Abstract
Extensive study has demonstrated that epilepsy occurs with greater frequency at certain times in the 24-h cycle. Although these findings implicate an overlap between the circadian rhythm and epilepsy, the molecular and cellular mechanisms underlying this circadian regulation are poorly understood. Because the 24-h rhythm is generated by the circadian molecular system, it is not surprising that this system comprised of many circadian genes is implicated in epilepsy. We summarized evidence in the literature implicating various circadian genes such as Clock, Bmal1, Per1, Rev-erbα, and Rorα in epilepsy. In various animal models of epilepsy, the circadian oscillation and the steady-state level of these genes are disrupted. The downstream pathway of these genes involves a large number of metabolic pathways associated with epilepsy. These pathways include pyridoxal metabolism, the mammalian target of rapamycin pathway, and the regulation of redox state. We propose that disruption of these metabolic pathways could mediate the circadian regulation of epilepsy. A greater understanding of the cellular and molecular mechanism of circadian regulation of epilepsy would enable us to precisely target the circadian disruption in epilepsy for a novel therapeutic approach.
Keywords: circadian, clock genes, epigenetic, epilepsy, metabolism
1 |. INTRODUCTION
Epilepsy is highly circadian in nature.1 This phenomenon has been studied and characterized extensively in human studies2 as well as animal studies utilizing different models of epilepsy.3,4 These studies show that different types of epilepsy present with distinct circadian signatures, depending on its seizure semiology and localization.2 Patients with epilepsy also present with disturbances in their sleep pattern and their sleep–wake cycle,5,6 which are behavioral outputs of the circadian rhythm. In patients with epilepsy, circadian rhythm has also been shown to influence autonomic response,7 hormonal rhythm,8 and response to antiseizure drugs.9 Despite the relatively well-characterized circadian nature of epilepsy, our understanding of the circadian mechanisms regulating seizures is incomplete at a cellular and molecular level.
In mammals, the circadian rhythm is generated and maintained by the circadian molecular system,10 comprised of several genes working in concert and generating rhythmic oscillations. Emerging studies in the field implicate some of these genes in epilepsy. However, a multitude of these circadian genes regulate a large number of physiological processes in mammals including bioenergetic pathways. Regulation of brain metabolism may be an important aspect of circadian transcription. This review starts by briefly reviewing the circadian molecular system and the various genes that make up the system. Evidence will then be presented that links the circadian molecular system and epilepsy in the context of circadian gene oscillation and the steady-state circadian expression level. Ultimately, three metabolic pathways will be discussed as potential downstream pathways to the circadian molecular system that links circadian rhythm and epilepsy.
2 |. CIRCADIAN MOLECULAR SYSTEM
The mammalian molecular circadian clock is a self-sustaining transcriptional feedback loop composed of interlocking positive and negative limbs of the feedback circuit (see Figure 1). Most of the genes involved are transcription factors, meaning that they control transcription of other genes through binding with a cis-regulatory element. The circadian system relies on this regulation through three main cis-regulatory elements within the promoter sequence of many circadian genes, which will be discussed below.
FIGURE 1.
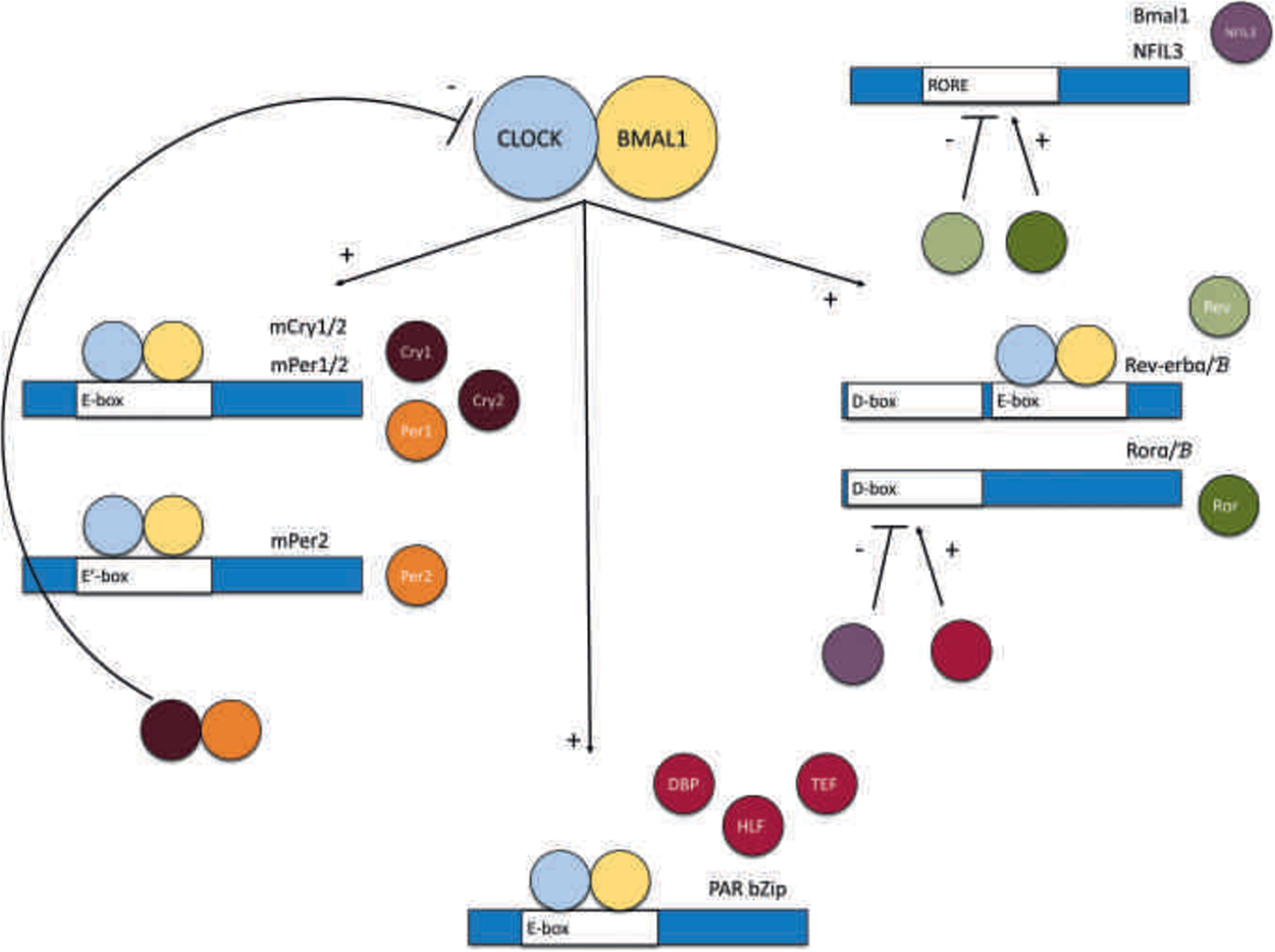
Components of the circadian molecular system. The circadian molecular system is made up of a transcriptional feedback loop defined by the interlocking positive and negative limbs. The positive limb is comprised of two core genes: Clock and Bmal1. CLOCK and BMAL1 form a heterodimer and bind to E-box sequences activating the three negative limbs of the system. The first negative limb is the Per and Cry genes. Upon translation, the PER and CRY proteins form a heterodimer and repress the transcription of the Clock and Bmal1 complex. The second negative limb is the Rev-erb and Ror genes. The proteins REV-ERB and ROR compete for the RORE sequence binding site, which is present in many genes. Upon binding, ROR induces the gene’s transcription, whereas REV-ERB inhibits the gene’s transcription. Those genes include Bmal1, a core component of the positive limb, thus creating this second negative feedback loop, and Nfil3, which will contribute in the feedback loop of the third negative limb. The third negative limb is made up of the PAR bZIP transcription factors, namely the genes Dbp, Hlf, and Tef. These PAR bZIP transcription factors compete with Nfil3 for D-box sequence binding. The PAR bZIP proteins bind to the D-box sequence to activate transcription, whereas NFIL3 represses transcription. The D-box sequence is present within the promoter sequence of Rev-erb and Ror genes. Thus, the PAR bZIP genes and Nfil3 create a third negative feedback loop and indirectly affect Bmal1 transcription through its regulation of Rev-erb and Ror transcription. Together, the positive limb and these three negative limbs form the circadian molecular system whose functional output is the circadian rhythm
The positive limb of this system is composed of two genes, also considered the core of the circadian molecular system, namely Clock and Bmal1.10 CLOCK and BMAL1 form a heterodimer complex that binds to an E-box sequence within the DNA, activating the transcription of many sets of different genes.10 Some sets of genes comprise the negative limb of the system, which eventually represses the transcription of Clock and Bmal1. The fluctuation or oscillation in the level of these genes as a result of this negative feedback loop is what creates the circadian rhythmicity.10 It is important to note here that Bmal1-knockout animals experienced loss of circadian rhythmicity.11 In contrast, Clock-knockout animals did not seem to experience loss of circadian rhythmicity.12 This appeared to be because of a Clock paralog, Npas2, that appears to be functionally redundant and can compensate for Clock in its absence by forming a heterodimer complex with Bmal1.12
There are three main components of the negative limb of the circadian molecular system, which we will briefly review here. The first set of genes are the Per (Per1 and Per2) and the Cry (Cry1 and Cry2) genes.10 The resulting PER1/2 and CRY1/2 proteins dimerize and inhibit the transcription of Clock and Bmal1 through histone deacetylation.10 The Per family of genes also includes Per3, which is a paralog of both Per1 and Per2, but appears not to be functional in circadian regulation.13 The other two components of the negative limbs are the Rev/Ror system and the PAR bZIP (the proline- and acid-rich [PAR] subfamily of basic region leucine-zipper [bZIP] transcription factors) system. These two negative limb components work together to generate another feedback loop for the purpose of Bmal1 regulation. This regulation is mediated by the competition between Rev-erbα/β (also called Nr1d1 or Nr1d2) and Rorα/β for their common binding site, the orphan receptor response element (RORE). The RORE binding site is present within many genes, importantly the circadian Bmal1 and Nfil3 genes (also called E4bp4).14 Upon binding, the ROR proteins initiate transcription of these genes, whereas the REV-ERB proteins inhibit it.14 Because REV-ERB and ROR differentially regulate the transcription of Bmal1, this creates a negative feedback loop that makes up the second component of the negative limb. In turn, the transcriptional regulation of Rev-erbα/β and Rorα/β is provided by the cis-elements D-box and E-box. As mentioned above, the E-box sequence is controlled by the CLOCK/BMAL1 complex, whereas the D-box integrates an additional layer of circadian regulation by the PAR bZIP system. The PAR bZIP transcription factors are comprised of three genes, namely, Dbp, Tef, and Hlf.15 Transcription of the PAR bZIP genes is initiated by the E-box binding of the Clock/Bmal1 complex. Upon transcription, the PAR bZIP proteins bind to the D-box sequence to activate transcription of the Rev and Ror genes, thus influencing circadian transcription in this manner.10 Transcription of Rev and Ror genes becomes a negative feedback loop, as one of its products of transcriptional activation through the RORE sequence, namely, the aforementioned Nfil3, is a transcriptional repressor of the D-box sequence.16 Therefore, the third component of the negative limb involves PAR bZIP proteins and NFIL3 that regulate the Rev and Ror gene transcription, which indirectly regulates the Bmal1 transcription.
