Abstract
Introduction:
PIK3CA is one of the most mutated oncogenes in solid tumors. In breast cancer (ER-positive, HER2-negative), these events represent a predictive biomarker of response to alpelisib. In glioblastomas (GBM), PIK3CA mutations were described as early constitutive events. Here, we investigated PIK3CA mutational profile across glioma molecular subgroups and its relevance during glioma recurrence. Furthermore, PIK3CA mutations’ effect in PI3K pathway, prognosis, and response to therapy was also explored.
Material and Methods:
Exons 10 and 21 of PIK3CA mutations were evaluated in 394 gliomas and 19 glioma recurrences from Instituto Português de Oncologia Lisboa Francisco Gentil (IPOLFG) and compared with The Cancer Genome Atlas (TCGA) data. TIMER2.0 and NetMHCpan4.1 were used to assess the immune-microenvironment contribution.
Results:
PIK3CA mutations were identified among all glioma subgroups, although with no impact on their stratification or prognosis. In both cohorts (IPOLFG and TCGA), PIK3CA mutation frequencies in IDH-mutant and IDH-wild-type GBM were similar (IPOLFG: 9% and 3%; TCGA: 8% and 2%). These mutations were not mutually exclusive with PTEN deletion and EGFR amplification. Despite their reduced frequency, we discovered PIK3CA mutations were maintained during glioma recurrence regardless of administered therapies. The immune microenvironment might not contribute to this phenotype as PIK3CA mutations did not influence immune cell infiltration.
Conclusions:
Despite the absence of a predominant effect in glioma stratification, PIK3CA mutations were maintained during glioma recurrence, possibly contributing to glioma cell survival, representing promising therapeutic targets in recurrent glioma. Nevertheless, understanding the potential synergistic effects between PIK3CA mutations, PTEN deletion, and EGFR amplification is pivotal to targeted therapies’ efficiency.
Keywords: PIK3CA mutations, gliomas, molecular subgroups, progression, recurrence, immune cell infiltrates, PI3K-Akt pathway
Introduction
Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) has been indicated as one of the most frequently mutated genes in solid tumors. 1 This gene encodes the p110α catalytic subunit of class IA PI3K lipid kinases, 2 enzymes that participate in the PI3K/Akt signaling pathway, regulating cell proliferation, angiogenesis, growth, motility, and survival. 3 Using new high-throughput molecular methodologies, PI3K-Akt overactivation has become recognized as one of the most important driver mechanisms of aggressiveness acquired by tumor cells. 2 At present, it is known that PI3K-Akt pathway abnormal activation is triggered mainly by 3 molecular events: PTEN loss of function/inactivation and EGFR or PIK3CA overactivation.4,5
In the last decades, PIK3CA mutations have been identified mainly in patients with breast (~35%), endometrial (~36%), bladder (>18%), and colorectal cancer (18%-38%).1,6-9 Exons 10 and 21 of PIK3CA are the main mutational hotspot regions found, with E542K, E545K, and H1047R being the most frequent mutations.9-11 However, discrepant frequencies of these alterations have been reported in glioblastoma (GBM), ranging from 5% to 30%.11-16 PIK3CA mutational analysis in less prevalent types of gliomas (oligodendrogliomas and astrocytomas) has been less explored.14,17 Importantly, the data available about the PIK3CA mutational profile in gliomas are based exclusively on histological criteria and could differ significantly when taking into account the 2016 World Health Organization (WHO) molecular classification of these tumors—GBM IDH-wildtype, GBM IDH-mutant, astrocytoma IDH-wildtype, astrocytoma IDH-mutant, and IDH-mutant and 1p/19q codeleted oligodendroglioma.18-20
In addition, PIK3CA activating mutations have been described as initial and clonal events found in all GBM surgical fragments, highlighting the potential relevance of these molecular alterations in glioma development. 16 Some studies have also stated that PIK3CA mutations might be associated with poor GBM patient outcomes16,21 and even earlier recurrence. 21
In fact, gliomas are the most common malignant brain tumors associated with a dismal prognosis, triggering several therapy resistance mechanisms that challenge patient treatment. 22 As the available therapeutic approaches for glioma are not efficient, 22 the potential inhibition of PI3K-Akt oncogenic signaling pathway should be further explored. Currently, the glioma molecular subgroup where PIK3CA mutations occur most frequently remains unclear, as well as their role in glioma stratification, prognosis, and aggressiveness.
PIK3CA has been highlighted as a promising therapeutic target in cancer.23-25 Recently, in breast cancer, PIK3CA mutations were associated with an increased sensitivity to the selective p110α inhibitor alpelisib.23,26 Consequently, alpelisib has been approved for treatment of advanced stage ER-positive, HER2-negative, and PIK3CA-mutated breast cancer, in combination with fulvestrant, leading to significantly prolonged progression-free survival (PFS).23,26 Alpelisib treatment should also be considered for patients with glioma, but it is necessary to clarify which molecular subgroups would benefit the most from the administration of these inhibitors.
In this study, we investigated the frequency and clinical relevance of PIK3CA mutations in exons 10 and 21 in glioma molecular subgroups, the significance of these mutations in glioma recurrence, and their relationship with other molecular alterations of the PI3K-Akt pathway and the immune microenvironment.
