Abstract
Viral RNAs are susceptible to co-translational RNA decay pathways mediated by the RNA helicase Upstream frameshift 1 (Upf1). Upf1 is a key component in nonsense-mediated decay (NMD), Staufen1-mediated mRNA decay (SMD), and structure-mediated RNA decay (SRD) pathways, among others. Diverse families of viruses have features that predispose them to Upf1 targeting, but have evolved means to escape decay through the action of cis- or trans-acting viral factors. Studies aimed at understanding how viruses are subjected to and circumvent NMD have increased our understanding of NMD target selection of host mRNAs. This review focuses on the knowledge gained from studying NMD in viral systems as well as related Upf1-dependent pathways and how these pathways restrict virus replication.
INTRODUCTION
Upf1 is an ATP-dependent RNA helicase that is required for several RNA quality control pathways and is highly conserved across eukaryotic systems [1–4]. Nonsense-mediated decay (NMD), the most extensively studied Upf1-dependent pathway, targets aberrant transcripts containing a premature termination codon (PTC) to prevent the production of truncated proteins with potentially detrimental effects [5, 6]. Phosphorylation of Upf1 is necessary for NMD [7–9] and inhibiting NMD results in increased steady-state levels of ~10% of cellular transcripts in multiple eukaryotic systems, demonstrating that NMD effectively regulates a considerable amount of gene expression post-transcriptionally [10–14]. mRNAs that do not contain PTCs make up a portion of cellular transcripts targeted by Upf1 and these transcripts often have upstream open reading frames (uORFs) [15] or introns in their 3’ untranslated region (3’ UTR) [16]. Upf1 predominantly binds 3’ UTRs and translocates over long distances using its RNA helicase activity, efficiently displacing bound proteins [17]. Transcriptome-wide mapping experiments revealed that Upf1 binding is enhanced in GC-rich regions, possibly as a result of reduced translocation [18–20]. The elevated levels of Upf1 binding to highly-structured, GC-rich 3’ UTRs regions [19] increases the likelihood that phosphorylated Upf1 will interact with a terminating ribosome and initiate mRNA decay [21]. The multi-cistronic organization of viral genomes predisposes viruses to NMD targeting since internal stop codons are perceived as PTCs by the host. Furthermore, viral 3’ UTRs are often GC-rich and highly structured for recruiting host factors [22] and/or components of the translation machinery [23], which also predisposes viral RNAs to NMD. As such, diverse families of viruses are targeted by NMD [24, 25], which the virus can evade through the presence of either specific cis-acting RNA sequences or viral proteins [26, 27]. In addition to NMD, Upf1 is essential for additional RNA degradation pathways that are initiated by various host factors (Fig. 1A) [21]. However, the roles of additional Upf1-dependent pathways in restricting viruses are poorly understood.