In general, the three components of the negative limb regulate circadian transcription by a combination of three main cis-regulatory elements, namely, the E-boxes, ROREs, and D-boxes, within the promoter and enhancer sequence of circadian regulated genes.10 It has been theorized that these three cis-elements collectively provide the necessary phase delay to cycle over a 24-h period: E-box in the subjective morning, D-box in the day, and RORE elements in the evening.10 It is also important to note that although not all of these circadian proteins are absolutely required for the circadian molecular clock, they serve to make the system more robust and precise.
One important molecular mechanism that contributes to the functioning of the circadian molecular system is epigenetic regulation. Epigenetic regulation does not change the DNA sequence but modifies the expression of the genes usually through mechanisms that change the structure of the nucleosome.17 The nucleosome refers to a unit composed of chromatin fibers where genomic DNA is packaged and wrapped around histones. These epigenetic mechanisms involve DNA methylation, histone modifications (such as histone tail acetylation, methylation, or phosphorylation), and chromatin remodeling.17 The circadian molecular system relies on epigenetic mechanisms for its daily function. An example of this was briefly mentioned before. The transcription feedback loop between PER/CRY and CLOCK/BMAL involves a rhythmic cycle of histone acetylation/deacetylation. CLOCK was shown to possess histone acetyltransferase activity,18 and acetylated histone H3 was found within the promoter sequence of Per1, Per2, and Cry1.19 As the Per1 and Per2 genes are translated, the PER protein complex also recruits SIN3 protein complex, which contains histone deacetylase protein (HDAC).20 This protein complex then deacetylates the histone that was acetylated by CLOCK upon transcription initiation, thus repressing the transcription of the Per genes.20 This rhythmic histone acetylation/deacetylation cycle adds another layer of regulation and corresponds to the rhythmic transcription feedback loop between CLOCK/BMAL and PER/CRY.10,20 Thus, the circadian molecular system is a molecular system that functions through direct feedback mechanisms by protein–protein interaction as well as epigenetic regulation.
3 |. CIRCADIAN GENE OSCILLATION IN EPILEPSY
As outlined above, circadian rhythm is generated through a rhythmic oscillation of a set of core circadian genes (see Figure 1). Given the established association between circadian rhythm and epilepsy, it would be expected that there is also a disruption in circadian gene oscillation in epilepsy, and that is the case. These disruptions have been described in multiple animal models of epilepsy, as will be detailed in subsequent sections below. The animal models used in these studies include models of temporal lobe epilepsy, such as the pilocarpine-induced and the kainic acid-induced status epilepticus models, where initial pharmacological induction of status epilepticus (the acute phase) and the kindling process (the silent phase) has resulted in network reorganization and subsequent spontaneous epileptogenesis (the chronic phase)21,22; models of transient brain insults, such as the electroconvulsive model of epilepsy, where seizure was induced acutely by electrical stimulation through ear clip electrodes23; and genetic animal models of epilepsy, such as the Kcna1-null mouse, which models epileptic patients with KCNA1 mutation.24
In the pilocarpine model, the hippocampus of epileptic rats lost the rhythmic oscillation of three transcripts, Per1, Per2, and Per3.25 Among those transcripts that still exhibited rhythmicity in this model, such as Bmal1, Cry1, and Cry2, the rhythmic oscillation was significantly shifted in phase, indicating a dysregulated circadian rhythm.25 Interestingly, Clock was reported to be arrhythmic in the hippocampus of both wild-type and epileptic rats of the pilocarpine model; therefore, their arrhythmicity was not significantly affected by the epileptic induction.25 Clock rhythmicity or lack thereof seems to be largely preserved in epilepsy animal models. In a different model, the electroconvulsive model, Clock rhythmicity in the rat’s frontal cortex was also not significantly different between wild-type and epileptic rats.26 This is in stark contrast to the other circadian transcripts studied in this model, such as Bmal1, Per2, Cry1, Cry2, Rev-erbα, and Rorα, all of which were significantly dysregulated.26
In a genetic model of epilepsy using the Kcna1-null mice, aberrant circadian gene oscillation was observed in the anterior hypothalamus.27 The study reported diminished oscillation in Clock, Per1, and Per2, whereas Bmal1 remained largely arrhythmic in both wild-type and epileptic mice.27 Interestingly, this study also examined changes in circadian gene oscillation in constant darkness (DD) condition and found that only Per1 exhibited any difference in oscillation under this condition.27 The DD condition is a tool used by chronobiologists to eliminate the effect of light entrainment on circadian gene oscillation to evaluate the endogenous circadian gene rhythm. The two aforementioned studies did not examine circadian gene oscillation in their epilepsy model under DD.25,26 This is an important factor to be considered when interpreting the results of these studies. In animal models of epilepsy, seizures have been found to occur in a diurnal circadian pattern that is maintained in phase even under DD.3,4 This implies that seizure occurrence is modulated by the endogenous circadian rhythm independent of the effect of light entrainment.
In addition, other factors also need to be considered when interpreting results on circadian gene oscillation. First, genes have different circadian oscillation patterns in different brain regions (see Table 1 for summary of the studies described above that illustrated this point). It is well established that circadian regulation of gene expression is remarkably tissue-specific.28,29 In the case of epilepsy, this increases the complexity even further, because circuit changes from a distantly located “nonepileptic” brain region may influence the circuit and manifestation of the epileptic foci.30 Consideration of these studies becomes difficult once examination of circadian changes extends beyond the epileptic foci to include more distant brain regions. Second, it is a well-characterized phenomenon that circadian rhythmicity at the mRNA level does not always result in rhythmicity of the mRNA or the protein at the steady-state level. This is thought to be due to the influences of other transcription factors as well as various posttranscriptional regulation that accompanies the positive arm of the circadian transcription.31 As a result, a change in the pattern of circadian gene oscillation in epilepsy may not result in a similar pattern of functional output. The changes in the functional output of the circadian system in epilepsy will be discussed more extensively in the next section below.
TABLE 1.
Summary of studies examining circadian gene oscillation under a 24-h light–dark cycle in epilepsy
| Study reference | Animal model | Brain region examined | Seizure profile | Sleep profile | Gene of interest | Effect on oscillation |
|---|---|---|---|---|---|---|
| Matos et al. 201825 | Rat pilocarpine model of temporal lobe epilepsy | Hippocampus | Diurnal, with peak in light phase | Fragmented, with increased slow-wave sleep and spikes interspersed throughout sleep-wake cycle | Clock | Unaffected (arrhythmic in both wild-type and epileptic rats) |
| Bmal1 | Rhythmicity maintained, but phase advanced and amplitude reduced | |||||
| Per1 | Rhythmicity loss | |||||
| Per2 | Rhythmicity loss | |||||
| Per3 | Rhythmicity loss | |||||
| Cry1 | Unaffected (rhythmic in both wild-type and epileptic rats) | |||||
| Cry2 | Unaffected (rhythmic in both wild-type and epileptic rats) | |||||
| Kim et al. 201826 | Rat acute ECS model | Frontal cortex | Seizure induced at same time throughout treatment | Not examined | Clock | Unaffected (rhythmic in both wild-type and epileptic rats) |
| Bmal1 | Rhythmicity maintained, but phase advanced and amplitude reduced | |||||
| Per1 | Rhythmicity maintained, but phase advanced | |||||
| Per2 | Rhythmicity loss | |||||
| Cry1 | Rhythmicity maintained, but phase delayed and amplitude reduced | |||||
| Cry2 | Rhythmicity maintained, but phase advanced and amplitude reduced | |||||
| Rev-erbα | Rhythmicity maintained, but phase advanced and amplitude increased | |||||
| Rorα | Rhythmicity maintained, but amplitude reduced | |||||
| Wallace et al. 201827 | Kcna1-null mice | Anterior hypothalamus | Diurnal, with peak in light phase | Fragmented, with reduced REM and NREM sleep | Clock | Rhythmicity maintained, but amplitude reduced |
| Bmal1 | Unaffected (arrhythmic in both wild-type and epileptic rats) | |||||
| Per1 | Rhythmicity maintained, but amplitude reduced | |||||
| Per2 | Rhythmicity maintained, but amplitude reduced |
Abbreviations: ECS, electroconvulsive seizure; NREM, non-REM; REM, rapid eye movement.
4 |. CIRCADIAN GENE AND PROTEIN EXPRESSION IN EPILEPSY
As mentioned above, changes in circadian gene oscillation vary across animal models of epilepsy. However, changes in circadian gene oscillation pattern do not necessarily translate to similar changes in steady-state mRNA and protein level.31 In most cases, they do translate to similar output. As an example, in the electroconvulsive model study mentioned above, some of the most significant change in the rat frontal cortex was in Rev-erbα, which showed a strong increase in gene oscillation and subsequently also an increase in steady-state mRNA and protein level.26 However, there are cases where changes in circadian oscillation does not translate to similar output. In that same study, there was no difference in the Per1 24-h rhythm, but both steady-state and protein levels of Per1 were decreased.26 Distinctions in steady-state level may prove to be more informative in understanding how the circadian system influences epilepsy rather than focusing on changes in circadian gene oscillation. In the case of Per1, for instance, this gene’s steady-state expression level has been extensively studied in the context of epilepsy. Per1 was demonstrated to be significantly upregulated in the mouse hippocampus following an acute epileptogenic insult with kainic acid or electroconvulsive shock.32 In contrast, repeated epileptogenic insult using electroconvulsive treatment resulted in reduced steady-state mRNA and protein level of Per1.26 When examining changes longitudinally, Per1 was found to be increased acutely post-status epilepticus and returned to normal level after kindling in the rat hippocampus of the pilocarpine model.25 Thus, PER1 may be a protein that is important in the acute phase of epileptogenesis and may serve as a biomarker of epileptic progression.
Similar to Per1, there are significant changes in the steady-state expression of Clock despite its circadian rhythmicity being only mildly affected across multiple models of epilepsy.25,26 At a functional level, Clock steady-state mRNA and protein levels showed a decrease in various animal models of epilepsy.25–27 Importantly, in resected neo-cortical samples from patients with focal epilepsy, Clock expression was also found to be significantly decreased.33 When Clock was knocked out in the forebrain excitatory neurons using the Emx-cre promoter, these animals developed spontaneous epileptic seizures with a phase timing related to their sleep–wake transition.33 This was one of two transgenic animal models of epilepsy related to circadian proteins. The other pertains to the PAR bZIP transcription factors, namely Dbp, Hlf, and Tef, three genes downstream of the CLOCK/BMAL1 complex (see Figure 1). Triple knockout of these three genes resulted in animals developing lethal audiogenic seizures.15 DBP by itself has been shown to be reduced in the hippocampus of rats in the kainate model.34 Thus, decrease in Clock expression and potentially its downstream targets seem to contribute strongly to epilepsy.
Because Clock appears to be an important circadian contributor to epilepsy, it would be remiss to not examine how Bmal1, the molecular binding partner of Clock, is affected in epilepsy. Similarly to Clock, Bmal1 expression was decreased in both the frontal cortex in the electroconvulsive mice26 and the hippocampus of the pilocarpine rat model.25 However, in the Kcna1-null mice, Bmal1 expression in the anterior hypothalamus was not significantly different to wild type.27 Studies in the Bmal1-knockout mouse have not yet reported an epileptic phenotype. However, Bmal1-knockout mouse was shown to exhibit reduced seizure threshold with electrical stimulation and reduced seizure endpoints.35 Although there is less evidence than for Clock, Bmal1 may also be implicated in epilepsy directly or indirectly through its association with Clock.
