Abstract
The family Solemoviridae includes viruses with icosahedral particles (26–34 nm in diameter) assembled on T=3 symmetry with a 4–6 kb positive-sense, monopartite, polycistronic RNA genome. Transmission of members of the genera Sobemovirus and Polemovirus occurs via mechanical wounding, vegetative propagation, insect vectors or abiotically through soil; members of the genera Polerovirus and Enamovirus are transmitted by specific aphids. Most solemoviruses have a narrow host range. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the family Solemoviridae, which is available at ictv.global/report/solemoviridae.
Keywords: ICTV Report, taxonomy, Solemoviridae
Abbreviations
CP, capsid protein; RdRP, RNA-directed RNA polymerase; sgRNA, subgenomic mRNA; VPg, virus protein genome-linked.
Virion
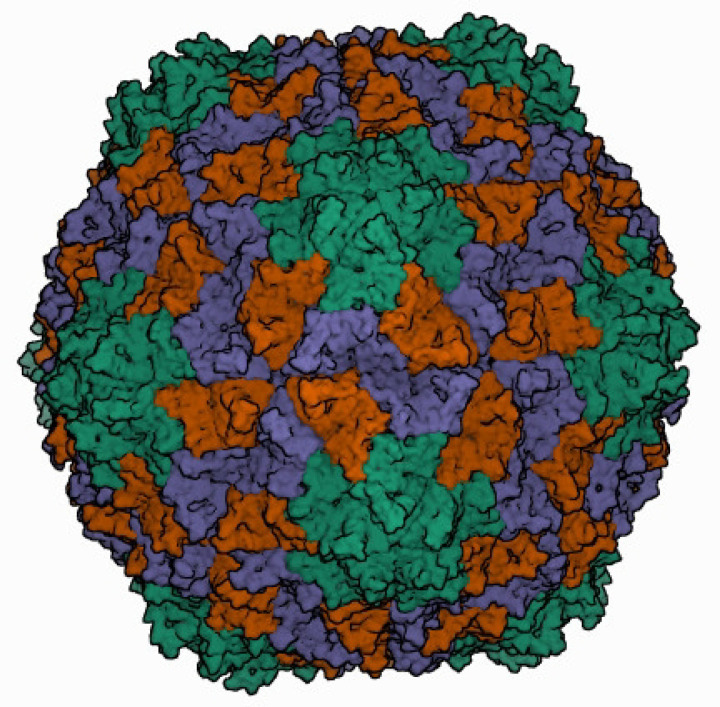
Icosahedral virions of 26–34 nm in diameter are composed of 180 monomers of viral capsid protein (CP) on a T=3 lattice symmetry (Table 1). CP monomers can adopt three conformations designated as A, B and C (Fig. 1) [1]. Sobemovirus capsids are stabilized by divalent cations, pH-dependent protein–protein interactions and salt bridges between protein and viral RNA. Polerovirus and enamovirus particles lack proof of bound metal ions [2]. The N-terminal random domain of CP monomers is rich in basic amino acids and regulates the stability, curvature and assembly of virions. The C-terminal shell domain possesses a jellyroll β-sandwich topology, common in most non-enveloped icosahedral viruses. Virions of poleroviruses and enamoviruses incorporate a small proportion of CP with a 25–50 kDa C-terminal extension of a readthrough domain that is associated with aphid transmission and virus particle stability.
Table 1.
Characteristics of members of the family Solemoviridae
|
Example: |
southern bean mosaic virus (DQ875594), species Southern bean mosaic virus, genus Sobemovirus |
|---|---|
|
Virion |
Non-enveloped icosahedral particles with T=3 symmetry, 20–34 nm in diameter, composed of 180 molecules of capsid protein |
|
Genome |
4–6 kb positive-sense, non-segmented RNA, with 5′-terminal VPg; there is no poly(A) tail |
|
Replication |
Cytoplasmic |
|
Translation |
From genomic and subgenomic RNAs via leaky scanning, stop codon readthrough and −1 ribosomal frameshifting |
|
Host range |
Plants (monocotyledons and dicotyledons) |
|
Taxonomy |
Realm Riboviria, kingdom Orthornavirae, phylum Pisuviricota, class Pisoniviricetes, order Sobelivirales; four genera, including >55 species |
Fig. 1.
Three-dimensional cryo-electron reconstruction of the particle of rice yellow mottle virus shown at 2.8 Å resolution. The icosahedral asymmetric unit contains three subunits: A (in green), B (in brown), and C (in blue). Image from the RCSB PDB (rcsb.org) of PDB ID 1F2N [1].
