Abstract
COVID-19 originated in Wuhan city, China, in 2019 erupted a global pandemic that had put down nearly 3 million lives and hampered the socio-economic conditions of all nations. Despite the available treatments, this disease is not being controlled totally and spreading swiftly. The deadly virus commences infection by hACE2 receptor and its co-receptors (DPP4) engagement with the viral spike protein in the lung alveolar epithelial cells, indicating a primary therapeutic target. The current research attempts to design an in-silico Bispecific antibody (BsAb) against viral spike glycoprotein and DPP4 receptors. Regdanvimab and Begelomab were identified to block the D614G mutated spike glycoprotein of SARS-CoV-2 and host DPP4 receptor, respectively. The designed BsAb was modified by using KIH (Knobs into Holes) and CrossMAb techniques to prevent heavy chain and light chain mispairings. Following the modifications, the site-specific molecular docking studies were performed, revealing a relatively higher binding affinity of BsAb with spike glycoprotein and DPP4 co-receptor than control BsAb. Also, for blocking the primary entry receptor, hACE2, an anti-viral peptide was linked to the Fc region of BsAb that blocks the hACE2 receptor by linker cleavage inside the infected host. Thus, the designed BsAb and anti-viral peptide therapy could be a promising triumvirate way to obstruct the viral entry by blocking the receptor engagement.
Keywords: SARS-CoV-2, Bispecific Antibody (BsAb), KIH, CrossMAb, Spike glycoprotein, DPP4, ACE2
1. Introduction
SARS-CoV-2, the seedbed of the pandemic initiated in 2020, reported the first case in Wuhan, China, in late December 2019 [1], and now it is swiftly scattering in the whole world, mainly affecting the United States, Italy, Spain, Germany, Iran, France, the United Kingdom, and India [2] . The world health organization (WHO) has declared COVID-19 disease as a pandemic on 11th March 2020 and issued a global emergency for public health. According to WHO, around the total number of confirmed cases by SARS-CoV-2 was reported till 20th August 2021 was 209,876,613 including 4,400,284 deaths, worldwide (WHO report 2019,https://covid19.who.int/). Ahead of time, the world had encountered zoonotic infection caused by viruses of the same family, notably Severe Acute Respiratory Syndrome (SARS) in 2002 [3] that caused 774 deaths and Middle East respiratory syndrome (MERS) in 2012, resulting in 858 deaths [4]. Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) is a beta coronavirus, member of the Coronaviridae family of the order Nidovirales [5], is a zoonotic virus resulting in pneumonia of the upper respiratory tract of the host causing respiratory or enteric mediated disease, and at times cause another disease like hepatitis and neurologic illness [6]. A total, seven types of coronaviruses have been identified to date- HCoV-229E, HCoV-NL63, HCoV- OC43, HCoV-KHU1, SARS-CoV-1, MERS-CoV and SARS-CoV-2. The sequence database studies indicate all seven humans CoVs originated from animals such as; SARS-CoV, MERS-CoV, HCoV-NL63, HCoV-229E are originated from bats HCoV-OC43, and HCoV-HKU1 are originated from rodents.
SARS-CoV-2 enters the human body through air droplets and initiates the viral life cycle by attaching RBD (Receptor binding domain) of the trimeric spike protein with the ACE2 receptor of the lungs [7]. DPP4 (dipeptidyl-peptidase 4) receptor was previously reported to be an entry receptor for MERS-CoV infection [8]. The viruses to enter into hosts require multiple transmembrane proteins apart from the primary receptor. The viruses of coronaviridae family can recognize the broad range of cell surface molecules and the designated receptors; these molecules are called co-receptors. DPP4 has been reported as a co-receptor of ACE2. At the time of virus entry, both SARS-CoV-2 and MERS-CoV interact with the identical residues of DPP4 that are K267 R336, R317, Q344 [9]. In Type-2 diabetic patients, there is an imbalance in the RAAS system, which causes the upregulation of DPP4 levels. These two factors cause heart failure and imbalance in the expression of ACE2 receptors. In the case of COVID-19, it seemed to affirm the fact that ACE2 receptor and DPP4 share a dynamic correlation and influence the lung inflammation and expressed strappingly in endothelium, kidney, lungs, in solid and hollow digestive organs (e.g., pancreas and small intestine) (Vankadari and Wilce, 2020).
COVID-19 experience was mild to moderate/severe respiratory illness, especially fever and cold [11]. Besides the host cell receptors and intermediates, the antigens involved in viral infection, replication, and other metabolic functions provide a wide array of targets for COVID-19 disease progression. The proposed treatment strategies include drug repurposings, such as the use of anti-viral (targeting polymerase and protease) [12], monoclonal antibodies (mAb) [13], immuno-modulators (targeting interleukin-6) [14] and interferon a/b [15]), and vaccines. Some nucleic acid-based vaccines, viral vector-based, and subunit vaccines are in clinical trials. FDA approves two mRNA-based vaccines, Moderna COVID-19 vaccine (mRNA-1273), Pfizer-BioNTech (BNT162b2), and one viral vector-based vaccine, Janssen COVID-19 vaccine (JNJ-78436735), for emergency use to prevent the SARS-CoV-2 infection. Two vaccines have been licensed for emergency use in India- Covishield (AZD1222/ ChAdOx1) and Covaxin (BBV152). Covishield is a non-replicating viral vector vaccine developed by AstraZeneca and Serum Institute of India, whereas Bharat Biotech Limited, India, manufactured Covaxin, an inactivated viral vaccine (https://www.cdc.gov/coronavirus/2019-ncov/vaccines/different-vaccines/janssen.html).
Monoclonal antibody therapy is another therapeutic strategy in the prevention of COVID-19 infection. In 1986, the first mAb muromonab-CD3 (ortho-clone OKT3) was approved by US-FDA, and now around 79 mAbs are in the market for the treatment of several diseases [16]. MAbs target pathogen surface antigens, proteins, and receptors in the host body to prevent the disease progress. For COVID-19, several authorized mAbs primarily target the spike protein of SARS-CoV-2 due to its role in host cell receptor engagement during viral entry.
