critical Care Medicine, Queen's University and Kingston Health Sciences Centre, Kingston, ON, Canada
9Wellcome-Wolfson Institute for Experimental Medicine, Queen's University Belfast, Belfast, Northern Ireland, UK
10Department of Intensive Care Medicine, Royal Victoria Hospital, Belfast, Northern Ireland, UK
11UCL Centre for Human Health and Performance, London, W1T 7HA, UK
12Clinical Research Centre at St Vincent's University Hospital, University College Dublin, Dublin, Ireland
13National Heart and Lung Institute, Imperial College London, London, UK
14Imperial College Healthcare NHS Trust:London,London,UK
15Heart Institute, University of Sao Paulo, Brazil
16MRC Human Genetics Unit, Institute of Genetics and Molecular Medicine, University of Edinburgh, Western General Hospital, Crewe Road, Edinburgh, EH4 2XU, UK
17Intensive Care National Audit & Research Centre, London, UK
18NIHR Health Protection Research Unit for Emerging and Zoonotic Infections, Institute of Infection, Veterinary and Ecological Sciences University of Liverpool, Liverpool, L69 7BE, UK
19Respiratory Medicine, Alder Hey Children's Hospital, Institute in The Park, University of Liverpool, Alder Hey Children's Hospital, Liverpool, UK
20Department of Intensive Care Medicine, Guy's and St. Thomas NHS Foundation Trust, London, UK
21Department of Medicine, University of Cambridge, Cambridge, UK
22NIHR Clinical Research Network (CRN), North West London Core Team, 3rd Floor Administrative Block South, Clock Tower, Hammersmith Hospital, Du Cane Road, London W12 0HS
23Cambridge University Hospitals NHS Foundation Trust, Hills Road, Cambridge, CB2 0QQ, UK
24Edinburgh Clinical Research Facility, Western General Hospital, University of Edinburgh, EH4 2XU, UK
25Biostatistics Group, State Key Laboratory of Biocontrol, School of Life Sciences, Sun Yat-sen University, Guangzhou, China
26Department of Infectious Diseases, Leiden University Medical Center, Leiden, The Netherlands
27Guys and St Thomas’ Hospital, London, UK
28Barts Health NHS Trust, London, UK
29James Cook University Hospital, Middlesbrough, UK
30Royal Stoke University Hospital, Staffordshire, UK
31North Middlesex University Hospital NHS trust, London, UK
32north Middlesex University Hospital NHS trust, London, UK
33The Royal Liverpool University Hospital, Liverpool, UK
34King's College Hospital, London, UK
35Charing Cross Hospital, St Mary's Hospital and Hammersmith Hospital, London, UK
36Nottingham University Hospital, Nottingham, UK
37John Radcliffe Hospital, Oxford, UK
38Kingston Hospital, Surrey, UK
39kingston Hospital, Surrey, UK
40Royal Infirmary of Edinburgh, Edinburgh, UK
41Queen Alexandra Hospital, Portsmouth, UK
42Morriston Hospital, Swansea, UK
43Addenbrooke's Hospital, Cambridge, UK
44BHRUT (Barking Havering) - Queens Hospital and King George Hospital, Essex, UK
45Royal Sussex County Hospital, Brighton, UK
46Queen Elizabeth Hospital, Birmingham, UK
47St George's Hospital, London, UK
48Stepping Hill Hospital, Stockport, UK
49Countess of Chester Hospital, Chester, UK
50Royal Blackburn Teaching Hospital, Blackburn, UK
51The Tunbridge Wells Hospital and Maidstone Hospital, Kent, UK
52Royal Gwent Hospital, Newport, UK
53Pinderfields General Hospital, Wakefield, UK
54Royal Berkshire NHS Foundation Trust, Berkshire, UK
55Broomfield Hospital, Chelmsford, UK
56Northumbria Healthcare NHS Foundation Trust, North Shields, UK
57Whiston Hospital, Prescot, UK
58Croydon University Hospital, Croydon, UK
59York Hospital, York, UK
60Heartlands Hospital, Birmingham, UK
61Ashford and St Peter's Hospital, Surrey, UK
62Barnet Hospital, London, UK
63East Surrey Hospital, Redhill, UK
64Ninewells Hospital, Dundee, UK
65Worthing Hospital, Worthing, UK and St Richard's Hospital, Chichester, UK
66Southampton General Hospital, Southampton, UK
67The Alexandra Hospital, Redditch and Worcester Royal Hospital, Worcester, UK
68Sandwell General Hospital and City Hospital, Birmingham, UK
69Blackpool Victoria Hospital, Blackpool, UK
70Royal Glamorgan Hospital, Pontyclun, UK
71The Royal Oldham Hospital, Manchester, UK
72Glasgow Royal Infirmary, Glasgow, UK