Genome
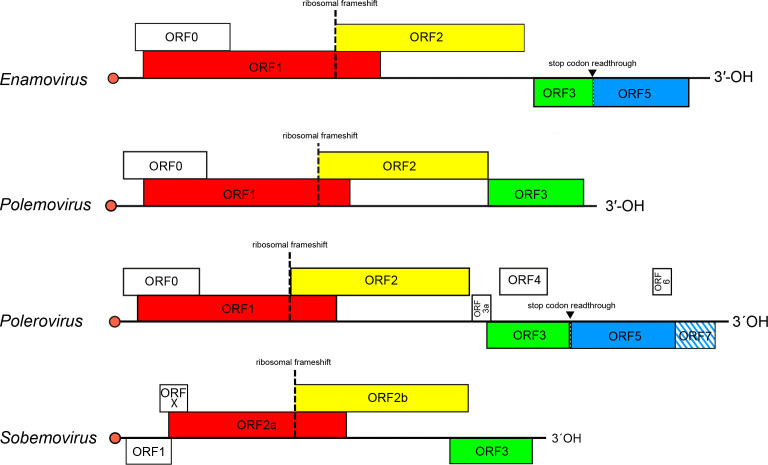
The genome comprises a polycistronic, positive-sense RNA molecule of 4–6 kb without a 3′-poly(A) tail. The genome 5′-end has a covalently attached virus protein genome-linked (VPg). The genome contains 4–10 open reading frames (ORFs) (Fig. 2). The 5′-proximal ORF encodes a non-conserved RNA silencing suppressor protein, followed by a polyprotein that is expressed by ribosomal leaky scanning and cleaved autocatalytically to different functional subunits (membrane anchor, serine protease, VPg and C-terminal domains). Expression of the viral RNA-directed RNA polymerase (RdRP) as an alternative C-terminal domain of the polyprotein is regulated by a −1 programmed ribosomal frameshift. A 3′-proximal ORF of solemoviruses encodes CP, expressed from a subgenomic mRNA (sgRNA). For polero- and enamoviruses, suppression of an amber stop codon of the CP gene leads to translation of readthrough domain protein from ORF5. Other ORFs are genus-specific.
Fig. 2.
Genome organization of the members of the family Solemoviridae. Red bullets indicate 5′-VPg. ORF reading frames are shown by their position above the genome in the order −1, 0 and +1 and encode the polyprotein (red), RdRP (yellow), CP (green) and readthrough domain (blue). The hatched area indicates an overlap between ORFs.
Replication
Replication occurs after synthesis of RdRP from genomic RNA. A conserved 5′-end sequence, 5′-ACAA(AA)−3′, and a stable 3′-end stem–loop are essential for template recognition. The VPg of cereal yellow dwarf virus RPV acts as a primer for full-length negative-sense strand RNA synthesis [3], whereas primer-independent replication has been demonstrated for Sesbania mosaic virus [4]. Sobemoviruses, polemoviruses and enamoviruses transcribe one sgRNA; poleroviruses generate up to three sgRNAs.
Pathogenicity
Infections can be symptomless or cause severe diseases. The family includes important plant pathogens with high economic impact, of which rice yellow mottle virus, potato leafroll virus, sugarcane yellow leaf virus and beet-infecting poleroviruses are the most devastating. Virulent infections induce chlorosis, phloem necrosis, stunting and sometimes sterility. Sobemoviruses infect different tissue types, whereas poleroviruses and enamoviruses are phloem-specific. Whereas sobemoviruses are mainly transmitted via mechanical wounds [5], transmission of poleroviruses and enamoviruses is by aphids in a persistent, circulative and non-propagative mode [6].
Taxonomy
Current taxonomy: ictv.global/taxonomy. The genera Sobemovirus and Polerovirus each include 20 or more species and the genus Enamovirus includes 5 species. The genus Polemovirus includes the species Poinsettia latent virus.
Resources
Full ICTV Report on the family Solemoviridae: ictv.global/report/solemoviridae.
Funding information
Production of this Profile, the ICTV Report, and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA). Solemoviridae SG cooperation was supported by G. F. PARROT programme.
Acknowledgements
Members of the ICTV Report Consortium are Stuart G. Siddell, Elliot J. Lefkowitz, Sead Sabanadzovic, Peter Simmonds, F. Murilo Zerbini, Donald B. Smith and Luisa Rubino.
Conflicts of interest
The authors declare that there are no conflicts of interest.
References
- 1.Qu C, Liljas L, Opalka N, Brugidou C, Yeager M, et al. 3D domain swapping modulates the stability of members of an icosahedral virus group. Structure. 2000;8:1095–1103. doi: 10.1016/s0969-2126(00)00508-6. [DOI] [PubMed] [Google Scholar]
- 2.Byrne MJ, Steele JFC, Hesketh EL, Walden M, Thompson RF, et al. Combining transient expression and cryo-EM to obtain high-resolution structures of luteovirid particles. Structure. 2019;27:1761–1770. doi: 10.1016/j.str.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osman TAM, Coutts RHA, Buck KW. In vitro synthesis of minus-strand RNA by an isolated cereal yellow dwarf virus RNA-dependent RNA polymerase requires VPg and a stem-loop structure at the 3’ end of the virus RNA. J Virol. 2006;80:10743–10751. doi: 10.1128/JVI.01050-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Govind K, Mäkinen K, Savithri HS. Sesbania mosaic virus (SeMV) infectious clone: possible mechanism of 3’ and 5’ end repair and role of polyprotein processing in viral replication. PLoS One. 2012;7:e31190. doi: 10.1371/journal.pone.0031190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sõmera M, Sarmiento C, Truve E. Overview on sobemoviruses and a proposal for the creation of the family Sobemoviridae . Viruses. 2015;7:3076–3115. doi: 10.3390/v7062761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brault V, Herrbach É, Reinbold C. Electron microscopy studies on luteovirid transmission by aphids. Micron. 2007;38:302–312. doi: 10.1016/j.micron.2006.04.005. [DOI] [PubMed] [Google Scholar]