Materials and Methods
Study design and biological samples
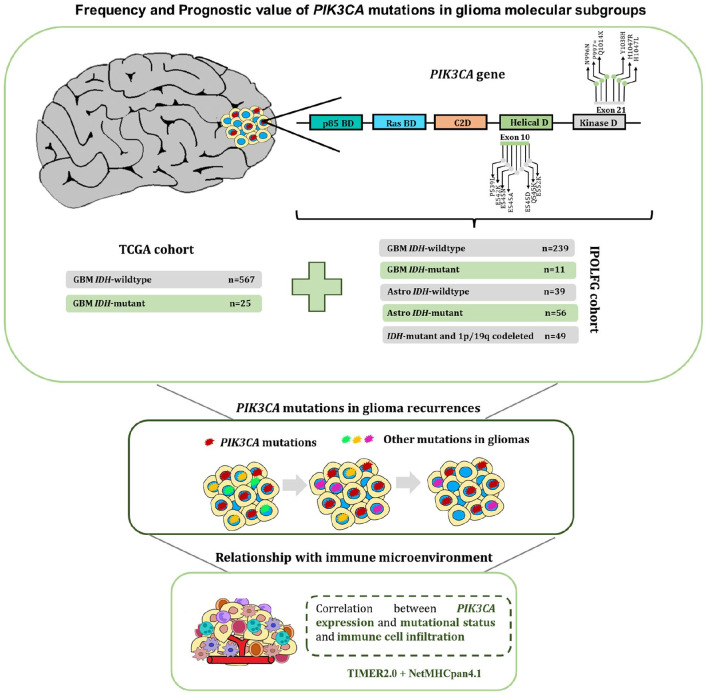
A detailed workflow of our study design is shown in Figure 1. The Instituto Português de Oncologia Lisboa Francisco Gentil (IPOLFG) cohort used in this study contains 394 glioma samples, previously reclassified and characterized according to the 2016 WHO classification. 20 Simultaneously, a dataset with 19 cases of glioma recurrences, corresponding to 19 primary and 23 matched recurrence samples, was analyzed to identify PIK3CA mutations. Tumor samples were received as fresh or paraffin-preserved tissue for DNA extraction, performed using the methodologies already described. 20 This study was previously approved by the IPOLFG Ethical Board Committee (UIC/1203), and written informed consent was obtained from all living patients.
Figure 1.
Study design and workflow used to investigate the frequency and relevance of PIK3CA mutations. The presence of PIK3CA mutations in exons 10 and 21 was assessed in 394 glioma samples from the IPOLFG cohort, classified according to the 2016 WHO molecular classification. Furthermore, PIK3CA mutational status was also assessed in 592 GBM samples from TCGA, divided into GBM IDH-wildtype and IDH-mutant. Then, 19 recurrence cases were evaluated to understand if PIK3CA mutations persisted throughout glioma progression. Finally, the relationship between PIK3CA expression and mutational status and the immune microenvironment was estimated using TIMER2.0 and NetMHCpan4.1.
GBM indicates glioblastomas; IPOLFG, Instituto Português de Oncologia de Lisboa Francisco Gentil; TCGA, The Cancer Genome Atlas; WHO, World Health Organization.
Genotyping
MGMT promoter methylation, PTEN deletion, and EGFR amplification were identified in diagnostic routine, as previously reported. 20 High-resolution chromosome comparative genome hybridization analysis was performed as previously described to evaluate PIK3CA copy number variations. 27
According to a current update, exons 9 and 20 of PIK3CA were renamed as exons 10 and 21, based on Ensembl Transcript ID: ENST00000263967.4, RefSeq: NM_006218.4. Both PIK3CA exons and R172 IDH2 hotspot mutations were evaluated by Sanger sequencing in the previously established glioma molecular subgroups.20,28 The primers and conditions used for polymerase chain reaction (PCR) amplification are listed in Supplementary Table 1. An additional set of primers was used to target a pseudogene with more than 95% of homology with exon 10 of PIK3CA, found in chromosome 22. 29
To determine the sequences of interest in PIK3CA and IDH2, ABI Prism 3130 Genetic Analyzer (Applied Biosystems, USA) was used following the protocol proposed by Big Dye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, USA), which can detect variants as low as 5% in a sample.
In silico analysis
In silico analysis was performed to predict the benign or pathogenic impact of new PIK3CA variants, not found in Ensembl, Catalogue of Somatic Mutations in Cancer (COSMIC), and the Human Gene Mutation Database (HGMB), using MutationTaster (http://www.mutationtaster.org), PolyPhen (http://genetics.bwh.harvard.edu/pph2), and Variant Effect Predictor (https://www.ensembl.org/info/docs/tools/vep/index.html).
In addition, the frequency of PIK3CA mutations in 567 GBM IDH-wild-type and 25 GBM IDH-mutant samples available in cBioPortal for Cancer Genomics (https://www.cbioportal.org/) was evaluated, hereafter referred to as The Cancer Genome Atlas (TCGA) cohort. Furthermore, the prognosis value of PIK3CA mutational status and the potential correlation between PIK3CA mutations, EGFR, and PTEN molecular alterations was determined.
The impact of PIK3CA expression, mutations, or amplification on the infiltration of distinct immune cell subsets in GBM and low-grade gliomas (LGG) was assessed using TIMER2.0. 30 As a complementary analysis, the correlation between PIK3CA expression or mutational status and the expression of various immune cell gene markers was also investigated using TIMER2.0. NetMHCpan4.1 algorithm was applied to determine whether PIK3CA mutations contribute to T-cell recruitment and activation, by estimating the binding affinity of wildtype and mutated 9 amino acid peptides to major histocompatibility complex (MHC) Class I molecules. 31
Statistical analysis
The primary endpoint used was overall survival (OS), defined as the time from glioma diagnosis to patient death or last follow-up. Survival analysis was done using Kaplan-Meier estimator and the log rank test for group comparison. Multivariable analysis was determined using Cox regression proportional hazard model. Fisher exact test was used to compare the prevalence of PIK3CA mutations across glioma molecular subgroups. The chi-square statistical test was used to analyze putative associations between PIK3CA mutations and PTEN and EGFR molecular alterations.
The differences between immune cell infiltrates and the differential expression of immune cell gene markers in GBM and LGG under distinct PIK3CA mutational status were determined by Wilcoxon rank sum test. Kruskal-Wallis H test was used to compare the immune infiltration distribution in PIK3CA-amplified GBM and LGG samples. Spearman rank correlation coefficients with purity adjustment were obtained when comparing PIK3CA expression with immune cell infiltration or the expression of immune cell markers in GBM and LGG cohorts. To compare the differences between the binding affinity of mutated and corresponding wild-type p110α peptides with MHC class I complexes, 2-way analysis of variance (ANOVA) was used followed by Bonferroni multiple comparisons test.
All tests were 2-sided, with a significance level of 5%. Statistical analysis and graphic representation were performed using IBM SPSS Statistics 21.0 and GraphPad Prism 8.4.3.