73St James's University Hospital and Leeds General Infirmary, Leeds, UK
74University Hospital North Durham, Darlington, UK and Darlington Memorial Hospital, Darlington, UK
75Fairfield General Hospital, Bury, UK
76Wythenshawe Hospital, Manchester, UK
77Royal Alexandra Hospital, Paisley, UK
78Good Hope Hospital, Birmingham, UK
79Tameside General Hospital, Ashton Under Lyne, UK
80Royal Derby Hospital, Derby, UK
81Medway Maritime Hospital, Gillingham, UK
82Royal Victoria Infirmary, Newcastle Upon Tyne, UK
83Poole Hospital, Poole, UK
84Bedford Hospital, Bedford, UK
85Queens Hospital Burton, Burton-On-Trent, UK
86North Manchester General Hospital, Manchester, UK
87Aberdeen Royal Infirmary, Aberdeen, UK
88Derriford Hospital, Plymouth, UK
89Manchester Royal Infirmary, Manchester, UK
90Salford Royal Hospital, Manchester, UK
91William Harvey Hospital, Ashford, UK
92Queen Elizabeth University Hospital, Glasgow, UK
93Bradford Royal Infirmary, Bradford, UK
94Bristol Royal Infirmary, Bristol, UK
95Norfolk and Norwich University hospital (NNUH), Norwich, UK
96Queen Elizabeth Hospital Gateshead, Gateshead, UK
97Sunderland Royal Hospital, Sunderland, UK
98Aintree University Hospital, Liverpool, UK
99Hull Royal Infirmary, Hull, UK
100Hull Royal Infirmary, Hull, Uk
101University College Hospital, London, UK
102Royal Devon and Exeter Hospital, Exeter, UK
103The Royal Papworth Hospital, Cambridge, UK
104Ipswich Hospital, Ipswich, UK
105Southmead Hospital, Bristol, UK
106Milton Keynes University Hospital, Milton Keynes, UK
107Royal Hampshire County Hospital, Hampshire, UK
108Queen Elizabeth Hospital, Woolwich, London, UK
109Great Ormond St Hospital and UCL Great Ormond St Institute of Child Health NIHR Biomedical Research Centre, London, UK
110Stoke Mandeville Hospital, Buckinghamshire, UK
111University Hospital of Wales, Cardiff, UK
112Basingstoke and North Hampshire Hospital, Basingstoke, UK
113Arrowe Park Hospital, Wirral, UK
114Chesterfield Royal Hospital Foundation Trust, Chesterfield, UK
115Musgrove Park Hospital, Taunton, UK
116Peterborough City Hospital, Peterborough, UK and Hinchingbrooke Hospital, Huntingdon, UK
117Royal Hallamshire Hospital and Northern General Hospital, Sheffield, UK
118Dumfries and Galloway Royal Infirmary, Dumfries, UK
119Royal Bolton Hospital, Bolton, UK
120Lister Hospital, Stevenage, UK
121Craigavon Area Hospital, County Armagh, NI
122Southport and Formby District General Hospital, Ormskirk, UK
123Calderdale Royal Hospital, Halifax, UK and Huddersfield Royal Infirmary, Huddersfield, UK
124Prince Charles Hospital, Merthyr Tydfil, UK
125Royal Bournemouth Hospital, Bournemouth, UK
126Royal Preston Hospital, Preston, UK
127Whittington Hospital, London, UK
128Princess Royal Hospital, Telford and Royal Shrewsbury Hospital, Shrewsbury, UK
129Macclesfield District General Hospital, Macclesfield, UK
130Royal Surrey County Hospital, Guildford, UK
131Hereford County Hospital, Hereford, UK
132University Hospital of North Tees, Stockton on Tees, UK
133Lincoln County Hospital, Lincoln, UK
134Royal Cornwall Hospital, Truro, UK
135Royal United Hospital, Bath, UK
136Royal Brompton Hospital, London, UK
137University Hospital Crosshouse, Kilmarnock, UK
138Basildon Hospital, Basildon, UK
139Glan Clwyd Hospital, Bodelwyddan, UK
140West Middlesex Hospital, Isleworth, UK
141Royal Lancaster Infirmary, Lancaster, UK
142Western General Hospital, Edinburgh, UK
143Chelsea & Westminster NHS Foundation Trust, London, UK
144The Queen Elizabeth Hospital, King's Lynn, UK
145King's Mill Hospital, Nottingham, UK
146Watford General Hospital, Watford, UK
147University Hospital Wishaw, Wishaw, UK
148Forth Valley Royal Hospital, Falkirk, UK
149George Eliot Hospital NHS Trust, Nuneaton, UK
150Barnsley Hospital, Barnsley, UK
151The Great Western Hospital, Swindon, UK
152Harefield Hospital, London, UK
153Rotherham General Hospital, Rotherham, UK
154Ysbyty Gwynedd, Bangor, UK
155Diana Princess of Wales Hospital, Grimsby, UK
156Russell's Hall Hospital, Dudley, UK
157Princess Royal Hospital, Haywards Heath, UK
158St Mary's Hospital, Newport, UK
159University Hospital Lewisham, London, UK