Finally, in the aforementioned electroconvulsive study, Rev-erbα showed the most significant change in its increase in gene oscillation pattern in line with an increase in steady-state mRNA and protein levels in the frontal cortex of epileptic mice.26 However, in contrast to this study, Rev-erbα was found to be reduced in the epileptic foci from temporal neocortex samples of patients with temporal lobe epilepsy.36 This study also demonstrated that Rev-erbα was decreased in the hippocampus and temporal neocortex of mice treated with pilocarpine both in the acute status epilepticus and the chronic phase.36 Thus, there is evidence implicating Rev-erbα in different models of epilepsy, albeit with varying results. In circadian regulation, Rev-erbα competes with Rorα and exerts opposing effects on Bmal1 regulation.14 Unsurprisingly, there is also some evidence implicating Rorα in various models of epilepsy. In the electroconvulsive study, Rorα expression was decreased in the frontal cortex of epileptic mice, opposite to the increased expression of Rev-erbα.26 In the hippocampus of the pilocarpine rat model, Rorα was also found to be decreased in both the acute and the silent phase of the model, but returned to a level comparable to control at the epileptic phase.37 In humans, Rorα mutations were identified in a subgroup of patients with genetic epilepsy related to intellectual disability and autism.38 Collectively, this evidence suggests the implication of the Rev-erbα and Rorα system in different types of epilepsy.
In summary, these studies implicated the genes that made up the circadian molecular system in epilepsy (pattern of changes summarized in Table 2 and illustrated in Figure 2). It is important to note that these genes often work together in the regulation of circadian transcription. The CLOCK/BMAL1 complex, for example, is the primary driver of circadian rhythm in the suprachiasmatic nucleus.10 Although their actions in a different brain region may be independent of one another, this has not been studied. Additionally, the other genes described above such as Per1, Rev-erbα, and Rorα are downstream targets of Clock and Bmal1. The implications of these genes in epilepsy are hard to interpret without weighing each within the context of CLOCK/BMAL1 regulation. To overcome this, studies are moving toward characterizing potential downstream targets or pathways of circadian regulation of epilepsy. Some of these targets and pathways will be described in the next section.
TABLE 2.
Summary of studies examining steady-state gene and protein expression of circadian genes of interest in various animal models of epilepsy
| Gene of interest | Study reference | Animal model of epilepsy | Brain region examined | mRNA level | Protein level |
|---|---|---|---|---|---|
| Clock | Matos et al. 201825 | Rat pilocarpine model | Hippocampus | Reduced | Not examined |
| Kim et al. 201826 | Rat acute ECS model | Frontal cortex | Unaffected | Reduced | |
| Wallace et al. 201827 | Kcna1-null mice | Anterior hypothalamus | Not examined | Unaffected | |
| Bmal1 | Matos et al. 201825 | Rat pilocarpine model | Hippocampus | Reduced | Not examined |
| Kim et al. 201826 | Rat acute ECS model | Frontal cortex | Reduced | Reduced | |
| Wallace et al. 201827 | Kcna1-null mice | Anterior hypothalamus | Not examined | Unaffected | |
| Per1 | Matos et al. 201825 | Rat pilocarpine model | Hippocampus | Increased acutely; returned to wild-type level on chronic phase | Not examined |
| Kim et al. 201826 | Rat acute ECS model | Frontal cortex | Reduced | Reduced | |
| Wallace et al. 201827 | Kcna1-null mice | Anterior hypothalamus | Not examined | Unaffected | |
| Eun et al. 201132 | Mouse acute kainic acid or ECS treatment | Hippocampus and neocortex | Increased acutely and then returned to wild-type level | Increased acutely | |
| Rev-erbα | Kim et al. 201826 | Rat acute ECS model | Frontal cortex | Increased | Increased |
| Yue et al. 202036 | Mouse pilocarpine model | Hippocampus and temporal neocortex | Not examined | Reduced | |
| Rorα | Kim et al. 201826 | Rat acute ECS model | Frontal cortex | Reduced | Reduced |
| Dbp | Klugmann et al. 200634 | Rat kainic acid model | Hippocampus | Reduced | Not examined |
Abbreviations: ECS, electroconvulsive seizure.
FIGURE 2.
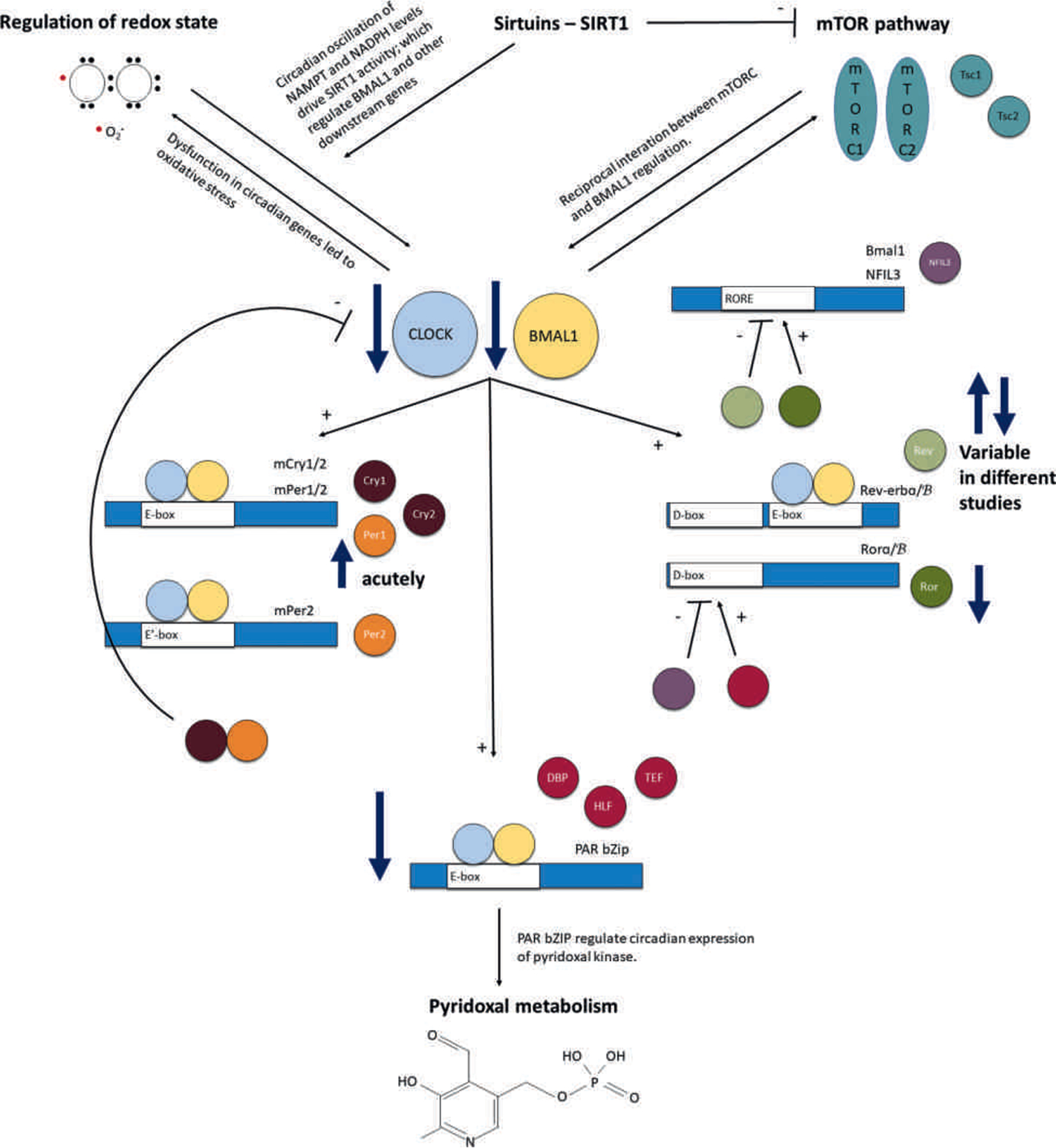
Summary of the changes in the circadian molecular system and its downstream regulation of metabolism in epilepsy. The circadian molecular system is intimately implicated in epilepsy. Several studies demonstrate evidence of reduced expression of the two core genes that make up the positive limb, namely Clock and Bmal1, in various animal models of epilepsy. There are also numerous changes in the negative limb of the system. Per1 was shown to be acutely upregulated in various studies on epileptic animals. Triple knockout animals of the PAR bZIP transcription factors experience epilepsy, implicating the loss of these transcription factors in epilepsy. Finally, changes have also been reported in the Rev-erb and Ror system. Ror was reported to be reduced in epileptic tissue, whereas there are conflicting results regarding Rev-erb, with some studies reporting increased and some decreased expression in models of epilepsy. We also propose that circadian regulation of brain metabolism may represent the link between the circadian molecular system and epilepsy. These metabolic pathways were pyridoxal metabolism, mammalian target of rapamycin (mTOR) pathway, regulation of redox state, and epigenetic regulation by sirtuins. The pyridoxal metabolism, specifically pyridoxal kinase—a rate-limiting enzyme—has been shown to be under regulation of the PAR bZIP transcription factors. Reciprocal interaction between the mTOR signaling and circadian molecular system transcription, particularly Bmal1, was discussed. Dysfunction in the circadian molecular system has led to oxidative stress and dysregulated cellular redox state. Finally, sirtuins directly inhibit mTOR and are regulated by the circadian rhythm of redox state as exemplified by nicotinamide phosphoribosyltransferase (NAMPT) and nicotinamide adenine dinucleotide phosphate (NADPH) oscillation. Thus, the circadian molecular system directly modulates most of these metabolic pathways while also receiving reciprocal input from some of these metabolic pathways. mTORC, mTOR complex
5 |. METABOLISM AS A DOWNSTREAM TARGET OF CIRCADIAN REGULATION OF EPILEPSY
Among the many physiological processes that are regulated by the circadian system, a large number relate to metabolism.39 Metabolism is tightly coupled to the circadian rhythm and is therefore modulated by the circadian molecular system. Metabolic pathways that may be implicated in circadian epilepsy will be discussed in more depth below.