Results
Frequency and prognostic impact of PIK3CA mutations in glioma molecular subgroups
Considering the lack of data about the frequency of PIK3CA mutations in exons 10 and 21 in glioma molecular subgroups, we evaluated the presence of these alterations in 394 diffuse glioma samples. A detailed workflow of this study is shown in Figure 1. PIK3CA mutations were most frequent in IDH-mutant and 1p/19q codeleted oligodendrogliomas (10%) and astrocytomas IDH-wildtype (10%) (Table 1). In contrast, the GBM IDH-wild-type subgroup, the most lethal, presented the lowest mutational frequency (3%).
Table 1.
PIK3CA mutations found in glioma molecular subgroups from the IPOLFG cohort.
| Glioma molecular subgroup | No. of samples | Frequency of PIK3CA mutations (%) | Exon | Nucleotide change | Amino acid change (no. of samples with mutation) |
|---|---|---|---|---|---|
| GBM, IDH-wildtype | 239 | 6/239 (3) | 21 10 10 21 |
c.3140A>G c.1633G>C c.1634A>C c.3112T>C a |
H1047R (n = 3) E545N (n = 1) E545A (n = 1) Y1038H (n = 1) |
| GBM, IDH-mutant | 11 | 1/11 (9) | 21 | c.3140A>G | H1047R (n = 1) |
| IDH-mutant and 1p/19q codeleted gliomas | 49 | 5/49 (10) | 10 10 10 21 21 |
c.1616C>T c.1635G>T c.1656G>A c.2991C>T c.2988T>C a |
P539L (n = 1) E545D (n = 1) E552K (n = 1) L997L (n = 1) N996N (n = 1) |
| Astrocytomas, IDH-wildtype | 39 | 4/39 (10) | 21 21 10 10 |
c.3140A>G c.3140A>T c.1624G>A c.1636C>A |
H1047R (n = 1) H1047L (n = 1) E542K (n = 1) Q546K (n = 1) |
| Astrocytomas, IDH-mutant | 56 | 3/56 (5) | 21 10 21 |
c.2965C>G c.1624G>A c.3040C>T a |
L989V (n = 1) E542K (n = 1) Q1014X (n = 1) |
Abbreviations: GBM, glioblastomas; IPOLFG, Instituto Português de Oncologia de Lisboa Francisco Gentil.
Unreported variant.
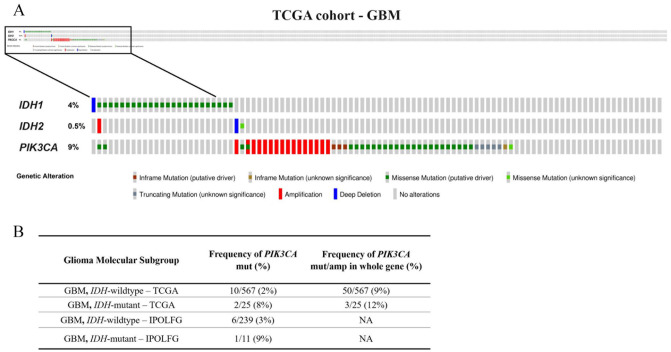
The frequency of PIK3CA mutations was higher in GBM IDH-mutant (9%) than GBM IDH-wildtype (3%) (Table 1), but without statistical significance (P = .273; Supplementary Table 2). Nevertheless, similar results were obtained in TCGA cohort when evaluating only PIK3CA hotspot exons: 8% in GBM IDH-mutant and 2% in GBM IDH-wildtype (Figure 2). Furthermore, when we analyzed the entire PIK3CA gene, an analysis that includes mutations and amplifications regardless of pathogenicity confirmation (described in Supplementary Table 3), these frequencies increased to 12% and 9%, respectively (Figure 1). Copy number variation analysis of TCGA data demonstrated that 16 GBM cases harbor a PIK3CA amplification, whereas in the IPOLFG cohort, no amplifications were detected (Supplementary Table 3 and Figure 2). This difference may be explained by the reduced sampling size of IPOLFG cohort and even by the distinct techniques used to assess copy number variations.
Figure 2.
Frequency of PIK3CA mutations and copy number variations in GBM molecular subgroups from TCGA cohort. (A) PIK3CA, IDH1, and IDH2 molecular alterations in GBM samples. Whole PIK3CA gene analysis showed that 9% of GBM cases contain molecular alterations. (B) Similarity between the frequency of PIK3CA mutations in exons 10 and 21 in GBM molecular subgroups from TCGA and IPOLFG cohorts. When considering PIK3CA mutations and amplifications throughout the whole gene, frequencies vary slightly in TCGA cohort.
GBM indicates glioblastomas; IPOLFG, Instituto Português de Oncologia de Lisboa Francisco Gentil; TCGA, The Cancer Genome Atlas.
Overall, in the IPOLFG cohort, we detected 19 PIK3CA mutations in the hotspot exons—5% of all glioma samples (Table 1). Although H1047R and E542K were the most common PIK3CA mutations detected, during this analysis, we also found 3 distinct undescribed variants (Table 1). Variant N996N was found 55 base pairs from the splice site, in IDH-mutant and 1p/19q codeleted oligodendrogliomas, and was estimated as pathogenic, likely to induce splice site changes, leading to the loss of amino acid sequences belonging to the helix. Unreported variant Y1038H in exon 21 was present in GBM IDH-wildtype subgroup and predicted to be pathogenic, with moderate impact on the protein encoded. Finally, Q1014X was found in the astrocytoma IDH-mutant subgroup and estimated as pathogenic due to a premature stop codon in position 3042 instead of 3207, resulting in a truncated p110 catalytic subunit lacking 55 amino acids.
