Abstract
Determination of hepatitis C virus (HCV) genotypes and subtypes has become increasingly important for the clinical management and prognosis of HCV infections. The aim of the present study was to assess the specificity and reliability of a newly developed, commercially available HCV genotyping kit (TRUGENE HCV 5′NC genotyping kit). This technique utilizes PCR fragments previously generated by the diagnostic Roche AMPLICOR HCV test, which are subsequently subjected to simultaneous PCR amplification and direct sequencing (CLIP sequencing) of the 5′ noncoding region (5′NCR). HCV isolates from 100 randomly chosen patients were genotyped by both the TRUGENE HCV 5′NC genotyping kit and DNA enzyme immunoassay (DEIA). Typing results obtained by both methods were in complete concordance in 91% of the cases. HCV RNA from the samples with discordant genotype assignment in both assays was additionally amplified with primers from the HCV core and NS5B regions. Phylogenetic analysis of the obtained sequences supported the results obtained from DEIA in six cases and CLIP sequencing in two cases. In the former six cases, the TRUGENE HCV 5′NC genotyping kit could not correctly differentiate between subtypes of genotypes 1 and 2 due to the high conservation of the 5′NCR. However, since there was not any misclassification between HCV genotypes 1 and non-1 types, the results obtained with this system are, in general, reliable and can be used in clinical practice. The TRUGENE HCV 5′NC genotyping kit in our hands proved to be a fast and convenient technique that might be an attractive option for HCV genotyping in laboratories already using the Roche AMPLICOR HCV test for diagnostic reverse transcription-PCR.
The hepatitis C virus (HCV) is a positive-stranded RNA virus that was identified as the main cause of posttransfusion non-A, non-B hepatitis (2). The genome of this virus is highly variable. So far, six major genotypes and more than 100 subtypes have been described (8, 19). Many studies have indicated an association between HCV genotype and both the responsiveness to alpha-interferon treatment and the degree of clinical progression of chronic HCV infection (4, 19, 28). Therefore, during the last few years, HCV genotyping has been implemented in clinical laboratory settings and there is an ongoing demand for the development of new, automated typing techniques that provide reliable results within a reasonable time frame (8, 16, 19). The use of a single amplification reaction for diagnostic HCV RNA detection including the possibility of genotype determination from these PCR products would be the most efficient way of HCV genotyping. In this study, we utilized PCR fragments previously generated by the Roche AMPLICOR HCV assay to assess the HCV subtype by subsequent PCR amplification and direct sequencing of the 5′ noncoding region (5′NCR) with a recently developed, and now commercially available, automated sequencing system.
MATERIALS AND METHODS
Specimens.
Randomly chosen sera from 100 HCV infected in- and outpatients (72 men and 28 women; mean age, 46 years [range, 18 to 87 years]) attending Essen University Hospital were included in this study. All patients tested positive for HCV antibodies by immunoassay (Sanofi Diagnostics Pasteur, Freiburg, Germany) and for HCV RNA by diagnostic reverse transcription (RT)-PCR (AMPLICOR; Roche Diagnostics, Mannheim, Germany). Sera were aliquoted and stored at −80°C until HCV typing.
TRUGENE HCV 5′NC genotyping kit.
The HCV subtype of all samples was determined by the newly developed TRUGENE HCV 5′NC genotyping kit (Visible Genetics Europe, Evry, France). This test uses the 244-bp fragment from the 5′NCR of HCV previously amplified by the diagnostic Roche AMPLICOR HCV kit (27). After purification with Chroma Spin 100 columns (Clontech Laboratories, Palo Alto, Calif.), the Roche amplicons were subjected to simultaneous PCR amplification and sequencing (CLIP sequencing), according to the manufacturer's instructions (instruction manual for TRUGENE HCV 5′NC genotyping kit, Visible Genetics, Toronto, Canada). Two characteristics of the CLIP reaction as a modification of the original coupled amplification and sequencing method by Ruano and Kidd (17) are noteworthy. (i) An engineered mutant of thermostable DNA polymerase is used which lacks 5′-3′ exonuclease activity and therefore produces uniform band intensities. (ii) Different far-red fluorescent dyes are linked to the two inward-facing CLIP primers, allowing a template to be sequenced in both directions in a single run (26). Automated sequencing of the 183-bp fragment (nucleotides [nt] 96 to 278 [numbering according to reference 2]) resulting from the CLIP reaction was performed on a MicroGene Clipper sequencer. This platform employs ultrathin (50-μm-thick) disposable gels (26). Each pair of forward and reverse sequences were combined and automatically aligned with reference sequences stored in the GeneLibrarian database in order to determine the HCV subtype and the closest isolate.
