Abstract
Introduction
Shiga toxin-producing Escherichia coli (STEC) is a zoonotic, foodborne gastrointestinal pathogen that has the potential to cause severe clinical outcomes, including haemolytic uraemic syndrome (HUS). STEC-HUS is the leading cause of renal failure in children and can be fatal. Over the last decade, STEC clonal complex 165 (CC165) has emerged as a cause of STEC-HUS.
Gap statement
There is a need to understand the pathogenicity and prevalence of this emerging STEC clonal complex in the UK, to facilitate early diagnosis, improve clinical management, and prevent and control outbreaks.
Aim
The aim of this study was to characterize CC165 through identification of virulence factors (VFs) and antimicrobial resistance (AMR) determinants in the genome and to integrate the genome data with the available epidemiological data to better understand the incidence and pathogenicity of this clonal complex in the UK.
Methodology
All isolates belonging to CC165 in the archives at the UK public health agencies were sequenced and serotyped, and the virulence gene and AMR profiles were derived from the genome using PHE bioinformatics pipelines and the Centre for Genomic Epidemiology virulence database.
Results
There were 48 CC165 isolates, of which 43 were STEC, four were enteropathogenic E. coli (EPEC) and one E. coli . STEC serotypes were predominately O80:H2 (n=28), and other serotypes included O45:H2 (n=9), O55:H9 (n=4), O132:H2 (n=1) and O180:H2 (n=1). All but one STEC isolate had Shiga toxin (stx) subtype stx2a or stx2d and 47/48 isolates had the eae gene encoding intimin involved in the intimate attachment of the bacteria to the human gut mucosa. We detected extra-intestinal virulence genes including those associated with iron acquisition (iro) and serum resistance (iss), indicating that this pathogen has the potential to translocate to extra-intestinal sites. Unlike other STEC clonal complexes, a high proportion of isolates (93%, 40/43) were multidrug-resistant, including resistance to aminoglycosides, beta-lactams, chloramphenicol, sulphonamides, tetracyclines and trimethoprim.
Conclusion
The clinical significance of this clonal complex should not be underestimated. Exhibiting high levels of AMR and a combination of STEC and extra-intestinal pathogenic E. coli (ExPEC) virulence profiles, this clonal complex is an emerging threat to public health.
Keywords: whole-genome sequencing, renal failure, complex STEC infection, haemolytic uraemic syndrome
Data Summary
FASTQ files are submitted to the National Center for Biotechnology Information (NCBI). All data can be found under Bioproject no. PRJNA315192 and strain-specific information is found in Table S1 (available in the online version of this article).
Introduction
The term haemolytic uraemic syndrome (HUS) was first used in the 1950s to describe the combination of clinical presentations of haemolytic anaemia, thrombocytopenia and renal failure [1]. Outbreaks of HUS led clinicians to suspect an infectious cause, and in 1985 Karmali et al. showed that Shiga toxin-producing Escherichia coli (STEC) could be isolated from the faecal specimens of a sub-set of HUS patients reporting a diarrhoeal prodrome [2]. Over the decades, other causes of non-infectious (atypical) HUS have been identified, but STEC remains the leading cause of HUS, especially in children [3].
Although Karmali et al. showed a number of different E. coli serotypes had the potential to produce Shiga toxin, a series of outbreaks of HUS caused by E. coli serotype O157:H7 during the 1980s established this serotype as the most common cause of STEC-HUS at that time, particularly in the UK and North America [4–6]. Subsequent studies showed that progression to STEC-HUS was most likely in patients infected with STEC harbouring the Shiga toxin (stx) type 2, in particular stx2a and stx2d [7]. The stx genes are bacteriophage-encoded and there is evidence that a wide range of E. coli serotypes other than O157:H7 have acquired the stx2a- (or stx2d) encoding phage [7].
Acquisition of stx by strains of E. coli that exhibit an attachment mechanism encoded by the locus of enterocyte effacement (LEE) result in a highly pathogenic strain capable of causing STEC-HUS [7]. The LEE encodes a Type III Secretion System (T3SS) including espA, espB, espF, tir and intimin that facilitates attachment to the human gut mucosa and microvillus effacement [8–10]. Intimin is encoded by the eae gene, which acts as a marker for the presence of the LEE. Non-LEE effectors include espC, espI, espJ, cif, nleA, nleB and nleC [8–10]. Certain STEC serotypes, including STEC O157:H7, harbour a plasmid-encoded enterohaemolysin (ehxA), a toxin (toxB) and serine protease (espP) [11].
Over the last 10 years, a novel serotype of STEC has emerged as a cause of STEC-HUS, STEC O80:H2 belonging to clonal complex 165 (CC165) [12–14]. This serotype has been isolated from patients with STEC-HUS presenting with multi-organ failure, extensive central nervous system involvement, and extensive thrombotic microangiopathy [12–14]. STEC O80:H2 has been described as a hybrid pathotype that combines the diarrhoeagenic E. coli virulence factors (VFs) stx, eae (intimin) and ehxA (enterohaemolysin), with extra-intestinal VFs [15].
The extra-intestinal virulence phenotype is attributed to the presence of a mosaic plasmid that harbours VFs that allow the translocation and survival of E. coli from the gut to extra-intestinal sites. This mosaic plasmid is closely related to the pathogenic plasmid pS88 found in extra-intestinal pathogenic E. coli (ExPEC), which harbours the genes iro (iron acquisition), iss (serum resistance) and hlyF (haemolysin) among many others such as sitABCD, iutA, iucC and ompTp [16, 17]. This mosaic plasmid found in O80:H2 has an additional resistance cassette which confers resistance to multiple classes of antibiotics (e.g. aminoglycosides, tetracyclines and β-lactams) [15].
The aim of this study was to review the data in the national enhanced STEC surveillance systems and whole genome sequencing data held by the public health agencies in the UK to investigate the incidence, and epidemiological and microbiological characteristics of this highly pathogenic strain of STEC.
Methods
Bacterial strains
The Gastrointestinal Bacteria Reference Unit (GBRU), Public Heath England (PHE), receives faecal specimens that test positive for STEC by PCR from patients with hospital or community-acquired cases of gastrointestinal disease or HUS, in England and Wales, when diagnostic labs are unable to isolate E. coli O157:H7 from the stool. In Scotland, faecal specimens from patients with symptoms of bloody diarrhoea and/or HUS are referred to the Scottish E. coli Reference Laboratory (SERL). STEC PCR and culture are performed at GBRU and SERL as previously described [18, 19].
