Abstract
The family Geminiviridae includes viruses with mono- or bipartite single-stranded, circular DNA genomes of 2.5–5.2 kb. They cause economically important diseases in most tropical and subtropical regions of the world. Geminiviruses infect dicot and monocot plants and are transmitted by insect vectors. DNA satellites are associated with some geminiviruses. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the family Geminiviridae which is available at ictv.global/report/geminiviridae.
Keywords: Geminiviridae, ICTV Report, taxonomy
Virion
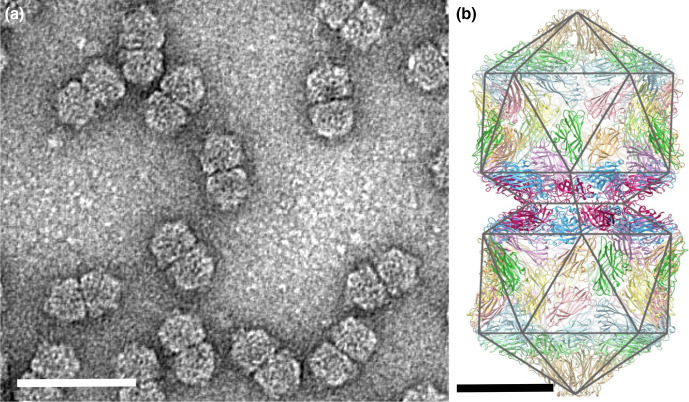
Geminiviruses have a unique particle morphology of twinned (geminate) icosahedra. For maize streak virus (genus Mastrevirus), virions are 22×38 nm, consisting of two incomplete icosahedra (T=1) containing 110 capsid protein (CP) subunits organized as 22 pentameric capsomers (Table 1, Fig. 1a) [1]. The structure of Ageratum yellow vein virus (genus Begomovirus) at 3.3 Å resolution shows that the N-terminus of CP adopts three different conformations essential for building the interface between geminate halves (Fig. 1b) [2].
Table 1.
Characteristics of members of the family Geminiviridae
|
Example: |
bean golden yellow mosaic virus (DNA-A: L01635, DNA-B: L01636), species Bean golden yellow mosaic virus, genus Begomovirus |
|---|---|
|
Virion |
Twinned (geminate) incomplete icosahedra, T=1, 22×38 nm with a single coat protein |
|
Genome |
2.5–5.2 kb of single-stranded, circular DNA, mono- or bipartite |
|
Replication |
Complementary strand synthesized in the nucleus by host replication factors; double-stranded circular molecules serve as templates for both transcription and replication; replication employs a rolling-circle mechanism and also a recombination-dependent mechanism |
|
Translation |
From transcribed mRNAs; members of some genera use transcript splicing |
|
Host range |
Plants (monocots and dicots) |
|
Taxonomy |
Realm Monodnaviria, kingdom Shotokuvirae, phylum Cressdnaviricota, class Repensiviricetes, order Geplafuvirales: >10 genera, >500 species |
Fig. 1.
(a) Purified particles of maize streak virus stained with uranyl acetate showing typical twinned quasi-isometric subunits. Bar, 50 nm. Reproduced with permission from [1]. (b) Complete atomic model for all 110 subunits in the capsid of Ageratum yellow vein virus, with a polyhedral cage showing the symmetry of the particle. The fivefold symmetry axis is vertical and through the centre of the particle in this view. Bar, 10 nm. CC BY 4.0 licence from [2].
Genome
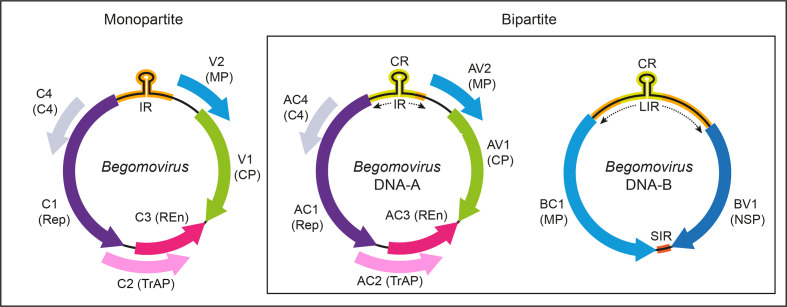
Viruses in most genera of the family have monopartite genomes, whereas those in the genus Begomovirus have mono- or bipartite genomes. The genomes of bipartite begomoviruses (Fig. 2) consist of two circular single-stranded components, DNA-A and DNA-B, each of 2.5–2.6 kb. The two components share approximately 200 bases of sequence (common region) that includes the replication origin. DNA-A encodes a capsid protein (ORF AV1, CP), a putative movement protein (ORF AV2, MP, absent in New World begomoviruses), a replication-associated protein (ORF AC1, Rep), a transcriptional activator (ORF AC2, TrAP), a replication enhancer (ORF AC3, REn) and the C4 protein (ORF AC4, C4). DNA-B encodes a nuclear shuttling protein (ORF BV1, NSP) and a movement protein (ORF BC1, MP). The genomes of monopartite begomoviruses (Fig. 2) resemble the DNA-A component of bipartite begomoviruses [3, 4]. The genome organization of members of each of the genera in the family Geminiviridae is shown in the ICTV Report (ictv.global/report/geminiviridae). DNA satellites are associated with many begomoviruses and some mastreviruses [4].