In the study described previously, the PAR bZIP triple knockout mice experienced lethal audiogenic seizures.15 These mice were found to have reduced expression of Pdxk,15 a gene encoding for pyridoxal kinase. Pyridoxal kinase is an enzyme involved in the conversion of pyridoxal to its active form, pyridoxal phosphate (PLP). PLP is a major cofactor of many different enzymes, including those catalyzing neurotransmitter metabolism such as glutamate decarboxylase (GAD).40 GAD is an enzyme that catalyzes the conversion of glutamate to γ-aminobutyric acid (GABA). It is vital in maintaining GABA homeostasis in the brain and depends largely on pyridoxal metabolism to regulate this pathway.40 Thus, pyridoxal metabolism directly feeds into GABA metabolism, a pathway relevant in epilepsy. A class of epilepsy directly associated with pyridoxal metabolism is called pyridoxine-dependent epilepsy. Pyridoxine-dependent epilepsy is an electroclinical syndrome in which seizures respond to pyridoxine (another form of pyridoxal) or PLP treatment.41 This epileptic encephalopathy was found to arise from mutations of varying genes involved in pyridoxal metabolism,42,43 but no known mutations in pyridoxal kinase have been described in human patients. However, enzymes involved in pyridoxal metabolism have been shown to be regulated in a circadian manner,29 just like pyridoxal kinase. Additionally, in these patients and in animal models based on human mutations, disruption in pyridoxal metabolism always led to reduced PLP level in the brain.15,43 Reduced PLP was also associated with reduced GABA concentration in the brain due to reduced activity of glutamate dehydrogenase.43
Whereas there is a large body of evidence suggesting that reduced PLP activity leads to seizure generation, some studies have reported that acute administration of PLP can exacerbate epileptic seizures in certain strains of mice44 and rats.45 In both these studies, the seizures were short-lasting and seemed to be associated also with acutely reduced availability of GABA.44,45 Therefore, pyridoxal metabolism appears to require tight regulation within a certain range in the brain to maintain GABA homeostasis. Regulated by the PAR bZIP transcription factors, the circadian oscillation of Pdxk is lower in the brain than in the liver, indicating the need for strict circadian regulation of brain PLP concentration.46 Interestingly, in the kainate model, where Dbp expression is reduced, overexpression of Dbp by an adenoviral vector actually worsened the seizures instead of rescuing them.34 This Dbp overexpression resulted in a moderate increase in Pdxk expression,34 which supports the hypothesis that tight circadian regulation of pyridoxal metabolism prevents epilepsy.
Another strong link between circadian system and epilepsy comes from the metabolic sensor and signaling pathway, mammalian target of rapamycin (mTOR). mTOR is the major nutrient sensing system that regulates cellular metabolism, protein synthesis, and cell autophagy.47 The mTOR system is composed of two distinct multiprotein complexes named mTORC1 and mTORC2.47 The mTOR complexes (mTORCs) are made up of numerous proteins, many of which have been implicated in epilepsy such as MTOR,48 TSC1 and TSC2,49 and DEPDC5.50 Mutations in these proteins cause epilepsy and collectively, they are called mTORopathies. Among the mTORopathies, tuberous sclerosis complex (TSC) is perhaps the most well-characterized, caused by mutations in TSC1 or TSC2. The gene products of TSC1 (called hamartin) and TSC2 (called tuberin) assemble the TSC1/2 complex, which physiologically suppresses mTOR by inhibiting Rheb, an essential activator of the mTORC.47 Patients with TSC present with epilepsy early in life and have the characteristic presence of “tubers,” tumorlike focal lesions all over the body including in brains.49 Children with TSC can present with sleep abnormalities such as increased incidences of night waking, parasomnias, severe difficult waking up early in the morning, and daytime sleepiness.5 These abnormalities were not significantly different between epileptic and nonepileptic subgroups, representing inherent sleep and circadian disruptions in TSC patients.5 In a polysomnography study on children with TSC, sleep architecture was found to be severely disrupted, with sleep fragmentation, a shorter total sleep time, reduced sleep efficiency, and decreased rapid eye movement sleep.51 Interestingly, in this study, nocturnal seizures were recorded in some patients and associated with a more severe sleep fragmentation.51 In an adult cohort of TSC patients, similar sleep abnormalities were found, namely insomnia and excessive daytime sleepiness, which, unlike in the pediatric group, was positively correlated with their seizure history and antiseizure drug use.6 To illustrate the scale of sleep abnormalities in TSC, a large natural history study of more than 2000 patients with TSC reported sleep abnormalities as the second most reported behavioral problem in about 40% of the patients.52 Thus, it is clear that patients with TSC present with chronic sleep abnormalities, regardless of their seizure history, which indicates an intrinsic circadian dysfunction. More importantly, nocturnal seizures have been recorded in these patient subgroups, which are characteristic of circadian epilepsy. This effect is not limited to disruptions in TSC1 or TSC2. Patients with mutations in genes associated with mTORopathies often present with chronic epilepsy related to sleep.53 Thus, there is strong clinical evidence implicating the mTORCs in the regulation of the circadian rhythm, particularly pertaining to the sleep–wake cycle.
At a cellular level, emerging evidence indicates reciprocal interaction between the mTOR system and the circadian system. In the study previously described, CLOCK expression was found to be reduced in patients with TSC.33 Animal models of TSC also demonstrated large changes in expression levels of circadian proteins, including increased CLOCK and BMAL1.54,55 There is a growing body of evidence to suggest that mTORC regulates circadian protein expression. The mTOR-effector kinase, S6K1, phosphorylated BMAL1 to associate with translational machinery and promote protein synthesis.56 Loss of PTEN function was shown to activate mTORC and lead to accumulation of BMAL1.57 On the other hand, circadian proteins also interact with and regulate mTORC. The circadian protein PER2 suppressed mTORC activity by recruiting TSC1 into the complex.58 Deficiency of BMAL1 activated the mTORC signaling pathway.59 These studies have collectively indicated the reciprocal interaction between the mTORC and the circadian molecular system. However, future studies should explore this interaction in the context of circadian epilepsy.
Next, it is prudent to discuss the implication of circadian regulation of redox state in the context of epilepsy. Redox state is a term used to describe the ratio of the oxidized and reduced form of a molecule in the cell.60 These molecules exist in interconvertible form between their oxidized and reduced forms, such as NAD+/NADH, NADP+/NADPH, and GSSG/GSH.60 Redox state needs to be maintained in constant homeostasis for proper cellular functioning. Unlike the reduced form of most molecules, the reduced form of oxygen and its derivatives, such as superoxide, are harmful to the cells and are collectively called free radicals or reactive oxygen species.61 Cellular defenses against the reactive oxygen species act by neutralizing these free radicals and are collectively called antioxidants.61 When the balance of reactive oxygen species and antioxidants is disrupted in favor of the radicals, a condition called oxidative stress occurs in which various constituents of a cell are damaged by uncontrolled radicals.61 Given how harmful oxidative stress is to the cell, unsurprisingly redox state is tightly regulated by many processes, including circadian regulation. The balance between the reduced and oxidized form of molecules in the cells oscillates in a circadian manner.62 Enzymes functioning as cellular antioxidants are also circadian regulated.63 More importantly, recent works have demonstrated that the cellular redox state also influences and feeds back into the circadian regulation.62,64
Importantly, in the brain, circadian regulation of redox state appears to influence neuronal excitability and may have implications in epilepsy. In the suprachiasmatic nucleus, circadian rhythm of redox state influences and regulates the circadian rhythm of neuronal excitability through a 24-h cycle.65 Recently, this correlated redox state and neuronal excitability cycle was also demonstrated in the hippocampal CA1.66 In the suprachiasmatic nucleus, redox state influences neuronal excitability by modulating potassium channel expression independent of transcriptional modulation.65 Circadian oscillation in neuronal potassium current has been reported and studied extensively.67 Furthermore, calcium channel and sodium channel conductance also oscillate in a circadian manner and contribute to neuronal excitability within the day–night cycle.68 Taken together, this circadian variation in ion channel conductance could possibly contribute toward the circadian rhythmicity of epilepsy. Looking at a bigger picture, oxidative stress has long been implicated in epilepsy.69 Dysfunction in the circadian molecular system has also been reported to result in oxidative stress. Mice with Bmal1 deletion experienced oxidative stress and neuronal oxidative damage, which subsequently resulted in neurodegeneration.70 Cells with mutations in the circadian genes experienced differential response to oxidative stress, indicating that the circadian genes regulate cellular survival and death signaling.71,72 Thus, regulation of cellular redox state and oxidative stress is a plausible link between circadian protein dysregulation and neuronal hyperexcitability in epilepsy.
Finally, one potential link between the circadian molecular system, its regulation of brain metabolism, and epilepsy is in the epigenetic regulation by sirtuin. Sirtuins are proteins belonging to the histone deacetylase family, which uniquely require NAD+ for their enzymatic activity.73 Due to this unique property, sirtuin deacetylase activity is tightly regulated by the cellular metabolic and redox state.73 One sirtuin in particular seems to be important in circadian epilepsy, namely SIRT1. In the Kcna1-null mice, circadian oscillation in Sirt1 was shown to be downregulated under 24-h light–dark cycle, which was associated with the dysfunctional oscillation of the other circadian genes.27 SIRT1 has been shown to be associated with CLOCK and exert regulation on the circadian molecular system through acetylation of BMAL1.74 SIRT1 also influences the expression of circadian genes downstream of Clock/Bmal1 such as Cry1 and Per2, and specifically promotes the deacetylation and degradation of Per2.75 SIRT1’s epigenetic activity was driven by the circadian oscillation in the activity of nicotinamide phosphoribosyltransferase, an enzyme involved in NAD+ metabolism, thus directly linking redox state regulation and the circadian molecular system.76 Finally, SIRT1 also interacts with the mTORC pathway by inhibiting mTORC activity, a pathway that has been associated with the promotion of neuronal outgrowth and survival.77,78 This promotion of neuronal survival by its deacetylase activity has been shown to be important in the context of an in vitro magnesium-free model of epilepsy and an in vivo kainate model of epilepsy.79,80 Hence, there is a strong precedent for the implication of SIRT1 in its metabolic and epigenetic modulation in epilepsy. Future study should address the gap and examine the direct contribution of SIRT1 to the circadian regulation of brain metabolism in epilepsy.
6 |. SYSTEMS BIOLOGY APPROACH TO UNDERSTANDING CIRCADIAN MODULATION OF METABOLISM
We have extensively discussed in the previous sections how the circadian molecular system regulates brain metabolic pathways potentially implicated in epilepsy, either directly or indirectly through epigenetic regulation (see Figure 2 for a summary of these potential regulations). Although a molecular approach is invaluable in understanding circadian epilepsy, an organismal systems biology perspective is equally important. The circadian molecular system is ubiquitous across all the organ systems. Thus, it integrates various systemic inputs as a feedback mechanism making circadian regulation of metabolism present in all of the human body.
Diet and gastrointestinal system function are major determinants of organismal metabolism. Feeding and fasting behavior is temporally regulated by the circadian system, providing a primary time cue for clock entrainment.81 Thus, diet and nutrition directly influence the circadian molecular system through metabolic modulation.81 There has been a multitude of evidence demonstrating dysfunction in the circadian molecular system due to dietary patterns, such as high-fat high-sugar diet82 and high-salt diet.83 On the other hand, dietary patterns, such as time-restricted feeding84 and ketogenic diet (to be discussed further in the section below), have been demonstrated to facilitate and restore proper circadian functioning in animal models of circadian dysfunction. There are few studies examining the impact of diets that cause circadian dysfunction on epilepsy, but preliminary results showed that they may increase seizure susceptibility.85 The ketogenic diet, on the other hand, which facilitates proper circadian functioning, has antiseizure properties. Adding another layer of complexity is the recent finding that the ketogenic diet’s antiseizure effect may be mediated by its effect on the gut microbiome.86 The ketogenic diet has been shown to promote the growth of specific species of gut microbiota that facilitated gamma glutamylation of amino acids that are associated with its antiseizure effect.86 Gut microbiome is significantly affected by dietary pattern and promotes a specific pattern of systemic metabolome.87 Coming full circle, there is evidence of a circadian diurnal pattern in the gut microbiome influenced by the feeding rhythm and dietary pattern.88 This diurnal pattern was associated with the diurnal rhythm of metabolites and promotion of systemic metabolic homeostasis.88 Metabolites generated by the gut microbiome, such as short-chain fatty acids like acetate and butyrate, have shown the capacity to entrain peripheral circadian clocks.89 When the host’s circadian rhythm is disrupted through light–dark cycle disruption or genetically by mutations in the circadian genes, gut microbiome composition has been found to be severely disrupted.90,91 Although it is unclear whether the bacteria themselves have their own intrinsic circadian clock, it is evident that gut microbiota were able to interact with and regulate the human host’s circadian molecular system. This subject is an interesting dynamic new field and warrants further study in the direct context of circadian epilepsy.