160Colchester General Hospital, Colchester, UK
161Queen Elizabeth the Queen Mother Hospital, Margate, UK
162Royal Albert Edward Infirmary, Wigan, UK
163Victoria Hospital, Kirkcaldy, UK
164Eastbourne District General Hospital, East Sussex, UK and Conquest Hospital, East Sussex, UK
165Cumberland Infirmary, Carlisle, UK
166New Cross Hospital, Wolverhampton, UK
167The Princess Alexandra Hospital, Harlow, UK
168Salisbury District Hospital, Salisbury, UK
169Dorset County Hospital, Dorchester, UK
170University College Dublin, St Vincent's University Hospital, Dublin, Ireland
171Glangwili General Hospital, Camarthen, UK
172Gloucestershire Royal Hospital, Gloucester, UK
173Yeovil Hospital, Yeovil, UK
174Leicester Royal Infirmary, Leicester, UK
175Royal Manchester Children's Hospital, Manchester, UK
176Royal Victoria Hospital, Belfast, NI
177Wrexham Maelor Hospital, Wrexham, Wales
178Walsall Manor Hospital, Walsall, UK
179Darent Valley Hospital, Dartford, UK
180Warrington General Hospital, Warrington, UK
181Warwick Hospital, Warwick, UK
182University Hospitals Coventry & Warwickshire NHS Trust, Coventry, UK
183University Hospital Monklands, Airdrie, UK
184Princess of Wales Hospital, Llantrisant, UK
185Northwick Park Hospital, London, UK
186Raigmore Hospital, Inverness, UK
187Royal Free Hospital, London, UK
188Scunthorpe General Hospital, Scunthorpe, UK
189West Cumberland Hospital, Whitehaven, UK
190Airedale General Hospital, Keighley, UK
191Birmingham Children's Hospital, Birmingham, UK
192Liverpool Heart and Chest Hospital, Liverpool, UK
193Pilgrim Hospital, Lincoln, UK
194Prince Philip Hospital, Lianelli, UK
195Furness General Hospital, Barrow-in-Furness, UK
196Scarborough General Hospital, Scarborough, UK
197Southend University Hospital, Westcliff-on-Sea, UK
198Alder Hey Children's Hospital, Liverpool, UK
199Torbay Hospital, Torquay, UK
200Borders General Hospital, Melrose, UK
201Kent & Canterbury Hospital, Canterbury, UK
202West Suffolk Hospital, Bury St Edmunds, UK
203James Paget University Hospital NHS Trust, Great Yarmouth, UK
204The Christie NHS Foundation Trust, Manchester, UK
205The Royal Marsden Hospital, London, UK
206University Hospital Hairmyres, East Kilbride, UK
207Withybush General Hospital, Pembrokeshire, Wales
208Ealing Hospital, Southall, UK
209North Devon District Hospital, Barnstaple, UK
210St John's Hospital Livingston, Livingston, UK
211Northampton General Hospital NHS Trust, Northampton, UK
212Harrogate and District NHS Foundation Trust, Harrogate, UK
213National Hospital for Neurology and Neurosurgery, London, UK
214Bronglais General Hospital, Aberystwyth, UK
215Golden Jubilee National Hospital, Clydebank, UK
216Homerton University Hospital Foundation NHS Trust, London UK
217Sheffield Children's Hospital, Sheffield, UK
218The Royal Alexandra Children's Hospital, Brighton, UK
Appendix 2: ISARIC4C Investigators
Consortium Lead Investigator: J Kenneth Baillie.
Chief Investigator: Malcolm G Semple.
Co-Lead Investigator: Peter JM Openshaw.
ISARIC Clinical Coordinator: Gail Carson.
Co-Investigator: Beatrice Alex, Petros Andrikopoulos, Benjamin Bach, Wendy S Barclay, Debby Bogaert, Meera Chand, Kanta Chechi, Graham S Cooke, Ana da Silva Filipe, Thushan de Silva, Annemarie B Docherty, Gonçalo dos Santos Correia, Marc-Emmanuel Dumas, Jake Dunning, Tom Fletcher, Christoper A Green, William Greenhalf, Julian L Griffin, Rishi K Gupta, Ewen M Harrison, Julian A Hiscox, Antonia Ying Wai Ho, Peter W Horby, Samreen Ijaz, Saye Khoo, Paul Klenerman, Andrew Law, Matthew R Lewis, Sonia Liggi, Wei Shen Lim, Lynn Maslen, Alexander J Mentzer, Laura Merson, Alison M Meynert, Shona C Moore, Mahdad Noursadeghi, Michael Olanipekun, Anthonia Osagie, Massimo Palmarini, Carlo Palmieri, William A Paxton, Georgios Pollakis, Nicholas Price, Andrew Rambaut, David L Robertson, Clark D Russell, Vanessa Sancho-Shimizu, Caroline J Sands, Janet T Scott, Louise Sigfrid, Tom Solomon, Shiranee Sriskandan, David Stuart, Charlotte Summers, Olivia V Swann, Zoltan Takats, Panteleimon Takis, Richard S Tedder, AA Roger Thompson, Emma C Thomson, Ryan S Thwaites, Lance CW Turtle, Maria Zambon.