Figure 1: Upf1-dependent decay and targeting of viral mRNAs.
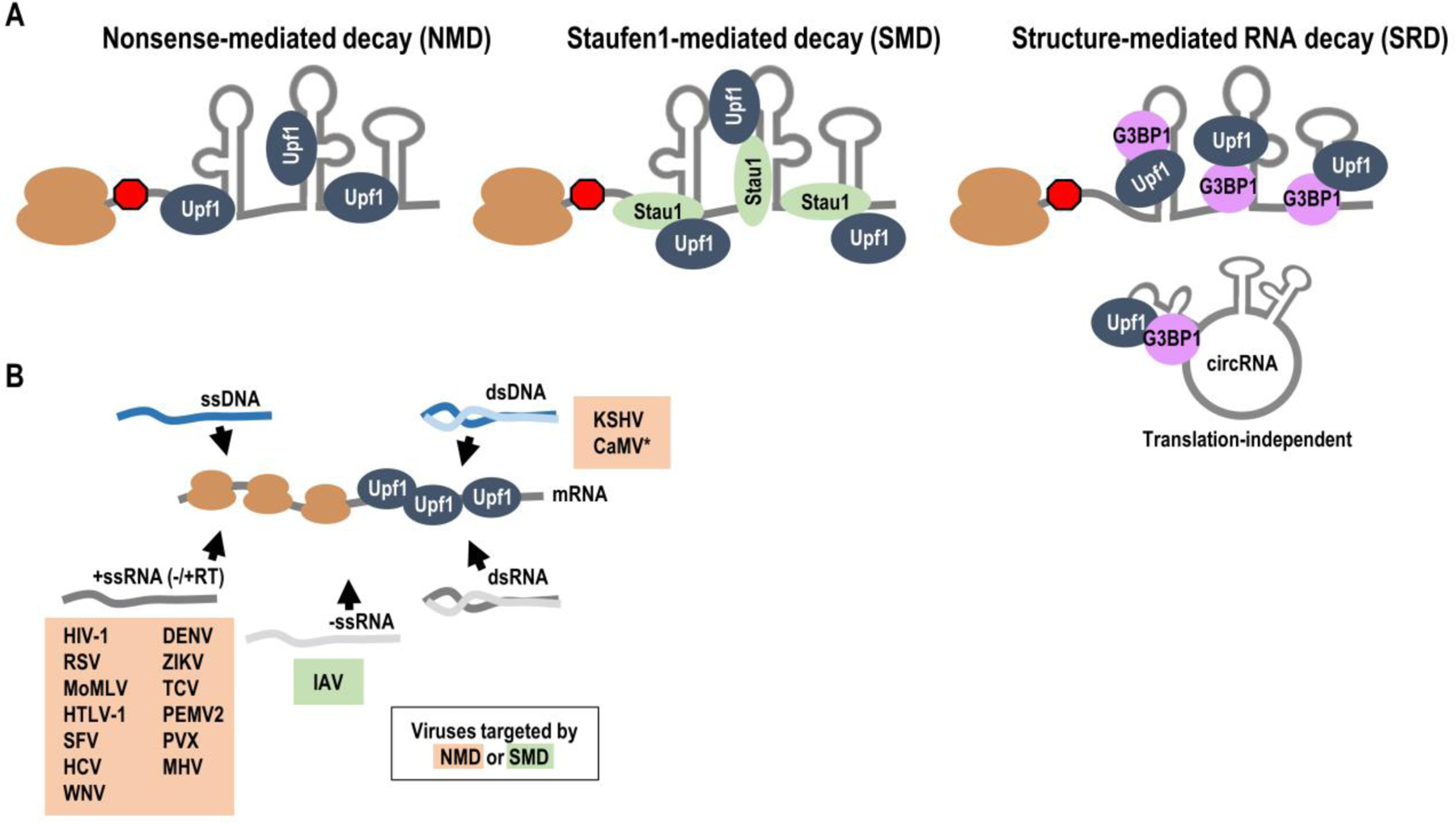
(A) Upf1 is essential for nonsense-mediated decay (NMD), Staufen1-mediated decay (SMD) and structure-mediated RNA decay (SRD). Upf1 associates with the 3’ UTRs of viral and host 3’ UTRs and can initiate NMD. Stau1 binds dsRNA structures in the 3’ UTRs of mRNAs and recruits Upf1 for subsequent SMD. G3BP1 binds highly structured regions in conjunction with Upf1 to target mRNAs or non-coding circRNAs for SRD. (B) All viruses produce mRNAs that are potential targets of Upf1. Most viruses that are known to be targeted by Upf1 are +ssRNA viruses. IAV (-ssRNA) is targeted by Upf1 and SMD. dsDNA viruses produce mRNAs that are targeted by NMD. *CaMV interferes with NMD, but direct evidence of susceptibility of viral RNAs towards NMD is lacking.
Diverse viruses are Upf1 targets for NMD
Both animal and plant viruses have been used for studying NMD. Studies using the retrovirus Rous sarcoma virus (RSV) were the first to demonstrate that Upf1 can target unspliced viral RNAs for NMD in a co-translational manner [28]. Interestingly, Upf1 is a component of the HIV-1 ribonucleoprotein (RNP) and positively regulates retrovirus replication (Table 1) [29–31]. The first evidence that NMD broadly inhibits RNA viruses came from studies demonstrating that NMD restricts Semliki forest virus (SFV) accumulation in mammalian cells [32] as well as Potato virus X (PVX) and Turnip crinkle virus (TCV) in plants [33]. Interestingly, the potyvirus Turnip mosaic virus was not targeted by NMD, which was attributed to its polyprotein expression strategy that prevents Upf1 accumulation along the viral mRNA [33]. These findings are in agreement with studies that demonstrate Upf1 is displaced by the translating ribosome and therefore only accumulates in 3’ UTRs [34]. NMD also targets several flaviviruses including Hepatitis C virus (HCV) [35] and Zika virus (ZIKV) [36], and the coronavirus Murine hepatitis virus (MHV) [37]. The DNA virus Kaposi’s sarcoma-associated herpesvirus (KSHV) was the first DNA virus determined to be targeted by Upf1 and NMD [38]. pre-mRNA splicing in the KSHV transcriptome results in retention of several introns downstream of termination codons that in turn predispose KSHV transcripts to NMD [39].