Another important systemic input in both the circadian system and epilepsy is the endocrine system. Hormones have been shown to interact with and influence seizure susceptibility in patients with epilepsy.92 An example of a hormone that regulates the circadian molecular system and is implicated in epilepsy is melatonin released by the pineal gland. Melatonin is secreted in a circadian fashion under direct regulation and innervation by the suprachiasmatic nucleus.93 Melatonin has been extensively characterized to be important for circadian regulation, especially for sleep–wake cycle.93,94 Patients with intractable epilepsy have been shown to have low melatonin levels.95 Importantly, although the melatonin circadian oscillation is unaffected, melatonin level was shown to be significantly reduced in both patients with diurnal and patients with nocturnal seizures.8 Melatonin supplementation in children with intractable epilepsy has shown variable success in reducing seizure burden and improving sleep comorbidities.96 In animal models of epilepsy, melatonin administration was able to reduce seizure frequency and attenuate neuronal cell death.97,98 Taken together, melatonin shows a promising antiseizure effect. The mechanism by which melatonin is antiseizure is still up for debate, but there is an interesting notion that melatonin is a potent metabolic modulator. Melatonin administration was able to upregulate antioxidant gene expression in neuronal cells in vitro and rat cortex in vivo.99 Melatonin and its metabolites have also been shown to possess free radical scavenging activities, acting as antioxidant molecules themselves.100 Furthermore, in multiple recent studies using ischemic–reperfusion injury as a model, melatonin treatment prevented cell death by activating the mTOR pathway and inhibiting mTORC-dependent autophagy.101 Interestingly, melatonin was able to attenuate oxidative stress-induced phosphorylation of mTORC, suggesting that its interaction with mTORC may be dependent on its modulation of the redox state.102 Whatever the mechanism may be, there is evidence that melatonin does interact with the mTORC. Finally, an early study showed that melatonin treatment significantly increased brain pyridoxal kinase activity.103 On the other hand, PLP was shown to inhibit an enzyme involved in the production of melatonin from its precursor, N-acetylserotonin.104 Thus, there appears to be a reciprocal interaction between melatonin and pyridoxal metabolism, but more study is needed to reexamine this interaction. Given the current evidence, it appears interesting that melatonin is able to interact with all three metabolic pathways that we discussed in earlier sections as potentially implicated in circadian epilepsy, namely pyridoxal metabolism, mTOR pathway, and redox regulation. Although all these studies were conducted in separate contexts, inferences drawn suggest that melatonin may contribute to circadian regulation of brain metabolism that is important in epilepsy.
7 |. THERAPEUTIC IMPLICATIONS: PRE-EXISTING ANTISEIZURE TREATMENT
Our understanding of the circadian aspect of epilepsy continues to evolve with more emerging studies in this field. One exciting implication of these studies is that chronotherapeutics could be a future direction for epilepsy management and treatment.105 At present, chronotherapeutics in epilepsy is limited to the use of antiseizure drugs treatment at the time of greatest seizure susceptibility based on the circadian timing of each patient’s seizures.105 Time-specific dosing of antiseizure drugs has led to better seizure freedom in some patients.9 This concept of differential dosing time of antiseizure drugs has also been validated in animal studies. Circadian variations have been shown in the pharmacokinetic profile of carbamazepine when injected at different times in rats106 and of valproic acid in mice.107 Interestingly, just like in human patients, these circadian variations in valproic acid pharmacokinetics correlate with differential antiseizure efficacy according to the circadian phase.108 Thus, there are multiple layers of evidence suggesting the importance of differential dosing of antiseizure drugs based on the circadian phenotype of the patients or animal models of epilepsy. This therapeutic strategy takes advantage of the circadian nature of epilepsy but does not actively correct the dysfunction in the circadian molecular system that could contribute to the pathogenic process. Thus, the next step would be working toward correcting circadian molecular system dysfunction as a novel therapeutic strategy in epilepsy.
There is already some evidence that pre-existing therapeutic strategies in epilepsy were able to influence the circadian system, either directly or indirectly. As mentioned earlier, the circadian system is heavily influenced by dietary input,81 including ketogenic diet. Ketogenic diet is a dietary therapy using a high-fat, low-carbohydrate diet ratio that puts the body in ketosis to generate ketone bodies.109 In the Kcna1-null mice, ketogenic diet has been shown to abolish the diurnal rhythmicity of the seizure in addition to effectively reducing seizure frequency.110 The Kcna1-null mice also exhibited atypical rest–activity rhythm, and feeding these mice the ketogenic diet restored the rhythm to a typical wild-type mice rhythm with significant peak activity during the dark phase.110 Together, these findings indicate that the ketogenic diet restored the circadian rhythm of epileptic mice in addition to providing effective seizure suppression. Ketogenic diet appears to be able to interact with and regulate the circadian molecular system. In the muscle, ketogenic diet could restore the transcription of the circadian gene Slc25a25, which was reduced in Bmal1-knockout and Clock-knockout mice.111 In the liver, ketogenic diet could rewire the peripheral clock and increase the amount of cycling transcript.112 Importantly, among the transcripts enriched by the ketogenic diet in the liver is Dbp,112 one of the PAR bZIP genes implicated in epilepsy. This enrichment was attributed to increased BMAL1 chromatin recruitment on the E-box promoter sequence of these genes.112 Ketogenic diet has been shown to influence chromatin structure and function through epigenetic mechanisms. Animal models of epilepsy have undergone general hypermethylation of hippocampal DNA, and in multiple studies, ketogenic diet could restore the DNA methylation status to control animals.113,114 In the context of neuroinflammation outside of epilepsy, ketogenic diet has been associated with other epigenetic changes such as β-hydroxybutyration and acetylation.115 Thus, there is evidence that ketogenic diet could influence the circadian molecular system through epigenetic modification. Although this hypothesis needs to be tested further, preliminary data indicate that this is the case. In the gut of mice fed the ketogenic diet, there is an inverse cyclic rhythmicity between the β-hydroxybutyrate level and histone deacetylation activity.112 This resulted in overall reduction of HDAC activity and an increase in acetylation of various epigenetic regulated circadian genes.112 Whether the epigenetic modulation of circadian genes is associated with its anti-seizure activity remains to be studied.
In addition to the ketogenic diet, existing antiseizure drugs may also interact with the circadian molecular system. Many of the clinically used antiseizure drugs have exerted epigenetic changes such as histone acetylation and DNA methylation on various target genes (for a comprehensive review, see Navarrete-Modesto et al.116). Valproic acid is of particular interest, as it exerted multiple epigenetic effects. Valproic acid has been shown in multiple studies to be a potent HDAC inhibitor.117,118 In addition to its acetylating effect, valproic acid also affects the DNA methylation of various genes.119,120 Although the epigenetic effect of valproic acid or any other antiseizure drug has not been described in circadian genes, it presents an interesting potential mechanism, considering how important epigenetic regulation is to the circadian molecular system. Valproic acid has been shown to directly interact with the circadian molecular system. Valproic acid treatment in vitro in multiple cell lines was able to upregulate transcription of various circadian genes, such as Bmal1, Cry1, Cry2, Per1, Per2, and Rev-erbα.121,122 Valproic acid was also able to phase-advance the rhythm of Per2 in vitro, thus affecting circadian gene oscillation.121 Therefore, valproic acid as an established antiseizure drug has a strong potential as a modulator of the circadian molecular system. Additional study is needed to characterize the specific effect of valproic acid and other antiseizure drugs in the context of correcting the circadian molecular dysfunction in epilepsy.
8 |. THERAPEUTIC IMPLICATIONS: INVESTIGATIONAL ANTISEIZURE DRUGS
Despite the many existing antiseizure treatments, one third of patients with epilepsy still have refractory epilepsy.123 Thus, there remains a need for investigational antiseizure drugs to be developed that could directly modify epileptogenesis. One interesting strategy for chronotherapeutic development could be to reevaluate and develop a new class of antiseizure treatment based on known mechanisms. In the previous section, melatonin was described as possessing antiseizure properties in both human patients96 and animal models of epilepsy.97,98 In addition to ongoing efforts to further study the clinical potential of melatonin as an antiseizure drug, studies have also examined the potential of agonists of melatonin receptor. Ramelteon, a selective agonist of melatonin receptor, significantly improved circadian rest–activity rhythm and positively modulated seizure diurnal rhythmicity and frequency in Kcna1-null mice.124 Another agonist of melatonin receptor MT1 and MT2, agomelatine, also demonstrated anticonvulsant effects in the mouse pentylenetetrazole (PTZ)-induced seizure and pilocarpine model.125 It is important to note that this study failed to find efficacy of agomelatine in other models of epilepsy, such as the strychnine-, electroshock-, and picrotoxin-induced seizure models.125 Finally, two other synthetic melatonin receptor agonists, Neu-P11 and Neu-P67, failed to demonstrate antiseizure effects in the mouse PTZ- and electroshock-induced seizure models.126 Thus, although melatonin and melatonin receptor agonists may be promising new antiseizure drugs, further study is needed to validate these findings.