Data, setting and source
England, Scotland and Wales operate a national enhanced STEC surveillance system for all STEC cases encompassing epidemiological data, reported through Enhanced Surveillance Questionnaires (ESQs) and microbiological data. This study includes all samples submitted to PHE and SERL between 2013 and 2020 belonging to CC165.
For STEC patients who had completed an ESQ, we extracted results relating to travel, clinical symptoms and food/animal exposures. STEC questionnaire data were linked to whole genome sequencing (WGS) data, including stx subtype, virulence profiling and antimicrobial resistance (AMR) detection.
WGS using Illumina technologies and data processing
Genomic DNA belonging to isolates from CC165 was extracted and sequenced as described in Dallman et al. [20]. SRA accession numbers were generated and are listed in Table S1.
Variant calling, SNP typing and phylogeny from Illumina data
The population structure of CC165 was investigated using SnapperDB v0.2.6 [21], which holds the called variant data relative to a CC165 reference genome (CP002729.1) for isolates sequenced at PHE. A whole genome alignment of all samples in CC165, mapped to reference CP002729, was produced. Gubbins v2.0.0 [22] was used on all samples within the alignment and an output that identified the recombinant regions was produced. SnapperDB v0.2.6 [21] was used again to re-extract the variants belonging to CC165, producing an alignment where a given variant position belonging to a minimum of 80% of the strains in the alignment and recombination regions were masked. IQTree v2.0.4 [23] was used to construct a maximum-likelihood phylogeny using the best-fit model and this was visualized in ITOL v5.7 [24].
Enterobase (v1.1.2) was used to generate a Minimum Spanning Tree (MSTree V2) based on the Achtman 7 Gene multilocus sequence type (MLST) scheme with 10469 strains included. Data selection involved genome uploads where ‘Species contains Escherichia coli ’ and ‘Lab contact contains Public Health England or GBRU’ between 2014 and 2021.
Virulence and AMR profiling using Illumina genome data
Virulence profile and AMR profiles were derived from the Illumina genome data as previously described [25–27], using the GeneFinder tool (https://github.com/phe-bioinformatics/gene_finder). The presence of each gene was confirmed when coverage and homology (CH) are over 85% compared to the reference gene. For the purpose of this study, genes where CH fell below 85% when compared to the reference were investigated further. The PHE virulence database was supplemented with variants of putative genes in E. coli virulence database v2020-05-29 from the Centre for Genomic Epidemiology (CGE) (https://bitbucket.org/genomicepidemiology/virulencefinder_db/src/master/virulence_ecoli.fsa).
Plasmid identification
The pO157 plasmid sequence (Accession AF074613.1) [28], as well as plasmid sequences corresponding to pR444_A and pR444_C (QBDM01000004.1 and QBDM01000002.1, ASM312350v1, BioProject PRJNA449634) [15] were extracted and curated into a custom database, designed for SRST2 [29]. SRST2 v0.2.0 was used to identify the presence of pR444_A, pR444_C and similarity to pO157 using Illumina FASTQ, with parameters for minimum coverage set at 40 and to report all consensus sequences.
Data deposition
Illumina FASTQ files are available from the National Center for Biotechnology Information (NCBI) Short Read Archive Bioproject no. PRJNA315192. The SRA (sequence read archive) accession numbers are given in Table S1.
Results
Epidemiology
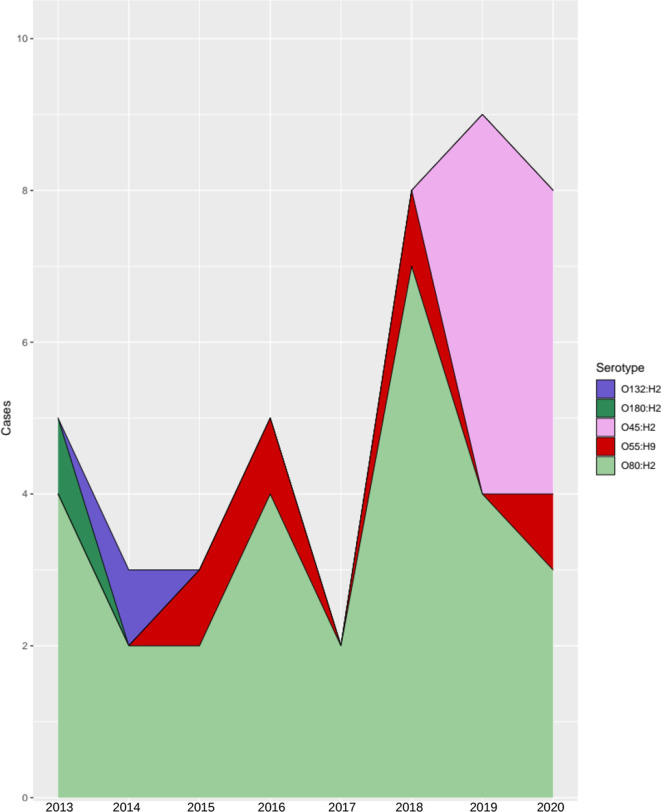
Since 2013, 43 isolates of STEC belonging to CC165 were detected in England (n=34), Scotland (n=5) and Wales (n=4). The majority of cases were female (26/43, 60%) and the age range was 0–85 years, with 30% (13/43) of cases under 5 years. Cases were reported throughout the year, with the highest number of cases reported in 2019 (Fig. 1). The maximum, minimum and median cases reported across the years for CC165 STEC were nine, three and five respectively. Geographical distribution in England revealed that London had the most cases (n=13), followed by the South East region (n=10) (Fig. S1). The remaining cases were distributed across the rest of the PHE centres, not surpassing three cases per PHE centre.
Fig. 1.
Yearly distribution of CC165 STEC cases in this dataset (n=43). The x-axis identifies the year of the case, and the y-axis gives the cumulative cases for the year. The stacked area graph shows the different serotypes detected in each year.
Of the 34 cases resident in England, an enhanced questionnaire was available for 32 cases. Fewer than half of the cases infected with CC165 reported either vomiting, nausea, abdominal pain and/or fever (Table 1). The majority of cases had diarrhoea (30/32, 94%) but only 5/32 (16%) reported bloody diarrhoea, compared to cases infected with STEC O157:H7 where 93–95 % had diarrhoea and 61–62% had bloody diarrhoea (PHE in-house data). There were 13/32 (41%) hospitalized cases, including 8/32 cases diagnosed with HUS (25%), of which half were female (4/8, 50%) and 6/8 (75%) were aged between 0 and 4 years. The two adults were 63 and 85 years old. Compared to STEC O157:H7, cases of CC165 STEC had a higher rate of hospitalisation (CC165: 41% vs O157: 34%) and progression to STEC-HUS (CC165: 25% vs O157: 3–5%) (PHE in-house data).