Table 1:
Pro-viral roles of Upf1 and associated decay factors
| Protein | Virus | Function | Refs. |
|---|---|---|---|
| Upf1 | HIV-1 | Required for efficient translation of pr55(Gag) | [29] |
| Upf1 | HIV-1 | Upf1 incorporates into viral RNPs to promote nuclear export of HIV-1 genomic RNA | [30] |
| Upf1 | HIV-1 | Upf1 helicase activity is required for HIV-1 reverse transcription and infectivity | [31] |
| Stau1 | Ebola | Stau1 facilitates interactions between viral RNA and replication machinery | [79] |
| Stau1 | EV-A71 | Promotes efficient viral protein translation by binding to 5’ UTR of viral RNAs | [80] |
| Stau1 | HIV-1 | Disassembles stress granules to promote efficient translation and trafficking of viral RNAs | [81] |
| Stau1 | IBDV, HCV | Stau1 binds viral dsRNA and interferes with type I interferon response and protein kinase R (PKR) | [82, 88] |
| Stau1 | HIV-1 | Component of HIV-1 RNP and regulates HIV-1 assembly and infectivity | [84–87] |
| Stau1 | IAV | Interacts with NS1 and viral ribonucleoprotein and is required for efficient replication | [89–91] |
| G3BP1 | SINV | Limits polyprotein expression, possibly by sequestering viral RNAs in nsP4 replication complexes | [107] |
| G3BP1 | SFV, CHIKV | nsP3 interacts with G3BP1 to sequester viral replication complexes and recruit translation factors | [108] |
| G3BP1 | MNV, HuNoV | Facilitates interactions between VPg, viral RNA, and translation machinery | [109] |
Virus evasion of NMD
Both cis-acting RNA sequences and trans-acting viral proteins can disrupt NMD and are summarized in Figure 2. Many viruses require ribosome recoding to produce C-terminally extended proteins and maximize coding potential [40, 41]. Viral ribosome recoding sequences (i.e., readthrough or frameshifting) just downstream from termination codons confer NMD-resistance in several plant viruses [42] as well as mammalian viruses like Moloney murine leukemia virus (MoMLV)[43, 44], likely as a result of ribosome displacement of Upf1 when the termination codon is by-passed [45].
Figure 2: Viruses disrupt NMD and SMD.

Both cis-acting RNA sequences and trans-acting viral proteins interfere with NMD. Flavivirus-mediated inhibition of NMD generally involves interfering with EJC components. SMD is disrupted by IAV NS1 blocking Upf1 association with Stau1.
Ribosomes terminating at the gag stop codon in retroviral mRNAs can leave a 3’ UTR that is greater than 7 kb in length, predisposing retroviruses to Upf1 targeting. However, NMD-resistance is conferred by RNA stability elements (RSEs; about 400 nt) located downstream of the gag stop codon, which are strongly conserved among retroviruses [46–49]. Human T-cell leukemia virus type 1 (HTLV-1) expresses Tax, a viral protein that directly binds Upf1 and INT6, a factor associated with eukaryotic initiation factor 3 (eIF3) and NMD [50]. Intricate studies have demonstrated that Tax binding to Upf1 inhibits Upf1 helicase activity and reduces RNA binding, thus effectively inhibiting NMD [51]. Upf1 activity is also antagonized by the SFV helicase and nsP3 in viruses in the Togaviridae [32]. Additionally, the HTLV-1 Rex protein has been proposed to protect both viral and host transcripts against NMD [52], however the mechanism remains unclear.
The NMD pathway is disrupted during HCV infection as a result of the viral core protein interfering with the interaction between Within bgcn homolog (WIBG) and the exon-junction complex (EJC) components Y14 and Magoh [35]. Y14, Magoh, and PYM1, an additional EJC component, are antiviral towards ZIKV, West Nile virus (WNV), and Dengue virus (DENV) as a result of Y14 directly binding viral RNA in a Magoh- and PYM1-dependent manner [53]. The WNV capsid inhibits interactions between PYM1, Magoh, and Y14, and WNV infection causes mis-localization of EJC components [53]. The ZIKV capsid interacts with nuclear Upf1 which leads to Upf1 degradation by the proteasome [36]. The targeting of cytoplasmic RNA viruses by the EJC is somewhat surprising since EJC proteins are largely associated with spliced mRNAs that form in the nucleus [54, 55]. However, studies in Drosophila have revealed that Y14 and Magoh can bind intronless transcripts [56] making it possible that EJC components could associate with viral RNAs promiscuously.
