Abstract
Background and Aim:
Infections with Campylobacter species have gained recognition as the most frequent cause of foodborne gastroenteritis globally. Their significance in South Africa is still an area of study interest. This study was, therefore, carried out to determine the occurrence of Campylobacter species in chickens from North West Province of South Africa as well as their antibiotic sensitivity status.
Materials and Methods:
A total of 2400 chicken fecal samples were collected and pooled to a total of 480 samples from five registered active poultry abattoirs in the Ngaka Modiri Molema District of North West Province, South Africa. Polymerase chain reaction (PCR) was used for the detection of Campylobacter spp. targeting the 16S rRNA gene while antibiotic sensitivity was determined using disk diffusion inhibition test.
Results:
After isolation, a total of 26 samples were confirmed to be harboring Campylobacter jejuni by PCR and sequencing. C. jejuni was found to be the only isolate detected in all the fecal samples tested. The study further demonstrated that C. jejuni infections were highest in the summer season (3%) followed by autumn and winter at 1%, while there were none detected in the spring. The isolated C. jejuni-positive samples on disk diffusion inhibition test displayed resistance to nalidixic acid, tetracycline, erythromycin, and ciprofloxacin at 98%, 80%, 83%, and 21%, respectively.
Conclusion:
C. jejuni isolated in this study is known to cause disease in humans, and thus its occurrence requires application of “One Health” strategy to reduce the spread of this zoonotic pathogen in South Africa.
Keywords: antibiotic resistance, Campylobacter jejuni, chickens, South Africa
Introduction
Foodborne diseases result from the consumption of food that is contaminated with pathogens which include bacteria, viruses, and parasites [1]. Outbreaks and sporadic cases of foodborne diseases are common worldwide [2]. One such infection is by bacteria of the genus Campylobacter which has gained recognition as the most frequent cause of global foodborne bacterial gastroenteritis overtaking those of Escherichia coli and Salmonella species in humans [3,4]. Campylobacter spp. which are commonly documented as the main contributors to foodborne diseases in humans are Campylobacter jejuni and Campylobacter coli [5,6].
Poultry is regarded as reservoir of Campylobacter spp., and it has been reported that up to a 100% of chickens at slaughter age may be infected [7]. However, chickens are considered as the main reservoir of Campylobacter spp. that leads to human campylobacteriosis, Campylobacter spp. can colonize chickens without causing any clinical signs in them [8]. In chickens, Campylobacter spp. are mostly found in the intestines, mainly in cecal and cloacal crypts [9,10]. However, the organism may be recovered from various organs, including the liver, small intestines, and gizzard [10]. As a result, it has become the major source of poultry meat and poultry product contamination that can lead to food safety concerns. Prevalence rates of Campylobacter spp. infection in slaughter age broiler flocks can be as high as 100% on some farm settings [11], thus making fecal contamination of poultry meat during slaughter a high-risk point in the transmission cycle of the disease to consumers.
Despite interest in studying this pathogen, a number of limitations still exist and the most important of which arise from the difficulty in culturing/and or isolating the organism. The types of Campylobacter spp. that can be isolated in the laboratory by culture procedures are influenced by the type of media used, culture method, and also the time and conditions from sample collection to culture [12]. It is difficult to detect and identify Campylobacter spp. due to its unique culture requirements as well as long incubation period [13,14]. However, different researchers have been using different methods to detect Campylobacter spp. including quantitative polymerase chain reaction (qPCR) [15], immunohistochemistry, fluorescence in situ hybridization [16], colorimetric aptasensor [17], and immunochromatographic assay [18].
Apart from knowing the diversity of Campylobacter spp. at slaughterhouses that may pose a risk to humans, there is also a growing concern about how this Campylobacter spp. respond to treatment with antimicrobial substances. The improper use of antimicrobial agents in food animals has resulted in the exposure and circulation of antimicrobial resistance bacteria, including antimicrobial-resistant Campylobacter [19,20]. Even though the resistance of Campylobactor spp. To antimicrobial agents has been reported worldwide, the situation seems to be graver and accelerating more rapidly in developing countries, where there is uncontrolled and widespread use of antibiotics [21]. It is important to document information on the occurrence as well as the development of antibiotic resistance by local isolates of Campylobacter spp. of each country.
Hence this study sought to characterize Campylobacter spp. found in feces of slaughter age broiler chickens at Ngaka Modiri Molema District Municipality of North West Province, South Africa, and further evaluated their antibiotic resistance status.
Materials and Methods
Ethical approval
The animal and human experimentation and animal care procedures ethical committee of NWU approved the study (Ethics number: NWU-00511-18-A5).