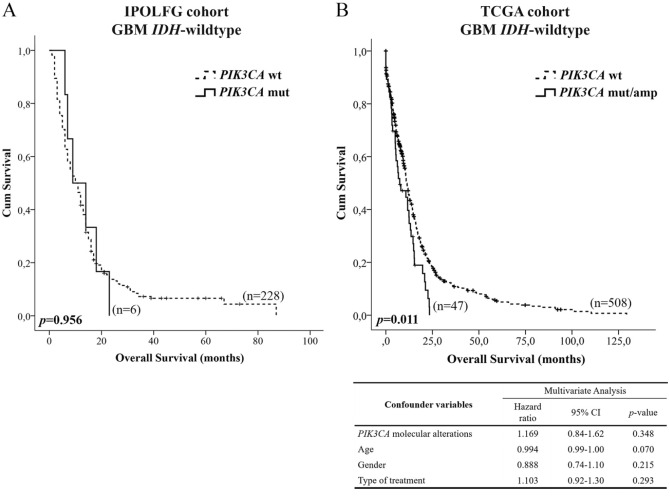
Then, we determined that PIK3CA mutations did not have prognostic value in the most representative molecular subgroup of the IPOLFG cohort, GBM IDH-wildtype (P = .956; Figure 3A). Nevertheless, the presence of PIK3CA mutations slightly decreased the median OS (10 months vs 9 months). This trend was confirmed using TCGA data, where we included all PIK3CA molecular alterations (Supplementary Table 3). According to the univariable analysis, these alterations were associated with shorter OS of GBM IDH-wild-type patients (P = .011; Figure 3B), with median OS being reduced from 11 to 7 months. However, multivariable analysis showed that PIK3CA molecular alterations were not independent prognostic factors (P = .348).
Figure 3.
Impact of PIK3CA molecular alterations in the overall survival of GBM IDH-wild-type patients. (A) Kaplan-Meier curve estimates the impact of PIK3CA mutations in the prognosis of patients included in the IPOLFG cohort. (B) Prognosis value of PIK3CA molecular alterations found in all exons was estimated using data from GBM IDH-wild-type patients available in TCGA cohort. Survival data were only available for 47 of the 53 cases containing PIK3CA molecular alterations. Multivariable analysis included 555 GBM IDH-wild-type samples.
GBM indicates glioblastomas; IPOLFG, Instituto Português de Oncologia de Lisboa Francisco Gentil; TCGA, The Cancer Genome Atlas.
In addition, we also evaluated the correlation between PIK3CA mutations in exons 10 and 21 and molecular alterations in other important players of the PI3K/Akt pathway, EGFR and PTEN, in both cohorts. EGFR amplification and PTEN deletion coexist with PIK3CA mutations in glioma molecular subgroups, although no significant associations were observed (Supplementary Tables 4 and 5). Furthermore, no significant association was found with MGMT methylation, a well-studied epigenetic alteration found in gliomas.
New PIK3CA polymorphism in glioma molecular subgroups
During PIK3CA mutational analysis, we detected a single-nucleotide polymorphism (SNP)—Rs45455192 (C>T), 55 nucleotides from the beginning of exon 10—that has never been described in gliomas. This SNP was detected in heterozygosity in the IPOLFG cohort in 18% of GBM IDH-wildtype, 27% of GBM IDH-mutant, 25% of IDH-mutant and 1p/19q codeleted oligodendrogliomas, 21% of astrocytomas IDH-wildtype, and 18% of astrocytomas IDH-mutant (Supplementary Table 6). For all glioma molecular subgroups analyzed, Rs45455192 was not associated with glioma patient prognosis (Supplementary Figure 1).
PIK3CA mutations in recurrent glioma
Recently, PIK3CA mutations were associated with earlier recurrence in patients with GBM 21 and reported as constitutive events shared by all GBM tumor mass. 16 However, these mutations were never investigated throughout glioma progression. Thus, we evaluated whether PIK3CA mutations persist throughout glioma recurrence or whether they are important events in glioma initiation that are lost during tumor recurrence.
We assessed 19 recurrent glioma cases. L989V and E545K, 2 reported mutations, were detected in 2 cases out of 19 (cases 2 and 9 depicted in Table 2), both in the primary tumor and in the recurrence samples. Clinical data indicate that both patients were treated with radiotherapy (RT) and/or chemotherapy (CT) (Supplementary Table 7), which might suggest that PIK3CA mutations are maintained throughout glioma recurrences regardless of the therapy administered.
Table 2.
PIK3CA mutational analysis in 19 recurrent glioma cases from the IPOLFG cohort.
| Initial diagnosis | First recurrence diagnosis | Second recurrence diagnosis | PIK3CA mutational analysis | |||
|---|---|---|---|---|---|---|
| Primary tumor | First recurrence | Second recurrence | ||||
| Case 1 (n = 2) |
Astrocytoma IDH-wildtype | GBM IDH-wildtype | NA | Wildtype | Wildtype | NA |
| Case 2 (n = 2) |
Astrocytoma IDH-mutant | GBM IDH-mutant | NA | Mutated (c.2965C>G) | Mutated (c.2965C>G) |
NA |
| Case 3 (n = 2) |
Astrocytoma IDH-mutant | GBM IDH-mutant | NA | Wildtype | Wildtype | NA |
| Case 4 (n = 3) |
Astrocytoma IDH-mutant | GBM IDH-mutant | GBM IDH-mutant | Wildtype | Wildtype | Wildtype |
| Case 5 (n = 2) |
Astrocytoma IDH-wildtype | GBM IDH-wildtype | NA | Wildtype | Wildtype | NA |
| Case 6 (n = 2) |
Astrocytoma IDH-wildtype | GBM IDH-wildtype | NA | Wildtype | Wildtype | NA |
| Case 7 (n = 2) |
Astrocytoma IDH-mutant | GBM IDH-mutant | NA | Wildtype | Wildtype | NA |
| Case 8 (n = 2) |
Astrocytoma IDH-wildtype | GBM IDH-wildtype | NA | Wildtype | Wildtype | NA |
| Case 9 (n = 3) |
GBM IDH-mutant | GBM IDH-mutant | GBM IDH-mutant | Mutated (c.1633G>A) |
Mutated (c.1633G>A) |
Mutated (c.1633G>A) |
| Case 10 (n = 2) |
GBM IDH-wildtype | GBM IDH-wildtype | NA | Wildtype | Wildtype | NA |
| Case 11 (n = 2) |
Astrocytoma IDH-wildtype | GBM IDH-wildtype | NA | Wildtype | Wildtype | NA |
| Case 12 (n = 2) |
Astrocytoma IDH-mutant | GBM IDH-mutant | NA | Wildtype | Wildtype | NA |
| Case 13 (n = 2) |
Astrocytoma IDH-mutant | GBM IDH-mutant | NA | Wildtype | Wildtype | NA |
| Case 14 (n = 2) |
Astrocytoma IDH-mutant | GBM IDH-mutant | NA | Wildtype | Wildtype | NA |
| Case 15 (n = 2) |
Astrocytoma IDH-mutant | GBM IDH-mutant | NA | Wildtype | Wildtype | NA |
| Case 16 (n = 2) |
Astrocytoma IDH-wildtype | GBM IDH-wildtype | NA | Wildtype | Wildtype | NA |
| Case 17 (n = 2) |
Astrocytoma IDH-wildtype | GBM IDH-wildtype | NA | Wildtype | Wildtype | NA |
| Case 18 (n = 3) |
Astrocytoma IDH-wildtype | Astrocytoma IDH-wildtype | GBM IDH-wildtype | Wildtype | Wildtype | Wildtype |
| Case 19 (n = 3) |
Astrocytoma IDH-mutant | Astrocytoma IDH-mutant | GBM IDH-mutant | Wildtype | Wildtype | Wildtype |
Abbreviations: GBM, glioblastomas; IPOLFG, Instituto Português de Oncologia de Lisboa Francisco Gentil; NA, not available.