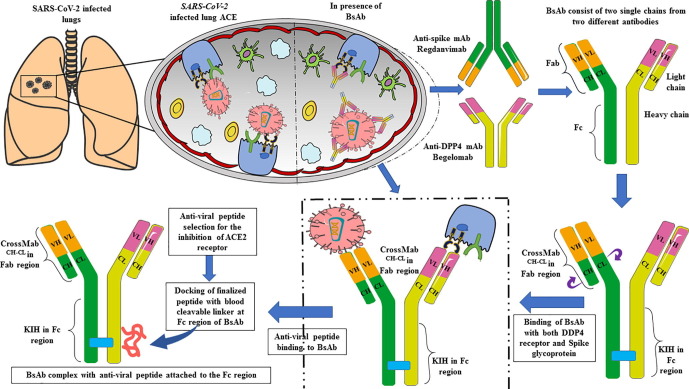
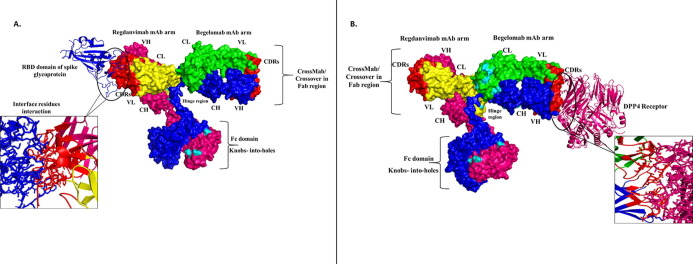
This study aimed to design a Bispecific Antibody (BsAb) conjugated with a peptide that could block all the possible targets employed during viral entry. The BsAb development approach to treat SARS-CoV-2 is more advantageous than traditional approaches because it targets two different entry receptors of SARS-CoV-2. The two mAbs selected for BsAb construction are; Regdanvimab (mAb that blocks spike protein), Begelomab (mAb that blocks the DPP4 co-receptor). The European Medical Agency (EMA) has approved Regdanvimab to treat COVID-19 (https://www.ema.europa.eu/en/news/ema-issues-advice-use-regdanvimab-treating-covid-19). Treatment with Regdanvimab has shown a reduced risk of infection in patients and cut off the demand for additional oxygen during infection. Whereas Begelomab, anti-CD26 mAb was reported against DPP4 receptor during COVID-19 infection and proven to treat COVID-19 patients with a severe diabetic condition (Galimberti et al., 2020). Both mAbs were modeled using the SWISS-MODEL followed by KIH mutation in the CH3 domain and Crossover (CrossMAb) mutation in the CH1 domain to prevent heavy and light chain mispairing, respectively. The anti-viral peptide was selected, followed by rigorous sorting based on different biophysical properties and evaluation of docking scores of each peptide with ACE2 receptor. The peptide with the highest docking score was conjugated to the Fc region of the BsAb using a blood cleavable linker. Thus, by this process, we have designed a bispecific antibody conjugated with anti-viral peptide to prevent the infection of SARS-CoV-2 via blocking its self spike protein and primary entry receptors, ACE2 and DPP4 (Fig. 1 ).
Fig. 1.
Graphical representation of designing and functioning of Bispecific antibody (BsAb) against SARS-CoV-2 and DPP4 receptor along with the inhibition of ACE-2 receptor with anti-viral peptide.
2. Methodology
2.1. Identification, selection, and homology modeling of SARS-CoV-2 spike glycoprotein
In search of an efficient therapy against COVID-19, we have shown the designing of a BsAb. Due to rapid mutation in the SARS-CoV-2 genome, with the help of various kinds of literature, we have found that spike glycoprotein is evolving very fast [17], [18]. This protein interacts with ACE2 and other co-receptors, for example, DPP4, to enter the host body. Spike glycoprotein is an attractive therapeutic option for the treatment against SARS-CoV-2. The RBD region present in the S1 subunit of spike glycoprotein is responsible for the binding with the ACE2 receptor [19]. This study selected the SARS-CoV-2 strain prevalent in Indian states and mutations in spike glycoprotein RBD region. After the identification of various mutations (D614G, Q271R, T723I, N440K, E484K, L54F, G1124V, R407I, A930V, Y144, E484Q, L5F, Q677H, S477N, N501Y, D138Y, P681H, Y453F, N439K) we have checked for mutation frequency. The spike glycoprotein with a high rate of mutation and responsible for enhanced infectivity was selected and modeled with the help of the SWISS-MODEL server (https://swissmodel.expasy.org/) [20]. This fully automated server is used to design the 3D structure of proteins and provides several user interactions such as; first approach mode, alignment mode, and project mode. After submitting the input file, this server gives different models based on parameters like template coverage, GMQE (Global Model Quality Estimate), QMEAN (Qualitative Model Energy ANalysis) [21].
2.2. Discerning ACE2 receptor/co-receptors and their homology modeling
After going through literature by Karunakaran et al., we have found that ACE2 alone is not responsible for viral entry; many other receptors are also responsible for the same [22]. We have utilized the STRING Database to identify the other spike glycoprotein receptors and their interaction (https://string-db.org/). This database aims to collect, score and integrate all available protein–protein interaction information [23]. STRING 11.0 version of the STRING database has the information of interacting proteins of around 5090 different organisms [23]. After comparing the obtained STRING data and available literature, we have found that apart from ACE2, DPP4 is mainly involved in viral entry and is associated with ACE2 as a co-receptor [10]. Further, we have retrieved the crystal structure of both ACE2 (PDB ID- 6VW1) and DPP4 (PDB ID-4L72) from RCBS (Research Collaboratory for Structural Bioinformatics) PDB (https://www.rcsb.org/). We found that both PDBs were complexed with some other compounds, so, after the removal of compounds, a 3D model was generated using the SWISS-MODEL server (https://swissmodel.expasy.org/). Further for the validation of all the generated models we have performed the Ramachandran Plot Analysis through ZLab Ramachandran Plot Server (https://zlab.umassmed.edu/bu/rama/).
2.3. Monoclonal antibodies selection for the designing of BsAb
BsAb comprises two different monoclonal antibodies that simultaneously bind with two different antigens. Recently, several antibodies have been reported against SARS-CoV-2; here, we have used Thera-SAbDab (therapeutic, structural antibody database) (http://opig.stats.ox.ac.uk/webapps/therasabdab) for the selection of antibodies. A WHO authorized database records all monoclonal antibodies and nanobodies against various targets and receptors and updates weekly with SAbDab (structural antibody database) [24]. SAbDab is a single platform containing multiple applications e.g. for Fv modeling ABodyBuilder (part of SAbPred (http://opig.stats.ox.ac.uk/webapps/newsabdab/sabpred/), for side-chain prediction PEARS, for epitope prediction EpiPred etc. We found that Regdanvimab (CT-P59), an efficient monoclonal antibody, was reported against the spike glycoprotein of SARS-CoV-2. Like other MAbs, it is still under consideration; nonetheless, few in vitro and in vivo studies have shown that this MAb effectively inhibits viral replication in the case of mutant spike glycoprotein [25]. Further, as per the term bispecific antibody, we have selected our second mAb, Begelomab [26], reported against the DPP4 receptor, which is used as an inhibitor against DPP4 in Type-2 diabetic conditions. Begelomab as an inhibitor for DPP4 receptor also helps reduce autophagy and reduce host-antiviral immune response [27].
2.4. Designing and modeling of BsAb via CrossMAb and Knobs into holes techniques
After selecting antibodies against their target, the next step is designing a bispecific antibody. For that, the sequence of VH, VL regions of Regdanvimab (CT-P59), and Begelomab antibodies were obtained directly from Thera-SAbDab Database, while for CH, CL sequence, model was generated by ABodyBuilder and obtained PDB IDs, 7CM4 and 5CMA, respectively from ABodyBuilder (http://opig.stats.ox.ac.uk/webapps/newsabdab/sabpred/abodybuilder/). From these PDB Ids on RCBS PDB, we took a sequence of CH, CL in FASTA format. Besides this, Fc (fragment crystalline) part was obtained from CT-P59 (PDB ID; 7CM4) antibody sequence. The Fc region for each antibody remains similar and constant for all species that bind to the Fc receptors present on the cell to activate the immune system. After designing the control BsAb, we have applied the CrossMAb and Knob into Hole (KIH) technique to overcome the light chain and heavy chain and mispairing, one-to-one. CrossMAb technique is first revealed to overcome light chain mispairing, based on the crossover/exchange of light or heavy chains in the Fab region. There are mainly three formats reported for CrossMAb designing: CrossMAbFab, CrossMAbVH-VL, and CrossMAbCH-CL. Several studies have reported more side products in CrossMAbFab and CrossMAb VH-VL than CrossMAbCH-CL [28] . However, the light chain mispairing problem can also be subdued by inverting the existing charge amino acid residues in both heavy and light chains [29]. Here, we have inverted the existing charged amino acid residues with oppositely charged residues for obtaining CrossMAbCH-CL. This inversion of charged residue leads to a striking interaction between oppositely charged residues, enabling correct light chain assembly. Whereas the KIH technique, a rational design strategy in antibody engineering for heteromerization in the Fc region, implies mainly subdue the heavy chain mispairing in the development of BsAb. This technique was applied in the Fc region of the heavy chain, where small amino acids (those have small side chain) got replaced with the larger ones (those have large side chain or aromatic side chain) to create a knob, and more considerable amino acid replaced with smaller ones to develop holes [30]. Further, this designed bispecific antibody was modeled using the SWISS-MODEL server.