Study period and location
The study was conducted from September 2017 to July 2018. This study was conducted around Mafikeng city in the Ngaka Modiri Molema district of the North West Province, South Africa (Figure-1). The province is the second-largest chicken producer in South Africa at 21.3% after Western Cape with 21.9% according to South African Poultry Association [http://www.sapoultry.co.za/pdf-news/sapa-survey-report-role-and-function-2014.pdf].
Figure-1.
Map of South Africa showing sampling area in the Ngaka Modiri Molema District of North West Province, South Africa [Source: https://showme.co.za/facts-about-south-africa/the-maps-of-south-africa].
Sample collection
A total of 2400 chicken fecal samples were collected randomly post-evisceration from the intestines of randomly selected broiler chickens at slaughterhouses. Feces were picked from the caeca/rectum and were placed into sterile fecal containers. Each container consisted of pooled feces from five different broiler chickens of the same farm, resulting in an overall total of 480 pooled samples (Table-1). All samples were placed on ice and immediately transported to the laboratory of the Centre for Animal Health Studies of North West University for analysis.
Table-1.
The number of samples collected and the total pooled samples.
| Season | Collected samples | Pooled samples |
|---|---|---|
| Summer | 600 | (600/5) 120 |
| Autumn | 600 | (600/5) 120 |
| Winter | 600 | (600/5) 120 |
| Spring | 600 | (600/5) 120 |
| Total | 2400 | (20,400/5) 480 |
Enrichment and culture of Campylobacter species
Fecal samples were cultured within an hour of collection onto a Campylobacter selective media to isolate and identify the bacteria for antimicrobial sensitivity testing [22]. Approximately 0.2 g of feces was added to 1 mL blood free Campylobacter selective broth and incubated at 5°C in a microaerophilic atmosphere for 48 h. The broth was then vortexed at 14,000 rpm for 3 min and plated onto Campylobacter blood-free selective agar base (Modified CCDA-Preston), which is supplemented with CCDA selective supplement SR0155E (Oxoid England). Inoculated plates were incubated at 41.5°C in the microaerophilic atmosphere for 48 h. Suspect colonies were subcultured on Campylobacter blood-free selective agar base (Modified CCDA-Preston) to get pure cultures.
Molecular analysis
The total genomic DNA of cultivated isolates was purified following Zymo Research Fungal/Bacterial DNA kit instructions (Zymo Research Corp., CA, USA). A final, 100 μL of DNA elution buffer was added to elute the DNA which was quantified using a Nanodrop (Bio-Rad Incorporated, CA, USA).
Conventional PCR was used to detect Campylobacter spp. in the chicken feces using universal 16S rRNA Campylobacter spp. primers, namely, the forward primer C412F: GGA TGA CAC ACT TTT CGG AGC and the reverse C1228R: CAT TGT AGC ACG TGT GTC. DNA extracted from chicken fecal samples was subjected to PCR according to conditions described by Bullman et al. [23] using Engine DYAD Peltier thermal cycler (Bio-Rad, USA). A total reaction volume of 25 μL containing 3 μL DNA template, 12.5 μL PCR Master Mix, 8.5 μL nucleus-free water, and 2 μL of primer mix (10 μM each) was used. Amplified PCR products were resolved on a 1.5% agarose gel stained with ethidium bromide and visualized with Syngene InGenius Bioimager (UK).
The PCR products were sent for sequencing at Inqaba Biotechnical Industries (Pty) Ltd., Pretoria, South Africa. The homology of partial sequences obtained was compared with the sequences from the NCBI GenBank using nucleotide Basic Local Alignment Search Tool (BLASTn) and nucleotide sequences with similarity above 95% were considered as accurate.
Antibiotic susceptibility testing of Campylobacter isolates
Phenotyping antibacterial susceptibility screening to ciprofloxacin (5 μg), nalidixic acid (30 μg), erythromycin (15 μg), and tetracycline (30 μg) was conducted as recommended by the World Health Organization (WHO) Advisory Group on Integrated Surveillance of Antimicrobial Resistance guidelines on foodborne bacteria. The test was performed using the Kirby–Bauer disk diffusion method and the results were interpreted using the Clinical and Laboratory Standards Institute (CLSI) guidelines [24]. Antibacterial susceptibility test was performed on Mueller-Hinton agar (Neogen Corporation, Lansing, MI, United States) that was supplemented with 10% sheep blood, according to CLSI guidelines. The plates were then sealed in an anaerobic jar 2.5L (Oxoid, UK) each containing gas generating sachet without catalyst in a microaerophilic incubator at 41°C for 24 h. The suspect colonies were subjected to Gram staining and examination under a light microscope for the identification of any potential Campylobacter spp. Antibiotic susceptibility was calculated by the zones of inhibition observed around each antibiotic disc in millimeters. Standard reference strains of Staphylococcus aureus (ATCC® 29213, Thermo Fischer, USA) and C. jejuni ATCC (33560) were used as quality controls [22,25].