Project Manager: Hayley Hardwick, Chloe Donohue, Fiona Griffiths, Wilna Oosthuyzen.
Project Administrator: Cara Donegan, Rebecca G. Spencer.
Data Analyst: Lisa Norman, Riinu Pius, Thomas M Drake, Cameron J Fairfield, Stephen R Knight, Kenneth A Mclean, Derek Murphy, Catherine A Shaw.
Data and Information System Manager: Jo Dalton, Michelle Girvan, Egle Saviciute, Stephanie Roberts, Janet Harrison, Laura Marsh, Marie Connor, Sophie Halpin, Clare Jackson, Carrol Gamble, Daniel Plotkin, James Lee.
Data Integration and Presentation: Gary Leeming, Andrew Law, Murray Wham, Sara Clohisey, Ross Hendry, James Scott-Brown.
Material Management: Victoria Shaw, Sarah E McDonald.
Patient Engagement: Seán Keating.
Outbreak Laboratory Staff and Volunteers: Katie A. Ahmed, Jane A Armstrong, Milton Ashworth, Innocent G Asiimwe, Siddharth Bakshi, Samantha L Barlow, Laura Booth, Benjamin Brennan, Katie Bullock, Benjamin WA Catterall, Jordan J Clark, Emily A Clarke, Sarah Cole, Louise Cooper, Helen Cox, Christopher Davis, Oslem Dincarslan, Chris Dunn, Philip Dyer, Angela Elliott, Anthony Evans, Lorna Finch, Lewis WS Fisher, Terry Foster, Isabel Garcia-Dorival, Philip Gunning, Catherine Hartley, Rebecca L Jensen, Christopher B Jones, Trevor R Jones, Shadia Khandaker, Katharine King, Robyn T. Kiy, Chrysa Koukorava, Annette Lake, Suzannah Lant, Diane Latawiec, Lara Lavelle-Langham, Daniella Lefteri, Lauren Lett, Lucia A Livoti, Maria Mancini, Sarah McDonald, Laurence McEvoy, John McLauchlan, Soeren Metelmann, Nahida S Miah, Joanna Middleton, Joyce Mitchell, Shona C Moore, Ellen G Murphy, Rebekah Penrice-Randal, Jack Pilgrim, Tessa Prince, Will Reynolds, P. Matthew Ridley, Debby Sales, Victoria E Shaw, Rebecca K Shears, Benjamin Small, Krishanthi S Subramaniam, Agnieska Szemiel, Aislynn Taggart, Jolanta Tanianis-Hughes, Jordan Thomas, Erwan Trochu, Libby van Tonder, Eve Wilcock, J. Eunice Zhang, Lisa Flaherty, Nicole Maziere, Emily Cass, Alejandra Doce Carracedo, Nicola Carlucci, Anthony Holmes, Hannah Massey.
Edinburgh Laboratory Staff and Volunteers: Lee Murphy, Sarah McCafferty, Richard Clark, Angie Fawkes, Kirstie Morrice, Alan Maclean, Nicola Wrobel, Lorna Donnelly, Audrey Coutts, Katarzyna Hafezi, Louise MacGillivray, Tammy Gilchrist.