2.5. Molecular docking of spike glycoprotein and DPP4 receptor with designed BsAb
After the bispecific antibody designing, molecular docking of spike glycoprotein and DPP4 receptor was performed with BsAb using H-DOCK server (http://hdock.phys.hust.edu.cn/). H-DOCK is an automated server that performs template-based and template-free site-specific docking [31] between protein–protein, protein-RNA, and protein-DNA. Here we have performed the site-specific docking of Regdanvimab (CT-P59) and Begelomab antibodies with spike glycoprotein and DPP4 receptor Fab arms, respectively. Here, spike glycoprotein and DPP4 work as ligand while both arms of BsAb work as receptors, so for the H-DOCK simulation, we have specified interacting residues in receptor and ligand such as in spike glycoprotein, residues of the RBD domain, which interacts with ACE2 (Rathod et al., 2020), was specified, whereas, in the DPP4 receptor, residues through which SARS-CoV-2 binds with DPP4 [9] were specified for site-specific docking.
2.6. Selection and screening of anti-viral peptide library to target ACE2 receptor
Anti-viral peptides have the colossal potentiality to inhibit virus replication by targeting various pathogen life cycle stages. ACE2 is the primary receptor that interacts with the RBD domain of spike glycoprotein to enter the human host. This receptor needs to be targeted to inhibit the entry of SARS-CoV-2 in humans [32]. For this, we have screened the anti-viral peptide library against the ACE2 receptor, which was available at Antiviral Peptide Database (AVPdb) (http://crdd.osdd.net/servers/avpdb). This database comprises anti-viral peptides against various viruses such as influenza, HCV (Hepatitis C virus), HSV (Herpes simplex virus), DENV (Dengue virus), SARS (Server acute respiratory syndrome), and many more. Those peptides reported to block the viral entry were selected [33]. This viral entry peptide library comprises 625 peptides. The sorting of these peptides was done on the basis of length and various physicochemical properties like allergenicity, antigenicity, toxicity, solubility, and stability. Allergenicity of each anti-viral peptide was predicted by using the Algpred server (http://www.imtech.res.in/raghava/algpred/); this server allows the prediction of allergenicity of any peptide by using amino acid sequence and based on similarity of known epitope with any region of the protein [34]. Antigenicity was predicted using VaxiJen 2.0 server (http://www.ddgpharmfac.net/vaxijen/), the first server that predicts the antigenicity in an independent orientation manner [35]. Toxicity was predicted using the ToxiNPred server (http://crdd.osdd.net/raghava/toxinpred/); this is a unique server used to predict the toxicity peptide in silico methods [36]. Solubility was predicted using peptide half-life prediction-HLP server (http://crdd.osdd.net/raghava/hlp/help.html), developed to predict peptide half-life. This server also predicts other physiochemical properties like charge, polarity, pK value, hydrophobicity, etc. [37]. The last parameter, stability, was checked using PepCalc.Com Server (https://pepcalc.com/peptide-solubility-calculator.php). This server calculates the peptide's molecular weight, extinction coefficient, net charge, iso-electric point, and water solubility [38]. After peptide library selection and screening, homology modeling of anti-viral peptides was made by using the PEP-FOLD3 server (http://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD3), this server generates native conformation of peptides having a length of 5 to 50 amino, within a few minutes [39].
2.7. Molecular docking of ACE2 receptor with anti-viral peptides
SARS-CoV-2 enters the cell through spike glycoprotein attachment with ACE2. Thus, ACE2 can also be considered a potential target to prevent the SARS-CoV-2 entry into the host cell with spike glycoprotein. Nowadays, peptides are being used as therapeutics to target ACE2; we performed molecular docking anti-viral peptides with ACE2. Before attaching anti-viral peptides with BsAb, molecular docking of ACE2 with all sorted anti-viral peptides was achieved (sorting based on various physicochemical properties). The site-specific docking was performed with the same server, i.e., the H-DOCK server. For site-specific docking, we have selected residues in ACE2 that interact with spike glycoprotein during SARS-CoV-2 infection, as Glu37 and Arg393. After docking, we got 130 models; however, the model with the highest binding energy was selected. Here, we have found that peptide AVP0800 has the highest binding energy score among all models, i.e.,–232.34.
2.8. Attachment of anti-viral peptide with designed BsAb by performing molecular docking
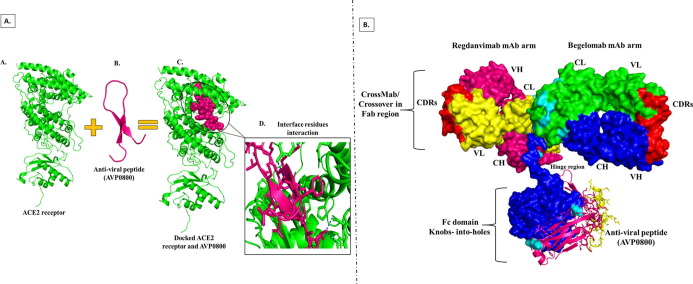
Although this designed antibody is bispecific, it will perform triple action and be a triumvirate. The best peptide docked with ACE2 receptor was attached to the Fc region of the designed bispecific antibody with the help of a blood cleavable linker [40]. Linkers provide stability and flexibility to the proteins; several linkers have been reported for connecting two or more proteins/peptides [41]. However, stable or rigid peptide linkers do not allow the separation of fusion proteins in vivo system. Cleavable linkers are comprised of disulfide bonds and cleaved by different proteases in blood; after cleavage, two fused proteins/peptides become separated to function individually. Thus, we have selected the Dithiocyclo peptide (linker sequence-CRRRRRREAEAC) (Xiaoying Chen et al., 2010), disulfide containing cleavable linker for the fusion of anti-viral peptide to the bispecific antibody; this linker has the cleavable sites which further separate the peptide and bispecific antibody and will act separately at their point of action [42]. We have also identified the cleavable sites of Dithiocyclopeptide with the help of the Peptide Cutter server (https://web.expasy.org/peptide_cutter/) and PROSPER server (http://lightning.med.monash.edu.au/PROSPER/), these servers are used for insilico identification of proteases substrates and protease cleavage sites [43]. After linker selection and docking of peptides with ACE2 receptor, the finalized peptide was selected for conjugation with designed BsAb. This peptide was attached or docked with a Dithiocyclo peptide linker to the BsAb; docking was done using the H-DOCK server. The Fc-binding site of Abs is the primary binding site, especially for the effector proteins and anti-viral peptides, whereas antigens bind to Fab region of Abs. Attachment of anti-viral peptides and other synthetic ligands at Fc-binding domain of Abs is highly desirable because it does not interfere with the antigen binding ability of immunoglobulin [44]. After going through this study, we have attached the selected anti-viral peptide between the CH2-CH3 domains of the Fc region of BsAb.