Results
A total of 480 samples were cultured on Campylobacter blood-free selective agar base (Modified CCDA-Preston). Colonies of typical Gram-negative curved rods were observed showing an “S” formation. Out of 480 samples, only 336 were suspected to be Campylobacter spp. The DNA extraction carried out from the identified colonies and PCR resulted in 70% (336/480) showing 816 bp Campylobacter spp.-like bands. However, after sequencing, only 26 PCR products generated nucleotide sequences which perfectly matched with C. jejuni using BLASTn and the sequences were submitted to the National Center for Biotechnology Information GenBank database (www.ncbi.nlm.nih.gov/BLAST) and assigned accession numbers, as shown in Table-2. The percentage of Campylobacter recovery was highest in summer and lowest in spring. The percentage positive, Campylobacter isolates, and seasons of fecal samples used in this study are summarized in Table-3.
Table-2.
Results of 26 isolates for 16S rRNA sequencing (PCR) and accession number.
| Samples ID | Sequence ID | Reference from NCBI database | Accession number in GenBank | Assigned accession number | Percentage similarity (%) |
|---|---|---|---|---|---|
| NWU1 | Seq1 | Campylobacter jejuni | LC382117 | MZ209102 | 100 |
| NWU2 | Seq2 | Campylobacter jejuni | CP028909 | MZ209103 | 99 |
| NWU3 | Seq3 | Campylobacter jejuni | KY559041 | MZ209104 | 99 |
| NWU4 | Seq4 | Campylobacter jejuni | CP022079 | MZ209105 | 99 |
| NWU5 | Seq5 | Campylobacter jejuni | KY559043 | MZ209106 | 99 |
| NWU6 | Seq6 | Campylobacter jejuni | CP028912 | MZ209107 | 100 |
| NWU7 | Seq7 | Campylobacter jejuni | CP028185 | MZ209108 | 99 |
| NWU8 | Seq8 | Campylobacter jejuni | CP022079 | MZ209109 | 99 |
| NWU9 | Seq9 | Campylobacter jejuni | LC382117 | MZ209110 | 99 |
| NWU10 | Seq10 | Campylobacter jejuni | MF872610 | MZ209111 | 99 |
| NWU11 | Seq11 | Campylobacter jejuni | CP028912 | MZ209112 | 99 |
| NWU12 | Seq12 | Campylobacter jejuni | CP022079 | MZ209113 | 99 |
| NWU13 | Seq13 | Campylobacter jejuni | KY559041 | MZ209114 | 99 |
| NWU14 | Seq14 | Campylobacter jejuni | CP022079 | MZ209115 | 99 |
| NWU15 | Seq15 | Campylobacter jejuni | CP028912 | MZ209116 | 99 |
| NWU16 | Seq16 | Campylobacter jejuni | KY559041 | MZ209117 | 99 |
| NWU17 | Seq17 | Campylobacter jejuni | CP023866 | MZ209118 | 98 |
| NWU18 | Seq18 | Campylobacter jejuni | KY559041 | MZ209119 | 99 |
| NWU19 | Seq19 | Campylobacter jejuni | CP028185 | MZ209120 | 99 |
| NWU20 | Seq20 | Campylobacter jejuni | MF872610 | MZ209121 | 99 |
| NWU21 | Seq21 | Campylobacter jejuni | CP028912 | MZ209122 | 98 |
| NWU22 | Seq22 | Campylobacter jejuni | CP028909 | MZ209123 | 99 |
| NWU23 | Seq23 | Campylobacter jejuni | CP022079 | MZ209124 | 99 |
| NWU24 | Seq24 | Campylobacter jejuni | KY559041 | MZ209124 | 99 |
| NWU25 | Seq25 | Campylobacter jejuni | CP028912 | MZ209126 | 100 |
| NWU26 | Seq26 | Campylobacter jejuni | MF872610 | MZ209127 | 100 |
PCR=Polymerase chain reaction
Table-3.
