Abstract
Emerging SARS-CoV-2 variants of concern (VOC) have been associated with enhanced transmissibility and immune escape. Next-generation sequencing (NGS) of the whole genome is the gold standard for variant identification for surveillance but is time-consuming and costly. Rapid and cost-effective assays that detect SARS-CoV-2 variants are needed. We evaluated Allplex SARS-CoV-2 Master Assay and Variants I Assay to detect HV69/70 deletion, Y144 deletion, E484K, N501Y, and P681H spike mutations in 248 positive samples collected in Kuala Lumpur, Malaysia, between January and May 2021. Spike variants were detected in 78/248 (31.5 %), comprising 60 VOC B.1.351 (beta) and 18 B.1.1.7 (alpha). With NGS as reference for 115 samples, the sensitivity for detecting the spike mutations was 98.7 % with the Master Assay and 100 % with the Variants I Assay. The emergence of beta variants correlated with increasing COVID-19 infections in Malaysia. The prevalence of alpha VOC and lineage B.1.466.2 was low. These assays detect mutations present in alpha, beta and gamma VOCs. Of the VOCs which have subsequently emerged, the assays should detect omicron (B.1.1.529) but not B.1.617.2 (delta). In conclusion, spike variant PCR assays can be used to rapidly monitor selected SARS-CoV-2 VOCs in resource-limited settings, but require updates as new variants emerge.
Keywords: COVID-19, SARS-CoV-2, Next-generation sequencing, RT-qPCR, Variants of concern, Malaysia
1. Introduction
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants pose an increased risk to global public health. The World Health Organization (WHO) has identified variants of interest (VOIs) with genetic changes that may lead to changes in virus transmission, disease severity, immune escape, diagnostic and treatment failure. Variants of concern (VOC) have evidence of increased virus transmission or virus virulence, or reduced effectiveness of public health measures, diagnostics, vaccines and therapeutics (World Health Organization, 2021). To date, WHO has designated four VOCs: B.1.1.7, B.1.351, P.1 and B.1.617.2 (also known as alpha, beta, gamma and delta variants, respectively). Notable spike (S) amino acid mutations are: HV69/70 deletion and P681H in B.1.1.7, K417N in B.1.351, and K417T in P.1. E484K and N501Y mutations are found in B.1.1.7, B.1.351 and P.1 (Janik et al., 2021). The most recent VOC B.1.617.2 contains L452R and P681R mutations (Planas et al., 2021). These S mutations could significantly change transmission dynamics as they increase transmissibility and facilitate immune escape (Sabino et al., 2021). Therefore, prompt variant monitoring is crucial to control outbreak spread.
Whole genome sequencing with next-generation sequencing (NGS) is the gold standard for detection of VOCs, but is less ideal in resource-limited settings as it is time-consuming, costly and requires extensive data processing (Armstrong et al., 2019). Direct PCR-based variant analysis of SARS-CoV-2 is rapid, inexpensive and could be utilised as an alternative to NGS (Kami et al., 2021). In this study, we investigated the emergence of VOCs in Malaysia using two multiplex RT-PCR commercial assays to identify VOC-defining mutations, with further validation using NGS.
2. Material and methods
We retrospectively examined 248 nasopharyngeal/oropharyngeal swabs (Ct values 12-39) from randomly selected COVID-19-positive patients admitted to University Malaya Medical Center (UMMC), a major teaching hospital covering the capital Kuala Lumpur and Selangor state, between 4 January and 28 May 2021. COVID-19 diagnosis was confirmed by real-time PCR using the WHO-recommended Berlin Charité protocol (Corman et al., 2020) and abTES COVID-19 qPCR I Kit (AITbiotech, Singapore). This study was approved by the institutional medical ethics committee (no. 2020730–8928). Informed consent for studies of archived and anonymised samples was not required.