3. Result
3.1. Selection and modeling of the mutated spike glycoprotein
For the bispecific antibody design, the targets need to be selected primarily. With the report of current cases of COVID-19 in India, we have found that the spike glycoprotein of SARS-CoV-2 is mainly responsible for alarming cases. Quite a few mutations were identified in the spike protein of SARS-CoV-2 in comparison to other proteins. To date, D614G is highly prevalent in Indian isolates and worldwide [45] and is responsible for augmenting viral transmission. According to Guruprasad et al., D614G mutation occurs 7859 times in spike glycoprotein, the highest among all mutations [46].
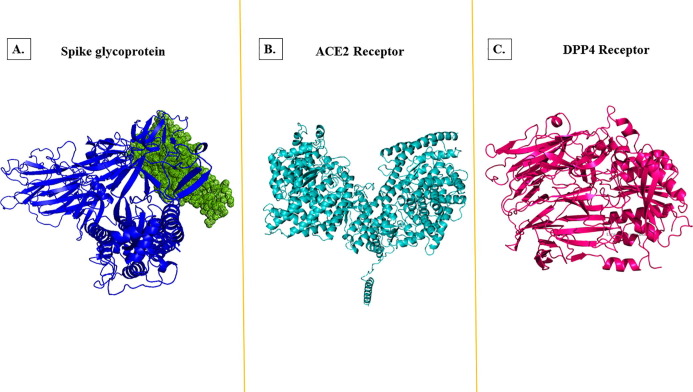
Consequently, we have selected the spike protein with D614G mutation as the target for bispecific antibody designing for this study (Supplementary sequence 1). The 3D structure of the mutated spike protein was generated by homology modeling. During homology modeling, we generated ten models, in which the model with the highest sequence similarity was selected for further molecular docking with BsAb (Fig. 3A).
Fig. 3.
Homology 3D modeling. (A.) Spike glycoprotein where dots in green color represent the receptor-binding domain. (B.) ACE2 (angiotensin-converting enzyme) and (C.) DPP4 (Dipeptidyl peptidase-4) receptor with the help of SWISS-MODEL. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Hit upon ACE2 receptor/co-receptors and their homology modeling
Interaction between the ACE2 receptor and its co-receptors was found with the help of the String database. The interconnection of receptors with each other was very interesting. We have found that around ten receptors- AGT (Angiotensinogen), AGTR1 (Type-1 Angiotensin II Receptor), AGTR2 (Type-2 Angiotensin II Receptor), DPP4 (Dipeptidyl Peptidase 4), PRCP (Lysosomal Pro-X carboxypeptidase), MME (Membrane metallo-endopeptidase), REN (Renin), MEP1A (Meprin A subunit alpha), MEP1B (Meprin A subunit beta), XPNPEP2 (Xaa-Pro aminopeptidase 2) are nicely interacting with ACE2 and helps in regulating its expression (Fig. 2 ). After analyzing the works of literature, we have discovered that the DPP4 receptor was reported as a primary entry receptor for SARS-CoV-1 [9]. Several other studies have shown that SARS-CoV-2 often utilizes the DPP4 receptor to enter the host. Type-2 diabetic patients are more susceptible to COVID-19 infection because they have an elevated level of soluble DPP4 protein. Consequently, we have selected the DPP4 receptor as a target of the bispecific antibody. The 3D model of both receptors has shown in Fig. 3 B and 3C. The validation of generated models ACE2, DPP4 and Spike glycoprotein was done using Ramachandran Plot Server and here we have found for all the models more than 95% residues were in favoured region. This denotes that the models are of good quality and are stable in nature (Supplementary Fig. 1A, B and C).
Fig. 2.
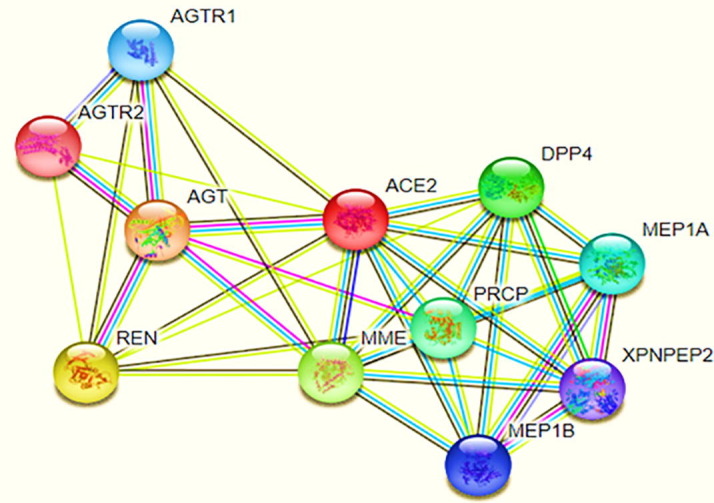
Interaction between ACE2 receptor with its associated co-receptors (Adopted from STRING DATABASE). From this figure, we analyzed the green color circle showing the closed neighborhood protein, red color circle showing protein fusions, the blue color circle is showing protein co-occurrence. In contrast, a purple color circle denoting experimentally determined proteins and black color lines shows the co-expression network of ACE2 with these ten receptor proteins. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Monoclonal antibodies selection for BsAb conniving
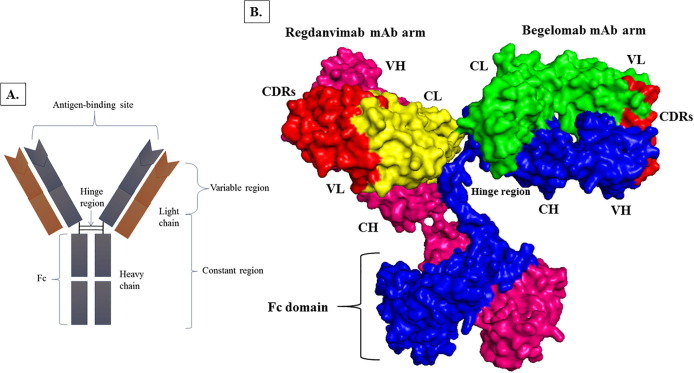
Several mAbs have been authorized as therapeutic in the treatment of SARS-CoV-2. This study focused on mAbs, which mainly targeted the RBD domain of spike glycoprotein and was used as an inhibitor against DPP4. Thus, Regdanvimab (CT-P59) mAb was selected against spike glycoprotein of D614G variant of SARS-CoV-2, and BegelomabmAb was selected against DPP4 co-receptor ACE2 for the designing of BsAb. After selecting mAbs, single arms for each antibody were selected and combined to design a BsAb (Fig. 4 ).
Fig. 4.