Local Principal Investigators: Kayode Adeniji, Daniel Agranoff, Ken Agwuh, Dhiraj Ail, Erin L. Aldera, Ana Alegria, Sam Allen, Brian Angus, Abdul Ashish, Dougal Atkinson, Shahedal Bari, Gavin Barlow, Stella Barnass, Nicholas Barrett, Christopher Bassford, Sneha Basude, David Baxter, Michael Beadsworth, Jolanta Bernatoniene, John Berridge, Colin Berry, Nicola Best, Pieter Bothma, David Chadwick, Robin Brittain-Long, Naomi Bulteel, Tom Burden, Andrew Burtenshaw, Vikki Caruth, David Chadwick, Duncan Chambler, Nigel Chee, Jenny Child, Srikanth Chukkambotla, Tom Clark, Paul Collini, Catherine Cosgrove, Jason Cupitt, Maria-Teresa Cutino-Moguel, Paul Dark, Chris Dawson, Samir Dervisevic, Phil Donnison, Sam Douthwaite, Andrew Drummond, Ingrid DuRand, Ahilanadan Dushianthan, Tristan Dyer, Cariad Evans, Chi Eziefula, Chrisopher Fegan, Adam Finn, Duncan Fullerton, Sanjeev Garg, Sanjeev Garg, Atul Garg, Effrossyni Gkrania-Klotsas, Jo Godden, Arthur Goldsmith, Clive Graham, Elaine Hardy, Stuart Hartshorn, Daniel Harvey, Peter Havalda, Daniel B Hawcutt, Maria Hobrok, Luke Hodgson, Anil Hormis, Michael Jacobs, Susan Jain, Paul Jennings, Agilan Kaliappan, Vidya Kasipandian, Stephen Kegg, Michael Kelsey, Jason Kendall, Caroline Kerrison, Ian Kerslake, Oliver Koch, Gouri Koduri, George Koshy, Shondipon Laha, Steven Laird, Susan Larkin, Tamas Leiner, Patrick Lillie, James Limb, Vanessa Linnett, Jeff Little, Mark Lyttle, Michael MacMahon, Emily MacNaughton, Ravish Mankregod, Huw Masson, Elijah Matovu, Katherine McCullough, Ruth McEwen, Manjula Meda, Gary Mills, Jane Minton, Mariyam Mirfenderesky, Kavya Mohandas, Quen Mok, James Moon, Elinoor Moore, Patrick Morgan, Craig Morris, Katherine Mortimore, Samuel Moses, Mbiye Mpenge, Rohinton Mulla, Michael Murphy, Megan Nagel, Thapas Nagarajan, Mark Nelson, Lillian Norris, Matthew K. O'Shea, Igor Otahal, Marlies Ostermann, Mark Pais, Carlo Palmieri, Selva Panchatsharam, Danai Papakonstantinou, Hassan Paraiso, Brij Patel, Natalie Pattison, Justin Pepperell, Mark Peters, Mandeep Phull, Stefania Pintus, Jagtur Singh Pooni, Tim Planche, Frank Post, David Price, Rachel Prout, Nikolas Rae, Henrik Reschreiter, Tim Reynolds, Neil Richardson, Mark Roberts, Devender Roberts, Alistair Rose, Guy Rousseau, Bobby Ruge, Brendan Ryan, Taranprit Saluja, Matthias L Schmid, Aarti Shah, Prad Shanmuga, Anil Sharma, Anna Shawcross, Jeremy Sizer, Manu Shankar-Hari, Richard Smith, Catherine Snelson, Nick Spittle, Nikki Staines, Tom Stambach, Richard Stewart, Pradeep Subudhi, Tamas Szakmany, Kate Tatham, Jo Thomas, Chris Thompson, Robert Thompson, Ascanio Tridente, Darell Tupper-Carey, Mary Twagira, Nick Vallotton, Rama Vancheeswaran, Lisa Vincent-Smith, Shico Visuvanathan, Alan Vuylsteke, Sam Waddy, Rachel Wake, Andrew Walden, Ingeborg Welters, Tony Whitehouse, Paul Whittaker, Ashley Whittington, Padmasayee Papineni, Meme Wijesinghe, Martin Williams, Lawrence Wilson, Sarah Cole, Stephen Winchester, Martin Wiselka, Adam Wolverson, Daniel G Wootton, Andrew Workman, Bryan Yates, Peter Young.
Appendix. Supplementary materials
References
- 1.WHO. WHO Coronavirus (COVID-19) Dashboard [Internet]. Available from: https://covid19.who.int/
- 2.Zheng Z., Peng F., Xu B., Zhao J., Liu H., Peng J., et al. Risk factors of critical & mortal COVID-19 cases: a systematic literature review and meta-analysis. J Infect. 2020;81(2):e16–e25. doi: 10.1016/j.jinf.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Q., Bastard P., Bolze A., Jouanguy E., Zhang S.-.Y., C.O.V.I.D. Human Genetic Effort, et al. Life-threatening COVID-19: defective interferons unleash excessive inflammation. Med (N Y). 2020;1(1):14–20. [DOI] [PMC free article] [PubMed]
- 4.Severe Covid-19 GWAS Group. D Ellinghaus, Degenhardt F., Bujanda L., Buti M., Albillos A., et al. Genomewide association study of severe COVID-19 with respiratory failure. N Engl J Med. 2020;383(16):1522–1534. doi: 10.1056/NEJMoa2020283. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu B.B., Gu D-Z, Yu J.N., Yang J., Shen W.Q. Association between ABO blood groups and COVID-19 infection, severity and demise: a systematic review and meta-analysis. Infect Genet Evol. 2020;84 doi: 10.1016/j.meegid.2020.104485. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370(6515) doi: 10.1126/science.abd4570. Oct 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.-.H., Zhang Y., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370(6515) doi: 10.1126/science.abd4585. Oct 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2020 doi: 10.1038/s41586-020-03065-y. Dec 11. [DOI] [PubMed] [Google Scholar]