Table 1.
Clinical presentation (n=32); England STEC cases where an ESQ was available
|
Symptom |
N |
% |
|---|---|---|
|
Diarrhoea |
30 |
94 |
|
Abdominal pain |
16 |
50 |
|
Vomiting |
14 |
44 |
|
Hospitalized |
13 |
41 |
|
Fever |
12 |
38 |
|
Nausea |
10 |
31 |
|
HUS |
8 |
25 |
|
Blood in stools |
5 |
16 |
|
Death |
0 |
0 |
ESQ data were available for none of the Welsh cases, and for 3/5 Scottish STEC cases. Of the three cases with extended data, 3/3 (100%) presented with bloody diarrhoea, 2/3 (67%) reported abdominal pain, and 1/3 (34%) reported nausea and vomiting. No cases were hospitalized overnight, but one case attended as a day case.
None of the cases resident in Scotland (0/3) reported travelling outside the UK. There were 10/32 England cases who reported having travelled outside of the UK, of which seven cases travelled to France, two cases reported travelling to Portugal and one travelled to Egypt.
ESQs for data from England revealed that 11/32 (34%) cases reported contact with dogs and 6/32 (19%) reported contact with cats. One case was reported for contact with each of the following: cattle, horses, poultry and deer. Three of 32 cases (9%) visited a farm, 4/32 cases (19%) walked on a farm where animals graze, 3/32 (9%) had contact with wildlife (bird feeder, possible deer droppings, pigeons or cats) and 4/32 (13%) had contact with soil, manure or sewage (Table 2). Food history of cases saw the most common food exposure was cooked poultry (18/32, 56% of cases), with raw vegetables following at 16/32 cases (50%) (Table 2).
Table 2.
Environmental and food exposures (n=32); England STEC cases where an ESQ was available
|
Environmental/food exposure |
N |
% |
|---|---|---|
|
Dog |
11 |
34 |
|
Cat |
6 |
19 |
|
Cattle |
1 |
3 |
|
Horse |
1 |
3 |
|
Poultry |
1 |
3 |
|
Deer |
1 |
3 |
|
Farm where animals graze |
6 |
19 |
|
Soil, manure or sewage |
4 |
13 |
|
Wildlife |
3 |
9 |
|
Farm |
3 |
9 |
|
Cooked poultry |
18 |
56 |
|
Raw vegetables |
16 |
50 |
|
Pasteurized milk |
17 |
53 |
|
Hard cheese |
15 |
47 |
|
Yoghurt |
14 |
44 |
|
Fish |
13 |
41 |
|
Cooked beef |
13 |
31 |
|
Ice cream |
8 |
25 |
|
Cooked pork |
7 |
22 |
|
Soft cheese |
7 |
22 |
|
Raw beef |
5 |
16 |
|
Cured meats |
5 |
16 |
|
Raw poultry |
4 |
13 |
Scottish enhanced surveillance data revealed that contact with cattle and/or cats was reported by three cases. Two of the three cases live on a farm. There were no common food exposures.
Phylogenetic analysis
We investigated the population structure (based on seven-gene MLST) of E. coli submitted to Enterobase by PHE between January 2014 and January 2021, highlighting the position of CC165 in the E. coli population (Fig. S2). CC165 in this data set comprises sequence type (ST) 301, ST165, ST189, ST382, ST1178 and ST6312.
There were 48 isolates belonging to CC165 in the PHE archive belonging to England, Wales and Scotland, comprising ST301 (n=43), ST189 (n=3), ST382 (n=1) and ST165 (n=1) (Table S1). The isolate belonging to ST165 was negative for both eae and stx. There were four isolates that had eae but were stx-negative and were designated enteropathogenic E. coli (EPEC). The isolates of EPEC belonged to ST189 (n=3) and ST382 (n=1). A phylogeny of CC165 was built with CP002729 as the reference. Three samples were excluded from the phylogeny by SnapperDB due to the mean depth coverage being below the cut-off threshold of 30. This resulted in 46 leaves in the tree, including the reference which was generated by aligning simulated reads to itself (Fig. 2). The sample belonging to ST382 (O unidentifiable:H5) was excluded from the phylogeny for the reasons stated, and therefore EPEC ST189 (O157:H26, n=3) and E. coli ST165 (O80:H19, n=1) belonged to the cluster designated as Cluster (C)1.
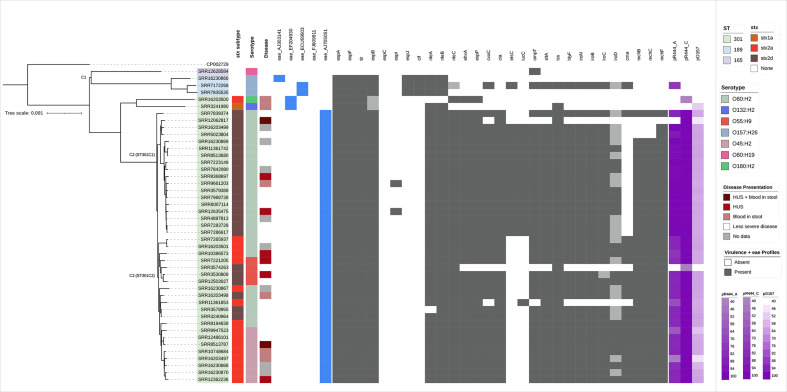
Fig. 2.
Population structure of CC165 in the UK (n=46) (three isolates excluded), constructed using SnapperDB and IQTree, and rooted at the midpoint. Sequence type, stx subtypes, severe clinical diagnosis and serotype are the first four colour strips mapped onto the phylogeny. The presence of variants of the eae gene is shown as blue for detected and white for not detected. VF profiles achieved from PHE pipelines supplemented with genes from CGE are annotated on the population structure of CC165 in the UK. Dark grey indicates detection, whereas light grey indicates the gene was not detected but coverage and homology values are high (>60). Purple on the right of the VF annotation indicates identity coverage of isolates to the three plasmid references, pR444_A, pR444_C and pO157. Shades of purple indicate coverage identity values, with light purple being 40.