Finally, novel small molecules have been identified that can modulate the circadian molecular system and are currently being investigated in the context of various neurological diseases.127,128 Some of these small molecules had effects on circadian period, either lengthening or shortening, whereas others modulated circadian phase by delaying it, advancing it, or enhancing its amplitude.129,130 Others had a more direct effects on the circadian molecular system through interacting with and regulating the negative limbs of the system. Agonists and inverse agonists of RORs and REV-ERBs as well as activators and inhibitors of CRYs have been identified.127,128 None of these small molecules have been extensively investigated in the context of epilepsy. However, if we speculate based on typical changes in these proteins in models of epilepsy, then one with potential to correct the circadian dysfunction in epilepsy would be the ROR agonists. One such candidate molecule is nobiletin, which was identified as a direct agonist of RORα.131 In a study using acute administration of PTZ, a convulsant, mice with oral nobiletin treatment experienced significantly reduced seizure burden.132 In other studies, nobiletin continued to demonstrate neuroprotective effects.133,134 Through its modulation of RORα, nobiletin has also been shown to induce positive metabolic changes, such as improvement of circadian glucose and lipid homeostasis, increased mitochondrial respiration, and reduced oxidative stress.131,135 Some of these metabolic changes could be important for circadian epilepsy, and nobiletin has the potential to directly address these dysfunctions. Thus, generally, there is precedent to further study and consider all other small molecule modifiers of the circadian clock as a potential novel class of antiseizure drugs. Importantly, further preclinical study should evaluate these potential chronotherapeutics in a systematic and consistent manner, because these findings have been made in multiple variable animal models of epilepsy and as previously demonstrated, findings of efficacy may be specific to a model of epilepsy.125
9 |. CONCLUSION
From many observations in human studies and research on animal models of epilepsy, it is clear that epileptic seizures may manifest with a circadian pattern. Although the mechanism is unclear, the circadian molecular system that generates the circadian rhythm is likely involved. These genes that oscillate in a circadian manner were largely found to be disrupted in various animal models of epilepsy (summarized in Figure 2). There is growing evidence that intrinsic disruption in the circadian molecular system can lead to epileptogenesis. This is supported by the genetic animal models with loss of circadian molecular proteins (namely CLOCK33 and PAR bZIP proteins15), where seizures were developed in vivo. However, there are also other data that support the idea that initial epileptic insult causes circadian disruptions. Numerous studies with pilocarpine and kainate models of epilepsy show that the initial acute epileptogenic insult with these drugs caused acute changes in the circadian molecular system (such as increased Per1 and Rev-erbα as well as reduced Rorα).32,36,37 Interestingly, in these animal models, where initial epileptic insult resulted in kindling during the silent phase and generation of spontaneous epilepsy, some of these changes persisted during the silent phase (such as the reduced Rorα),37 some persisted throughout and up to the spontaneous epilepsy phase (such as the increased Rev-erbα),36 and some returned to normal after the acute phase (such as the increased Per1).25 Thus, these changes in the circadian molecular system could either be an acute response to the epileptic insult or a change that contributes during the kindling process to the generation of a spontaneously epileptic network. It appears that there is a bidirectional linkage between epilepsy and circadian dysfunction. Circadian dysfunction can lead to the formation of an epileptic network, and epileptic insult can lead to acute circadian dysfunction. Another interesting question is whether epilepsy, independent of seizure activity, induces changes in the circadian biology. Although we acknowledge that epilepsy has systemic effects, the presence of seizures generated in the brain still constitutes a large part of what makes up the diagnosis of epilepsy.136 This question is difficult to address in animal models, as it is not possible to develop an animal model of epilepsy that does not have any seizure activity. In most animal models, however, studies with treatment of antiseizure drugs have shown improvement in the circadian dysfunction that tends to correlate with seizure improvement.110,132 If we examine human studies, most studies have shown that the severity of the patient’s sleep dysfunction is usually associated with the severity of their seizures.6,51 Thus, the current evidence suggests that seizure activity in an epilepsy phenotype is tied with the circadian dysfunction. Correcting the circadian dysfunction can result in improvement of the seizure activity and vice versa.
Without a full picture, we propose that metabolic dysfunction may bridge the gap between circadian dysfunction and epilepsy. We present evidence from the literature suggesting three downstream pathways of the circadian molecular system, namely, pyridoxal metabolism, mTORC signaling, and regulation of redox state as the basis for the missing link between circadian regulation and epilepsy. Circadian regulation appears to influence all three metabolic pathways; while only two of them, namely, mTORC signaling and redox regulation, have been shown to feed back into the circadian molecular system through the effect of sirtuin. In addition to the studies that directly examined these three metabolic pathways in the context of circadian epilepsy, various other systems that interact with the circadian molecular system appear to support the notion that these metabolic pathways are involved in circadian epilepsy. As mentioned above, epigenetic regulation through the sirtuins, and particularly Sirt1,27 interacts with mTORC78 and cellular redox signaling.74 Evidence from another system that supports this notion is the endocrine system through the release of melatonin. Melatonin has been shown to interact with all three metabolic pathways, namely, by interacting with pyridoxal kinase,103 activating the mTOR pathway,101 and being a potent antioxidant (thus, regulating the cellular redox state).100 Melatonin has shown potential antiseizure properties in both animal and human studies,96–98 the mechanism of which is unknown. It is possible that melatonin could modulate these metabolic pathways important in circadian epilepsy and exert its antiseizure properties in such a manner. Taken together, all this evidence supports the notion that circadian regulation of brain metabolism is an important pathway that could underlie circadian epilepsy. Future studies in this budding field should address this hypothesis directly. Work emerging from this field could pave the way to a new class of antiseizure drugs that target the circadian molecular system or the circadian modulation of brain metabolism.
Key Points.
Epilepsy is circadian in nature and is associated with disruptions in the circadian molecular system
Circadian oscillation of various genes that make up the circadian molecular system is highly disrupted in animal models of epilepsy
Functional output of the system (mRNA and protein levels of the circadian genes) is significantly affected in epilepsy animal models
We propose three metabolic pathways (pyridoxal metabolism, mTOR, and redox state) as links between the circadian system and epilepsy
Studies in the field are working toward development of chronotherapeutics by targeting these novel pathways in epilepsy
ACKNOWLEDGMENTS
F.C. and J.L. are supported by CURE and NIH/NINDS R01 NS104428-01. We would like to acknowledge all lab members who participated in constructive discussion of the manuscript, particularly Adam Friedberg and Carin Papendorp, who provided critical feedback on the manuscript.
Funding information
Citizens United for Research in Epilepsy; National Institute of Neurological Disorders and Stroke, Grant/Award Number: RO1 NS104428-01
Footnotes
CONFLICT OF INTEREST
Neither of the authors has any conflict of interest to disclose.
REFERENCES
- 1.Khan S, Nobili L, Khatami R, Loddenkemper T, Cajochen C, Dijk DJ, et al. Circadian rhythm and epilepsy. Lancet Neurol. 2018; 17: 1098–108. [DOI] [PubMed] [Google Scholar]
- 2.Loddenkemper T, Vendrame M, Zarowski M, Gregas M, Alexopoulos AV, Wyllie E, et al. Circadian patterns of pediatric seizures. Neurology. 2011; 76 (2): 145–53. [DOI] [PubMed] [Google Scholar]
- 3.Wright S, Wallace E, Hwang Y, Maganti R. Seizure phenotypes, periodicity, and sleep–wake pattern of seizures in Kcna-1 null mice. Epilepsy Behav. 2016; 55: 24–9. [DOI] [PubMed] [Google Scholar]
- 4.Quigg M, Clayburn H, Straume M, Menaker M, Bertram EH III. Effects of circadian regulation and rest—activity state on spontaneous seizures in a rat model of limbic epilepsy. Epilepsia. 2000; 41: 502–9. [DOI] [PubMed] [Google Scholar]
- 5.Trickett J, Heald M, Oliver C, Richards C. A cross-syndrome cohort comparison of sleep disturbance in children with Smith-Magenis syndrome, Angelman syndrome, autism spectrum disorder and tuberous sclerosis complex. J Neurodev Disord. 2018; 10 (1): 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Eeghen AM, Numis AI, Staley BA, Therrien SE, Thibert RL, Thiele EA. Characterizing sleep disorders of adults with tuberous sclerosis complex: a questionnaire-based study and review. Epilepsy Behav. 2011; 20: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Persson H, Kumlien E, Ericson M, Tomson T. Circadian variation in heart-rate variability in localization-related epilepsy. Epilepsia. 2007; 48: 917–22. [DOI] [PubMed] [Google Scholar]
- 8.Yalýn Ö, Arman F, Erdoğan F, Kula M. A comparison of the circadian rhythms and the levels of melatonin in patients with diurnal and nocturnal complex partial seizures. Epilepsy Behav. 2006; 8: 542–6. [DOI] [PubMed] [Google Scholar]
- 9.Thome-Souza S, Klehm J, Jackson M, Kadish NE, Manganaro S, Fernández IS, et al. Clobazam higher-evening differential dosing as an add-on therapy in refractory epilepsy. Seizure. 2016; 40: 1–6. [DOI] [PubMed] [Google Scholar]
- 10.Buhr ED, Takahashi JS. Molecular components of the mammalian circadian clock. Handb Exp Pharmacol. 2013;(217): 3–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000; 103: 1009–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeBruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, Reppert SM. A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron. 2006; 50: 465–77. [DOI] [PubMed] [Google Scholar]
- 13.Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001; 30: 525–36. [DOI] [PubMed] [Google Scholar]
- 14.Guillaumond F, Dardente H, Giguère V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. 2005; 20: 391–403. [DOI] [PubMed] [Google Scholar]
- 15.Gachon F, Fonjallaz P, Damiola F, Gos P, Kodama T, Zakany J, et al. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 2004; 18 (12): 1397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 2001; 15 (8): 995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010; 28: 1057–68. [DOI] [PubMed] [Google Scholar]
- 18.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK Is a histone acetyltransferase. Cell. 2006; 125: 497–508. [DOI] [PubMed] [Google Scholar]
- 19.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003; 421 (6919): 177–82. [DOI] [PubMed] [Google Scholar]