Seasonal patterns of isolated Campylobacter spp. in chicken feces.
| Season | Collected samples | Total pooled samples | Positive samples | Campylobacter spp. (%) | Campylobacter jejuni (%) |
|---|---|---|---|---|---|
| Summer | 600 | (600/5) 120 | 144/480 | 14/480 (3) | 14/480 (3) |
| Autumn | 600 | (600/5) 120 | 96/480 | 6/480 (1) | 6/480 (1) |
| Winter | 600 | (600/5) 120 | 96/480 | 6/480 (1) | 6/480 (1) |
| Spring | 600 | (600/5) 120 | 0 | 0 | 0 |
| Total | 2400 | (2400/5) 480 | 336/480 | 26/480 (5) | 26/480 (5) |
Antibiotic susceptibility
The antibiotic susceptibility profiles of C. jejuni showed a high percentage resistance to nalidixic acid at 98%, tetracycline at 80%, erythromycin at 83%, and the lowest percentage resistance was ciprofloxacin at 21%. The antibiotics and concentrations used in this study, as well as the interpretation of results obtained, are listed in Figure-2. Multidrug resistance to more than 2 classes of antibiotics was found in 4 (15%) isolates.
Figure-2.
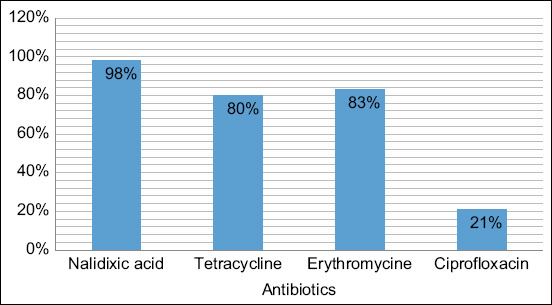
Antimicrobial resistance of Campylobacter isolated from fecal samples of broiler chickens.
Discussion
This study investigated Campylobacter spp. diversity in slaughter age chickens. C. jejuni (7.7%) was the only Campylobacter spp. isolated. This is a very significant finding since this is an important zoonotic infection under the “One health” paradigm, as it causes disease to humans which is acquired through chicken product consumption. The study, therefore, highlights that C. jejuni is the dominant species of the foodborne bacteria of the genus Campylobacter occurring in Mafikeng. This observation may, however, also mean that the method of isolation and/or sample processing we used was not adequate enough to pick up the other Campylobacter species in the samples collected. This can be seen from the number of Campylobacter-like bands that were seen after PCR on the gel, but which were not identified as such after sequencing. It is known that the time it takes to collect the sample and final transportation to the laboratory greatly affects the yield of Campylobacter diverse species.
Poultry meat and products have been considered a major source of C. jejuni for humans [26] and this study has also confirmed this likely source of transmission. The contamination of the meat and products with fecal matter containing C. jejuni, a known human pathogen, occurs during the slaughter process, especially at the time of degutting. The consumption of undercooked poultry meat and its products can thus be a major cause of human infections [27]. Furthermore, cross-contamination in the kitchen from contaminated meat to other food items, especially those that will not be cooked, is also considered a major pathway of transmission [27,28]. Although this study did not explore that route of transmission in the kitchen, the presence of contamination of the carcasses at slaughter makes this cross-contamination very possible.
A study by Richardson et al. [29] conducted in Soweto, South Africa, reported the relative ease of C. jejuni acquisition by families. This was attributed to the fact that poultry meat and meat products are highly consumed and more available because they are cheap and are also farmed in the backyards in different communities [29]. In Kwazulu-Natal Province, 43% of C. jejuni was confirmed [30]. In the study conducted by Richardson et al. [29], C. jejuni was recovered in 86% of fowl feces, pet dog feces, and bovine intestine (another popular food source). A previous study by Samie et al. [31] reported prevalence of 10.2% and 6.5% for C. jejuni and C. coli, respectively, from stool samples of hospital patients and asymptomatic pupils in Venda, Limpopo Province, South Africa. Another study conducted by Lastovica [32] in Cape Town, Western Cape Province, South Africa, detected 40%, 7.7%, and 24.6% of C. jejuni, C. coli, and Campylobacter concisus infections from diarrheagenic stools of children using culture methods.
In the current study, 3% (n=14) of total C. jejuni was isolated and confirmed during summer, 1% (n=6) in autumn, and 1% (n=6) in winter and there was no detection of C. jejuni during spring. Reports from other studies indicate that infections of Campylobacter are mostly sporadic and occur during warmer months of the summer and autumn. Various countries have indicated seasonal patterns to the pathogen, such as New Zealand and Australia [33]. In South Africa, there are limited data on the seasonality of Campylobacter spp. infections. This is probably because South Africa, as a developing country, lacks resources to investigate the pathogen on a continuous basis. This statement is supported by Coker et al. [34], who stated that public health infrastructure contributes to the lack of data in developing countries. Plats-Mills and Kosek [35] speculated that a lack of data in this regard could be due to the fact that the temperature variations in some developing countries are not as extreme as in developed countries. Nonetheless, the fact that Campylobacter is difficult to isolate is the most likely reason for the lack of data. However, the findings in the present study seem to indicate that the summer and warmer months have a higher risk because C. jejuni was isolated in more chicken feces during that period. The study area (North West Province) is a semi-desert area with extreme temperatures during summer. Another reason could be that there is less ventilation in summer, so chickens tend to be more stressed and shed more bacteria during this season.