Studies using plant viruses have uncovered additional cis- and trans-acting viral elements that can thwart NMD. The long-distance p26 movement protein from Pea enation mosaic virus 2 (PEMV2) confers resistance to both viral and non-viral 3’ UTRs [57]. RNA-seq analyses revealed that both an NMD inhibitor and PEMV2 infection resulted in a high degree of overlap between down- and up-regulated transcripts, suggesting that PEMV2 severely impairs the NMD pathway [57]. The transactivator protein (TAV) from Cauliflower mosaic virus (CaMV), a DNA virus, disrupts the VARICOSE decapping complex and confers NMD resistance to host transcripts, but it remains unknown if CaMV transcripts are themselves subjected to NMD [58]. A short unstructured region (USR) immediately downstream of the TCV subgenomic RNA termination codon confers NMD resistance to both viral and non-viral NMD targets [42]. Introducing a 2-nt mutation downstream of the USR that forces the USR to form a stable hairpin abolishes NMD protection, demonstrating that the unstructured RNA is inherently NMD-resistant.
Viral RNA sequences provide insight into NMD target selection
The discovery of cis-acting viral RNA sequences that confer NMD-resistance has shed light on how cellular transcripts with NMD-inducing features evade NMD surveillance. The RSV RSE has long been studied for its role in conferring stability and NMD-resistance [49, 59]. Polypyrimidine tract binding protein 1 (PTBP1) binds polypyrmidine tracts within the RSV RSE [60], promoting Upf1 dissociation from NMD targets [61]. Nearly 200 human NMD targets are protected by PTBP1 and these transcripts are enriched for polypyrimidine hexamers downstream of their termination codons that bind PTBP1 and confer NMD-resistance [60]. Similar findings have revealed that hnRNP L protects cellular transcripts with long 3’ UTRs against NMD by binding to 3’ UTRs with CA repeats [62].
Interestingly, highly unstructured RNA sequences immediately downstream of the termination codon, as is found in TCV, are over 4-fold enriched in NMD-resistant human transcripts when compared to target transcripts [19, 42]. Another study identified several NMD-inhibiting termination-proximal cis elements in human mRNAs that were enriched for A/U nucleotides (~70%) in the first 200-nt downstream of the stop codon and lack stable secondary structures [63].
In Arabidopsis thaliana, alternative splicing generates three eukaryotic release factor 1 (eRF1) transcripts, where eRF1–1 contains an NMD-inducing intron in the 3’ UTR [64]. Interestingly, the eRF1–1 termination codon is followed by a 6-nt sequence (CAAUCA) that is similar to the Tobacco mosaic virus (TMV) ribosome readthrough signal [64, 65] and readthrough of this codon confers NMD-resistance and contributes to autoregulation of eRF1 protein levels [66]. A human genome-wide study found that the TMV readthrough signal was heavily favored among transcripts that undergo ribosome readthrough when treated with aminoglycosides and become NMD-resistant [67]. Collectively, these examples demonstrate how cis-acting viral sequences that confer NMD-resistance can provide insight into cellular NMD target selection, a process that is not fully understood [6].
Timing and efficiency of virus inhibition by NMD
Suppressing Upf1 expression results in increased virus accumulation across eukaryotic systems. However, the ability of Upf1 and NMD to limit virus replication in a biologically significant way at the organismal level remains unknown. NMD has been proposed to mainly occur during the pioneer round of mRNA translation [68], however multiple studies have shown that NMD can occur by chance during all rounds of translation [69–71]. Using single-molecule imaging and tracking individual ribosomes on an NMD target, Hoek et. al. (2019) demonstrated that approximately 10% of terminating ribosomes induce NMD [71]. This inefficiency of NMD activation presents a challenge for host cells to use NMD to eliminate viral RNAs. NMD may be most effective at restricting virus replication during early stages of infection when few copies of the viral RNAs are present. After an infection is established, the 10% overall efficiency of NMD is unlikely to appreciably limit virus replication since the cell is overcome with viral RNAs. In support of this conjecture, MHV is most efficiently inhibited by NMD in the first five hours of infection before expression of the viral N protein that inhibits NMD [37].
Additional Upf1-mediated decay pathways
Staufen1-mediated mRNA decay (SMD) requires that the double-stranded RNA binding protein Stau1 bind the 3’ UTRs of target mRNAs [72, 73]. Stau1 then recruits Upf1 to the 3’ UTRs of target mRNAs for subsequent target degradation [74]. Stable 3’ UTR stem-loop structures or intermolecular base-pairing between an mRNA 3’ UTR and long non-coding RNA (lncRNA) are sufficient to recruit Stau1 [75]. Knockdown of Stau1 results in the upregulation of 1% of human transcripts demonstrating that SMD, like NMD, can also regulate gene expression post-transcriptionally [76]. The efficiencies of NMD and SMD are inversely correlated with one another since Upf1 is required for both pathways but both Stau1 and the NMD factor Upf2 share binding sites within Upf1 [77, 78]. Whereas Stau1 plays a pro-viral role in multiple virus lifecycles (see Table 1) [79–91], there is currently limited evidence for Stau1 and SMD targeting viral RNAs. Importantly, Cho et. al. found that Influenza A virus (IAV) NS1 binding to Stau1 prevents Stau1 from interacting with Upf1 and initiating SMD decay of IAV RNAs [92]. Transcriptome-wide identification of Stau1 binding sites has revealed striking similarities to Upf1 where Stau1 preferentially binds long 3’ UTRs containing GC-rich secondary structures absent of specific sequence motifs [93–95]. Although most studies to date have focused on NMD-targeting of viruses, the similarities and intertwined functions of Stau1 and Upf1 suggest that SMD could also restrict accumulation in diverse families of viruses.