C2 comprised the isolates that harbour the stx-encoding prophage (STEC) and was divided into two sub-clusters, ST301C1 and ST301C2. ST301C1 comprised isolates belonging to serotype O80:H2 (n=18), whereas ST301C2 contained isolates belonging to three different serotypes, O55:H9 (n=4), O45:H2 (n=8) and O80:H2 (n=9). Two isolates of ST301 (serotypes O180:H2 and O132:H2) did not cluster with ST301C1 or ST301C2. Fig. 1 also describes the emergence of CC165 STEC serotypes from 2013 to 2020, with O45:H2 first emerging into the population in 2019.
STEC virulence factors
As described by Blanco et al., and Cointe et al. [15, 30], isolates of O80:H2 are characterized by a unique variant of the eae gene, eaeXi (AJ705051). Isolates of CC165 were examined for eae, using variants from the CGE database. There were 41 isolates of CC165 that harboured the eaeXi variant, which were all isolates in C2, and two isolates that were excluded from the tree, which includes serotypes O80:H2, O55:H9 and O45:H2. The remaining six isolates that had eae detected harboured four other variants (Fig. 2).
There were 43 isolates, all ST301, that harboured stx2d (n=25), stx2a (n=17) and stx1a (n=1) (Table S1). Isolates that were positive for stx2a belonged to four serotypes: O45:H2 (n=9), O80:H2 (n=6), O55:H9 (n=1) and O180:H2 (n=1). Isolates that harboured stx2d belonged to two different serotypes: O80:H2 (n=22) and O55:H9 (n=3) (Table S1). Of the eight cases that were diagnosed with HUS, four were infected with isolates that had stx2a and four had stx2d.
Fig. 2 shows the distribution of VFs for each isolate across the population structure of CC165.
As expected, LEE-encoded T3SS proteins (espA, espB, espF and tir) were present in all eae-positive isolates [8, 31]. We also interrogated the genomes to determine whether non-LEE-encoded effectors, espC, espI, espJ, cif, nleA, nleB and nleC, were present [10, 32, 33]. For the majority of samples, nleA and nleB were present in the eae-positive isolates whereas nleC was only confirmed in isolates with stx2a or stx2d. Using the 85% coverage and homology criteria, espC was not detected in any of the isolates of CC165, but CH values <10% for all but one isolate were detected (ST2). The gene espC is not considered essential in attaching and effacing lesion formation [32]. The presence of espI was confirmed in two STEC isolates, although lower CH values for espI were detected for other isolates (ST2). The genes espJ and cif were detected in 3/4 EPEC isolates, but not in those that had acquired stx (Fig. 2).
In STEC O157:H7, the pO157 plasmid harbours toxB, katP, ehxA and espP [11, 28, 34–37]. In our dataset, both ehxA (an enterohaemolysin) and espP (an extracellular serine protease associated with the LEE [35]) were detected in all STEC isolates that harboured stx2a or stx2d, with one exception (n=41). The gene toxB was detected in one EPEC isolate (ST2) and katP was not detected in any of the isolates in this dataset.
VFs associated with the pO157 plasmid were present in our dataset, including the genes ehxA and espP. However. coverage against the pO157 reference was between 45 and 64 % in 41 STEC isolates and one EPEC (Fig. 2). Our analysis also included the pR444_C 95 kb plasmid identified by Cointe et al. [15]. The results did not reveal much variation between STEC clusters, with coverage ranging between 99.5 and 100% with two exceptions, in relation to the reference pR444_C (Fig. 2).
Extra-intestinal VFs
The plasmid-encoded haemolysin, hlyF, is located on the pS88-like plasmid [15, 17, 38] and was detected in most STEC (38/43) and one EPEC (1/4) (total=39/48). Other genes encoded on the pS88-like plasmid, as described by Cointe et al. [15, 17], were investigated. The genes cvaC, cia, ompT and sitA appear to be present in both ST301C1 and ST301C2 (Fig. 2). Conversely, etsC and iucC were present mostly in ST301C1 (Fig. 2). All but one STEC isolate (n=42) and one EPEC isolate had iss (43/48). The gene iss is a serum resistance-associated protein and is also encoded on the pS88 or pS88-like plasmid [17, 39].
The iro gene cluster is responsible for extra-intestinal iron aggregation commonly associated with ExPEC and the genes are encoded on a plasmid [17, 40–42]. The genes iroB, iroC and iroN were detected in most STEC isolates as well as one EPEC (Fig. 2). The presence of iroD was lower, with some STEC isolates below the threshold for detection (Fig. 2, ST2).
The genes cma, mchB, mchC and mchF are associated with microcin, a natural bactericidal antibiotic, with cma suggested to be encoded on a plasmid [15, 38, 43, 44]. The genes mchB, mchC and mchF were detected in all isolates that belonged to ST301C2. In ST301C1, mchB and mchC were present in 15/18 isolates while mchF was present in 17/18. (Fig. 2). The gene cma was detected in 23/48 isolates, mainly ST301C2 (19/21) and four isolates outside the cluster including one EPEC (Fig. 2, ST2).
The presence of a pS88-like plasmid (pR444_A [15]) across CC165 was investigated, with genes that are encoded on the pS88 plasmid being found in most STEC isolates. The majority of ST301C1 had over 90% coverage for a pS88-like plasmid. Variation against the reference pR444_A in ST301C2 was detected, with C2 coverage ranging from 44 to 86% (Fig. 2). The coverage correlating to this plasmid, as well as the presence of genes that are typically encoded on the pS88 plasmid suggest the presence of a pS88-like plasmid in STEC isolates and one EPEC (Fig. 2).
Antimicrobial resistance
Eighty-eight per cent (42/48) of isolates in CC165 were multidrug-resistant (MDR), defined as resistant to three or more antimicrobial classes (Fig. 3). Phenotypic AMR profiling was not conducted, as studies have previously assessed the relationship of genotype and phenotype in non-O157 STEC isolates [27].
Fig. 3.
AMR profiles achieved from the PHE pipeline supplemented with trimethoprim genes dfrA34, dfrA35 and dfrA36 from CGE, annotated on the phylogeny of CC165 in the UK. The blue box indicates a resistance gene was detected. In the case of fluoroquinolone, the blue box indicates reduced susceptibility.