- 20.Duong HA, Robles MS, Knutti D, Weitz CJ. A molecular mechanism for circadian clock negative feedback. Science. 2011; 332: 1436–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curia G, Longo D, Biagini G, Jones RS, Avoli M. The pilocarpine model of temporal lobe epilepsy. J Neurosci Methods. 2008; 172 (2): 143–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lévesque M, Avoli M. The kainic acid model of temporal lobe epilepsy. Neurosci Biobehav Rev. 2013; 37: 2887–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang SS, Jeong HG, Chung HJ. Electroconvulsive seizures in rats and fractionation of their hippocampi to examine seizure-induced changes in postsynaptic density proteins. J Vis Exp. 2017;(126):56016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smart SL, Lopantsev V, Zhang CL, Robbins CA, Wang H, Chiu SY, et al. Deletion of the KV1.1 potassium channel causes epilepsy in mice. Neuron. 1998; 20: 809–19. [DOI] [PubMed] [Google Scholar]
- 25.Matos HC, Koike BDV, Pereira WDS, de Andrade TG, Castro OW, Duzzioni M, et al. Rhythms of core clock genes and spontaneous locomotor activity in post-status epilepticus model of mesial temporal lobe epilepsy. Front Neurol. 2018; 9: 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim SH, Park HG, Jeong SH, Kang UG, Ahn YM, Kim YS. Electroconvulsive seizure alters the expression and daily oscillation of circadian genes in the rat frontal cortex. Psychiatry Investig. 2018; 15: 717–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallace E, Wright S, Schoenike B, Roopra A, Rho JM, Maganti RK. Altered circadian rhythms and oscillation of clock genes and sirtuin 1 in a model of sudden unexpected death in epilepsy. Epilepsia. 2018; 59: 1527–39. [DOI] [PubMed] [Google Scholar]
- 28.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002; 109 (3): 307–20. [DOI] [PubMed] [Google Scholar]
- 29.Mure LS, Le HD, Benegiamo G, Chang MW, Rios L, Jillani N, et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science. 2018; 359: eaao0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Streng ML, Krook-Magnuson E. Excitation, but not inhibition, of the fastigial nucleus provides powerful control over temporal lobe seizures. J Physiol. 2020; 598: 171–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurley JM, Loros JJ, Dunlap JC. Circadian oscillators: around the transcription-translation feedback loop and on to output. Trends Biochem Sci. 2016; 41: 834–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eun B, Kim HJ, Kim SY, Kim TW, Hong ST, Choi KM, et al. Induction of Per1 expression following an experimentally induced epilepsy in the mouse hippocampus. Neurosci Lett. 2011; 498: 110–3. [DOI] [PubMed] [Google Scholar]
- 33.Li P, Fu X, Smith NA, Ziobro J, Curiel J, Tenga MJ, et al. Loss of CLOCK results in dysfunction of brain circuits underlying focal epilepsy. Neuron. 2017; 96 (2): 387–401. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klugmann M, Leichtlein CB, Symes CW, Klaussner BC, Brooks AI, Young D, et al. A novel role of circadian transcription factor DBP in hippocampal plasticity. Mol Cell Neurosci. 2006; 31: 303–14. [DOI] [PubMed] [Google Scholar]
- 35.Gerstner JR, Smith GG, Lenz O, Perron IJ, Buono RJ, Ferraro TN. BMAL1 controls the diurnal rhythm and set point for electrical seizure threshold in mice. Front Syst Neurosci. 2014; 8: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yue J, He J, Wei Y, Shen K, Wu K, Yang X, et al. Decreased expression of Rev-Erbα in the epileptic foci of temporal lobe epilepsy and activation of Rev-Erbα have anti-inflammatory and neuroprotective effects in the pilocarpine model. J Neuroinflamm. 2020; 17 (1): 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rocha AKAdA, de Lima E, Amaral FGD, Peres R, Cipolla-Neto J, Amado D. Pilocarpine-induced epilepsy alters the expression and daily variation of the nuclear receptor RORα in the hippocampus of rats. Epilepsy Behav. 2016; 55: 38–46. [DOI] [PubMed] [Google Scholar]
- 38.Guissart C, Latypova X, Rollier P, Khan TN, Stamberger H, McWalter K, et al. Dual molecular effects of dominant RORA mutations cause two variants of syndromic intellectual disability with either autism or cerebellar ataxia. Am J Hum Genet. 2018; 102: 744–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eckel-Mahan K, Sassone-Corsi P. Metabolism control by the circadian clock and vice versa. Nat Struct Mol Biol. 2009; 16: 462–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Percudani R, Peracchi A. A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 2003; 4: 850–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Karnebeek CDM, Tiebout SA, Niermeijer J, Poll-The BT, Ghani A, Coughlin CR, et al. Pyridoxine-dependent epilepsy: an expanding clinical spectrum. Pediatr Neurol. 2016; 59: 6–12. [DOI] [PubMed] [Google Scholar]
- 42.Scharer G, Brocker C, Vasiliou V, Creadon-Swindell G, Gallagher RC, Spector E, et al. The genotypic and phenotypic spectrum of pyridoxine-dependent epilepsy due to mutations in ALDH7A1. J Inherit Metab Dis. 2010; 33: 571–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darin N, Reid E, Prunetti L, Samuelsson L, Husain Ralf A, Wilson M, et al. Mutations in PROSC disrupt cellular pyridoxal phosphate homeostasis and cause vitamin-B6-dependent epilepsy. Am J Hum Genet. 2016; 99: 1325–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Norris DK, Murphy RA, Chung SH. Alteration of amino acid metabolism in epileptogenic mice by elevation of brain pyridoxal phosphate. J Neurochem. 1985; 44: 1403–10. [DOI] [PubMed] [Google Scholar]
- 45.Kouyoumdjian JC, Ebadi M. Anticonvulsant activity of muscimol and gamma-aminobutyric acid against pyridoxal phosphate-induced epileptic seizures. J Neurochem. 1981; 36: 251–7. [DOI] [PubMed] [Google Scholar]
- 46.Schibler U The daily rhythms of genes, cells and organs. EMBO Rep. 2005; 6: S9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009; 122: 3589–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim JK, Cho J, Kim SH, Kang HC, Kim DS, Kim VN, et al. Brain somatic mutations in MTOR reveal translational dysregulations underlying intractable focal epilepsy. J Clin Invest. 2019; 129 (10): 4207–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Curatolo P, Maria BL. Tuberous sclerosis. Handb Clin Neurol. 2013; 111: 323–31. [DOI] [PubMed] [Google Scholar]
- 50.Ishida S, Picard F, Rudolf G, Noé E, Achaz G, Thomas P, et al. Mutations of DEPDC5 cause autosomal dominant focal epilepsies. Nat Genet. 2013; 45: 552–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruni O, Cortesi F, Giannotti F, Curatolo P. Sleep disorders in tuberous sclerosis: a polysomnographic study. Brain Dev. 1995; 17: 52–6. [DOI] [PubMed] [Google Scholar]
- 52.de Vries PJ, Belousova E, Benedik MP, Carter T, Cottin V, Curatolo P, et al. TSC-associated neuropsychiatric disorders (TAND): findings from the TOSCA natural history study. Orphanet J Rare Dis. 2018; 13: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baldassari S, Picard F, Verbeek NE, van Kempen M, Brilstra EH, Lesca G, et al. The landscape of epilepsy-related GATOR1 variants. Genet Med. 2019; 21: 398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lipton JO, Boyle LM, Yuan ED, Hochstrasser KJ, Chifamba FF, Nathan A, et al. Aberrant proteostasis of BMAL1 underlies circadian abnormalities in a paradigmatic mTOR-opathy. Cell Rep. 2017; 20 (4): 868–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramanathan C, Kathale ND, Liu D, Lee C, Freeman DA, Hogenesch JB, et al. mTOR signaling regulates central and peripheral circadian clock function. PLoS Genet. 2018; 14: e1007369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lipton JO, Yuan ED, Boyle LM, Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, et al. The circadian protein BMAL1 regulates translation in response to S6K1-mediated phosphorylation. Cell. 2015; 161 (5): 1138–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsumoto CS, Almeida LO, Guimarães DM, Martins MD, Papagerakis P, Papagerakis S, et al. PI3K-PTEN dysregulation leads to mTOR-driven upregulation of the core clock gene BMAL1 in normal and malignant epithelial cells. Oncotarget. 2016; 7: 42393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu R, Dang F, Li P, Wang P, Xu Q, Liu Z, et al. The circadian protein Period2 suppresses mTORC1 activity via recruiting Tsc1 to mTORC1 complex. Cell Metab. 2019; 29 (3): 653–67. e6. [DOI] [PubMed] [Google Scholar]
- 59.Khapre RV, Kondratova AA, Patel S, Dubrovsky Y, Wrobel M, Antoch MP, et al. BMAL1-dependent regulation of the mTOR signaling pathway delays aging. Aging. 2014; 6: 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001; 30 (11): 1191–212. [DOI] [PubMed] [Google Scholar]
- 61.Sies H, Berndt C, Jones DP. Oxidative stress. Annu Rev Biochem. 2017; 86: 715–48. [DOI] [PubMed] [Google Scholar]
- 62.Milev NB, Reddy AB. Circadian redox oscillations and metabolism. Trends Endocrinol Metab. 2015; 26: 430–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sani M, Sebai H, Ghanem-Boughanmi N, Boughattas NA, Ben-Attia M. Circadian (about 24-hour) variation in malondialdehyde content and catalase activity of mouse erythrocytes. Redox Rep Commun Free Radical Res. 2015; 20: 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Putker M, O’ Neill JS. Reciprocal control of the circadian clock and cellular redox state—a critical appraisal. Mol Cells. 2016; 39: 6–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang TA, Yu YV, Govindaiah G, Ye X, Artinian L, Coleman TP, et al. Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science. 2012; 337: 839–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Naseri Kouzehgarani G, Bothwell MY, Gillette MU. Circadian rhythm of redox state regulates membrane excitability in hippocampal CA1 neurons. Eur J Neurosci. 2020; 51: 34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Itri JN, Vosko AM, Schroeder A, Dragich JM, Michel S, Colwell CS. Circadian regulation of a-type potassium currents in the suprachiasmatic nucleus. J Neurophysiol. 2010; 103: 632–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bothwell MY, Gillette MU. Circadian redox rhythms in the regulation of neuronal excitability. Free Radic Biol Med. 2018; 119: 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pearson-Smith JN, Patel M. Metabolic dysfunction and oxidative stress in epilepsy. Int J Mol Sci. 2017; 8: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Musiek ES, Lim MM, Yang G, Bauer AQ, Qi L, Lee Y, et al. Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. J Clin Invest. 2013; 123: 5389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tamaru T, Hattori M, Ninomiya Y, Kawamura G, Varès G, Honda K, et al. ROS stress resets circadian clocks to coordinate pro-survival signals. PLoS One. 2013; 8: e82006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Magnone MC, Langmesser S, Bezdek AC, Tallone T, Rusconi S, Albrecht U. The mammalian circadian clock gene per2 modulates cell death in response to oxidative stress. Front Neurol. 2014; 5: 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li X, Kazgan N. Mammalian sirtuins and energy metabolism. Int J Biol Sci. 2011; 7: 575–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008; 134: 329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008; 134 (2): 317–28. [DOI] [PubMed] [Google Scholar]
- 76.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009; 324: 654–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guo W, Qian L, Zhang J, Zhang W, Morrison A, Hayes P, et al. Sirt1 overexpression in neurons promotes neurite outgrowth and cell survival through inhibition of the mTOR signaling. J Neurosci Res. 2011; 89: 1723–36. [DOI] [PubMed] [Google Scholar]
- 78.Ghosh HS, McBurney M, Robbins PD. SIRT1 negatively regulates the mammalian target of rapamycin. PLoS One. 2010; 5: e9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang S, Yang X, Lin Y, Qiu X, Li H, Zhao X, et al. Cellular NAD depletion and decline of SIRT1 activity play critical roles in PARP-1-mediated acute epileptic neuronal death in vitro. Brain Res. 2013; 1535: 14–23. [DOI] [PubMed] [Google Scholar]
- 80.Liu D, Li S, Gong L, Yang Y, Han Y, Xie M, et al. Suppression of microRNA-141 suppressed p53 to protect against neural apoptosis in epilepsy by SIRT1 expression. J Cell Biochem. 2019; 120: 9409–20. [DOI] [PubMed] [Google Scholar]