Viral RNA was tested using Allplex SARS-CoV-2 Master Assay (Seegene, South Korea) that screens for SARS-CoV-2 envelope (E), nucleoprotein (N), RNA-dependent RNA polymerase (RdRp), S gene, and S mutations (any of the HV69/70 deletion, Y144 deletion, E484K, N501Y, and P681H) following manufacturer's instructions. Samples identified with S mutations and 38 non-variant samples (selected randomly) were then subjected to Allplex SARS-CoV-2 Variants I Assay (Seegene, South Korea) that detects SARS-CoV-2 RdRp gene, HV69/70 deletion, E484K and N501Y mutations. PCR was performed with a CFX96 Touch Real-Time PCR Detection System (Bio-Rad, USA) and results were analysed with Seegene Viewer V1.0 software (Seegene). The results allow the provisional assignment of cases as possible VOCs (B.1.1.7 or B.1.351/P.1) or VOIs (B.1.525 or B.1.526).
For validation, the 77 identified spike variants and 38 non-variant samples were subjected to NGS following the ARTIC nCoV-2019 sequencing protocol V3 (Quick, 2020), making a total of 115 samples sequenced. Briefly, reverse transcription of viral RNA was performed using First Strand Synthesis System (Invitrogen, USA) with random hexamers. Whole genome amplification was conducted using two pools of nCoV-2019/V3 primer sets (Quick, 2020; Chong et al., 2020). Libraries were prepared and sequencing was performed using MinION Mk1C (Nanopore, United Kingdom). The cut-off was set to Q30 to trim the sequence read using BBDuk plugin in Geneious Prime 2020 (Biomatters, New Zealand). After mapping to the reference genome Wuhan-Hu-1 (GenBank accession number MN908947), consensus sequences were obtained using Geneious Prime 2020. Lineage groups were determined using the Phylogenetic Assignment of Named Global Outbreak (PANGO) Lineages (www.pangolin.cog-uk.io). All generated sequences were deposited into GISAID. The epidemiology of the VOCs was analysed with other complete genome sequences from Malaysia available in the GISAID database (www.gisaid.org) as of 25 July 2021. Malaysian COVID-19 case numbers were obtained from the Ministry of Health (https://kpkesihatan.com/).
3. Results
All 248 cases were confirmed positive for SARS-CoV-2 E, N, RdRp, and S gene, and 77 (31.0 %) of these samples were identified as spike variants with the Allplex SARS-CoV-2 Master Assay (Table 1 ). These 77 spike variants and 38 randomly selected samples which were negative for spike variants (total n = 115) were then tested using Allplex SARS-CoV-2 Variants I Assay and NGS. All 77 spike variants had detectable spike mutations using the Variants I Assay, comprising 18 with HV69/70 deletion and N501Y mutation confirmed as lineage B.1.1.7 using NGS, and 59 positive for E484K and N501Y mutations confirmed as B.1.351 by NGS (Table 1). One of the 38 samples testing negative for spike variants with the Master Assay was also found to be B.1.351 using the Variants I Assay and NGS. With the Master Assay, Ct values of the E/N/RdRp and S variants were very similar with average deviation of 1.2 cycles. Similarly, the Variants I Assay showed similar Ct values of RdRp and HV69/70 deletion/E484K/N501Y, with average deviation of 1.0 cycles. This reflects the high consistency of both assays.
Table 1.
Proportions of variants of concern (VOCs) and non-VOC defined by Allplex assays and NGS of samples from 4 January to 28 May 2021.
| Months | Allplex Master Assay (n = 248) |
Allplex Variants I Assay + NGS (n = 115) |
||
|---|---|---|---|---|
| VOC | Non-VOC | HV69/70 del + N501Y (B.1.1.7) | E484 K + N501Y (B.1.351) | |
| January | 0/22 (0 %) | 22/22 (100 %) | 0/22 (0 %) | 0/22 (0 %) |
| February | 3/30 (10 %) | 27/30 (90 %) | 0/3 (0 %) | 3/3 (100 %) |
| March | 3/46 (6.5 %) | 43/46 (93.5 %) | 2/3 (66.7 %) | 1/3 (33.3 %) |
| April | 68/145 (46.9 %) | 77/145 (53.1 %) | 16/82 (19.5 %) | *53/82 (64.6 %) |
| May | 3/5 (60.0 %) | 2/5 (40.0 %) | 0/5 (0 %) | 3/5 (60.0 %) |
| Total | 77/248 (31.0 %) | 171/248 (69.0 %) | 18/115 (15.7 %) | 60/115 (52.2 %) |
An additional B.1.351 case was detected with the Allplex Variants I Assay and NGS, but missed with the Master Assay.