In this study, C. jejuni was found to be resistant to nalidixic acid at 98%, tetracycline at 80%, erythromycin at 83%, and ciprofloxacin at 21%. Studies in other countries also show that Campylobacter spp. have developed resistance to antibiotics, specifically to macrolide antibiotics and fluoroquinolones, globally [36]. In another study in South Africa, ciprofloxacin resistance in clinical C. jejuni isolates from commercial chicken increased from 1.4% to 79% in 14 years between 1998 and 2011 [37]. Furthermore, C. jejuni isolated from our study was 80% resistant to tetracycline. According to Basardien [37], tetracycline resistance in C. jejuni isolated in commercial chicken increased from 14.2% to 86% between the years 1998 and 2011.
In the present study, C. jejuni isolates were 93% resistant to erythromycin. Macrolide antibiotics, including erythromycin, are considered the first drug of choice for human campylobacteriosis cases [38-40]. The resistance of clinical C. jejuni isolated in commercial chicken to erythromycin has increased from 3.4% and 97% between the years 1998 and 2012 [37]. However, the recent study conducted by Wieczorek et al. [41] reported 0% resistance to erythromycin for Campylobacter isolates in Poland. Erythromycin is generally used as the first drug of choice for treating Campylobacter gastroenteritis [4,20].
Investigation of the 26 C. jejuni isolates for antibiotic resistance revealed that 80% of the isolates were resistant to tetracycline, which is an antibiotic used against both Gram-negative and Gram-positive bacteria, including some other atypical and non-infectious microorganisms through inhibition of protein synthesis in these harmful agents [42,43]. Tetracycline is one of the most commonly used drugs in the livestock industry and it is, therefore, not surprising that one would find the levels of resistance revealed by our study.
The lowest resistance was noticed for ciprofloxacin at 21%. Similar results were obtained from different studies conducted by Geissler et al. [44] and Cody et al. [45], which reported 9% and 25% ciprofloxacin resistance by C. jejuni in the United States and Ireland, respectively.
In this study, multidrug resistance patterns were also observed among the isolates. Our findings do not stand alone since similar findings were observed in Poland, where 321 (60.9%) Campylobactor spp. Isolates from poultry were showing multidrug resistance [41], confirming that there is a disturbing increase in the emergence of multidrug-resistant Campylobacter [36,46,47]. Once again this observation of development of antibiotic resistance is of major concern, which is listed by the WHO as an item that requires “One Health” approach (https://www.who.int/news-room/q-a-detail/one-health).
This study is one of the many that has highlighted increased antibiotic resistance build-up in South Africa because of high levels of antibiotic use as therapeutic agents and growth promoters in the South African poultry industry. This has an impact on human health as antimicrobials are used to treat infections that cause morbidities and mortalities. Increased antibiotic resistance decreases the effectiveness of these drugs and results in human health being compromised [48].
Conclusion
This study successfully isolated Campylobacter spp. using direct DNA extraction from chicken feces and broth enrichment approach. The common species occurring in the study area is C. jejuni and its highest prevalence was during summer. This suggests that a high risk of contracting campylobacteriosis due to C. jejuni could possibly be during this season and chicken meat may be an important source. This study also revealed that the species of C. jejuni isolated from chicken feces had an increased prevalence of antibiotic resistance to ciprofloxacin, nalidixic acid, tetracycline, and erythromycin, which are used in the therapeutic treatment of humans that have acquired serious cases of campylobacteriosis. Detection of zoonotic C. jejuni and its development of antibiotic resistance highlights the need for “One Health” approach to combat the spread of these bacteria from the environment, animals, and humans in Mafikeng and South Africa as a whole.
Authors’ Contributions
MS and RVN: Conceived and designed the study and provided reagents and materials. KM: Performed the experiments and wrote the first draft. TAR: Supervised the molecular analyses. OMMT: Analyzed the data, interpreted the results, and reviewed and edited the manuscript. All authors read and approved the final manuscript.
Acknowledgments
The authors are thankful to bursaries from North-West University and FoodBev SETA of South Africa for providing necessary facilities for the study. The authors did not receive any funds for this study.
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published map and institutional affiliation.