Thirty-nine of 48 (81%) isolates had AMR determinants known to confer resistance to beta-lactams, with 87% (34/39) isolates harbouring blaTEM-1. Forty-two of 48 (88%) isolates had a mutation at position 83[S-L] in gyrA, known to confer reduced susceptibility to ciprofloxacin [27] (fluoroquinolones).
Resistance to aminoglycoside was determined by the presence of strA-strB, ant2′-Ia, aph6-Id or aad variants; 32/48 isolates (67%) held one or more of these determinants. Trimethoprim resistance was present in 40/48 isolates, with dfrA36 being most common (n=25/40, 63%), followed by dfrA-5 (n=15). We noted a striking difference in gene presence between ST301C1 and ST301C2, with C2 predominantly harbouring dfrA36.
There were 36/48 isolates with tetA, with one isolate also harbouring tetM. Forty-one of 48 isolates had sul-1, sul-2 or both, with one isolate also having sul-3. Chloramphenicol resistance was held by 27/48 isolates, attributed to catA-1 (n=3), floR (n=9), catA-1 and floR (n=14), and catA-1, floR and cml-1 (n=1).
Isolate SRR3574263 that had sul1, 2 and 3 also had blaCTX-M-32 , tetA and tetM, the three chloramphenicol genes, dfrA-1 and dfrA36, macrolide resistance ermB, the gyrase mutation and aadA-23, aph6-Id, strA-strB, aadA-3, aac3-IIa and ant2''-Ia, a total of 20 AMR determinants, from the eight antibiotic classes investigated.
Discussion
Although overall the number of cases remains low compared to those infected with STEC O157:H7 (CC11) or STEC O26:H11 (CC29), here we present evidence that strains of STEC belonging to CC165 are emerging as a cause of STEC-HUS in the UK. While this complex is not present at high levels in the UK population, we did see higher levels in certain regions. The majority of cases occurred in London and the South East of England, although this is probably due the higher number of hospitals using commercial PCR for the detection of gastrointestinal pathogens in this region.
CC11 and CC29 are dominated by one serotype, STEC O157:H7 and STEC O26:H11, respectively, whereas CC165 is composed of at least four different STEC serotypes. While STEC O80:H2 was the dominate serotype within CC165, STEC O45:H2, STEC O55:H9, STEC O132:H2 and STEC O180:H2 were also reported in this complex in the UK.
As previously described in other countries, the hospitalization rate and proportion of cases developing HUS in the UK was higher for cases infected with STEC belonging to CC165 compared to STEC O157:H7, although the rate of bloody diarrhoea was lower in CC165 STEC. Symptoms of CC165 STEC that are comparable to STEC O157:H7 were diarrhoea, nausea, abdominal pain and vomiting.
Additionally, the diagnosis of HUS was not exclusive to the O80:H2 serotype, with the serotypes O55:H9 and O45:H2 being associated with the progression of HUS in this dataset. A lower proportion of cases reported bloody diarrhoea compared to cases of STEC O157:H7 and this may confound diagnosis and be a contributing factor in the atypical STEC-HUS presentation described by colleagues elsewhere [13].
With so few cases in the UK to date, it is difficult to determine whether strains belonging to CC165 have become endemic in the UK, or whether they are associated with travellers’ diarrhoea, imported food products or animal migration. Recent data suggest that serogroup O80:H2 has become the second most frequent STEC serogroup isolated in France, after STEC O157:H7 [45, 46], and a subset of UK cases reported travelling to France prior to the onset of symptoms. Similar to this dataset, STEC isolates belonging to CC165, such as serotype O80:H2, O55:H9 and O45:H2 that harbour extra-intestinal virulence factors, have caused gastrointestinal symptoms in humans in Switzerland [47, 48], the Netherlands [13, 49] and Italy [49], supporting the conclusion that this hybrid pathotype is not limited to one serotype.
Analysis of recent exposures from animal contact and outdoor activities did not reveal any common exposures, but there was reported contact with animals and farmland, which has also been reported in STEC O157:H7 over the past 10 years. STEC O80:H2 has been isolated from cattle in Spain [30] and Belgium [16]. These data suggest that like STEC O157:H7 and STEC O26:H11, CC165 STEC has a zoonotic reservoir, and is likely to be transmitted to humans via the food chain and/or contact with animals. To date, there have been very few cattle surveys screening for non-O157 STEC in the UK. Monitoring the prevalence of CC165 STEC along with other non-O157 in cattle and other ruminants would facilitate our understanding of the source and route of transmission in the UK. The prevalence and severity of this serogroup make it essential to establish effective typing technology used for monitoring the transmission of CC165 STEC strains throughout the food chain and environment.
Unusually for STEC, isolates belonging to CC165 are characteristically MDR with the majority of isolates exhibiting resistance to eight different classes of antibiotic [27, 50]. Although antibiotic treatment of STEC is contraindicated because certain antibiotics may induce toxin release, which enhances the risk of progression to HUS, patients are often treated when they first present to healthcare centres because the clinical picture often mimics sepsis (diarrhoea, confusion, hypotension, leukocytosis and acute renal failure). Moreover, there is an argument that once HUS has developed, antimicrobial therapy may shorten the course of illness and improve clinical outcomes [51, 52].
In 2020, a study by Gentle et al., [27] investigated the correlation between genotypic prediction and phenotypic expression of AMR in STEC. The authors noted a discrepancy between isolates of CC165 that were phenotypically resistant to trimethoprim but had no AMR genes known to confer resistance to trimethoprim. These isolates were included in this study and we detected dfrA36. This gene, known to confer resistance to trimethoprim, was first characterized by Wüthrich et al. [53] in 2019, but it first appeared in this dataset in 2013 and has been circulating in the population since.
Azithromycin is an antimicrobial used for STEC treatment when appropriate, but the high levels of macrolide resistance within this dataset suggest this treatment would not be effective. The high levels of resistance in CC165 are concerning, particularly relating to the emergence of previously undetected genes, and should treatment be deemed appropriate for STEC infections, it is important that clinicians are aware of the MDR profile of strains belonging to CC165.