DEIA.
All samples were also typed by DNA enzyme immunoassay (DEIA) (Gen-Eti-K DEIA; Sorin Biomedica, Saluggia, Italy), as described previously in full detail (24). In brief, a fragment from the HCV core region was amplified by nested RT-PCR, and the resulting amplicons were hybridized to type- or subtype-specific oligonucleotide probes adsorbed to microwell plates. The double-stranded DNA hybrid thus formed was detected by DEIA.
Amplification and direct sequencing of HCV core and NS5B fragments.
HCV RNA from the samples with discordant genotyping results by CLIP sequencing and DEIA were subjected to additional RT-PCR with primers from the core and NS5B regions. The full details of both procedures are given elsewhere (5, 25). The PCR products obtained were sequenced directly from both directions.
Phylogenetic analysis.
Sequences of the 183-bp HCV 5′NCR (nt 96 to 278 [numbering according to reference 2]), 216-bp core (nt 461 to 676 [numbering according to reference 2]), and 156-bp HCV NS5B (nt 8353 to 8508 [numbering according to reference 2]) DNA fragments from those samples with discrepant typing results by the TRUGENE HCV 5′NC genotyping kit and DEIA were subjected to phylogenetic analysis using the PHYLIP software package, version 3.5c (7). Distances between pairs of sequences were estimated with the DNADIST program. Phylogenetic trees were constructed by the unweighted pair group method using arithmetic averages on the previous sets of pairwise distance. Significance of the grouping was assured by bootstrap resampling (1,000 replicates). The following sequences from the GenBank were included in the analysis (accession numbers given in parentheses): subtype 1a (AF011751 and M62321); subtype 1b (AF165057 and D10934); subtype 1c (D14853); subtype 2a (AF177036 and D00944); subtype 2b (D10077, D10649, D10988, and D45877); subtype 2c (AF041329, L23457, L23458, and L38337); subtype 2i (X76411); subtype 3a (D17763 and D28917); subtype 3b (D10080, D11443, and D49374); subtype 4 (L23470, L29625, and Z29446). The partial HCV 5′NCR, core, and NS5B sequences obtained from those samples which gave initially discrepant typing results by the TRUGENE HCV 5′NC genotyping kit and DEIA have been deposited in GenBank under accession numbers AF245282 to AF245290 (HCV 5′NCR), AF233701 to AF23379 (HCV core), and AF233693 to AF233699 (HCV NS5B).
RESULTS
An HCV type and subtype could be determined for all 100 samples with the TRUGENE HCV 5′NC genotyping kit, whereas one sample was not typeable by DEIA. The results obtained with the two tests were in complete agreement in 91% of the cases. A different genotype assignment was achieved in two cases (1b versus 3a and 1b versus 4) and divergent HCV 1 and 2 subtypes were determined for six isolates (Table 1).
TABLE 1.
Comparison of HCV typing results obtained by GEN-ETI-K DEIA and TRUGENE HCV 5′NC genotyping kita
| DEIA HCV subtype | No. of isolates with HCV subtype according to TRUGENE HCV 5′NC genotyping kit
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 1a | 1b | 1c | 2a/c | 2b | 3a | 4 | 5 | |
| 1a | 14 | 1 | 1 | |||||
| 1b | 2 | 25 | 1 | 1 | ||||
| 2a | 5 | |||||||
| 2b | 2 | 9 | ||||||
| 2c | 4 | |||||||
| 3a | 28 | |||||||
| 4 | 5 | |||||||
| 5 | 1 | |||||||
| Not typeable | 1 | |||||||
Discrepant genotype and subtype assignments are indicated in boldface type.