References
- 1.Tauxe R.V, Doyle M.P, Kuchenmüller T, Schlundt J, Stein C.E. Evolving public health approaches to the global challenge of foodborne infections. Int. J. Food Microbiol. 2010;139(1):S16–S28. doi: 10.1016/j.ijfoodmicro.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 2.Scallan E, Hoekstra R.M, Angulo F.J, Tauxe R.V, Widdowson M.A, Roy S.L, Jones J.L, Griffin P.M. Foodborne illness acquired in the United States major pathogens. Emerg. Infect. Dis. 2011;17(1):7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Man S.M. The clinical importance of emerging Campylobacter species. Nat. Rev. Gastroenterol. Hepatol. 2011;8(12):669–685. doi: 10.1038/nrgastro.2011.191. [DOI] [PubMed] [Google Scholar]
- 4.El-Naenaeey E.S, El-Hamid A, Khalifa E. Foodborne Campylobacter species:Taxonomy, isolation, virulence attributes and antimicrobial resistance. Zagazig Vet. J. 2020;48(4):414–432. [Google Scholar]
- 5.He Y, Yao X, Gunther N.W, Xie Y, Tu S.I, Shi X. Simultaneous detection and differentiation of Campylobacter jejuni, C. coli, and C. lari in chickens using a multiplex real-time PCR assay. Food Anal. Methods. 2010;3(4):321–329. [Google Scholar]
- 6.Nur-Aziera-Aina C.M.N, Nasir N.S.M, Zaidah A.R. Detection of Campylobacter jejuni among commercial broiler chickens in east-coast Malaysia. J. World Poult. Res. 2020;10(2):367–370. [Google Scholar]
- 7.Jacobs-Reitsma W.F, van de Giessen A.W, Bolder N.M, Mulder R.W.A. Epidemiology of Campylobacter spp. at two Dutch broiler farms. Epidemiol. Infect. 1995;114(3):413–421. doi: 10.1017/s0950268800052122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Habib I, Berkvens D, de Zutter L, Dierick K, van Huffel X, Speybroeck N, Geeraerd A.H, Uyttendaele M. Campylobacter contamination in broiler carcasses and correlation with slaughterhouses operational hygiene inspection. Food Microbiol. 2012;29(1):105–112. doi: 10.1016/j.fm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Berndtson E, Danielsson-Tham M.L, Engvall A. Campylobacter incidence on a chicken farm and the spread of Campylobacter during the slaughter process. Int. J. Food Microbiol. 1996;32(1-2):35–47. doi: 10.1016/0168-1605(96)01102-6. [DOI] [PubMed] [Google Scholar]
- 10.Sahin O, Morishita T.Y, Zhang Q. Campylobacter colonization in poultry:Sources of infection and modes of transmission. Anim. Health Res. Rev. 2002;3(2):95–105. doi: 10.1079/ahrr200244. [DOI] [PubMed] [Google Scholar]
- 11.Sahin O, Kassem I.I, Shen Z, Lin J, Rajashekara G, Zhang Q. Campylobacter in poultry:Ecology and potential interventions. Avian Dis. 2015;59(2):185–200. doi: 10.1637/11072-032315-Review. [DOI] [PubMed] [Google Scholar]
- 12.Fonseca B.B, Fernandez H, Rossi D.A. Campylobacter spp, Related Organisms in Poultry:Pathogen-Host Interactions, Diagnosis and Epidemiology. 1st ed. Cham: Springer International Publishing; 2016. [Google Scholar]
- 13.Brandl M.T, Haxo A.F, Bates A.H, Mandrell R.E. Comparison of survival of Campylobacter jejuni in the phyllosphere with that in the rhizosphere of spinach and radish plants. Appl. Environ. Microbiol. 2004;70(2):1182–1189. doi: 10.1128/AEM.70.2.1182-1189.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chon J.W, Seo K.H, Kim B, Jeong D, Song K.Y. Advanced methods for isolating from and confirming Campylobacter spp. in milk and dairy products. J. Dairy Sci. Biotechnol. 2020;38(3):121–133. [Google Scholar]
- 15.Kingsbury J.M, Soboleva T.K. Evaluation of culture-based and molecular detection methods for Campylobacter in New Zealand raw cows'milk. J. Appl. Microbiol. 2020;130(2):478–492. doi: 10.1111/jam.14798. [DOI] [PubMed] [Google Scholar]
- 16.Dutta N, Banga H.S, Deshmukh S, Devi G. Localization and detection of Campylobacter jejuni using bio-molecular techniques like immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) in raw chicken meat. J. Entomol. Zool. Stud. 2020;8(4):467–470. [Google Scholar]
- 17.Kim Y.J, Kim H.S, Chon J.W, Kim D.H, Hyeon J.Y, Seo K.H. New colorimetric aptasensor for rapid on-site detection of Campylobacter jejuni and Campylobacter coli in chicken carcass samples. Anal. Chim. Acta. 2018;1029:78–85. doi: 10.1016/j.aca.2018.04.059. [DOI] [PubMed] [Google Scholar]
- 18.He D, Wu Z, Cui B, Xu E, Jin Z. Establishment of a dual mode immunochromatographic assay for Campylobacter jejuni detection. Food Chem. 2019;289:708–713. doi: 10.1016/j.foodchem.2019.03.106. [DOI] [PubMed] [Google Scholar]