The relationship between immune cell infiltration and PIK3CA in diffuse gliomas
It is known that distinct glioma molecular profiles are associated with the recruitment of different immune subsets, which can justify variations in tumor aggressiveness and therapy resistance.32-35 Knowing that PIK3CA mutations seem to be maintained throughout glioma progression, we decided to explore the relationship between the immune microenvironment and the PIK3CA gene to understand whether this interplay can influence glioma aggressiveness.
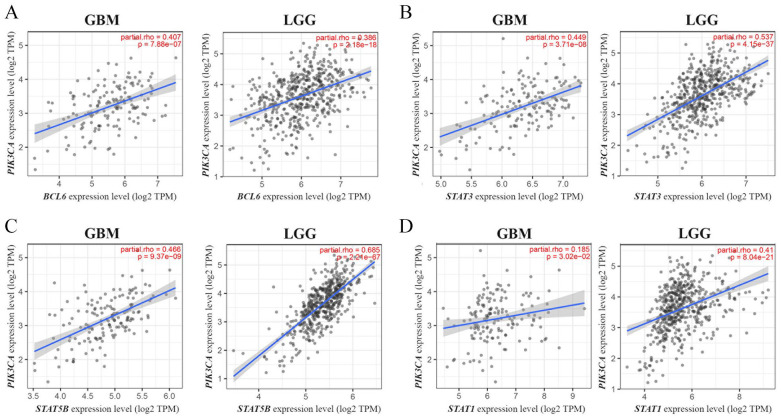
Infiltration levels for various types of immune cells were assessed according with PIK3CA expression and mutational and amplification status in GBM and LGG samples, using TIMER2.0 (Supplementary Tables 8, 9, and 10). The results obtained show no significant associations with the infiltration of the immune subsets analyzed. We also explored the interaction of PIK3CA expression and mutational status with the expression of various immune cell markers, in GBM and LGG (Supplementary Tables 11 and 12). Only BCL6, STAT3, STAT5B, and STAT1 showed a significant positive correlation with PIK3CA expression levels, the latter only in LGG samples (Figure 4). Finally, no differences were detected between the binding affinity of wildtype and mutated p110α peptides to MHC Class I molecules (Supplementary Figure 2), according to the NetMHCpan4.1 algorithm.
Figure 4.
Correlation between PIK3CA expression and the expression of the immune cell gene markers BCL6 (A), STAT3 (B), STAT5B (C), and STAT1 (D), in GBM and LGG samples from TCGA. Data were obtained from the TIMER2.0 resource, including P values and Spearman’s rank correlation coefficients with purity adjustment (partial rho). GBM indicates glioblastomas; LGG, low-grade gliomas; TCGA, The Cancer Genome Atlas.
These results seem to indicate that PIK3CA mutational status is not associated with immune cell infiltration, although a possible crosstalk between PI3K/Akt and JAK-STAT pathways was suggested.
Discussion
PIK3CA somatic activating mutations have been found in various cancer types, but their impact in patient prognosis and response to therapy is still under investigation. 1 Considering that PIK3CA mutations are described as predictive biomarkers of a better response to alpelisib treatment in breast cancer, 23 their potential as biomarkers should be further explored in other contexts. Currently, there is an urgent need for new biomarkers to improve the clinical management of patients with glioma, as these neoplasms are highly invasive and lethal, and effective therapies are nonexistent. 22 To understand if PIK3CA mutations could constitute good biomarkers in glioma, we evaluated, for the first time, the frequency and clinical relevance of mutations in exons 10 and 21 of PIK3CA in 5 well-characterized molecular subgroups of glioma previously defined.20,36
The overall frequency of PIK3CA mutations in the IPOLFG glioma cohort was low (5%), with the highest percentage belonging to the IDH-mutant and 1p/19q codeleted glioma and astrocytoma IDH-wild-type subgroups (10%). However, these frequencies might be slightly underestimated, as only exons 10 and 21 of PIK3CA were analyzed and not the entire gene. Overall, PIK3CA mutations were distributed among all glioma molecular subgroups, suggesting that their individual presence is not associated with the differences in subgroup aggressiveness. Therefore, these events alone cannot help to stratify patients with glioma according to their diagnosis. Only one other study has calculated the frequency of PIK3CA mutations in IDH-mutant 1p/19q codeleted gliomas. 17 TCGA Research Network 17 analyzed the entire PIK3CA gene in a more robust cohort and determined that 20% (17/84) of IDH-mutant 1p/19q codeleted LGG harbored PIK3CA mutations. These results highlight the importance of PIK3CA mutations in this subgroup.