- 9.Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S., et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85(9):4122–4134. doi: 10.1128/JVI.02232-10. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84(24):12658–12664. doi: 10.1128/JVI.01542-10. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol. 2011;85(2):873–882. doi: 10.1128/JVI.02062-10. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. 16e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA. 2020 doi: 10.1073/pnas.2003138117. May 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin B., Ferguson C., White J.T., Wang S., Vessella R., True L.D., et al. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59(17):4180–4184. Sep 1. [PubMed] [Google Scholar]
- 15.Limburg H., Harbig A., Bestle D., Stein D.A., Moulton H.M., Jaeger J., et al. TMPRSS2 is the major activating protease of influenza a virus in primary human airway cells and influenza B virus in human type II pneumocytes. J Virol. 2019;93(21) doi: 10.1128/JVI.00649-19. Nov 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Afar D.E., Vivanco I., Hubert R.S., Kuo J., Chen E., Saffran D.C., et al. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 2001;61(4):1686–1692. Feb 15. [PubMed] [Google Scholar]
- 17.Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020 doi: 10.15252/embj.20105114. Apr 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaarala M.H., Porvari K.S., Kellokumpu S., Kyllönen A.P., Vihko P.T. Expression of transmembrane serine protease TMPRSS2 in mouse and human tissues. J Pathol. 2001;193(1):134–140. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH743>3.0.CO;2-T. Jan. [DOI] [PubMed] [Google Scholar]
- 19.Zhou P., Yang X.-.L., Wang X.-.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bilinska K., Jakubowska P., Von Bartheld C.S., Butowt R. Expression of the SARS-CoV-2 ENTRY PROTEINs, ACE2 and TMPRSS2, in cells of the olfactory epithelium: identification of cell types and trends with age. ACS Chem Neurosci. 2020;11(11):1555–1562. doi: 10.1021/acschemneuro.0c00210. 03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuler B.A., Habermann A.C., Plosa E.J., Taylor C.J., Jetter C., Negretti N.M., et al. Age-determined expression of priming protease TMPRSS2 and localization of SARS-CoV-2 in lung epithelium. J Clin Invest. 2021;131(1) doi: 10.1172/JCI140766. Jan 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection. J Virol. 2019;15(6):93. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F., Han M., Dai P., Xu W., He J., Tao X., et al. Distinct mechanisms for TMPRSS2 expression explain organ-specific inhibition of SARS-CoV-2 infection by enzalutamide. Nat Commun. 2021;12(1):866. doi: 10.1038/s41467-021-21171-x. Feb 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peacock T.P., Goldhill D.H., Zhou J., Baillon L., Frise R., Swann O.C., et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol. 2021 doi: 10.1038/s41564-021-00908-w. Apr 27. [DOI] [PubMed] [Google Scholar]
- 25.Kawase M., Shirato K., van der Hoek L., Taguchi F., Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J Virol. 2012;86(12):6537–6545. doi: 10.1128/JVI.00094-12. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mykytyn A.Z., Breugem T.I., Riesebosch S., Schipper D., van den Doel P.B., Rottier R.J., et al. SARS-CoV-2 entry into human airway organoids is serine protease-mediated and facilitated by the multibasic cleavage site. Elife. 2021;10 doi: 10.7554/eLife.64508. Jan 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y., Vedantham P., Lu K., Agudelo J., Carrion R., Nunneley J.W., et al. Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res. 2015;116:76–84. doi: 10.1016/j.antiviral.2015.01.011. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann M., Hofmann-Winkler H., Smith J.C., Krüger N., Arora P., Sørensen L.K., et al. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine. 2021 doi: 10.1016/j.ebiom.2021.103255. Mar 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ClinicalTrial. gov [Internet]. Available from: https://clinicaltrials.gov/ct2/results?cond=&term=camostat&cntry=&state=&city=&dist=
- 30.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45(D1):D158–D169. doi: 10.1093/nar/gkw1099. Jan 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021 doi: 10.1038/s41586-021-03819-2. Jul 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ittisoponpisan S., Islam S.A., Khanna T., Alhuzimi E., David A., Sternberg M.J.E. Can predicted protein 3D structures provide reliable insights into whether missense variants are disease associated? J Mol Biol. 2019;431(11):2197–2212. doi: 10.1016/j.jmb.2019.04.009. May 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaser R., Adusumalli S., Leng S.N., Sikic M., Ng P.C. SIFT missense predictions for genomes. Nat Protoc. 2016;11(1):1–9. doi: 10.1038/nprot.2015.123. Jan. [DOI] [PubMed] [Google Scholar]