Designing of BsAb. (A.) Antibody structure (B.) BsAb designing; comprising two single arms of Regdanvimab and BegelomabMAbs. In the Regdanvimab arm, yellow denotes the light chain, hot pink represents the heavy chain, and red signifies the CDRs.In the Begelomab arm, green epitomizes the light chain, blue represents the heavy chain, and red characterizes the CDRs. In the Fc region, hot pink and blue colors represent the heavy chain of RegdanvimabmAb and BegelomabmAb. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Designing and modeling of BsAb by applying KIH and CrossMAb techniques
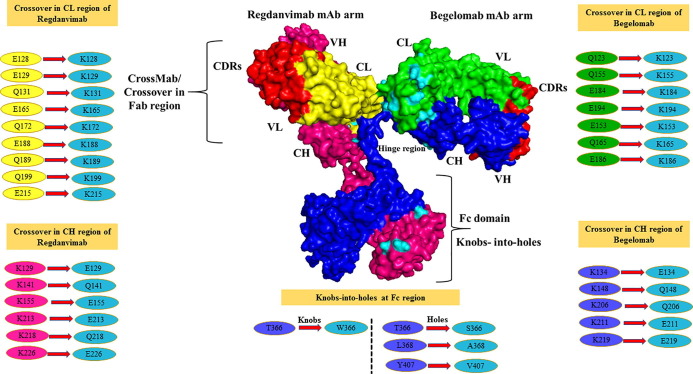
For the designing of BsAb, KIH and CrossMab technique was applied. As mentioned in the methodology section, the KIH technique was applied to subdue heavy chain mispairing in the Fc region. For knob creation, we have introduced Tryptophan (Trp) in place of Threonine (Thr) at 366th position in one side of the heavy chain in the Fc region, which developed as T366W mutation. According to Regula et al. study, we have created holes by introducing small amino acids at the place of more prominent amino acids on another side of the heavy chain in the Fc region. For this, we have introduced small amino acids at the place of larger amino acids as, Serine in place of Threonine at 366th position (T366S), Alanine at the place of Leucine at 368th position (L368A), Valine at the place of Tyrosine at 407th position (Y407V) and create a hole in opposition of knob (created on the heavy chain in the Fc region) [29].
Further, CrossMAb was applied by taking the same study as a reference; firstly, we have replaced the existing charged residues in the CH-CL interfaces in both arms (heavy and light chain) of BsAb with their opposite charged residues (negative charge residues replaced with positively charged residues and vice-versa). We have identified the negative charge residues at positions E128, E129, Q131, E165, Q172, E188, Q189, Q199, E215 in the light chain (CL), these residues replaced by oppositely (positive) charged residue as Lys (K). In heavy chain (CH), positive charge residues identified at positions K129, K141, K155, K113, K218, K226, for inversion replaced with oppositely charged residues (negative) as (Glu) E and (Gln) Q. On the other hand, negative charge residues were also identified at positions Q123, E153, Q155, Q165, E184, E186, E194 in the light chain (CL) of Begelomab mAb and replaced with Lys (K). In the heavy chain (CH) of Begelomab mAb, positive charge residues were identified at positions K134, K148, K206, K211, K219 and replaced with oppositely charged residues E (Glu) and Q (Gln) (Supplementary sequences 2 and 3). The designed BsAb with all the modifications has shown in Fig. 5 .
Fig. 5.
Representation of a BsAb comprising Regdanvimab and BegelomabMAbs. In both the arms of mAbs, cyan color denotes the Crossover (CrossMAb) CH-CL changes, whereas cyan color represents the KIH modifications in the Fc region. All the amino acid residue modifications have shown in circle form. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. Molecular docking analysis of spike glycoprotein and DPP4 receptor with BsAb
Spike glycoprotein was docked with the single-arm of Regdanvimab (CT-P59), whereas the DDP4 receptor was docked with the other arm, i.e., Begelomab. Further, molecular docking of modified BsAb (CrossMAbCH-CL and KIH) was done compared to control BsAb (without KIH or CrossMAbCH-CL). Here, we have performed separate dockings, firstly of spike glycoprotein (mainly RBD region) with CDRs of the Regdanvimab (CT-P59) arm (Fig. 6 A). When set for docking, the H-DOCK server gave ten models, in that we have selected that model which had high docking score in comparison to control. The score of the unmodified Regdanvimab arm was −208.05, whereas the modified Regdanvimab arm with spike glycoprotein was −227.37, which was the highest among all ten models.
Fig. 6.
Molecular docking of BsAb with its target. (A) Spike Glycoprotein docking with CDRs of Regdanvimab arm of BsAbs and their interaction has shown in the box. (B) DPP4 docking with CDRs of the Begelomab arm of BsAb, and their interaction has shown in stick form in the box as interface residue interaction.
Further, the same procedure is done for docking of the Begelomab arm with the DPP4 receptor; the DPP4 receptor is docked with CDRs of the Begelomab arm (Fig. 6B). With docking data, the highest binding score of the unmodified Begelomab arm was 188.13, whereas the highest score of the modified Begelomab arm with DPP4 receptor was −193.63. With this data, it can be concluded that after introducing KIH and CrossMAbCH-CL techniques, the binding affinity of spike glycoprotein and DPP4 with their specific single-arm mAb got enhanced compared to control.
3.6. Selection and assessment of anti-viral peptide library to target ACE2 receptor and their homology modeling
During SARS-CoV-2 infection, ACE2 interacts with the RBD region of the spike protein. This interaction via small molecules like peptides, neutralizing antibodies, and nanobodies can be an attractive therapeutic option for treating COVID-19 [47], [48]. We have selected an anti-viral peptide library comprised of 625 peptides and is responsible for inhibiting the viral entry. These peptides were sorted based on their length, allergenicity, antigenicity, toxicity, solubility, and stability of peptides. The first parameter we applied was the length; peptides had 15 amino acids selected. After that, peptides that were non-allergenic and antigenic were selected. The peptides having antigenicity scores > 0.5 were selected. Further peptides which were non-toxic, water-soluble, and are highly stable were selected after sorting by these properties. We had thirteen peptides left at last (Table 1 ). Further homology modeling of all thirteen peptides was done to perform the next step.
Table 1.
List of anti-viral peptides predicted from AVPdb.