In addition, PIK3CA mutations were not considered independent prognostic factors of OS in the IPOLFG or the TCGA cohorts. A recent study published by Tanaka et al 21 reported that PIK3CA hotspot mutations are associated with shorter PFS in patients with GBM IDH-wildtype independently of confounder variables, but they found no significant association with OS, similar to the results obtained in our study. Further research in other cohorts is therefore crucial to better understand the role of PIK3CA mutations in the prognosis of patients with glioma.
Even though PIK3CA mutations did not significantly impact glioma diagnosis and prognosis, we are concerned about the harmful effect that these mutations could have when combined with other molecular alterations that dysregulate the PI3K-Akt pathway. Class I PI3K is responsible for the phosphorylation of phosphatidylinositol 4,5 bisphosphate (PIP2) to phosphatidylinositol-3,4,5-triphosphate (PIP3) when activated by upstream receptor tyrosine kinases (RTKs). PIP3 accumulates in the plasma membrane and activates protein kinase B (PKB; also known as Akt) through phosphatidylinositol dependent kinase (PDK) recruitment. 37 Akt plays a central role as mediator of this signaling pathway, being responsible for the phosphorylation of several downstream targets. Mechanistic target of rapamycin (mTOR) is one of the most relevant downstream targets of this signaling pathway, triggering cell death inhibition and promoting cell survival, growth, and angiogenesis.2,38 Phosphatase and tensin homologue deleted on chromosome 10 (PTEN) antagonizes PI3K activity by dephosphorylating PIP3 back to PIP2, making it a strict negative regulator of PI3K activity. 37 Considering that in diffuse large B-cell lymphoma, PTEN deletion and PIK3CA mutations are mutually exclusive events, 39 we analyzed whether there is some correlation between PIK3CA mutations, EGFR amplification, and PTEN deletion in gliomas. None of these events are mutually exclusive in glioma subgroups. Despite not being significantly correlated, it would be interesting to evaluate whether there is a cooperative impact of these molecular alterations in glioma aggressiveness and response to therapy. This cooperative effect may compromise the individual targeting of p110α, PTEN, or EGFR, due to compensatory mechanisms potentiated by molecular alterations present in other genes belonging to this pathway. For example, in lung adenocarcinomas, cumulative PIK3CA mutations and EGFR mutations are associated with poorer outcomes compared with tumors with EGFR mutations, suggesting that the mutual effect between these molecular alterations promotes tumor aggressiveness. 40 Furthermore, concomitant alterations in PIK3CA and PTEN genes were correlated with an additive or synergistic effect in PI3K activation in a model of endometrial cancer. 6 Therefore, the putative mutual effect between these molecular alterations should be further assessed in gliomas.
We also found a SNP, described for the first time in molecular subgroups of glioma in this study, that did not demonstrate any correlation with patient prognosis. This polymorphism was described in human oral squamous carcinoma 41 and also in a sample of pancreaticobiliary adenocarcinoma. 42 In the future, it could be interesting to analyze the association between this polymorphism and glioma risk to understand whether it is a germinal or somatic alteration.
Another important question addressed in this study was the role of PIK3CA mutations among glioma progression. Here, for the first time, we detected that, when present, PIK3CA variants are in the primary tumor and in the respective recurrences. As these patients were submitted to RT and/or CT, these variants seem to be maintained during glioma recurrences independently of the therapy administered. However, we have only reported 2 PIK3CA mutated recurrence cases, and thus, further studies are needed to validate whether PIK3CA mutations provide any adaptative tumor advantage, contributing to therapy resistance. Recently, PIK3CA mutations were proposed as a biomarker to monitor CT resistance in colorectal cancer. 43 They have also been associated with paclitaxel, anthracycline, and anti-HER2 therapy resistance in patients with breast cancer.44-46 In cervical cancer cells, E545K mutation confers radio resistance through β-catenin signaling pathway activation. 47 Therefore, it is of the utmost importance to clarify the role of PIK3CA mutations in glioma progression and response to therapy.
Recently, some studies have highlighted the importance of exploring the relationship between specific glioma molecular profiles and immune cell infiltration, which can have a considerable impact on tumor aggressiveness.32-35 However, no study thus far has examined the association between PIK3CA mutations and the remodeling of glioma’s immune microenvironment. Hence, we sought to better understand this relationship and how it might affect glioma aggressiveness and therapy resistance. Overall, our results suggest that PIK3CA mutations do not heavily influence glioma immunogenicity. It is known that, when compared with other tumors, central nervous system tumors usually display lower infiltration of immune cells. 48 Similar to our results, a recent study found that, in triple negative breast cancer, PIK3CA mutations were not associated with the density of tumor-infiltrating lymphocytes. 49 However, other studies seem to highlight an association between PI3K/Akt activation and immune suppression in breast, prostate, and bladder cancers.50-52 Further research should be performed to better understand these dynamics in glioma.
We did find a positive correlation between PIK3CA expression and distinct markers for different immune cell populations, BCL6, STAT3, STAT5B, and STAT1, in glioma. These results indicate a strong crosstalk between the PI3K/Akt and the JAK-STAT pathways in glioma, independent from their roles in immune cell function. Constitutive activation of STATs is thought to contribute to oncogenesis and has been found in gliomas. 53 STAT3 overactivation, which can be induced by phosphorylation via EGFR, has been linked with shorter OS in patients with GBM. 54 Likewise, STAT5B was indicated as a putative therapeutic target for patients with GBM, being an inductor of GBM cell growth, cell cycle progression, invasion, and migration. 55 In the future, it could be interesting to explore if alterations in both the PI3K/Akt and the JAK-STAT pathway could have a synergistic effect in increasing glioma aggressiveness.
Conclusions
Overall, PIK3CA mutations were identified among all glioma molecular subgroups, with higher prevalence in IDH-mutant and 1p/19q codeleted oligodendrogliomas and astrocytomas IDH-wildtype. In addition, PIK3CA molecular alterations were not independent prognostic factors in GBM IDH-wild-type patients and were not mutually exclusive with EGFR amplification or PTEN deletion. Despite low frequency of these mutations in glioma molecular subgroups, they constitute early events maintained during tumor progression regardless of the therapy administered, possibly hinting at a role in glioma cell survival. Thus, even though PIK3CA mutations alone might not be a useful biomarker in glioma patient stratification, these results highlight PIK3CA as a potential promising therapeutic target in glioma recurrent cases.