NGS showed that all 115 samples sequenced had 94.8–99.6 % similarity to the reference genome with mean coverage ranging between 18 to 1986, and the PANGO lineages of all consensus sequences could be determined. NGS confirmed the 37 non-variant samples testing negative for spike variants with both the Master and Variants I Assay, and identified lineages B.1 (1, 2.7 %), B.1.221 (1, 2.7 %), B.1.466.2 (13, 35.1 %), and B.1.524 (22, 59.5 %). Of note, the 242–244 deletion in the spike gene of B.1.351 samples overlapped with forward primer 74 from the nCoV-2019/V3 primer sets from ARTIC, resulting in very low coverage. Sequences (n = 107) with at least 98.6 % similarity to the reference genome and mean coverage above 63 were submitted to GISAID with accession numbers EPI_ISL_3246352-EPI_ISL_3246430, EPI_ISL_3492535, EPI_ISL_2769407- EPI_ISL_1769413, EPI_ISL_2769415- EPI_ISL_2769423, EPI_ISL_2769425- EPI_ISL_2769427, EPI_ISL_2769436, EPI_ISL_4056144- EPI_ISL_4056146, EPI_ISL_4056163, EPI_ISL_4056167, and EPI_ISL_2784329. Overall, the sensitivity for detecting SARS-CoV-2 was 100 % with the Master Assay (248/248) and Variants I Assay (115/115) compared to the reference Berlin Charité and abTES COVID-19 qPCR I Kit initially used to screen samples. There was a total of 78 samples with the specified spike mutations confirmed by NGS, and sensitivity for detecting these was 98.7 % (77/78) with the Master Assay (which missed a single B.1.351 sample with a high Ct value of 35.62) and 100 % (78/78) with the Variants I Assay. The specificity for both assays are 100 % (37/37). Overall, the Allplex assays were highly sensitive.
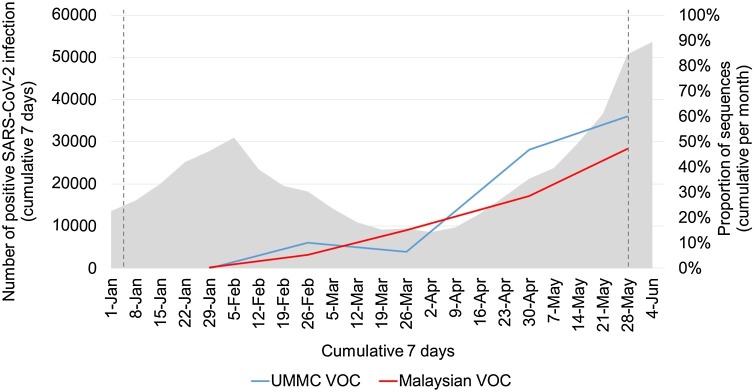
Next, to determine the prevalence of VOC, we plotted the relative proportions of VOC detected in UMMC and from reported Malaysian SARS-CoV-2 sequences (including B.1.617.2) in GISAID since January 2021 (Fig. 1 ). The proportion of VOC increased over time, to 60.0 % in UMMC and 47.2 % in Malaysia by end of May 2021, accompanied by a steep rise in nationwide cases. GISAID data showed B.1.1.7 prevalence was low from January to April, ranging between 0.4 % to 1.9 %, and was not detected in May. B.1.351 made up the majority of VOCs in Malaysia, rapidly increasing from 4.6 % in February to 35.8 % in May 2021. VOC P.1 was not reported in Malaysia. This national VOC data is broadly similar to our UMMC data. Although we did not detect B.1.617.2 in UMMC during the study period, nationally a low rate was detected in April (0.8 %) which increased to 11.4 % in May.