The majority of UK isolates in CC165 belonged to STEC O80:H2 and, as described in previous studies in France, Spain and the Netherlands, had stx2d and eae. Strains of STEC harbouring both stx2d and eae are rare in the UK but have been associated with severe clinical outcomes, including HUS, elsewhere [54, 55]. STEC O80:H2 has been described as causing severe extra-renal manifestations such as protracted seizures, myocardial damage, transaminitis, bacteraemia and peritonitis [12–14]. The rate of hospitalization in cases in England from this study was 40%, with 25% of cases having complications of HUS, but the drawback of this study is that there was lack of detailed clinical data and outcomes. We also detected STEC O80:H2 harbouring both stx2a and eae, which is the virulence gene combination most commonly seen in non-O157 STEC-causing HUS in the UK (https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/732577/non-O157_STEC_Evidence_Base.pdf). Previous studies have shown that the presence of stx2a and stx2d is significantly associated with severe clinical manifestations [56].
Like STEC O157:H7 and STEC O26:H11, two other STEC serotypes that cause HUS, isolates in this study also had a combination of non-LEE-encoding effectors and ehxA. In contrast to other STEC serotypes, they also carried a suite of virulence factors encoded on a pS88-like plasmid more commonly found in extra-intestinal E. coli that may facilitate survival outside the gut. The pO157 plasmid was not found in our isolates, although two VFs (ehxA and espP) characteristic of pO157 were detected, demonstrating the dynamic nature of genes on mobile genetic elements. By integrating the virulence gene profile with the phylogeny, we observed loss and gain of genes within the CC165 population. Specifically, acquisition of the stx-encoding phage corresponded to the loss of phage-encoded espJ and cif, where three of the four EPEC harboured espJ and cif. We also observed the loss or gain of individual plasmid-encoded genes, rather than the loss of the entire plasmid. We theorize that this is due to either two separate acquisition events of two highly similar plasmids into the STEC cluster, or a selective pressure from the environment that induced gene loss between the STEC subclusters, ST301C1 and ST301C2.
Here, we present the emergence of this new clonal complex in the UK. There is evidence of domestically acquired infection as the majority of cases do not report travel outside the UK, but it is not clear whether the source is imported food or whether STEC belonging to CC165 is endemic in the UK cattle population. There is also evidence of the pathogenic potential of this clonal complex and that clinical management may be challenging due to the complex presentation of patients and the multidrug resistance exhibited by the pathogen. The lack of comprehensive microbiological and/or epidemiological surveillance of STEC other than serotype O157:H7 in the UK continues to be a concern. There is a need to expand the implementation of methods capable of detecting all STEC serotypes, specifically PCR targeting stx at the local hospital level. Studies focusing on the detection of STEC O157:H7 from food and animal samples should be extended to include all STEC to better understand the zoonotic reservoir and transmission routes of this pathogen.
Supplementary Data
Funding information
This study was funded by the National Institute for Health Research (NIHR) Health Protection Research Unit in Gastrointestinal Infections, a partnership between Public Health England, the University of Liverpool and the University of Warwick. The views expressed are those of the author(s) and not necessarily those of the NIHR, Public Health England or the Department of Health and Social Care.
Author contributions
Conceptualization: T.J.D., C.J. Methodology: E.V.R., T.J.D., C.J. Software: E.V.R., T.J.D. Validation: E.V.R. Formal Analysis: E.V.R., B.V. Investigation: E.V.R. Resources: R.S., L.B., A.S.P., L.A., A.H., T.J.D., C.J. Data Curation: T.J.D., C.J. Writing – Original Draft Preparation: E.V.R., C.J. Writing – Review and Editing: E.V.R., B.V., R.S., L.B., A.S.P., L.A., A.H., L.B., G.G., N.M., T.J.D., C.J. Visualization: E.V.R., B.V. Supervision: G.G., L.B., N.M., T.J.D., C.J. Project Administration: N.M., T.J.D., C.J. Funding: N.M., T.J.D., C.J.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AMR, antimicrobial resistance; CC, clonal complex; CGE, Centre for Genomic Epidemiology; CH, coverage and homology; EPEC, enteropathogenic E. coli; ESQ, enhanced surveillance questionnaire; ExPEC, extra-intestinal pathogenic E. coli; GBRU, Gastrointestinal Bacteria Reference Unit; HUS, haemolytic uraemic syndrome; LEE, locus of enterocyte effacement; MDR, multi-drug-resistant; MLST, multi-locus sequence typing; PHE, Public Health England; SERL, Scottish E. coli Reference Laboratory; ST, sequence type; STEC, shiga toxin-producing Escherichia coli; T3SS, type 3 secretion system; VF, virulence factor; WGS, whole genome sequencing.
One supplementary table and three supplementary figures are available with the online version of this article.
References
- 1.Gasser C, Gautier E, Steck A, Siebenmann RE, Oechslin R. Hemolytic-uremic syndrome: bilateral necrosis of the renal cortex in acute acquired hemolytic anemia. Schweiz Med Wochenschr. 1955;85:905–909. [PubMed] [Google Scholar]
- 2.Karmali MA, Petric M, Lim C, Fleming PC, Arbus GS, et al. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli . J Infect Dis. 1985;151:775–782. doi: 10.1093/infdis/151.5.775. [DOI] [PubMed] [Google Scholar]
- 3.Sheerin NS, Glover E. Haemolytic uremic syndrome: diagnosis and management. F1000Res. 2019;8:F1000 Faculty Rev-1690. doi: 10.12688/f1000research.19957.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell BP. A multistate outbreak of Escherichia coli O157:H7-associated bloody diarrhea and hemolytic uremic syndrome from hamburgers. The Washington experience. JAMA. 1994;272:1349–1353. doi: 10.1001/jama.272.17.1349. [DOI] [PubMed] [Google Scholar]
- 5.Pennington TH. E. coli O157 outbreaks in the United Kingdom: past, present, and future. Infect Drug Resist. 2014;7:211–222. doi: 10.2147/IDR.S49081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams NL, Byrne L, Smith GA, Elson R, Harris JP, et al. Shiga Toxin-Producing Escherichia coli O157, England and Wales, 1983-2012. Emerg Infect Dis. 2016;22:590–597. doi: 10.3201/eid2204.151485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.EFSA BIOHAZ Panel. Koutsoumanis K, Allende A, Alvarez‐Ordóñez A, Bover‐Cid S, et al. Pathogenicity assessment of Shiga toxin‐producing Escherichia coli (STEC) and the public health risk posed by contamination of food with STEC. EFS2. 2020;18:e05967. doi: 10.2903/j.efsa.2020.5967. [DOI] [Google Scholar]