In the future, clarifying not only the impact of PIK3CA mutations in response to alpelisib treatment in cell models but also the potential role of other molecular alterations might have on the efficacy of p110α targeted therapies will be essential for their success in glioma.
Supplemental Material
Supplemental material, sj-docx-1-onc-10.1177_11795549211068804 for PIK3CA Mutations in Diffuse Gliomas: An Update on Molecular Stratification, Prognosis, Recurrence, and Aggressiveness by Cheila Brito, Ana Tomás, Ana Azevedo, Susana Esteves, Manuela Mafra, Lúcia Roque and Marta Pojo in Clinical Medicine Insights: Oncology
Acknowledgments
The authors thank Liga Portuguesa Contra o Cancro—Núcleo Regional do Sul and the Neurology Department of Instituto Português de Oncologia Lisboa Francisco Gentil.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by Liga Portuguesa Contra o Cancro—Núcleo Regional Sul (LPCC-NRS)—Terry Fox grant 2018/2019. Marta Pojo was supported by LPCC-NRS. iNOVA4Health—UIDB/04462/2020 and UIDP/04462/2020, a program financially supported by Fundação para a Ciência e Tecnologia/Ministério da Ciência, Tecnologia e Ensino Superior, through national funds is acknowledged.
Author Contributions: MP and LR were responsible for concept and design. CB, AT, AA, and MM were responsible for experiments and procedures. Data analysis was done by CB, AT, SE, MP, and LR. CB, AT, and MP were responsible for writing the article. All authors have sufficiently contributed to the work performed. All authors read and approved the final manuscript.
ORCID iDs: Ana Tomás  https://orcid.org/0000-0003-4710-2605
https://orcid.org/0000-0003-4710-2605
Marta Pojo  https://orcid.org/0000-0002-4036-6731
https://orcid.org/0000-0002-4036-6731
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Millis SZ, Jardim DL, Albacker L, et al. Phosphatidylinositol 3-kinase pathway genomic alterations in 60,991 diverse solid tumors informs targeted therapy opportunities. Cancer. 2019;125:1185–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15:7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Katso R, Okkenhaug K, Ahmadi K, White S, Timms J, Waterfield MD. Cellular function of phosphoinositide 3-kinases: implications for development, immunity, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2001;17:615–675. [DOI] [PubMed] [Google Scholar]
- 4. Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang J, Nie J, Ma X, Wei Y, Peng Y, Wei X. Targeting PI3K in cancer: mechanisms and advances in clinical trials. Mol Cancer. 2019;18:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oda K, Stokoe D, Taketani Y, McCormick F. High frequency of coexistent mutations of PIK3CA and PTEN genes in endometrial carcinoma. Cancer Res. 2005;65:10669–10673. [DOI] [PubMed] [Google Scholar]
- 7. Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang Y, Kwok-Shing Ng P, Kucherlapati M, et al. A pan-cancer proteogenomic atlas of PI3K/AKT/mTOR pathway alterations. Cancer Cell. 2017;31:820–832.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ligresti G, Militello L, Steelman LS, et al. PIK3CA mutations in human solid tumors: role in sensitivity to various therapeutic approaches. Cell Cycle. 2009;8:1352–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. [DOI] [PubMed] [Google Scholar]
- 12. Hartmann C, Bartels G, Gehlhaar C, Holtkamp N, von Deimling A. PIK3CA mutations in glioblastoma multiforme. Acta Neuropathol. 2005;109:639–642. [DOI] [PubMed] [Google Scholar]
- 13. Verhaak RGW, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR and NF1. Cancer Cell. 2010;17:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Broderick DK, Di C, Parrett TJ, et al. Mutations of PIK3CA in anaplastic oligodendrogliomas, high-grade astrocytomas, and medulloblastomas. Cancer Res. 2004;64:5048–5050. [DOI] [PubMed] [Google Scholar]
- 15. Brennan CW, Verhaak RGW, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee JK, Wang J, Sa JK, et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet. 2017;49:594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. The Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iuchi T, Sugiyama T, Ohira M, et al. Clinical significance of the 2016 WHO classification in Japanese patients with gliomas. Brain Tumor Pathol. 2018;35:71–80. [DOI] [PubMed] [Google Scholar]
- 19. Tabouret E, Nguyen AT, Dehais C, et al. Prognostic impact of the 2016 WHO classification of diffuse gliomas in the French POLA cohort. Acta Neuropathol. 2016;132:625–634. [DOI] [PubMed] [Google Scholar]
- 20. Brito C, Azevedo A, Esteves S, et al. Clinical insights gained by refining the 2016 WHO classification of diffuse gliomas with: EGFR amplification, TERT mutations, PTEN deletion and MGMT methylation. BMC Cancer. 2019;19:968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tanaka S, Batchelor TT, Iafrate AJ, et al. PIK3CA activating mutations are associated with more disseminated disease at presentation and earlier recurrence in glioblastoma. Acta Neuropathol Commun. 2019;7:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Velásquez C, Mansouri S, Mora C, et al. Molecular and clinical insights into the invasive capacity of glioblastoma cells. J Oncol. 2019;2019:1740763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. André F, Ciruelos E, Rubovszky G, et al. Alpelisib for PIK3CA-mutated, hormone receptor–positive advanced breast cancer. N Engl J Med. 2019;380:1929–1940. [DOI] [PubMed] [Google Scholar]
- 24. Vasan N, Razavi P, Johnson JL, et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kα inhibitors. Science. 2019;366:714–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Juric D, Rodon J, Tabernero J, et al. Phosphatidylinositol 3-kinase α–selective inhibition with alpelisib (BYL719) in PIK3CA-altered solid tumors: results from the first-in-human study. J Clin Oncol. 2018;36:1291–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Markham A. Alpelisib: first global approval. Drugs. 2019;79:1249–1253. [DOI] [PubMed] [Google Scholar]