| S.No | Anti-viral peptides | Peptide Sequence | Target | Length | Allergenicity | Antigenicity | Toxicity | Solubility | Stability |
|---|---|---|---|---|---|---|---|---|---|
| 1. | AVP0691 | AAPTGDPKPKKNKKP | Viral entry | 15 | Non-allergenic | ANTIGEN (0.9266) | Non-toxic | Good water solubility | HIGH (3.121) |
| 2. | AVP0724 | DDHETDMELKPANAA | Viral entry | 15 | Non-allergenic | ANTIGEN (1.2953) | Non-toxic | Good water solubility | HIGH (1.097) |
| 3. | AVP0725 | DMELKPANAATRTSR | Viral entry | 15 | Non-allergenic | ANTIGEN (1.5144) | Non-toxic | Good water solubility | HIGH (1.127) |
| 4. | AVP0727 | TRTSRGWHTTDLKYN | Viral entry | 15 | Non-allergenic | ANTIGEN (1.3233) | Non-toxic | Good water solubility | HIGH (1.178) |
| 5. | AVP0739 | YGYREGSHTEHTTYA | Viral entry | 15 | Non-allergenic | ANTIGEN (0.7556) | Non-toxic | Good water solubility | HIGH (1.538) |
| 6. | AVP0771 | EQSRKPPNPTPPPPG | Viral entry | 15 | Non-allergenic | ANTIGEN (0.8679) | Non-toxic | Good water solubility | HIGH (2.645) |
| 7. | AVP0800 | EPCTVGHRRYFTFGG | Viral entry | 15 | Non-allergenic | ANTIGEN (1.0104) | Non-toxic | Good water solubility | HIGH (2.314) |
| 8. | AVP0807 | IDLNITMLEDHEFVP | Viral entry | 15 | Non-allergenic | ANTIGEN (1.3527) | Non-toxic | Good water solubility | HIGH (1.800) |
| 9. | AVP0810 | RHEIKDSGLLDYTEV | Viral entry | 15 | Non-allergenic | ANTIGEN (1.0570) | Non-toxic | Good water solubility | HIGH (3.706) |
| 10. | AVP0813 | QRRNQLHDLRFADID | Viral entry | 15 | Non-allergenic | ANTIGEN (1.8815) | Non-toxic | Good water solubility | HIGH (1.036) |
| 11. | AVP1075 | RRKKAAVALLPAVLL | Viral entry | 15 | Non-allergenic | ANTIGEN (0.8598) | Non-toxic | Good water solubility | HIGH (2.083) |
| 12. | AVP1091 | RRKKLPAVLLALLAP | Viral entry | 15 | Non-allergenic | ANTIGEN (0.8218) | Non-toxic | Good water solubility | HIGH (3.493) |
| 13. | AVP1853 | FKCRRWQWRMKKLGA | Viral entry | 15 | Non-allergenic | ANTIGEN (1.3904) | Non-toxic | Good water solubility | HIGH (2.319) |
3.7. Anti-viral peptide molecular docking with ACE2 receptor
Before initiating docking, proteases cleavable site on Dithiocyclopeptide linker was identified (by PEPTIDE CUTTER and PROSPER server) between E (Glu) and A (Ala) residues (CRRRRRRE AEAC). Thus, the linker AEAC sequence will remain attached with peptide after cleavage by proteases, so we modeled peptides with the AEAC sequence and docked them with the ACE2 receptor. For this purpose, an H-DOCK server was used, which provided ten models for each peptide. For site-specific docking, we have selected residues Glu37 and Arg39in ACE2, as these residues mainly interact with spike glycoprotein during SARS-CoV-2infection. After docking, we got 130 models; however, the model with the highest binding energy was selected. Here, we have found that peptide AVP0800 has the highest binding energy score among all models, i.e., –232.34. Therefore, to inhibit the ACE2 receptor interaction with spike protein, peptide AVP0800 was attached with BsAb with the help of a blood cleavable linker. This peptide has shown constricted interaction with both selected residues, Glu37 and Arg393. Here, we have shown only polar contacts of R393 and N394 of ACE2 with AVP0800 peptide (Fig. 7 A).
Fig. 7.
Docking of an anti-viral peptide with ACE2 and BsAb. (A) Peptide-ACE2 docking structures embedded with ACE2 and their interface interactions. (a) Represents the ACE2 receptor (b) Represents the AVP0800 anti-viral peptide (c) Represent the docked complex of ACE2 (cyan color) and AVP0800 anti-viral peptide (hot pink color) (d) Represents the polar interaction at the interface between ACE2 and AVP0800. (B) Attachment of peptide between CH2-CH3 Domains of the Fc region of BsAb. In the Fc region, hot pink and blue colors represent the heavy chain of RegdanvimabmAb and BegelomabmAb, respectively, and the yellow color represents the anti-viral peptide. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.8. Molecular docking analysis of AVP0800 peptide with BsAb
At last, we attached peptide with the help of disulfide containing cleavable linker between CH2-CH3 domains at the Fc region of BsAb. As we mentioned in the methodology part, this linker has cleavable sites that further separate the peptide and bispecific antibody; they will act separately at their point of action. Site-specific docking was performed by selecting residues as Lys (K), Gly (G), Ser (S), etc., between CH2 and CH3 domain at the Fc region [49]. In this, we also got ten models based on different docking scores. We have selected the first model with the highest docking score −205.16. The docked model of BsAb and peptide and has shown in Fig. 7B.
4. Discussion
COVID-19 is an alarming situation that displayed a constant death rate each since the pandemic. Due to the higher rate of transmissibility and higher mortality, developing an effective therapeutic strategy is an utmost requirement in public health concerns, promising treatment against the SARS-CoV-2 virus and eliminating the COVID-19. Monoclonal antibody therapy is considered an effective strategy for inhibiting the virus-receptor engagement or the host cell components. mAbs could be developed to inhibit viral entry (targeting viral surface antigen or host receptors), inhibit viral replication or transcription, or inhibit host immune cells that promote inflammation. BsAbs are constructed by joining two mAbs that target a pathogen and host cell or two host cells. BsAb engineering has various applications in NCDs or infectious disease prevention and therapeutics. BsAb engineering is a significant challenge in immunotherapeutics and needs scientific, systematic efforts and expertise in bioinformatics.
Here, we have aimed to engineer a BsAb that prevent SARS-CoV-2 infection in human employing insilico approach. The BsAb was constructed by combining existing mAbs, Regdanvimab (CT-P59), and Begelomab, targeting the spike glycoprotein of SARS-CoV-2 and DPP4 co-receptor, respectively. An anti-viral peptide was conjugated at the FC region of BsAb with the help of a blood protease cleavable linker to block the ACE2 receptor of the host cell. This BsAb was designed to inhibit viral entry by blocking all possible targets, i.e., spike glycoprotein of the virus, entry promoting receptors, ACE2 and DPP4. Hence, BsAb development is considered a promising strategy in preventing SARS-CoV-2 infection compared to mAb treatment. ACE2 receptor is utilized for SARS-CoV-2 entry by the interaction of the primary receptor with spike glycoprotein. Higher expression of DPP4 observed in Diabetic patients also elevates the SARS-CoV-2 infection as it is the co-receptor of the ACE2 receptor. Sequence retrieval of mAbs, ACE2 receptor, and DPP4 co-receptor was done from different databases. Anti-viral peptides were utilized to increase the half-life and stability of BsAb in further in vivo studies. A blood cleavable linker was used to attach anti-viral peptide with BsAb, which could ensure intrinsic action of the anti-viral peptide and BsAb once entered into the human circulation. In-vitro and in-vivo BsAb synthesis could generate few side products due to two antibody light or heavy chain mispairing. KIH and CrossMAb techniques were applied to overcome the heavy chain and light chain mispairing that could reduce the number of combinations yet to be generated. The heavy chain and light chain pairing of two antibodies expressed in a cell could theoretically result in 16 different combinations consisting of one BsAband, the others being non-specific or monospecific antibodies. Introduction of KIH mutations in the CH3 domain reduces the combinations to 4, and further introduction of CrossMAb technique in CH1 produces single combination efficient to inhibit the viral entry as evident from the docking scores. The modeled structures were docked with respective receptors that showed significant docking scores of DPP4, spike glycoprotein, and anti-viral peptide with BsAb, indicating this BsAb could efficiently inhibit SARS-CoV-2 entry. Hence, these in-silico studies affirm the bispecific antibody development that could be an efficient therapeutic in the future against COVID-19.