- 81.Potter GD, Cade JE, Grant PJ, Hardie LJ. Nutrition and the circadian system. Br J Nutr. 2016; 116: 434–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blancas-Velazquez A, la Fleur SE, Mendoza J. Effects of a free-choice high-fat high-sugar diet on brain PER2 and BMAL1 protein expression in mice. Appetite. 2017; 117: 263–9. [DOI] [PubMed] [Google Scholar]
- 83.Oike H, Nagai K, Fukushima T, Ishida N, Kobori M. High-salt diet advances molecular circadian rhythms in mouse peripheral tissues. Biochem Biophys Res Commun. 2010; 402: 7–13. [DOI] [PubMed] [Google Scholar]
- 84.Chaix A, Lin T, Le HD, Chang MW, Panda S. Time-restricted feeding prevents obesity and metabolic syndrome in mice lacking a circadian clock. Cell Metab. 2019; 29 (2): 303–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alzoubi KH, Hasan ZA, Khabour OF, Mayyas FA, Al Yacoub ON, Banihani SA, et al. The effect of high-fat diet on seizure threshold in rats: role of oxidative stress. Physiol Behav. 2018; 196: 1–7. [DOI] [PubMed] [Google Scholar]
- 86.Olson CA, Vuong HE, Yano JM, Liang QY, Nusbaum DJ, Hsiao EY. The gut microbiota mediates the anti-seizure effects of the ketogenic diet. Cell. 2018; 173 (7): 1728–41. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tang Z-Z, Chen G, Hong Q, Huang S, Smith HM, Shah RD, et al. Multi-omic analysis of the microbiome and metabolome in healthy subjects reveals microbiome-dependent relationships between diet and metabolites. Front Genet. 2019; 10: 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015; 17: 681–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee J, d’Aigle J, Atadja L, Quaicoe V, Honarpisheh P, Ganesh BP, et al. Gut microbiota-derived short-chain fatty acids promote poststroke recovery in aged mice. Circ Res. 2020; 127 (4): 453–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Voigt RM, Forsyth CB, Green SJ, Mutlu E, Engen P, Vitaterna MH, et al. Circadian disorganization alters intestinal microbiota. PLoS One. 2014; 9: e97500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Voigt RM, Summa KC, Forsyth CB, Green SJ, Engen P, Naqib A, et al. The circadian clock mutation promotes intestinal dysbiosis alcoholism. Clin Exp Res. 2016; 40: 335–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Taubøll E, Sveberg L, Svalheim S. Interactions between hormones and epilepsy. Seizure. 2015; 28: 3–11. [DOI] [PubMed] [Google Scholar]
- 93.Stehle JH, von Gall C, Korf HW. Melatonin: a clock-output, a clock-input. J Neuroendocrinol. 2003; 15: 383–9. [DOI] [PubMed] [Google Scholar]
- 94.Dijk DJ, Cajochen D. Melatonin and the circadian regulation of sleep initiation, consolidation, structure, and the sleep EEG. J Biol Rhythms. 1997; 12: 627–35. [DOI] [PubMed] [Google Scholar]
- 95.Bazil CW, Short D, Crispin D, Zheng W. Patients with intractable epilepsy have low melatonin, which increases following seizures. Neurology. 2000; 55: 1746–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Coppola G, Iervolino G, Mastrosimone M, La Torre G, Ruiu F, Pascotto A. Melatonin in wake-sleep disorders in children, adolescents and young adults with mental retardation with or without epilepsy: a double-blind, cross-over, placebo-controlled trial. Brain Dev. 2004; 26: 373–6. [DOI] [PubMed] [Google Scholar]
- 97.de Lima E, Soares JM, del Carmen Sanabria Garrido Y, Gomes Valente S, Priel MR, Chada Baracat E, et al. Effects of pine-alectomy and the treatment with melatonin on the temporal lobe epilepsy in rats. Brain Res. 2005; 1043 (1–2): 24–31. [DOI] [PubMed] [Google Scholar]
- 98.Petkova Z, Tchekalarova J, Pechlivanova D, Moyanova S, Kortenska L, Mitreva R, et al. Treatment with melatonin after status epilepticus attenuates seizure activity and neuronal damage but does not prevent the disturbance in diurnal rhythms and behavioral alterations in spontaneously hypertensive rats in kainate model of temporal lobe epilepsy. Epilepsy Behav. 2014; 31: 198–208. [DOI] [PubMed] [Google Scholar]
- 99.Kotler M, Rodríguez C, Sáinz RM, Antolín I, Menéndez-Peláez A. Melatonin increases gene expression for antioxidant enzymes in rat brain cortex. J Pineal Res. 1998; 24: 83–9. [DOI] [PubMed] [Google Scholar]
- 100.Galano A, Tan DX, Reiter RJ. On the free radical scavenging activities of melatonin’s metabolites, AFMK and AMK. J Pineal Res. 2013; 54: 245–57. [DOI] [PubMed] [Google Scholar]
- 101.Chen WR, Liu HB, Chen YD, Sha Y, Ma Q, Zhu PJ, et al. Melatonin attenuates myocardial ischemia/reperfusion injury by inhibiting autophagy via an AMPK/mTOR signaling pathway. Cell Physiol Biochem. 2018; 47: 2067–76. [DOI] [PubMed] [Google Scholar]
- 102.Kimball SR, Abbas A, Jefferson LS. Melatonin represses oxidative stress-induced activation of the MAP kinase and mTOR signaling pathways in H4IIE hepatoma cells through inhibition of Ras. J Pineal Res. 2008; 44: 379–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Antón-Tay F, Sepulveda J, González S. Increase of brain pyridoxal phosphokinase activity following melatonin administration. Life Sci. 1970; 9: 1283–8. [DOI] [PubMed] [Google Scholar]
- 104.Nir I, Hirschmann N, Sulman FG. Inhibition of pineal hydroxyindole-O-methyl transferase by pyridoxal-5’-phosphate. Biochem Pharmacol. 1976; 25: 581–3. [DOI] [PubMed] [Google Scholar]
- 105.Sánchez Fernández I, Loddenkemper T. Chronotherapeutic implications of cyclic seizure patterns. Nat Rev Neurol. 2018; 14: 696–7. [DOI] [PubMed] [Google Scholar]
- 106.Bruguerolle B, Valli M, Bouyard L, Jadot G, Bouyard P. Circadian effect on carbamazepine kinetics in rat. Eur J Drug Metabol Pharmacokinet. 1981; 6: 189–93. [DOI] [PubMed] [Google Scholar]
- 107.Ben-Cherif W, Dridi I, Aouam K, Ben-Attia M, Reinberg A, Boughattas NA. Chronotolerance study of the antiepileptic drug valproic acid in mice. J Circadian Rhythms. 2012; 10: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Khedhaier WBC, Dridi I, Aouam K, Ben-Attia M, Reinberg A, Boughattas NA. Circadian variation in anticonvulsant activity of valproic acid in mice. Biomed Pharmacother. 2017; 95: 25–30. [DOI] [PubMed] [Google Scholar]
- 109.Keene DL. A systematic review of the use of the ketogenic diet in childhood epilepsy. Pediatr Neurol. 2006; 35: 1–5. [DOI] [PubMed] [Google Scholar]
- 110.Fenoglio-Simeone KA, Wilke JC, Milligan HL, Allen CN, Rho JM, Maganti RK. Ketogenic diet treatment abolishes seizure periodicity and improves diurnal rhythmicity in epileptic Kcna1-null mice. Epilepsia. 2009; 50: 2027–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nakao R, Shimba S, Oishi K. Ketogenic diet induces expression of the muscle circadian gene Slc25a25 via neural pathway that might be involved in muscle thermogenesis. Sci Rep. 2017; 7: 2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tognini P, Murakami M, Liu Y, Eckel-Mahan KL, Newman JC, Verdin E, et al. Distinct circadian signatures in liver and gut clocks revealed by ketogenic diet. Cell Metabol. 2017; 26 (3): 523–38. [DOI] [PubMed] [Google Scholar]
- 113.Kobow K, Kaspi A, Harikrishnan KN, Kiese K, Ziemann M, Khurana I, et al. Deep sequencing reveals increased DNA methylation in chronic rat epilepsy. Acta Neuropathol. 2013; 126: 741–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lusardi TA, Akula KK, Coffman SQ, Ruskin DN, Masino SA, Boison D. Ketogenic diet prevents epileptogenesis and disease progression in adult mice and rats. Neuropharmacol. 2015; 99: 500–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dąbek A, Wojtala M, Pirola L, Balcerczyk A. Modulation of cellular biochemistry, epigenetics and metabolomics by ketone bodies. Implications of the ketogenic diet in the physiology of the organism and pathological states. Nutrients. 2020; 12: 788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Navarrete-Modesto V, Orozco-Suárez S, Feria-Romero IA, Rocha L. The molecular hallmarks of epigenetic effects mediated by antiepileptic drugs. Epilepsy Res. 2019; 149: 53–65. [DOI] [PubMed] [Google Scholar]
- 117.Göttlicher M, Minucci S, Zhu P, Krämer OH, Schimpf A, Giavara S, et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001; 20: 6969–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Krämer OH, Zhu P, Ostendorff HP, Golebiewski M, Tiefenbach J, Peters MA, et al. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003; 1 (22): 3411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Milutinovic S, D’Alessio AC, Detich N, Szyf M. Valproate induces widespread epigenetic reprogramming which involves demethylation of specific genes. Carcinogenesis. 2007; 28: 560–71. [DOI] [PubMed] [Google Scholar]
- 120.Wolters JEJ, van Breda SGJ, Caiment F, Claessen SM, de Kok T, Kleinjans JCS. Nuclear and mitochondrial DNA methylation patterns induced by valproic acid in human hepatocytes. Chem Res Toxicol. 2017; 16 (30): 1847–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Johansson A-S, Brask J, Owe-Larsson B, Hetta J, Lundkvist GBS. Valproic acid phase shifts the rhythmic expression of PERIOD2::LUCIFERASE. J Biol Rhythms. 2011; 26: 541–51. [DOI] [PubMed] [Google Scholar]
- 122.Griggs CA, Malm SW, Jaime-Frias R, Smith CL. Valproic acid disrupts the oscillatory expression of core circadian rhythm transcription factors. Toxicol Appl Pharmacol. 2018; 339: 110–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Younus I, Reddy DS. A resurging boom in new drugs for epilepsy and brain disorders. Expert Rev Clin Pharmacol. 2018; 11: 27–45. [DOI] [PubMed] [Google Scholar]
- 124.Fenoglio-Simeone K, Mazarati A, Sefidvash-Hockley S, Shin D, Wilke J, Milligan H, et al. Anticonvulsant effects of the selective melatonin receptor agonist ramelteon. Epilepsy Behav. 2009; 16: 52–7. [DOI] [PubMed] [Google Scholar]
- 125.Aguiar CC, Almeida AB, Araújo PV, Vasconcelos GS, Chaves EM, do Vale OC, et al. Anticonvulsant effects of agomelatine in mice. Epilepsy Behav. 2012; 24: 324–8. [DOI] [PubMed] [Google Scholar]
- 126.Mosińska P, Socała K, Nieoczym D, Laudon M, Storr M, Fichna J, et al. Anticonvulsant activity of melatonin, but not melatonin receptor agonists, Neu-P11 and Neu-P67, in mice. Behav Brain Res. 2016; 307: 199–207. [DOI] [PubMed] [Google Scholar]
- 127.He B, Chen Z. Molecular targets for small-molecule modulators of circadian clocks. Curr Drug Metabol. 2016; 17: 503–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cha HK, Chung S, Lim HY, Jung JW, Son GH. Small molecule modulators of the circadian molecular clock with implications for neuropsychiatric diseases. Front Mol Neurosci. 2018; 11: 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chen Z, Yoo SH, Park YS, Kim KH, Wei S, Buhr E, et al. Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc Natl Acad Sci U S A. 2012; 109 (1): 101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen Z, Yoo SH, Takahashi JS. Small molecule modifiers of circadian clocks. Cell Mol Life Sci. 2013; 70: 2985–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.He B, Nohara K, Park N, Park Y-S, Guillory B, Zhao Z, et al. The small molecule nobiletin targets the molecular oscillator to enhance circadian rhythms and protect against metabolic syndrome. Cell Metabol. 2016; 23: 610–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yang B, Wang J, Zhang N. Effect of nobiletin on experimental model of epilepsy. Transl Neurosci. 2018; 9: 211–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nakajima A, Ohizumi Y, Yamada K. Anti-dementia activity of nobiletin, a citrus flavonoid: a review of animal studies. Clin Psychopharmacol Neurosci. 2014; 12: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jeong KH, Jeon MT, Kim HD, Jung UJ, Jang MC, Chu JW, et al. Nobiletin protects dopaminergic neurons in the 1-methyl-4-phenylpyridinium-treated rat model of Parkinson’s disease. J Med Food. 2015; 18: 409–14. [DOI] [PubMed] [Google Scholar]
- 135.Nohara K, Mallampalli V, Nemkov T, Wirianto M, Yang J, Ye Y, et al. Nobiletin fortifies mitochondrial respiration in skeletal muscle to promote healthy aging against metabolic challenge. Nat Commun. 2019; 10 (1): 3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fisher RS, Acevedo C, Arzimanoglou A, Bogacz A, Cross JH, Elger CE, et al. ILAE official report: A practical clinical definition of epilepsy. Epilepsia. 2014; 55: 475–82. [DOI] [PubMed] [Google Scholar]