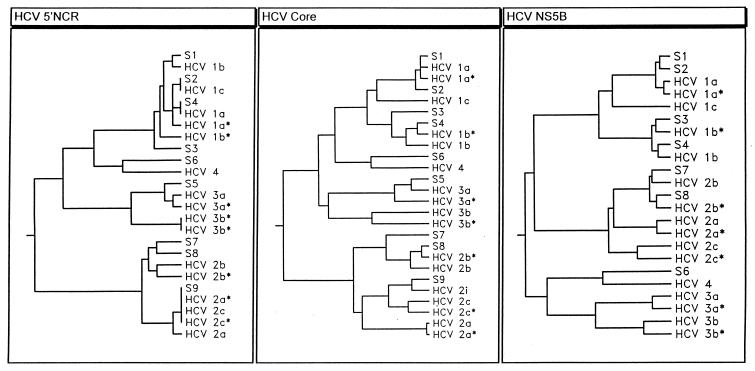
For those samples that gave discrepant results with the TRUGENE HCV 5′NC genotyping kit and DEIA, a phylogenetic analysis of the 5′NCR sequences was performed to check for the reliability of HCV subtype determinations by means of the GeneLibrarian database. Identical results were obtained in six cases. Samples S3, S7, and S8, however, turned out to be HCV subtypes 1b and 2b in phylogenetic analysis, whereas typing by the GeneLibrarian database indicated the presence of subtypes 1a and 2a/c, respectively (Fig. 1; Table 2). Direct sequencing and analysis of the less-conserved HCV core and NS5B fragments revealed that the TRUGENE HCV 5′NC genotyping kit provided the correct genotype assessment (genotypes 3a and 4) for samples S5 and S6. Regarding the discrepant subtyping results (samples S1 to S4, S7, and S8), DEIA was correct in all six cases. Sample S9, which could not be typed by DEIA and yielded subtype 2a/c by the TRUGENE HCV 5′NC genotyping kit, belonged to the rare subtype 2i (Fig. 1; Table 2).
FIG. 1.
Phylogenetic trees of samples S1 to S9 with discordant HCV typing results by DEIA (GEN-ETI-K DEIA) and TRUGENE HCV 5′NC genotyping kit, derived from an analysis of partial 5′NCR (nt 96 to 278), HCV core (nt 461 to 676), and NS5B (nt 8353 to 8508) sequences. As subtype prototypes, sequences from the GenBank were included. Asterisks are used for discrimination of different isolates from the same HCV subtype. The respective accession numbers are given in the text.
TABLE 2.
Final HCV subtype assignment based on results of HCV core and NS5B sequencing for samples that yielded discrepancies in other assays
| Sample | HCV subtype according to:
|
Interpretation | |||
|---|---|---|---|---|---|
| DEIA | TRUGENE 5′NC HCV genotyping kit | Direct sequencing
|
|||
| Core | NS5B | ||||
| S1 | 1a | 1b | 1a | 1a | 1a |
| S2 | 1a | 1c | 1a | 1a | 1a |
| S3 | 1b | 1a | 1b | 1b | 1b |
| S4 | 1b | 1a | 1b | 1b | 1b |
| S5 | 1b | 3a | 3a | No amplicon | 3a |
| S6 | 1b | 4 | 4 | 4 | 4 |
| S7 | 2b | 2a/c | 2b | 2b | 2b |
| S8 | 2b | 2a/c | 2b | 2b | 2b |
| S9 | Not typeable | 2a/c | 2i | Not done | 2i |
DISCUSSION
Nucleotide sequence analysis is the current “gold standard” for identifying different HCV genotypes and subtypes but is generally regarded as not practical for routine clinical laboratory settings (8, 19). Therefore, a variety of surrogate HCV typing procedures has been proposed during recent years, mainly based upon amplification of the viral sequences by PCR, using either type-specific primers (15), analysis of PCR products by hybridization with genotype-specific probes (21, 25), or restriction fragment length polymorphism (1, 22). The underlying assumption of all these assays that the region (e.g., 5′NCR, core, or NS5B) analyzed is representative of the whole HCV genome is generally supported by the very consistent typing results which have been obtained so far using assays based on sequence analysis of different regions of the HCV genome (8, 19).