- 27. Roque L, Rodrigues R, Pinto A, Moura-Nunes V, Soares J. Chromosome imbalances in thyroid follicular neoplasms: a comparison between follicular adenomas and carcinomas. Genes Chromosomes Cancer. 2003;36:292–302. [DOI] [PubMed] [Google Scholar]
- 28. Campos C, Fragoso S, Luís R, et al. High-throughput sequencing identifies 3 novel susceptibility genes for hereditary melanoma. Genes. 2020;11:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baker CL, Vaughn CP, Samowitz WS. A PIK3CA pyrosequencing-based assay that excludes pseudogene interference. J Mol Diagn. 2012;14:56–60. [DOI] [PubMed] [Google Scholar]
- 30. Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–W514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cejalvo T, Gargini R, Segura-Collar B, et al. Immune profiling of gliomas reveals a connection with IDH1/2 mutations, tau function and the vascular phenotype. Cancers. 2020;12:3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bunse L, Pusch S, Bunse T, et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat Med. 2018;24:1192–1203. [DOI] [PubMed] [Google Scholar]
- 34. Wang Q, Hu B, Hu X, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2017;32:42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jeanmougin M, Håvik AB, Cekaite L, et al. Improved prognostication of glioblastoma beyond molecular subtyping by transcriptional profiling of the tumor microenvironment. Mol Oncol. 2020;14:1016–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. [DOI] [PubMed] [Google Scholar]
- 37. Jiang N, Dai Q, Su X, Fu J, Feng X, Peng J. Role of PI3K/AKT pathway in cancer: the framework of malignant behavior. Mol Biol Rep. 2020;47:4587–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li X, Wu C, Chen N, et al. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget. 2016;7:33440–33450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Abubaker J, Bavi PP, Al-Harbi S, et al. PIK3CA mutations are mutually exclusive with PTEN loss in diffuse large B-cell lymphoma. Leukemia. 2007;21:2368–2370. [DOI] [PubMed] [Google Scholar]
- 40. Eng J, Woo KM, Sima CS, et al. Impact of concurrent PIK3CA mutations on response to EGFR tyrosine kinase inhibition in EGFR-mutant lung cancers and on prognosis in oncogene-driven lung adenocarcinomas. J Thorac Oncol. 2015;10:1713–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kostakis GC, Papadogeorgakis N, Koumaki V, Kamakari S, Koumaki D, Alexandridis C. Absence of hotspot mutations in exons 9 and 20 of the PIK3CA gene in human oral squamous cell carcinoma in the Greek population. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2010;109:e53–e58. [DOI] [PubMed] [Google Scholar]
- 42. Weiss GA, Rossi MR, Khushalani NI, et al. Evaluation of phosphatidylinositol-3-kinase catalytic subunit (PIK3CA) and epidermal growth factor receptor (EGFR) gene mutations in pancreaticobiliary adenocarcinoma. J Gastrointest Oncol. 2013;4:20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang Q, Shi YL, Zhou K, et al. PIK3CA mutations confer resistance to first-line chemotherapy in colorectal cancer. Cell Death Dis. 2018;9:739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guo S, Loibl S, von Minckwitz G, Darb-Esfahani S, Lederer B, Denkert C. PIK3CA H1047R mutation associated with a lower pathological complete response rate in triple-negative breast cancer patients treated with anthracycline-taxane–based neoadjuvant chemotherapy. Cancer Res Treat. 2020;52:689–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mosele F, Stefanovska B, Lusque A, et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann Oncol. 2020;31:377–386. [DOI] [PubMed] [Google Scholar]
- 46. Loibl S, von Minckwitz G, Schneeweiss A, et al. PIK3CA mutations are associated with lower rates of pathologic complete response to anti-human epidermal growth factor receptor 2 (HER2) therapy in primary HER2-overexpressing breast cancer. J Clin Oncol. 2014;32:3212–3220. [DOI] [PubMed] [Google Scholar]
- 47. Jiang W, Wu Y, He T, et al. Targeting of β-catenin reverses radioresistance of cervical cancer with the PIK3CA E545K mutation. Mol Cancer Ther. 2020;19:337–347. [DOI] [PubMed] [Google Scholar]
- 48. Sampson JH, Gunn MD, Fecci PE, Ashley DM. Brain immunology and immunotherapy in brain tumours. Nat Rev Cancer. 2020;20:12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boissière-Michot F, Chabab G, Mollevi C, et al. Clinicopathological correlates of γδ T cell infiltration in triple-negative breast cancer. Cancers. 2021;13:765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. An Y, Adams JR, Hollern DP, et al. Cdh1 and Pik3ca mutations cooperate to induce immune-related invasive lobular carcinoma of the breast. Cell Rep. 2018;25:702–714.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Borcoman E, De La Rochere P, Richer W, et al. Inhibition of PI3K pathway increases immune infiltrate in muscle-invasive bladder cancer. Oncoimmunology. 2019;8:e1581556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Crane CA, Panner A, Murray JC, et al. PI(3) kinase is associated with a mechanism of immunoresistance in breast and prostate cancer. Oncogene. 2009;28:306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Swiatek-Machado K, Kaminska B. STAT signaling in glioma cells. In: Baranska J, ed. Glioma Signaling. Vol 1202. Cham: Springer; 2020:203–222. [DOI] [PubMed] [Google Scholar]
- 54. Birner P, Toumangelova-Uzeir K, Natchev S, Guentchev M. STAT3 tyrosine phosphorylation influences survival in glioblastoma. J Neurooncol. 2010;100:339–343. [DOI] [PubMed] [Google Scholar]
- 55. Liang QC, Xiong H, Zhao ZW, et al. Inhibition of transcription factor STAT5b suppresses proliferation, induces G1 cell cycle arrest and reduces tumor cell invasion in human glioblastoma multiforme cells. Cancer Lett. 2009;273:164–171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-onc-10.1177_11795549211068804 for PIK3CA Mutations in Diffuse Gliomas: An Update on Molecular Stratification, Prognosis, Recurrence, and Aggressiveness by Cheila Brito, Ana Tomás, Ana Azevedo, Susana Esteves, Manuela Mafra, Lúcia Roque and Marta Pojo in Clinical Medicine Insights: Oncology