Recently, structure-mediated RNA decay (SRD) was demonstrated to selectively degrade transcripts with highly-structured 3’ UTRs in a Upf1- and Ras GTPase-activating protein-binding protein 1 (G3BP1)-dependent manner [96]. G3BP1 and Upf1 interact via base-paired RNA sequences that do not overlap with Stau1 binding sites as determined by CLIP-seq [95, 96]. G3BP1 forms stress granule cores during stress leading to sequestration and translational repression of cellular transcripts [97–100]. G3BP1 has several pro-viral (Table 1) and anti-viral roles in virus lifecycles. G3BP1 inhibits Sendai virus and Vesicular stomatitis virus by facilitating the RIG-I antiviral response [101, 102], G3BP1 sequesters HIV-1 RNA transcripts [103], and G3BP1 is specifically targeted and cleaved by the viral proteases from several picornaviruses [104–106]. Interestingly, SRD targets non-coding circular RNAs (circRNAs) in addition to mRNAs [96] opening the possibility that non-coding viral RNAs could be targeted by Upf1 and SRD.
CONCLUSIONS
The necessity of all viruses to either translate their genomes or antigenomes or produce mRNAs predisposes all viruses to Upf1-targeting (Fig. 1B) and future work will undoubtedly identify additional families of viruses subjected to Upf1-mediated decay. Most research to date has focused on the role of NMD in virus lifecycles, and the identification of cis- and trans-acting viral factors that interfere with NMD has broadened our understanding of NMD itself. Global inhibition of the NMD pathway has been observed during plant and animal virus infections and may contribute to pathogenesis. However, future work is needed to determine if viruses manipulate host gene expression through NMD-inhibition as a means to shape the transcriptome in a way that favors virus replication. Upf1’s involvement in additional pathways like SMD or SRD suggest that viruses are likely affected by additional Upf1-dependent pathways and additional studies dissecting the involvement of individual pathways in virus replication is needed.
HIGHLIGHTS.
Upf1 targets viral RNAs for degradation through multiple pathways
Viruses disrupt Upf1-mediated decay in different way through cis- and trans-acting factors
Viral systems have yielded insight into Upf1 target selection within host cells
ACKNOWLEDGEMENTS
This work was funded by NIH award F32 GM119235 and NIH training grant (T32 AI 125186-3) to JPM, University of Missouri-Kansas City institutional start-up funds to JPM, and NSF award MCB-1818229 to AES
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors state that they have no conflict of interests regarding the work in this manuscript
REFERENCES AND RECOMMENDED READING
Recent papers of special and outstanding interest have been highlighted as:
* of special interest
** of outstanding interest
REFERENCES
- 1.Kim YK, Maquat LE. UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA 2019;25(4):407–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Causier B, Li Z, De Smet R, Lloyd JPB, Van de Peer Y, Davies B. Conservation of Nonsense-Mediated mRNA Decay Complex Components Throughout Eukaryotic Evolution. Scientific reports. 2017;7(1):16692-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lloyd JPB. The evolution and diversity of the nonsense-mediated mRNA decay pathway. F1000Research. 2018;7:1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imamachi N, Tani H, Akimitsu N. Up-frameshift protein 1 (UPF1): multitalented entertainer in RNA decay. Drug discoveries & therapeutics. 2012;6(2):55–61. [PubMed] [Google Scholar]
- 5.Kurosaki T, Popp MW, Maquat LE. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nature reviews Molecular cell biology. 2019;20(7):406–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kishor A, Fritz SE, Hogg JR. Nonsense-mediated mRNA decay: The challenge of telling right from wrong in a complex transcriptome. Wiley interdisciplinary reviews RNA. 2019;10(6):e1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durand S, Franks TM, Lykke-Andersen J. Hyperphosphorylation amplifies UPF1 activity to resolve stalls in nonsense-mediated mRNA decay. Nature communications. 2016;7:12434-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, et al. Phosphorylation of hUPF1 Induces Formation of mRNA Surveillance Complexes Containing hSMG-5 and hSMG-7. Mol Cell. 2003;12(5):1187–200. [DOI] [PubMed] [Google Scholar]
- 9.Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JWB, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133(2):314–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt SA, Foley PL, Jeong D-H, Rymarquis LA, Doyle F, Tenenbaum SA, et al. Identification of SMG6 cleavage sites and a preferred RNA cleavage motif by global analysis of endogenous NMD targets in human cells. Nucleic acids research. 2015;43(1):309–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lelivelt MJ, Culbertson MR. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol 1999;19(10):6710–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drechsel G, Kahles A, Kesarwani AK, Stauffer E, Behr J, Drewe P, et al. Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell. 2013;25(10):3726–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. Rna 2005;11(10):1530–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nature genetics. 2004;36(10):1073–8. [DOI] [PubMed] [Google Scholar]