- 35.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.González-Pérez A., López-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88(4):440–449. doi: 10.1016/j.ajhg.2011.03.004. Apr 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. Jul 1Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R., et al. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat Commun. 2020;11(1):3523. doi: 10.1038/s41467-020-17409-9. Jul 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edie S., Zaghloul N.A., Leitch C.C., Klinedinst D.K., Lebron J., Thole J.F., et al. Survey of human chromosome 21 gene expression effects on early development in Danio rerio. G3 (Bethesda) 2018;8(7):2215–2223. doi: 10.1534/g3.118.200144. Jul 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou W., Nielsen J.B., Fritsche L.G., Dey R., Gabrielsen M.E., Wolford B.N., et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet. 2018;50(9):1335–1341. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. Sep 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.COVID-19 host genetics initiative. Mapping the human genetic architecture of COVID-19. Nature. 2021 doi: 10.1038/s41586-021-03767-x. Jul 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10(5) doi: 10.1371/journal.pgen.1004383. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.GTEx Consortium Laboratory, data analysis &coordinating center (LDACC)—analysis working group, statistical methods groups—analysis working group, enhancing GTEx (eGTEx) groups, NIH common fund, NIH/NCI, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–213. doi: 10.1038/nature24277. Oct 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aruffo A., Bowen M.A., Patel D.D., Haynes B.F., Starling G.C., Gebe J.A., et al. CD6-ligand interactions: a paradigm for SRCR domain function? Immunol Today. 1997;18(10):498–504. doi: 10.1016/s0167-5699(97)01130-4. Oct. [DOI] [PubMed] [Google Scholar]
- 46.Freeman M., Ashkenas J., Rees D.J., Kingsley D.M., Copeland N.G., Jenkins N.A., et al. An ancient, highly conserved family of cysteine-rich protein domains revealed by cloning type I and type II murine macrophage scavenger receptors. Proc Natl Acad Sci USA. 1990;87(22):8810–8814. doi: 10.1073/pnas.87.22.8810. Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Resnick D., Pearson A., Krieger M. The SRCR superfamily: a family reminiscent of the Ig superfamily. Trends Biochem Sci. 1994;19(1):5–8. doi: 10.1016/0968-0004(94)90165-1. Jan. [DOI] [PubMed] [Google Scholar]
- 48.The National Genomic Research Library v5.1, Genomics England [Internet]. Available from: https://doi.org/10.6084/m9.figshare.4530893.v6 2020.
- 49.Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pöhlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2014;88(2):1293–1307. doi: 10.1128/JVI.02202-13. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto N., Bauer G. Apparent difference in fatalities between Central Europe and East Asia due to SARS-COV-2 and COVID-19: four hypotheses for possible explanation. Med Hypotheses. 2020;144 doi: 10.1016/j.mehy.2020.110160. Aug 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang F., Huang S., Gao R., Zhou Y., Lai C., Li Z., et al. Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discov. 2020;6(1):83. doi: 10.1038/s41421-020-00231-4. Nov 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Breining P., Frølund A.L., Højen J.F., Gunst J.D., Staerke N.B., Saedder E., et al. Camostat mesylate against SARS-CoV-2 and COVID-19-Rationale, dosing and safety. Basic Clin Pharmacol Toxicol. 2021;128(2):204–212. doi: 10.1111/bcpt.13533. Feb. [DOI] [PubMed] [Google Scholar]
- 53.Sakr Y., Bensasi H., Taha A., Bauer M., Ismail K., the UAE-Jena Research Group Camostat mesylate therapy in critically ill patients with COVID-19 pneumonia. Intensive Care Med. 2021;47(6):707–709. doi: 10.1007/s00134-021-06395-1. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gunst J.D., Staerke N.B., Pahus M.H., Kristensen L.H., Bodilsen J., Lohse N., et al. Efficacy of the TMPRSS2 inhibitor camostat mesilate in patients hospitalized with COVID-19-a double-blind randomized controlled trial. EClinicalMedicine. 2021;35 doi: 10.1016/j.eclinm.2021.100849. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Foipan dropped as potential coronavirus treatment [Internet]. Available from: https://www.thepharmaletter.com/in-brief/brief-foipan-dropped-as-potential-coronavirus-treatment
- 56.Wilson S., Greer B., Hooper J., Zijlstra A., Walker B., Quigley J., et al. The membrane-anchored serine protease, TMPRSS2, activates PAR-2 in prostate cancer cells. Biochem J. 2005;388(Pt 3):967–972. doi: 10.1042/BJ20041066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lam D.K., Dang D., Flynn A.N., Hardt M., Schmidt B.L. TMPRSS2, a novel membrane-anchored mediator in cancer pain. Pain. 2015;156(5):923–930. doi: 10.1097/j.pain.0000000000000130. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.D'Andrea M.R., Derian C.K., Leturcq D., Baker S.M., Brunmark A., Ling P., et al. Characterization of protease-activated receptor-2 immunoreactivity in normal human tissues. J Histochem Cytochem. 1998;46(2):157–164. doi: 10.1177/002215549804600204. Feb. [DOI] [PubMed] [Google Scholar]