In this study, we have combined diagnostic HCV RNA detection with the widely distributed Roche AMPLICOR HCV assay and direct sequencing of the already available HCV 5′NCR PCR product by a newly developed commercial system (26; TRUGENE HCV 5′NC genotyping kit manual). Following this approach, HCV genotyping including analysis of the sequence data was possible in about 5 h. Genotyping by DEIA, besides requiring an additional nested PCR with primers from the HCV core region, requires an overnight incubation with the biotinylated hybridization probes and, therefore, cannot be accomplished in less than 15 h. Comparison of the results obtained by the TRUGENE HCV 5′NC genotyping kit with those of the already well-established DEIA (24) indicated an identical HCV subtype assignment in 91% of all cases. Direct sequencing of the 183-bp 5′NCR fragment in our study, however, was not able to completely resolve all existing HCV genotypes and subtypes. This failure in three cases was attributable to an inadequate subtype assignment by the current version of the GeneLibrarian module, as shown by phylogenetic analysis of the obtained 5′NCR sequences. Possibilities to improve the overall performance of the TRUGENE system might, therefore, comprise the inclusion of more HCV 5′NCR sequences in the GeneLibrarian database, the modification of the general algorithm used for sequence analysis, and the search for modes of HCV subtype assignment other than by a simple alignment of sequences. The remaining discrepancies recorded in this study between TRUGENE HCV 5′NC genotyping kit and DEIA could not be resolved by a more-sophisticated sequence analysis system and are the result of the high sequence conservation of HCV 5′NCR (13). For instance, subtypes 1a and 1b in the 5′NCR fragment analyzed by TRUGENE HCV 5′NC genotyping kit differ in only 1 nt in position 99, and subtype 2b only differs from 2a by 2 nt at positions 124 and 164. A reliable discrimination of subtypes 2a and 2c isolates is also not possible based upon sequence analysis of the 5′NCR alone (20). Consequently, failures in correct assignment of all HCV subtypes similar to those observed in this study had already been demonstrated in several comparative evaluations of HCV genotyping procedures based solely on the analysis of the 5′NCR (3, 9–12, 14, 18, 21, 23), indicating that less-conserved parts of the HCV genome, such as the core (used by DEIA) or NS5B, are more suited for the identification of all existing HCV subtypes.
These inherent disadvantages of the 5′NCR for HCV typing are, however, not crucial for clinical purposes. From the point of view of a clinician, currently it is not necessary to know precisely the subtype of the HCV strain present but to achieve a reliable discrimination between HCV genotypes 1 and non-1 types. According to the most-recent therapeutic recommendations, highly viremic patients chronically infected with HCV genotype 1 isolates should be treated with alpha-interferon and ribavirin for at least 1 year, whereas for infections with non-1 types an initial course of 6 months is sufficient (6). Since with the TRUGENE HCV 5′NC genotyping kit no misclassifications between HCV genotypes 1 and non-1 types occurred, the typing results obtained with this system are, in general, reliable for clinical practice.
Taken together, the newly developed TRUGENE HCV 5′NC genotyping kit and the MicroGene Clipper sequencing platform turned out to be a convenient analytical system that provides clinically valid HCV typing results in about 5 h. The assay might be an attractive option for HCV genotyping in laboratories that already use the Roche AMPLICOR HCV test for diagnostic RT-PCR.
ACKNOWLEDGMENTS
We are grateful to T. Teckentrupp for skillful technical assistance.
This study was supported in part by a grant to the German National Reference Centre for Hepatitis C.