- 19.Aarestrup F.M, Engberg J. Antimicrobial resistance of thermophilic Campylobacter. Vet. Res. 2001;32(3-4):311–321. doi: 10.1051/vetres:2001127. [DOI] [PubMed] [Google Scholar]
- 20.Szczepanska B, Andrzejewska M, Spica D, Klawe J.J. Prevalence and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from children and environmental sources in urban and suburban areas. BMC Microbiol. 2017;17(1):80. doi: 10.1186/s12866-017-0991-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hart C.A, Kariuki S. Antimicrobial resistance of developing countries. BMJ. 1998;317(7159):647–650. doi: 10.1136/bmj.317.7159.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simango C. Antimicrobial susceptibility of Campylobacter species. S. Afr. J. Epidemiol. Infect. 2013;28(3):139–142. [Google Scholar]
- 23.Bullman S, O'Leary J, Corcoran D, Sleator R.D, Lucey B. Molecular-based detection of non-culturable and emerging campylobacteria in patients presenting with gastroenteritis. Epidemiol. Infect. 2012;140(4):684–688. doi: 10.1017/S0950268811000859. [DOI] [PubMed] [Google Scholar]
- 24.Huq M, Gonis G, Istivan T. Development and evaluation of a multiplex PCR for the detection of Campylobacter concisus and other Campylobacter spp. from gastroenteritis cases. J. Med. Microbiol. 2014;4(1):29–37. [Google Scholar]
- 25.Jonker A, Picard J.A. Antimicrobial susceptibility in thermophilic Campylobacter species isolated from pigs and chickens in South Africa. J. S. Afr. Vet. Assoc. 2010;81(4):228–236. doi: 10.4102/jsava.v81i4.153. [DOI] [PubMed] [Google Scholar]
- 26.Igwaran A, Okoh A.I. Human campylobacteriosis:A public health concern of global importance. Heliyon. 2019;5(11):e02814. doi: 10.1016/j.heliyon.2019.e02814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagenaar J.A, French N.P, Havelaar A.H. Preventing Campylobacter at the source:Why is it so difficult? Clin. Infect. Dis. 2013;57(11):1600–1606. doi: 10.1093/cid/cit555. [DOI] [PubMed] [Google Scholar]
- 28.Uçar A, Yilmaz M.V, Çakıroğlu F.P. Food safety-problems and solutions. In:Significance, Prevention and Control of Food Related Diseases. Ch. 1. InTech, EU, Croatia. 2016 [Google Scholar]
- 29.Richardson N.J, Koornhof H.J, Bokkenheuser V.D, Mayet Z, Rosen E.U. Age related susceptibility to Campylobacter jejuni infection in a high prevalence population. Arch. Dis. Child. 1983;58(8):616–619. doi: 10.1136/adc.58.8.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pillay S, Amoako D.G, Abia A.L, Somboro A.M, Shobo C.O, Perrett K, Bester L.A, Essack S.Y. Characterisation of Campylobacter spp. isolated from poultry in KwaZulu-Natal, South Africa. Antibiotics. 2020;9(2):42. doi: 10.3390/antibiotics9020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samie A, Obi C.L, Barrett L.J, Powell S.M, Guerrant R.L. Prevalence of Campylobacter species, Helicobacter pylori and Arcobacter species in stool samples from the Venda region, Limpopo, South Africa:Studies using molecular diagnostic methods. J. Infect. 2007;54(6):558–566. doi: 10.1016/j.jinf.2006.10.047. [DOI] [PubMed] [Google Scholar]
- 32.Lastovica A.J. Emerging Campylobacter spp.:The tip of the iceberg. Clin. Microbiol. Newsl. 2006;28(7):9–56. [Google Scholar]
- 33.McCarthy N.D, Gillespie I.A, Lawson A.J, Richardson J, Neal K.R, Hawtin P.R, Maiden M.C.J, O'Brien S.J. Molecular epidemiology of human Campylobacter jejuni shows association between seasonal and international patterns of disease. Epidemiol. Infect. 2012;140(12):2247–2255. doi: 10.1017/S0950268812000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coker A.O, Isokpehi R.D, Thomas B.N, Amisu K.O, Obi C.L. Human campylobacteriosis in developing countries1. Emerg. Infect. Dis. 2002;8(3):237. doi: 10.3201/eid0803.010233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Platts-Mills J.A, Kosek M. Update on the burden of Campylobacter in developing countries. Curr. Opin. Infect. Dis. 2014;27(5):444–450. doi: 10.1097/QCO.0000000000000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahimi E, Ameri M. Antimicrobial resistance patterns of Campylobacter spp. isolated from raw chicken, turkey, quail, partridge, and ostrich meat in Iran. Food Control. 2011;22(8):1165–1170. [Google Scholar]
- 37.Basardien L. Molecular Characterization of Campylobacter Isolates from FreeRange and Commercial Chicken in South Africa. University of the Western Cape, South Africa. 2012. Available from: http://www.etd.uwc.ac.za/xmlui/handle/11394/5068?. Retrieved on 12-02-2021.