The anticipated in-vitro or in-vivo mode of action of this BsAb is initiated by cleavage of the linker followed by releasing the peptide that inhibits the ACE2 receptor. The separated BsAb blocks the Spike glycoprotein and DPP4 co-receptor in parallel while present in proximity, and the Fc of BsAb binds to the Fc receptor of NK cells to initiate ADCC (Antibody-dependent cellular cytotoxicity). Further, in vitro or in vivo studies generate bispecific antibodies by fusion of two antibody-producing cell lines that combine heavy (H) and light(L) chains of two different antibodies [50] to produce diverse combinations of H and L chains of both antibodies. In-vitroBsAb combinations with mutations in the Fc region, i.e., BsAb with heterodimeric heavy chains, can be eluted by protein A chromatography [51] as they exhibit decreased affinity for protein A in contrast to homodimeric Fc and displayed retention of Fc-mediated effector functions, such as ADCC [52]. The CrossMAbCH1-CL showed better side-product combinations, as evident from the bispecific CrossMAb against VEGF and Ang-2 [53]. Hence, the BsAb in vitro studies are anticipated to show promising results to block the viral entry and perform the Fc mediated effector functions that could significantly reduce the risk of SARS-CoV-2 infection progression. This research could open new doors for developing a novel and effective therapeutic strategy for COVID-19 besides other infectious diseases.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
RO is thankful to Central University of Rajasthan for providing a university fellowship. VKP is thankful to the Central University of Rajasthan for providing lab facilities.
Author contribution
Protocol designed by RO, KG, TSR, AM, VKP.
Methodology performed by RO, KG, TSR, VKP.
The manuscript was written by RO, KG, TSR, VKP
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.humimm.2022.01.004.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.J. Lee, G. Chowell, E. Jung, A dynamic compartmental model for the Middle East respiratory syndrome outbreak in the Republic of Korea: A retrospective analysis on control interventions and superspreading events, J. Theor. Biol. (2020). [DOI] [PMC free article] [PubMed]
- 5.Yang Y., Xiao Z., Ye K., He X., Sun B., Qin Z., Yu J., Yao J., Wu Q., Bao Z., Zhao W. SARS-CoV-2: characteristics and current advances in research. Virol. J. 2020;17:1–17. doi: 10.1186/s12985-020-01369-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., Qu Y., Li F., Lv Q.i., Wang W., Xue J., Gong S., Liu M., Wang G., Wang S., Song Z., Zhao L., Liu P., Zhao L.i., Ye F., Wang H., Zhou W., Zhu N.a., Zhen W., Yu H., Zhang X., Guo L.i., Chen L., Wang C., Wang Y., Wang X., Xiao Y., Sun Q., Liu H., Zhu F., Ma C., Yan L., Yang M., Han J., Xu W., Tan W., Peng X., Jin Q.i., Wu G., Qin C. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583(7818):830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 7.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein. BioRxiv. 2020 doi: 10.1101/2020.02.19.956581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyerholz D.K., Lambertz A.M., McCray P.B. Dipeptidyl Peptidase 4 Distribution in the Human Respiratory Tract Implications for the Middle East Respiratory Syndrome. Am. J. Pathol. 2016;186(1):78–86. doi: 10.1016/j.ajpath.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y.u., Zhang Z., Yang L.i., Lian X., Xie Y., Li S., Xin S., Cao P., Lu J. The MERS-CoV Receptor DPP4 as a Candidate Binding Target of the SARS-CoV-2 Spike. IScience. 2020;23(6):101160. doi: 10.1016/j.isci.2020.101160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vankadari N., Wilce J.A. Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26, Emerg. Microbes Infect. 2020;9:601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A Novel Coronavirus from Patients with Pneumonia in China. N. Engl. J. Med. 2019;382(2020):727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.M. Angel, crossm Compounds with Therapeutic Potential against Novel, (2020) 1–7.
- 13.J. Xu, S. Zhao, T. Teng, A.E. Abdalla, W. Zhu, Systematic Comparison of Two Animal-to-Human Transmitted Human Coronaviruses : SARS-CoV-2 and, (2020). https://doi.org/10.3390/v12020244. [DOI] [PMC free article] [PubMed]
- 14.R. Fleischmann, M.C. Genovese, Y. Lin, G.S. John, J.A. Maldonado-cocco, M. Stanislav, A.J. Kivitz, Original article Long-term safety of sarilumab in rheumatoid arthritis : an integrated analysis with up to 7 years ’, (2020) 292–302. https://doi.org/10.1093/rheumatology/kez265. [DOI] [PMC free article] [PubMed]
- 15.Hu W., Yen Y., Singh S., Kao C., Wu-hsieh B.A. SARS-CoV Regulates Immune Function-Related. 2012;25:277–288. doi: 10.1089/vim.2011.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu R.M., Hwang Y.C., Liu I.J., Lee C.C., Tsai H.Z., Li H.J., Wu H.C. Development of therapeutic antibodies for the treatment of diseases. J. Biomed. Sci. 2020;27:1–30. doi: 10.1186/s12929-019-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galimberti S., Baldini C., Baratè C., Ricci F., Balducci S., Grassi S., Ferro F., Buda G., Benedetti E., Fazzi R., Baglietto L., Lucenteforte E., Di A., Petrini M. The CoV-2 outbreak: how haemotologists could help to fight Covid-19. Pharmacol. Res. 2020 doi: 10.1016/j.phrs.2020.104866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gómez C.E., Perdiguero B., Esteban M. Emerging sars-cov-2 variants and impact in global vaccination programs against sars-cov-2/covid-19. Vaccines. 2021;9:1–13. doi: 10.3390/vaccines9030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rathod S.B., Prajapati P.B., Punjabi L.B., Prajapati K.N., Chauhan N., Mansuri M.F. Peptide modelling and screening against human ACE2 and spike glycoprotein RBD of SARS - CoV - 2. Silico Pharmacol. 2020;8(1) doi: 10.1007/s40203-020-00055-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., De Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.K. Karunakaran, M. Ganapathiraju, Interactome of SARS-CoV-2 / nCoV19 modulated host proteins with computationally predicted PPIs, Res. Sq. (2020). https://doi.org/10.21203/rs.3.rs-28592/v1.