REFERENCES
- 1.Buoro S, Pizzighella S, Boschetto R, Pellizari L, Cusan N, Bonaguro R, Mengoli C, Caudai C, Paluda M, Valensin P E, Palù G. Typing of hepatitis C virus by a new method based on restriction fragment length polymorphism. Intervirology. 1999;42:1–8. doi: 10.1159/000024953. [DOI] [PubMed] [Google Scholar]
- 2.Choo Q-L, Richman K, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby A, Barr P J, Weiner A J, Bradley D W, Kuo G, Houghton M. Genetic organization and diversity of hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cretel E, Gallian P, Obadia Y, Rousseau S, De Micco P, De Lamballerie X. Analysis of hepatitis C virus isolates using molecular and serological typing techniques. Acta Virol. 1997;41:269–275. [PubMed] [Google Scholar]
- 4.Davis G L, Lau J N. Factors predictive of a beneficial response to therapy of hepatitis C. Hepatology. 1997;26(Suppl. 3):122S–127S. doi: 10.1002/hep.510260721. [DOI] [PubMed] [Google Scholar]
- 5.Enomoto N, Takada A, Nakao T, Date T. There are two major types of hepatitis C virus in Japan. Biophys Biochem Res Commun. 1990;170:1021–1025. doi: 10.1016/0006-291x(90)90494-8. [DOI] [PubMed] [Google Scholar]
- 6.European Association for the Study of the Liver. EASL international consensus conference on hepatitis C. Consensus statement. J Hepatol. 1999;30:956–961. [PubMed] [Google Scholar]
- 7.Felsenstein J. PHYLIP (Phylogenetic Inference Package). Seattle: University of Washington; 1993. [Google Scholar]
- 8.Forns X, Bukh J. Methods for determining hepatitis C virus genotype. Viral Hep Rev. 1998;4:1–19. [Google Scholar]
- 9.Forns X, Maluenda M D, Lopez-Labrador F X, Ampurdanes S, Olmedo E, Costa J, Simmonds P, Sanchez-Tapias J M, Jimenez De Anta M T, Rhodes J. Comparative study of three methods for genotyping hepatitis C virus strains in samples from Spanish patients. J Clin Microbiol. 1996;34:2516–2521. doi: 10.1128/jcm.34.10.2516-2521.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Germer J J, Rys P N, Thorvilson J N, Persing D H. Determination of hepatitis C virus genotype by direct sequence analysis of products generated with the Amplicor test. J Clin Microbiol. 1999;37:2625–2630. doi: 10.1128/jcm.37.8.2625-2630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giannini C, Thiers V, Nousbaum J-B, Stuyver L, Maertens G, Bréchot C. Comparative analysis of two assays for genotyping hepatitis C virus based on genotype-specific primers or probes. J Hepatol. 1995;23:246–253. [PubMed] [Google Scholar]
- 12.Le Pogam S, Dubois F, Christen R, Raby C, Cavicchini A, Goudeau A. Comparison of DNA enzyme immunoassay and line probe assays (Inno-LiPA I and II) for hepatitis C virus genotyping. J Clin Microbiol. 1998;36:1461–1463. doi: 10.1128/jcm.36.5.1461-1463.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Major M E, Feinstone M. The molecular virology of hepatitis C. Hepatology. 1997;25:1527–1538. doi: 10.1002/hep.510250637. [DOI] [PubMed] [Google Scholar]
- 14.O'Brien C B, Henzel B S, Wolfe L, Gutekunst K, Moonka D. cDNA sequencing of the 5′ noncoding region (5′NCR) to determine hepatitis C genotypes in patients with chronic hepatitis C. Dig Dis Sci. 1997;42:1087–1093. doi: 10.1023/a:1018813825486. [DOI] [PubMed] [Google Scholar]
- 15.Okamoto H, Sugiyama Y, Okada S, Kurai K, Akahane Y, Sugai Y, Tanaka T, Sato K, Tsuda F, Miyakawa Y, Mayumi M. Typing hepatitis C virus by polymerase chain reaction with type-specific primers: application to clinical surveys and tracing infectious sources. J Gen Virol. 1992;73:673–679. doi: 10.1099/0022-1317-73-3-673. [DOI] [PubMed] [Google Scholar]