- 15.Nyikó T, Sonkoly B, Mérai Z, Benkovics AH, Silhavy D. Plant upstream ORFs can trigger nonsense-mediated mRNA decay in a size-dependent manner. Plant molecular biology. 2009;71(4–5):367–78. [DOI] [PubMed] [Google Scholar]
- 16.Kertész S, Kerényi Z, Mérai Z, Bartos I, Pálfy T, Barta E, et al. Both introns and long 3’-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res 2006;34(21):6147–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiorini F, Bagchi D, Le Hir H, Croquette V. Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities. Nature communications. 2015;6(1):7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurt JA, Robertson AD, Burge CB. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res 2013;23(10):1636–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imamachi N, Salam KA, Suzuki Y, Akimitsu N. A GC-rich sequence feature in the 3’ UTR directs UPF1-dependent mRNA decay in mammalian cells. Genome Res 2017;27(3):407–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colombo M, Karousis ED, Bourquin J, Bruggmann R, Mühlemann O. Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways. RNA 2017;23(2):189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavysh D, Neu-Yilik G. UPF1-Mediated RNA Decay-Danse Macabre in a Cloud. Biomolecules. 2020;10(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyde JL, Chen R, Trobaugh DW, Diamond MS, Weaver SC, Klimstra WB, et al. The 5’ and 3’ ends of alphavirus RNAs--Non-coding is not non-functional. Virus research. 2015;206:99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon AE, Miller WA. 3’ cap-independent translation enhancers of plant viruses. Annual review of microbiology. 2013;67:21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hogg JR. Viral Evasion and Manipulation of Host RNA Quality Control Pathways. Journal of virology. 2016;90(16):7010–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balistreri G, Bognanni C, Mühlemann O. Virus Escape and Manipulation of Cellular Nonsense-Mediated mRNA Decay. Viruses. 2017;9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popp MW, Cho H, Maquat LE. Viral subversion of nonsense-mediated mRNA decay. RNA 2020. [DOI] [PMC free article] [PubMed]
- 27.Leon K, Ott M. An ‘Arms Race’ between the Nonsense-mediated mRNA Decay Pathway and Viral Infections. Seminars in cell & developmental biology. 2020. [DOI] [PMC free article] [PubMed]
- 28.LeBlanc JJ, Beemon KL. Unspliced Rous sarcoma virus genomic RNAs are translated and subjected to nonsense-mediated mRNA decay before packaging. Journal of virology. 2004;78(10):5139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ajamian L, Abrahamyan L, Milev M, Ivanov PV, Kulozik AE, Gehring NH, et al. Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation. RNA 2008;14(5):914–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ajamian L, Abel K, Rao S, Vyboh K, García-de-Gracia F, Soto-Rifo R, et al. HIV-1 Recruits UPF1 but Excludes UPF2 to Promote Nucleocytoplasmic Export of the Genomic RNA. Biomolecules. 2015;5(4):2808–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serquiña AK, Das SR, Popova E, Ojelabi OA, Roy CK, Göttlinger HG. UPF1 is crucial for the infectivity of human immunodeficiency virus type 1 progeny virions. Journal of virology. 2013;87(16):8853–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balistreri G, Horvath P, Schweingruber C, Zünd D, McInerney G, Merits A, et al. The host nonsense-mediated mRNA decay pathway restricts Mammalian RNA virus replication. Cell Host Microbe 2014;16(3):403–11. [DOI] [PubMed] [Google Scholar]
- 33.Garcia D, Garcia S, Voinnet O. Nonsense-mediated decay serves as a general viral restriction mechanism in plants. Cell Host Microbe 2014;16(3):391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurosaki T, Maquat LE. Rules that govern UPF1 binding to mRNA 3’ UTRs. Proc Natl Acad Sci U S A. 2013;110(9):3357–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramage HR, Kumar GR, Verschueren E, Johnson JR, Von Dollen J, Johnson T, et al. A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay. Mol Cell. 2015;57(2):329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fontaine KA, Leon KE, Khalid MM, Tomar S, Jimenez-Morales D, Dunlap M, et al. The Cellular NMD Pathway Restricts Zika Virus Infection and Is Targeted by the Viral Capsid Protein. mBio 2018;9(6). *Study demonstrates that nuclear Upf1 is degraded by the ZIKV capsid protein, a novel method of NMD interference.