- 22.Gadanec L.K., McSweeney K.R., Qaradakhi T., Ali B., Zulli A., Apostolopoulos V. Can SARS-CoV-2 virus use multiple receptors to enter host cells? Int. J. Mol. Sci. 2021;22:1–35. doi: 10.3390/ijms22030992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J., Von Mering C. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raybould M.I.J., Marks C., Lewis A.P., Shi J., Bujotzek A., Taddese B., Deane C.M. Thera-SAbDab: The Therapeutic Structural Antibody Database. Nucleic Acids Res. 2020;48:D383–D388. doi: 10.1093/nar/gkz827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim C., Ryu D.-K., Lee J., Kim Y.-I., Seo J.-M., Kim Y.-G., Jeong J.-H., Kim M., Kim J.-I., Kim P., Bae J.S., Shim E.Y., Lee M.S., Kim M.S., Noh H., Park G.-S., Park J.S., Son D., An Y., Lee J.N., Kwon K.-S., Lee J.-Y., Lee H., Yang J.-S., Kim K.-C., Kim S.S., Woo H.-M., Kim J.-W., Park M.-S., Yu K.-M., Kim S.-M., Kim E.-H., Park S.-J., Jeong S.T., Yu C.H., Song Y., Gu S.H., Oh H., Koo B.-S., Hong J.J., Ryu C.-M., Park W.B., Oh M.-D., Choi Y.K., Lee S.-Y. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 2021;12(1) doi: 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galimberti S., Baldini C., Baratè C., Ricci F., Balducci S., Grassi S., Ferro F., Buda G., Benedetti E., Fazzi R., Baglietto L., Lucenteforte E., Di Paolo A., Petrini M. The CoV-2 outbreak: how hematologists could help to fight Covid-19. Pharmacol. Res. 2020;157:104866. doi: 10.1016/j.phrs.2020.104866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valencia I., Peiró C., Lorenzo Ó., Sánchez-Ferrer C.F., Eckel J., Romacho T. DPP4 and ACE2 in Diabetes and COVID-19: Therapeutic Targets for Cardiovascular Complications? Front. Pharmacol. 2020;11:1–14. doi: 10.3389/fphar.2020.01161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H., Saxena A., Sidhu S.S., Wu D. Fc engineering for developing therapeutic bispecific antibodies and novel scaffolds. Front. Immunol. 2017;8:1–15. doi: 10.3389/fimmu.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J.T. Regula, S. Imhof-Jung, M. Mølhøj, J. Benz, A. Ehler, A. Bujotzek, W. Schaefer, C. Klein, Variable heavy-variable light domain and Fab-arm CrossMabs with charged residue exchanges to enforce correct light chain assembly, Protein Eng. Des. Sel. 31 (2018) 289–299. https://doi.org/10.1093/protein/gzy021. [DOI] [PMC free article] [PubMed]
- 30.Ridgway J.B.B., Presta L.G., Carter P. “Knobs-into-holes” engineering of antibody C(H)3 domains for heavy chain heterodimerization. Protein Eng. 1996;9(7):617–621. doi: 10.1093/protein/9.7.617. [DOI] [PubMed] [Google Scholar]
- 31.Yan Y., Tao H., He J., Huang S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020;15(5):1829–1852. doi: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- 32.Akachar J., Bouricha E.M., Hakmi M., Belyamani L., El Jaoudi R., Ibrahimi A. Identifying epitopes for cluster of differentiation and design of new peptides inhibitors against human SARS-CoV-2 spike RBD by an in-silico approach. Heliyon. 2020;6(12):e05739. doi: 10.1016/j.heliyon.2020.e05739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qureshi A., Thakur N., Tandon H., Kumar M. AVPdb: A database of experimentally validated anti-viral peptides targeting medically important viruses. Nucleic Acids Res. 2014;42(D1):D1147–D1153. doi: 10.1093/nar/gkt1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saha S., Raghava G.P.S. AlgPred: Prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res. 2006;34(Web Server):W202–W209. doi: 10.1093/nar/gkl343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.BMC Bioinformatics, antigens and subunit vaccines (2021) 1–2. [DOI] [PMC free article] [PubMed]
- 36.Gupta S., Kapoor P., Chaudhary K., Gautam A., Kumar R., Raghava G.P.S., Patterson R.L. In silico approach for predicting toxicity of peptides and proteins. PLoS One. 2013;8(9):e73957. doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma A., Singla D., Rashid M., Raghava G.P.S. Designing of peptides with desired half-life in intestine-like environment. BMC Bioinform. 2014;15:1–8. doi: 10.1186/1471-2105-15-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amit C., Muralikumar S., Janaki S., Lakshmipathy M., Therese K.L., Umashankar V., Padmanabhan P., Narayanan J. Designing and enhancing the antifungal activity of corneal specific cell penetrating peptide using gelatin hydrogel delivery system. Int. J. Nanomed. 2019;14:605–622. doi: 10.2147/IJN.S184911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamiable A., Thévenet P., Rey J., Vavrusa M., Derreumaux P., Tufféry P. PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016;44(W1):W449–W454. doi: 10.1093/nar/gkw329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kishimoto S., Nakashimada Y., Yokota R., Hatanaka T., Adachi M., Ito Y. Site-Specific Chemical Conjugation of Antibodies by Using Affinity Peptide for the Development of Therapeutic Antibody Format. Bioconjug. Chem. 2019;30(3):698–702. doi: 10.1021/acs.bioconjchem.8b0086510.1021/acs.bioconjchem.8b00865.s001. [DOI] [PubMed] [Google Scholar]
- 41.Lu J., Jiang F., Lu A., Zhang G.e. Linkers having a crucial role in antibody–drug conjugates. Int. J. Mol. Sci. 2016;17(4):561. doi: 10.3390/ijms17040561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiaoying Chen W.-C.-S., Bai Y., Zaro J.L. Design of an in vivo cleavable disulfide linker in recombinant fusion protein. Biotechniques. 2010;49:513–518. doi: 10.2144/000113450.Design. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song J., Tan H., Perry A.J., Akutsu T., Webb G.I., Whisstock J.C., Pike R.N., Srinivasan N. PROSPER: An Integrated Feature-Based Tool for Predicting Protease Substrate Cleavage Sites. PLoS One. 2012;7(11):e50300. doi: 10.1371/journal.pone.0050300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choe W., Durgannavar T., Chung S. Fc-binding ligands of immunoglobulin G: An overview of high affinity proteins and peptides. Materials (Basel) 2016;9(12):994. doi: 10.3390/ma9120994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.U. States, S.E. Galloway, P. Paul, D.R. Maccannell, M.A. Johansson, Emergence of SARS-CoV-2 B . 1 . 1 . 7 Lineage —, 70 (2021) 95–99. [DOI] [PMC free article] [PubMed]
- 46.Guruprasad L. Human SARS CoV-2 spike protein mutations. Proteins Struct. Funct. Bioinform. 2021;89:569–576. doi: 10.1002/prot.26042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6(1) doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Lu L.u. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.AR and YI. Satoka Mori, Arisa Abe, Naoto Ishikawa, A novel site-specific chemical conjugation of IgG antibodies by affinity peptide for immunoassays, J. Biochem. (2020). https://doi.org/10.1093/jb/mvaa084. [DOI] [PubMed]
- 50.Suresh M.R., Cuello A.C., Milstein C. Bispecific monoclonal antibodies from hybrid hybridomas. Methods Enzymol. 1986;121:210–228. doi: 10.1016/0076-6879(86)21019-8. [DOI] [PubMed] [Google Scholar]
- 51.H. Lindhofer, R. Mocikat, B. Steipe, S. Thierfelder, Preferential species-restricted heavy/light chain pairing in rat/mouse quadromas. Implications for a single-step purification of bispecific antibodies., J. Immunol. 155 (1995) 219 LP – 225. [PubMed]
- 52.Shatz W., Chung S., Li B., Marshall B., Tejada M., Phung W., Sandoval W., Kelley R.F., Scheer J.M. Knobs-into-holes antibody production in mammalian cell lines reveals that asymmetric afucosylation is sufficient for full antibody-dependent cellular cytotoxicity. MAbs. 2013;5(6):872–881. doi: 10.4161/mabs.26307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kloepper J., Riedemann L., Amoozgar Z., Seano G., Susek K., Yu V., Dalvie N., Amelung R.L., Datta M., Song J.W., Askoxylakis V., Taylor J.W., Lu-Emerson C., Batista A., Kirkpatrick N.D., Jung K., Snuderl M., Muzikansky A., Stubenrauch K.G., Krieter O., Wakimoto H., Xu L., Munn L.L., Duda D.G., Fukumura D., Batchelor T.T., Jain R.K. Ang-2/VEGF bispecific antibody reprograms macrophages and resident microglia to anti-tumor phenotype and prolongs glioblastoma survival. Proc. Natl. Acad. Sci. U. S. A. 2016;113(16):4476–4481. doi: 10.1073/pnas.1525360113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.