- 16.Ross R S, Viazov S, Renzing-Köhler K, Roggendorf M. Changes in the epidemiology of hepatitis C in Germany: shift in the predominance of hepatitis C subtypes. J Med Virol. 2000;60:122–125. [PubMed] [Google Scholar]
- 17.Ruano G, Kidd K K. Coupled amplification and sequencing of genomic DNA. Proc Natl Acad Sci USA. 1991;88:2815–2819. doi: 10.1073/pnas.88.7.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seme K, Poljak M, Dovic P, Koren S. Comparative evaluation of four genotyping methods for hepatitis C virus. Folia Biol (Praha) 1997;43:219–224. [PubMed] [Google Scholar]
- 19.Simmonds P. Variability of the hepatitis C virus genome. In: Reesink H W, editor. Hepatitis C virus. Current studies in hematology and blood transfusion. Vol. 62. Freiburg, Germany: Karger; 1998. pp. 38–63. [DOI] [PubMed] [Google Scholar]
- 20.Smith D B, Mellor J, Jarvis L M, Davidson F, Kolberg J, Urdea M, Yap P L, Simmonds P. Variation of the hepatitis C virus 5′ non-coding region: implications for secondary structure, virus detection and typing. J Gen Virol. 1995;76:1749–1761. doi: 10.1099/0022-1317-76-7-1749. [DOI] [PubMed] [Google Scholar]
- 21.Stuyver L, Wyseur A, van Arnhem W, Lunel F, Laurent-Puig P, Pawlotsky J M, Kleter B, Bassit L, Nkengasong J, van Dorn L J. Hepatitis C virus genotyping by means of 5′UR/core line probe assay and molecular analysis of untypeable samples. Virus Res. 1995;38:137–157. doi: 10.1016/0168-1702(95)00052-r. [DOI] [PubMed] [Google Scholar]
- 22.Thiers V, Jaffredo F, Tuveri R, Chodan N, Bréchot C. Development of a simple restriction fragment length polymorphism (RFLP) based assay for HCV genotyping and comparative analysis with genotyping and serotyping results. J Virol Methods. 1997;65:9–17. doi: 10.1016/s0166-0934(96)02162-3. [DOI] [PubMed] [Google Scholar]
- 23.Vatteroni M, Maggi F, Morrica A, Fornai C, Giorgi M, Pistello M, Bendinelli M. Comparative evaluation of five rapid methods for identifying subtype 1b and 2c hepatitis C virus isolates. J Virol Methods. 1997;66:187–194. doi: 10.1016/s0166-0934(97)00054-2. [DOI] [PubMed] [Google Scholar]
- 24.Viazov S, Zibert A, Ramakrishnan K, Widell A, Cavicchini A, Schreier E, Roggendorf M. Typing of hepatitis C virus isolates by DNA enzyme immunoassay. J Virol Methods. 1994;48:81–92. doi: 10.1016/0166-0934(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 25.Viazov S, Kuzin S, Paladi N, Tchernovetsky M, Isaeva E, Mazhul L, Vasychova F, Widell A, Roggendorf M. Hepatitis C virus genotypes in different regions of the former Soviet Union (Russia, Moldova, and Uzbekistan) J Med Virol. 1997;53:36–40. [PubMed] [Google Scholar]
- 26.Yager T D, Baron L, Batra R, Bouevitch A, Chan D, Chan K, Darasch S, Gilchrist R, Izmailov A, Lacroix J-M, Marchelleta K, Renfrew J, Rushlow D, Steinbach E, Ton C, Waterhouse P, Zaleski H, Dunn J M, Stevens J. High performance DNA sequencing, and the detection of mutations and polymorphisms, on the Clipper sequencer. Electrophoresis. 1999;20:1280–1300. doi: 10.1002/(SICI)1522-2683(19990101)20:6<1280::AID-ELPS1280>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 27.Young K K, Resnick R M, Myers T W. Detection of hepatitis C virus RNA by a combined reverse transcription-polymerase chain reaction. J Clin Microbiol. 1993;31:882–886. doi: 10.1128/jcm.31.4.882-886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zein N N, Rakela J, Krawitt E L, Reddy K R, Tominaga T, Persing D H. Hepatitis C virus genotypes in the United States: epidemiology, pathogenicity, and response to interferon therapy. Ann Intern Med. 1996;125:634–639. doi: 10.7326/0003-4819-125-8-199610150-00002. [DOI] [PubMed] [Google Scholar]