- 59.Cocks T.M., Fong B., Chow J.M., Anderson G.P., Frauman A.G., Goldie R.G., et al. A protective role for protease-activated receptors in the airways. Nature. 1999;398(6723):156–160. doi: 10.1038/18223. Mar 11. [DOI] [PubMed] [Google Scholar]
- 60.Reed C.E., Kita H. The role of protease activation of inflammation in allergic respiratory diseases. J Allergy Clin Immunol. 2004;114(5):997–1008. doi: 10.1016/j.jaci.2004.07.060. Novquiz 1009. [DOI] [PubMed] [Google Scholar]
- 61.Heuberger D.M., Schuepbach R.A. Protease-activated receptors (PARs): mechanisms of action and potential therapeutic modulators in PAR-driven inflammatory diseases. Thromb J [Internet]. 2019;17 doi: 10.1186/s12959-019-0194-8. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6440139/ Mar 29 [cited 2021 Feb 3]Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su X., Camerer E., Hamilton J.R., Coughlin S.R., Matthay M.A. Protease-activated receptor-2 activation induces acute lung inflammation by neuropeptide-dependent mechanisms. J Immunol. 2005;175(4):2598–2605. doi: 10.4049/jimmunol.175.4.2598. Aug 15. [DOI] [PubMed] [Google Scholar]
- 63.Matsushima R., Takahashi A., Nakaya Y., Maezawa H., Miki M., Nakamura Y., et al. Human airway trypsin-like protease stimulates human bronchial fibroblast proliferation in a protease-activated receptor-2-dependent pathway. Am J Physiol Lung Cell Mol Physiol. 2006;290(2):L385–L395. doi: 10.1152/ajplung.00098.2005. Feb. [DOI] [PubMed] [Google Scholar]
- 64.Chokki M., Yamamura S., Eguchi H., Masegi T., Horiuchi H., Tanabe H., et al. Human airway trypsin-like protease increases mucin gene expression in airway epithelial cells. Am J Respir Cell Mol Biol. 2004;30(4):470–478. doi: 10.1165/rcmb.2003-0199OC. Apr. [DOI] [PubMed] [Google Scholar]
- 65.David A., Khanna T., Beykou M., Hanna G., Sternberg M.J. Structure, function and variants analysis of the androgen-regulated TMPRSS2, a drug target candidate for COVID-19 infection. BioRxiv. 2020 May. [Google Scholar]
- 66.Monticelli M., Mele B.H., Andreotti G., Cubellis M.V., Riccio G. Why does SARS-CoV-2 hit in different ways? Host genetic factors can influence the acquisition or the course of COVID-19. Eur J Med Genet. 2021;64(6) doi: 10.1016/j.ejmg.2021.104227. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zarubin A., Stepanov V., Markov A., Kolesnikov N., Marusin A., Khitrinskaya I., et al. Structural variability, expression profile, and pharmacogenetic properties of TMPRSS2 gene as a potential target for COVID-19 therapy. Genes (Basel) 2020;12(1):E19. doi: 10.3390/genes12010019. Dec 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vargas-Alarcón G., Posadas-Sánchez R., Ramírez-Bello J. Variability in genes related to SARS-CoV-2 entry into host cells (ACE2, TMPRSS2, TMPRSS11A, ELANE, and CTSL) and its potential use in association studies. Life Sci. 2020;260 doi: 10.1016/j.lfs.2020.118313. Nov 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hou Y., Zhao J., Martin W., Kallianpur A., Chung M.K., Jehi L., et al. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020;18(1):216. doi: 10.1186/s12916-020-01673-z. Jul 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paniri A., Hosseini M.M., Akhavan-Niaki H. First comprehensive computational analysis of functional consequences of TMPRSS2 SNPs in susceptibility to SARS-CoV-2 among different populations. J Biomol Struct Dyn. 2021;39(10):3576–3593. doi: 10.1080/07391102.2020.1767690. Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Senapati S., Kumar S., Singh A.K., Banerjee P., Bhagavatula S. Assessment of risk conferred by coding and regulatory variations of TMPRSS2 and CD26 in susceptibility to SARS-CoV-2 infection in human. J Genet. 2020;99(1):53. doi: 10.1007/s12041-020-01217-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sah P., Fitzpatrick M.C., Zimmer C.F., Abdollahi E., Juden-Kelly L., Moghadas S.M., et al. Asymptomatic SARS-CoV-2 infection: a systematic review and meta-analysis. Proc Natl Acad Sci U S A. 2021;118(34) doi: 10.1073/pnas.2109229118. Aug 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Full summary-level data in support of the findings of this study are available for download from https://genomicc.org/data. Individual level data can be analysed by qualified researchers in the ISARIC 4C/GenOMICC data analysis platform by application at https://genomicc.org/data. BioBank data and Genomics England data are available to registered researchers at https://www.ukbiobank.ac.uk/ and https://www.genomicsengland.co.uk/. The COVID-19 Host Genetics Initiative2 (COVID-19hg) summary statistics are available at https://www.COVID-19hg.org/.
Supplementary Information is available for this paper.