Fig. 2.
Genome organization of begomoviruses. The ORFs ([A]V1, [A]V2, [A]C1, [A]C2, [A]C3, [A]C4, BV1 and BC1) and their protein products (CP, capsid protein; MP, movement protein; Rep, replication-associated protein; TrAP, transcriptional activator protein; REn, replication enhancer protein; C4, C4 protein; NSP, nuclear shuttle protein) are shown. ORF [A]V2 is not present in New World begomoviruses. IR, intergenic region; LIR, long intergenic region; SIR, short intergenic region; CR, common region. The hairpin which includes the origin of replication is indicated in the IR/LIR.
Replication
Complementary-sense DNA synthesis to produce dsDNA depends solely on host factors. Virus ssDNA synthesis is initiated by cleavage of the virion-sense strand by Rep in the 5′-TAATATTAC-3′ sequence conserved in most geminiviruses within the intergenic region (IR)/ long intergenic region (LIR). Geminiviruses do not encode a DNA polymerase, relying on host factors recruited during the early stages of replication. Coding regions in both strands diverge from the IR/LIR, and transcription is bi-directional. Geminiviruses use multiple overlapping transcripts for gene expression [3, 4]. The begomovirus tomato yellow leaf curl virus replicates in the salivary glands of whiteflies [5].
Taxonomy
Current taxonomy: ictv.global/taxonomy. The family includes multiple genera collectively including >500 species, the most diverse genera being Begomovirus (>440 species) and Mastrevirus (>40 species). Host are dicots except for maldoviruses and mastreviruses (dicots and monocots) and eragroviruses (monocots). Vectors are whiteflies (begomoviruses), aphids (capulaviruses), treehoppers (grabloviruses, topocuviruses), leafhoppers (becurtoviruses, curtoviruses, mastreviruses, mulcrileviruses, turncurtoviruses), or unknown (citlodaviruses, eragroviruses, maldoviruses, opunviruses, topileviruses).
Resources
Full ICTV Report on the family Geminiviridae: ictv.global/report/geminiviridae.
Funding information
Production of this summary, the online chapter, and associated resources was funded by a grant from the Wellcome Trust (WT108418AIA).
Acknowledgements
Members of the ICTV Report Consortium are Stuart G. Siddell, Elliot J. Lefkowitz, Sead Sabanadzovic, Peter Simmonds, F. Murilo Zerbini, Donald B. Smith and Luisa Rubino.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: CP, capsid protein; IR, intergenic region; LIR, long intergenic region; MP, movement protein; NSP, nuclear shuttling protein; REn, replication enhancer; Rep, replication-associated protein; TrAP, transcriptional activator.
References
- 1.Zhang W, Olson NH, Baker TS, Faulkner L, Agbandje-McKenna M, et al. Structure of the maize streak virus geminate particle. Virology. 2001;279:471–477. doi: 10.1006/viro.2000.0739. [DOI] [PubMed] [Google Scholar]
- 2.Hesketh EL, Saunders K, Fisher C, Potze J, Stanley J, et al. The 3.3 Å structure of a plant geminivirus using cryo-EM. Nat Commun. 2018;9:2369. doi: 10.1038/s41467-018-04793-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rojas MR, Hagen C, Lucas WJ, Gilbertson RL. Exploiting chinks in the plant’s armor: evolution and emergence of geminiviruses. Annu Rev Phytopathol. 2005;43:361–394. doi: 10.1146/annurev.phyto.43.040204.135939. [DOI] [PubMed] [Google Scholar]
- 4.Navas-Castillo J, Fiallo-Olivé E. In: Encyclopedia of Virology. Zuckerman M, Bamford D, editors. Vol. 3. Oxford: Academic Press; 2021. Geminiviruses (Geminiviridae) pp. 411–419. vol. [Google Scholar]
- 5.He Y-Z, Wang Y-M, Yin T-Y, Fiallo-Olivé E, Liu Y-Q, et al. A plant DNA virus replicates in the salivary glands of its insect vector via recruitment of host DNA synthesis machinery. Proc Natl Acad Sci U S A. 2020;117:16928–16937. doi: 10.1073/pnas.1820132117. [DOI] [PMC free article] [PubMed] [Google Scholar]