Fig. 1.
Daily reported infections (grey) in Malaysia and proportions of SARS-CoV-2 VOC in Malaysian sequences from UMMC (blue) and GISAID (red). UMMC data is from 4 January to 28 May 2021 and the period covered by this study is shown by the dotted lines. GISAID data is from 1 January to 28 May 2021. Genomes were classified as either VOC (B.1.1.7, B.1.351, B.1.617.2) or non-VOC (all other lineages). VOC P.1 was not reported in Malaysia during this period. UMMC, University Malaya Medical Centre; GISAID, Global initiative in sharing all influenza data; VOC, variant of concern.
4. Discussion
The B.1.1.7 VOC emerged in September 2020 in the UK while B.1.351 emerged later in December 2020 in South Africa. Since then, both variants have quickly spread to multiple countries (Galloway et al., 2021; Pearson et al., 2021). This study was carried out during the emergence of B.1.1.7 and B.1.351 in the capital Kuala Lumpur and surrounding state Selangor in early 2021. These were two of the worst-affected regions, which together contributed 39 % of infections reported in Malaysia during the study period. As UMMC serves Kuala Lumpur and Selangor, we showed that our cohort could reflect the VOC proportions circulating in Malaysia.
Malaysia first reported B.1.1.7 on 28 December 2020, from a traveller returning from UK (Ong, 2021), and B.1.351 in early February 2021 (Ministry of Health Malaysia, 2021a). B.1.1.7 prevalence remained low, while B.1.351 has been replacing other lineages since February 2021 and became the predominant VOC in both UMMC (Table 1) and reported SARS-CoV-2 sequences in Malaysia (GISAID). The emergence and establishment of B.1.351 in the presence of B.1.1.7 is suggestive of natural selection of a more transmissible virus (Adam, 2021), which likely explains the steep increase in overall infections since early April 2021. Another notable variant under alerts for further monitoring by WHO, B.1.466.2, was mainly reported in east Malaysia (Sam et al., 2021). As we also confirmed B.1.466.2 in 13/37 non-variant cases in UMMC, this suggests that this lineage may have contributed to new infections along with B.1.351.
Genomic surveillance and rapid variant detection are important to understand local epidemiology. We showed that the Allpex SARS-CoV-2 Master and Variants I assays have high sensitivity in detecting selected spike mutations present in VOCs including B.1.1.7 and B.1.351, which is comparable to current diagnostic screening assays of known VOCs (Vogels et al., 2021; Volz et al., 2021). However, new important variants such as B.1.617.1 (kappa), B.1.617.2 (delta) and B.1.617.3 have subsequently emerged, with mutations such as L452R and P681R that are not detected by these assays, necessitating additional panels. The VOC B.1.617.2 was first reported in Malaysia in late April (CodeBlue, 2021), and this is a more transmissible variant than B.1.1.7 and B.1.351 which has rapidly driven new waves in countries such as USA and India (Adam, 2021). More recently, the newly described VOC B.1.1.529 (omicron) was first reported in Malaysia in November 2021 (Ministry of Health Malaysia, 2021b). As omicron carries the spike mutations HV69/70 deletion, Y144 deletion, N501Y, and P681H, both Master and Variants I assays should detect it. In resource-limited settings, these multiplex variant PCR assays are useful as rapid and relatively cheaper screening tools to identify samples for further NGS. However, manufacturers need to be responsive to emergence of key new variants, and NGS continues to be the gold standard.
Author statement
Jolene Yin Ling Fu: Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing - Original draft. Yoong Min Chong: Conceptualization, Formal analysis, Investigation, Methodology, Visualization. I-Ching Sam: Conceptualization, Investigation, Resources, Writing – review & editing. Yoke Fun Chan: Conceptualization, Supervision, Resources, Writing – review & editing.