- 37. Wada M, Lokugamage KG, Nakagawa K, Narayanan K, Makino S. Interplay between coronavirus, a cytoplasmic RNA virus, and nonsense-mediated mRNA decay pathway. Proc Natl Acad Sci U S A. 2018;115(43):E10157–e66.30297408 *First study demonstrating coronaviruses are targeted by NMD and MHV is most susceptible to NMD in the early stages of infection before the N protein is synthesized. N protein disrupts NMD targeting and protects viral RNAs.
- 38. Zhao Y, Ye X, Shehata M, Dunker W, Xie Z, Karijolich J. The RNA quality control pathway nonsense-mediated mRNA decay targets cellular and viral RNAs to restrict KSHV. Nature communications. 2020;11(1):3345. **First study to demonstrate that DNA viruses produce transcripts that are subjected to NMD. Since all viruses must produce mRNAs that would presumably be accessible to Upf1, this study lays the groundwork for discovering additional classes of viruses that are targeted by NMD.
- 39.Ye X, Zhao Y, Karijolich J. The landscape of transcription initiation across latent and lytic KSHV genomes. PLoS Pathog 2019;15(6):e1007852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miras M, Miller WA, Truniger V, Aranda MA. Non-canonical Translation in Plant RNA Viruses. Frontiers in plant science. 2017;8:494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Firth AE, Brierley I. Non-canonical translation in RNA viruses. J Gen Virol 2012;93(Pt 7):1385–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. May JP, Yuan X, Sawicki E, Simon AE. RNA virus evasion of nonsense-mediated decay. PLoS Pathog 2018;14(11):e1007459.30452463 *The authors identified an unstructured RNA sequence immediately following a subgenomic RNA stop codon that confers NMD-resistance. Interestingly, similar unstructured sequences are enriched among human NMD-resistant transcripts.
- 43.Baker SL, Hogg JR. A system for coordinated analysis of translational readthrough and nonsense-mediated mRNA decay. PLoS One. 2017;12(3):e0173980–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang X, Zhu Y, Baker SL, Bowler MW, Chen BJ, Chen C, et al. Structural basis of suppression of host translation termination by Moloney Murine Leukemia Virus. Nature communications. 2016;7:12070-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zünd D, Gruber AR, Zavolan M, Mühlemann O. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3’ UTRs. Nature structural & molecular biology. 2013;20(8):936–43. [DOI] [PubMed] [Google Scholar]
- 46.Weil JE, Beemon KL. A 3’ UTR sequence stabilizes termination codons in the unspliced RNA of Rous sarcoma virus. RNA 2006;12(1):102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Withers JB, Beemon KL. The structure and function of the rous sarcoma virus RNA stability element. Journal of cellular biochemistry. 2011;112(11):3085–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Withers JB, Beemon KL. Structural features in the Rous sarcoma virus RNA stability element are necessary for sensing the correct termination codon. Retrovirology. 2010;7:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quek BL, Beemon K. Retroviral strategy to stabilize viral RNA. Curr Opin Microbiol 2014;18:78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mocquet V, Neusiedler J, Rende F, Cluet D, Robin JP, Terme JM, et al. The human T-lymphotropic virus type 1 tax protein inhibits nonsense-mediated mRNA decay by interacting with INT6/EIF3E and UPF1. Journal of virology. 2012;86(14):7530–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fiorini F, Robin JP, Kanaan J, Borowiak M, Croquette V, Le Hir H, et al. HTLV-1 Tax plugs and freezes UPF1 helicase leading to nonsense-mediated mRNA decay inhibition. Nature communications. 2018;9(1):431. **Using magnetic tweezers, the authors revealed how the HTLV-1 Tax protein causes global NMD inhibition by blocking Upf1 helicase activity and limititng RNA association.
- 52.Nakano K, Ando T, Yamagishi M, Yokoyama K, Ishida T, Ohsugi T, et al. Viral interference with host mRNA surveillance, the nonsense-mediated mRNA decay (NMD) pathway, through a new function of HTLV-1 Rex: implications for retroviral replication. Microbes and infection. 2013;15(6–7):491–505. [DOI] [PubMed] [Google Scholar]
- 53. Li M, Johnson JR, Truong B, Kim G, Weinbren N, Dittmar M, et al. Identification of antiviral roles for the exon-junction complex and nonsense-mediated decay in flaviviral infection. Nat Microbiol 2019;4(6):985–95.30833725 **Defined antiviral roles for exon-junction complex (EJC) components targeting flaviviruses. Although EJCs usually associate with RNAs in the nucleus, this study demonstrates that strictly cytoplasmic viral RNAs.