- 8.Stevens MP, Frankel GM. The locus of enterocyte effacement and associated virulence factors of enterohemorrhagic Escherichia coli . Microbiol Spectr. 2014;2:EHEC–0007. doi: 10.1128/microbiolspec.EHEC-0007-2013. [DOI] [PubMed] [Google Scholar]
- 9.Donnenberg MS, Tzipori S, McKee ML, O’Brien AD, Alroy J, et al. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J Clin Invest. 1993;92:1418–1424. doi: 10.1172/JCI116718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cepeda-Molero M, Berger CN, Walsham ADS, Ellis SJ, Wemyss-Holden S, et al. Attaching and effacing (A/E) lesion formation by enteropathogenic E. coli on human intestinal mucosa is dependent on non-LEE effectors. PLoS Pathog. 2017;13:e1006706. doi: 10.1371/journal.ppat.1006706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt H, Beutin L, Karch H. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect Immun. 1995;63:1055–1061. doi: 10.1128/iai.63.3.1055-1061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soysal N, Mariani-Kurkdjian P, Smail Y, Liguori S, Gouali M, et al. Enterohemorrhagic Escherichia coli hybrid pathotype O80:H2 as a new therapeutic challenge. Emerg Infect Dis. 2016;22:1604–1612. doi: 10.3201/eid2209.160304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wijnsma KL, Schijvens AM, Rossen JWA, Kooistra-Smid AMDM, Schreuder MF, et al. Unusual severe case of hemolytic uremic syndrome due to Shiga toxin 2d-producing E. coli O80:H2. Pediatr Nephrol. 2017;32:1263–1268. doi: 10.1007/s00467-017-3642-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mariani-Kurkdjian P, Lemaître C, Bidet P, Perez D, Boggini L, et al. Haemolytic-uraemic syndrome with bacteraemia caused by a new hybrid Escherichia coli pathotype. New Microbes New Infect. 2014:127–131. doi: 10.1002/nmi2.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cointe A, Birgy A, Mariani-Kurkdjian P, Liguori S, Courroux C, et al. Emerging multidrug-resistant hybrid pathotype shiga toxin-producing Escherichia coli O80 and related strains of clonal complex 165, Europe. Emerg Infect Dis. 2018;24:2262–2269. doi: 10.3201/eid2412.180272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Rauw K, Thiry D, Caljon B, Saulmont M, Mainil J, et al. Characteristics of Shiga toxin producing- and enteropathogenic Escherichia coli of the emerging serotype O80:H2 isolated from humans and diarrhoeic calves in Belgium. Clin Microbiol Infect. 2019;25:e5-111.e8. doi: 10.1016/j.cmi.2018.07.023. [DOI] [PubMed] [Google Scholar]
- 17.Peigne C, Bidet P, Mahjoub-Messai F, Plainvert C, Barbe V, et al. The plasmid of Escherichia coli strain S88 (O45:K1:H7) that causes neonatal meningitis is closely related to avian pathogenic E. coli plasmids and is associated with high-level bacteremia in a neonatal rat meningitis model. Infect Immun. 2009;77:2272–2284. doi: 10.1128/IAI.01333-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkins C, Ling CL, Ciesielczuk HL, Lockwood J, Hopkins S, et al. Detection and identification of bacteria in clinical samples by 16S rRNA gene sequencing: comparison of two different approaches in clinical practice. J Med Microbiol. 2012;61:483–488. doi: 10.1099/jmm.0.030387-0. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins C, Perry NT, Godbole G, Gharbia S. Evaluation of chromogenic selective agar (CHROMagar STEC) for the direct detection of Shiga toxin-producing Escherichia coli from faecal specimens. J Med Microbiol. 2020;69:487–491. doi: 10.1099/jmm.0.001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dallman TJ, Greig DR, Gharbia SE, Jenkins C. Phylogenetic context of Shiga toxin-producing Escherichia coli serotype O26:H11 in England. Microb Genom. 2019 doi: 10.1099/mgen.0.000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dallman T, Ashton P, Schafer U, Jironkin A, Painset A, et al. SnapperDB: a database solution for routine sequencing analysis of bacterial isolates. Bioinformatics. 2018;34:3028–3029. doi: 10.1093/bioinformatics/bty212. [DOI] [PubMed] [Google Scholar]
- 22.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, et al. Corrigendum to: IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:2461. doi: 10.1093/molbev/msaa131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashton PM, Perry N, Ellis R, Petrovska L, Wain J, et al. Insight into Shiga toxin genes encoded by Escherichia coli O157 from whole genome sequencing. PeerJ. 2015;3:e739. doi: 10.7717/peerj.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chattaway MA, Dallman TJ, Gentle A, Wright MJ, Long SE, et al. Whole genome sequencing for public health surveillance of shiga toxin-producing Escherichia coli other than serogroup O157. Front Microbiol. 2016;7:258. doi: 10.3389/fmicb.2016.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gentle A, Day MR, Hopkins KL, Godbole G, Jenkins C. Antimicrobial resistance in Shiga toxin-producing Escherichia coli other than serotype O157 : H7 in England, 2014-2016. J Med Microbiol. 2020;69:379–386. doi: 10.1099/jmm.0.001146. [DOI] [PubMed] [Google Scholar]
- 28.Burland V, Shao Y, Perna NT, Plunkett G, Sofia HJ, et al. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 1998;26:4196–4204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:11–90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanco M, Blanco JE, Mora A, Dahbi G, Alonso MP, et al. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eae-xi) J Clin Microbiol. 2004;42:645–651. doi: 10.1128/JCM.42.2.645-651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaytán MO, Martínez-Santos VI, Soto E, González-Pedrajo B. Type three secretion system in attaching and effacing pathogens. Front Cell Infect Microbiol. 2016;6:129. doi: 10.3389/fcimb.2016.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elliott SJ, Sperandio V, Girón JA, Shin S, Mellies JL, et al. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli . Infect Immun. 2000;68:6115–6126. doi: 10.1128/IAI.68.11.6115-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong ARC, Pearson JS, Bright MD, Munera D, Robinson KS, et al. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol Microbiol. 2011;80:1420–1438. doi: 10.1111/j.1365-2958.2011.07661.x. [DOI] [PubMed] [Google Scholar]