Research funding
This study was funded by Seegene Inc, South Korea (grant number IF050-2021), and the Fundamental Research Grant Scheme from the Ministry of Education Malaysia (grant number FRGS/1/2020/SKK0/UM/02/5). The funders had no role in study design, collection, analysis and interpretation of data, writing of the report, and the decision to submit the article for publication.
Declaration of Competing Interest
This study was part-funded by a grant to YFC and ICS from Seegene Inc, South Korea.
Acknowledgements
None.
Data availability
Data will be made available on request.
References
- Adam What scientists know about new, fast-spreading coronavirus variants. Nature. 2021;594(7861):19–21. doi: 10.1038/d41586-021-01390-4. [DOI] [PubMed] [Google Scholar]
- Armstrong, et al. Pathogen genomics in public health. N. Engl. J. Med. 2019;381(26):2569–2580. doi: 10.1056/NEJMsr1813907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong Y.M., et al. SARS-CoV-2 lineage B.6 was the major contributor to early pandemic transmission in Malaysia. PLoS Negl. Trop. Dis. 2020;14(11) doi: 10.1371/journal.pntd.0008744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CodeBlue . 2021. Malaysia Reports First Local Covid-19 Case with Indian Variant.https://codeblue.galencentre.org/2021/05/17/malaysia-reports-first-local-covid-19-case-with-indian-variant/ (Accessed 25 December 2021) [Google Scholar]
- Corman, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway, et al. Emergence of SARS-CoV-2 B.1.1.7 lineage - United States, December 29, 2020-January 12, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70(3):95–99. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janik, et al. The emerging concern and interest SARS-CoV-2 variants. Pathogens. 2021;10(6):633. doi: 10.3390/pathogens10060633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami, et al. Rapid and simultaneous identification of three mutations by the Novaplex SARS-CoV-2 variants I assay kit. J. Clin. Virol. 2021;141 doi: 10.1016/j.jcv.2021.104877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministry of Health Malaysia . 2021. Situation and Information Update on the COVID-19 Spike Mutation in Malaysia - 1 April 2021.http://covid-19.moh.gov.my/semasa-kkm/2021/04/maklumat-terkini-mutasi-protein-spike-di-malaysia (Accessed 25 December 2021) [Google Scholar]
- Ministry of Health Malaysia . 2021. Current Developments of Omicron’s New Variant (B.1.1.529) and Control and Preventive Measures in Malaysia – 3 December 2021.https://covid-19.moh.gov.my/semasa-kkm/2021/12/kenyataan-media-kkm-perkembangan-semasa-omicron-b11529-dan-langkah-kawalan-dan-pencegahan-03122021 (Accessed 25 December 2021) [Google Scholar]
- Ong . 2021. UK-Variant of Covid-19 has Reached Malaysia, Dr Noor Hisham Confirms.https://www.malaymail.com/news/malaysia/2021/01/11/uk-variant-of-covid-19-has-reached-malaysia-dr-noor-hisham-confirms/1939483 (Accessed 25 December 2021) [Google Scholar]
- Pearson, et al. 2021. Estimates of Severity and Transmissibility of Novel SARS-CoV-2 Variant 501Y.V2 in South Africa.https://cmmid.github.io/topics/covid19/sa-novel-variant.html (Accessed 25 December 2021) [Google Scholar]
- Planas, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596(7871):276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Quick . 2020. nCoV-2019 Sequencing Protocol v3 (LoCost) V.3.https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye (Accessed 25 December 2021) [Google Scholar]
- Sabino, et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397(10273):452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sam, et al. Changing predominant SARS-CoV-2 lineages drives successive COVID-19 waves in Malaysia, February 2020 to March 2021. J. Med. Virol. 2021 doi: 10.1002/jmv.27441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels, et al. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol. 2021;19(5) doi: 10.1371/journal.pbio.3001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz, et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593(7858):266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- World Health Organization . 2021. Tracking SARS-CoV-2 Variants.https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (Accessed 25 December 2021) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.