- 38.European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC) The European Union one health 2018 zoonoses report. EFSA J. 2019;17(12):e05926. doi: 10.2903/j.efsa.2019.5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Giannatale E, Calistri P, Di Donato G, Decastelli L, Goffredo E, Adriano D, Mancini M.E, Galleggiante A, Neri D, Antoci S, Marfoglia C. Thermotolerant Campylobacter spp. in chicken and bovine meat in Italy:Prevalence, level of contamination and molecular characterization of isolates. PLoS One. 2019;14(12):e0225957. doi: 10.1371/journal.pone.0225957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luangtongkum T, Jeon B, Han J, Plummer P, Logue C.M, Zhang Q. Antibiotic resistance in Campylobacter:Emergence, transmission, and persistence. Future Microbiol. 2009;4(2):189–200. doi: 10.2217/17460913.4.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wieczorek K, Szewczyk R, Osek J. Prevalence, antimicrobial resistance, and molecular characterization of Campylobacter jejuni and C. coli isolated from retail raw meat in Poland. Vet. Med. Czech. 2012;57(6):293–299. [Google Scholar]
- 42.Abdi-Hachesoo B, Khoshbakht R, Sharifiyazdi H, Tabatabaei M, Hosseinzadeh S, Asasi K. Tetracycline resistance genes in Campylobacter jejuni and C. coli isolated from poultry carcasses. Jundishapur J. Microbiol. 2014;7(9):e12129. doi: 10.5812/jjm.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamidian M, Sanaei M, Azimi-Rad M, Tajbakhsh M, Dabiri H, Zali M.R. Fla-typing, RAPD analysis, isolation rate and antimicrobial resistance profile of Campylobacter jejuni and Campylobacter coli of human origin collected from hospitals in Tehran, Iran. Ann. Microbiol. 2011;61(2):315–321. [Google Scholar]
- 44.Geissler A.L, Bustos Carrillo F, Swanson K, Patrick M.E, Fullerton K.E, Bennett C, Barrett K, Mahon B.E. Increasing Campylobacter infections, outbreaks, and antimicrobial resistance in the United States, 2004-2012. Clin. Infect. Dis. 2017;65(10):1624–1631. doi: 10.1093/cid/cix624. [DOI] [PubMed] [Google Scholar]
- 45.Cody A.J, Clarke L, Bowler I.C, Dingle K.E. Ciprofloxacin-resistant campylobacteriosis in the UK. Lancet. 2010;376(9757):1987. doi: 10.1016/S0140-6736(10)62261-1. [DOI] [PubMed] [Google Scholar]
- 46.Moore J.E, Barton M.D, Blair I.S, Corcoran D, Dooley J.S, Fanning S, Kempf I, Lastovica A.J, Lowery C.J, Matsuda M, McDowell D.A. The epidemiology of antibiotic resistance in Campylobacter. Microbes. Infect. 2006;8(7):1955–1966. doi: 10.1016/j.micinf.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 47.Bester A.C, Roniger M, Oren Y.S, Im M.M, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach D.S, Kerem B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145(3):435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Angulo F.J, Collignon P, Powers J.H, Chiller T.M, Aidara-Kane A, Aarestrup F.M. World Health Organization ranking of antimicrobials according to their importance in human medicine:A critical step for developing risk management strategies for the use of antimicrobials in food production animals. Clin. Infect. Dis. 2009;49(1):132–141. doi: 10.1086/599374. [DOI] [PubMed] [Google Scholar]