- 34.Brunder W, Schmidt H, Karch H. KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7. Microbiology (Reading) 1996;142 (Pt 11):3305–3315. doi: 10.1099/13500872-142-11-3305. [DOI] [PubMed] [Google Scholar]
- 35.Brunder W, Schmidt H, Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol Microbiol. 1997;24:767–778. doi: 10.1046/j.1365-2958.1997.3871751.x. [DOI] [PubMed] [Google Scholar]
- 36.Makino K, Ishii K, Yasunaga T, Hattori M, Yokoyama K, et al. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res. 1998;5:1–9. doi: 10.1093/dnares/5.1.1. [DOI] [PubMed] [Google Scholar]
- 37.Tatsuno I, Horie M, Abe H, Miki T, Makino K, et al. toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect Immun. 2001;69:6660–6669. doi: 10.1128/IAI.69.11.6660-6669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson TJ, Johnson SJ, Nolan LK. Complete DNA sequence of a ColBM plasmid from avian pathogenic Escherichia coli suggests that it evolved from closely related ColV virulence plasmids. J Bacteriol. 2006;188:5975–5983. doi: 10.1128/JB.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cyoia PS, Rodrigues GR, Nishio EK, Medeiros LP, Koga VL, et al. Presence of virulence genes and pathogenicity islands in extraintestinal pathogenic Escherichia coli isolates from Brazil. J Infect Dev Ctries. 2015;9:1068–1075. doi: 10.3855/jidc.6683. [DOI] [PubMed] [Google Scholar]
- 40.Sorsa LJ, Dufke S, Heesemann J, Schubert S. Characterization of an iroBCDEN gene cluster on a transmissible plasmid of uropathogenic Escherichia coli: evidence for horizontal transfer of a chromosomal virulence factor. Infect Immun. 2003;71:3285–3293. doi: 10.1128/IAI.71.6.3285-3293.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caza M, Lépine F, Milot S, Dozois CM. Specific roles of the iroBCDEN genes in virulence of an avian pathogenic Escherichia coli O78 strain and in production of salmochelins. Infect Immun. 2008;76:3539–3549. doi: 10.1128/IAI.00455-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feldmann F, Sorsa LJ, Hildinger K, Schubert S. The salmochelin siderophore receptor IroN contributes to invasion of urothelial cells by extraintestinal pathogenic Escherichia coli in vitro . Infect Immun. 2007;75:3183–3187. doi: 10.1128/IAI.00656-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baquero F, Lanza VF, Baquero M-R, Del Campo R, Bravo-Vázquez DA. Microcins in enterobacteriaceae: peptide antimicrobials in the eco-active intestinal chemosphere. Front Microbiol. 2019;10:2261. doi: 10.3389/fmicb.2019.02261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodríguez E, Gaggero C, Laviña M. The structural gene for microcin H47 encodes a peptide precursor with antibiotic activity. Antimicrob Agents Chemother. 1999;43:2176–2182. doi: 10.1128/AAC.43.9.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ingelbeen B, Bruyand M, Mariani-Kurkjian P, Le Hello S, Danis K, et al. Emerging Shiga-toxin-producing Escherichia coli serogroup O80 associated hemolytic and uremic syndrome in France, 2013-2016: Differences with other serogroups. PLoS One. 2018;13:e0207492. doi: 10.1371/journal.pone.0207492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruyand M, Mariani-Kurkdjian P, Le Hello S, King L-A, Van Cauteren D, et al. Paediatric haemolytic uraemic syndrome related to Shiga toxin-producing Escherichia coli, an overview of 10 years of surveillance in France, 2007 to 2016. Euro Surveill. 2019;24 doi: 10.2807/1560-7917.ES.2019.24.8.1800068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fierz L, Cernela N, Hauser E, Nüesch-Inderbinen M, Stephan R. Characteristics of shigatoxin-producing Escherichia coli strains isolated during 2010-2014 from human infections in Switzerland. Front Microbiol. 2017;8:1471. doi: 10.3389/fmicb.2017.01471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nüesch-Inderbinen M, Cernela N, Wüthrich D, Egli A, Stephan R. Genetic characterization of Shiga toxin producing Escherichia coli belonging to the emerging hybrid pathotype O80:H2 isolated from humans 2010-2017 in Switzerland. Int J Med Microbiol. 2018;308:534–538. doi: 10.1016/j.ijmm.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 49.Gigliucci F, van Hoek AHAM, Chiani P, Knijn A, Minelli F, et al. Genomic characterization of hlyF -positive Shiga toxin–producing Escherichia coli, Italy and the Netherlands, 2000–2019. Emerg Infect Dis. 2021;27:853–861. doi: 10.3201/eid2703.203110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Day M, Doumith M, Jenkins C, Dallman TJ, Hopkins KL, et al. Antimicrobial resistance in Shiga toxin-producing Escherichia coli serogroups O157 and O26 isolated from human cases of diarrhoeal disease in England, 2015. J Antimicrob Chemother. 2017;72:145–152. doi: 10.1093/jac/dkw371. [DOI] [PubMed] [Google Scholar]
- 51.Menne J, Nitschke M, Stingele R, Abu-Tair M, Beneke J, et al. Validation of treatment strategies for enterohaemorrhagic Escherichia coli O104:H4 induced haemolytic uraemic syndrome: case-control study. BMJ. 2012;345 doi: 10.1136/bmj.e4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nitschke M, Sayk F, Härtel C, Roseland RT, Hauswaldt S, et al. Association between azithromycin therapy and duration of bacterial shedding among patients with Shiga toxin-producing enteroaggregative Escherichia coli O104:H4. JAMA. 2012;307:1046–1052. doi: 10.1001/jama.2012.264. [DOI] [PubMed] [Google Scholar]
- 53.Wüthrich D, Brilhante M, Hausherr A, Becker J, Meylan M, et al. A novel trimethoprim resistance gene, dfrA36, characterized from Escherichia coli from calves. mSphere. 2019;4 doi: 10.1128/mSphere.00255-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bielaszewska M, Friedrich AW, Aldick T, Schürk-Bulgrin R, Karch H. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin Infect Dis. 2006;43:1160–1167. doi: 10.1086/508195. [DOI] [PubMed] [Google Scholar]
- 55.Delannoy S, Mariani-Kurkdjian P, Bonacorsi S, Liguori S, Fach P. Characteristics of emerging human-pathogenic Escherichia coli O26:H11 strains isolated in France between 2010 and 2013 and carrying the stx2d gene only. J Clin Microbiol. 2015;53:486–492. doi: 10.1128/JCM.02290-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Byrne L, Adams N, Jenkins C. Association between shiga toxin-producing Escherichia coli O157:H7 stx gene subtype and disease severity, England, 2009-2019. Emerg Infect Dis. 2020;26:2394–2400. doi: 10.3201/eid2610.200319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.