- 54.Saulière J, Murigneux V, Wang Z, Marquenet E, Barbosa I, Le Tonquèze O, et al. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nature structural & molecular biology. 2012;19(11):1124–31. [DOI] [PubMed] [Google Scholar]
- 55.Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 2001;20(17):4987–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choudhury SR, Singh AK, McLeod T, Blanchette M, Jang B, Badenhorst P, et al. Exon junction complex proteins bind nascent transcripts independently of pre-mRNA splicing in Drosophila melanogaster. Elife 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. May JP, Johnson PZ, Ilyas M, Gao F, Simon AE. The Multifunctional Long-Distance Movement Protein of Pea Enation Mosaic Virus 2 Protects Viral and Host Transcripts from Nonsense-Mediated Decay. mBio 2020;11(2):e00204–20.32156817 *Plant virus movement protein confers protection against NMD to a significant portion of cellular transcripts. Significant overlap of differentially expressed transcripts between an NMD inhibitor and PEMV2 infection was observed, suggesting that virus infection broadly inhibits NMD.
- 58. Lukhovitskaya N, Ryabova LA. Cauliflower mosaic virus transactivator protein (TAV) can suppress nonsense-mediated decay by targeting VARICOSE, a scaffold protein of the decapping complex. Scientific reports. 2019;9(1):7042.31065034 *First study to demonstrate that a virus can interfere with the late-stages of NMD by disrupting the functions of VARICOSE and the decapping complex.
- 59.Barker GF, Beemon K. Rous sarcoma virus RNA stability requires an open reading frame in the gag gene and sequences downstream of the gag-pol junction. Mol Cell Biol 1994;14(3):1986–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ge Z, Quek BL, Beemon KL, Hogg JR. Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway. Elife 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fritz SE, Ranganathan S, Wang CD, Hogg JR. The RNA-binding protein PTBP1 promotes ATPase-dependent dissociation of the RNA helicase UPF1 to protect transcripts from nonsense-mediated mRNA decay. The Journal of biological chemistry. 2020;295(33):11613–25.32571872 *Work by Fritz et. al. demonstrates that PTBP1 displaces Upf1 from NMD targets. The RSV RNA stability element (RSE) is enriched with PTBP1 binding sites and this study provides a mechanism of action for the NMD-resistant RSE.
- 62.Kishor A, Ge Z, Hogg JR. hnRNP L-dependent protection of normal mRNAs from NMD subverts quality control in B cell lymphoma. EMBO J 2019;38(3):e99128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toma KG, Rebbapragada I, Durand S, Lykke-Andersen J. Identification of elements in human long 3’ UTRs that inhibit nonsense-mediated decay. RNA 2015;21(5):887–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chapman B, Brown C. Translation termination in Arabidopsis thaliana: characterisation of three versions of release factor 1. Gene 2004;341:219–25. [DOI] [PubMed] [Google Scholar]
- 65.Skuzeski JM, Nichols LM, Gesteland RF, Atkins JF. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J Mol Biol 1991;218(2):365–73. [DOI] [PubMed] [Google Scholar]
- 66.Nyikó T, Auber A, Szabadkai L, Benkovics A, Auth M, Mérai Z, et al. Expression of the eRF1 translation termination factor is controlled by an autoregulatory circuit involving readthrough and nonsense-mediated decay in plants. Nucleic Acids Res 2017;45(7):4174–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wangen JR, Green R. Stop codon context influences genome-wide stimulation of termination codon readthrough by aminoglycosides. Elife 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a Pioneer Round of mRNA Translation: mRNAs Subject to Nonsense-Mediated Decay in Mammalian Cells Are Bound by CBP80 and CBP20. Cell. 2001;106(5):607–17. [DOI] [PubMed] [Google Scholar]
- 69.Durand S, Lykke-Andersen J. Nonsense-mediated mRNA decay occurs during eIF4F-dependent translation in human cells. Nature structural & molecular biology. 2013;20(6):702–9. [DOI] [PubMed] [Google Scholar]
- 70.Rufener SC, Mühlemann O. eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nature structural & molecular biology. 2013;20(6):710–7. [DOI] [PubMed] [Google Scholar]
- 71. Hoek TA, Khuperkar D, Lindeboom RGH, Sonneveld S, Verhagen BMP, Boersma S, et al. Single-Molecule Imaging Uncovers Rules Governing Nonsense-Mediated mRNA Decay. Mol Cell. 2019;75(2):324–39.e11.31155380 **Hoek et al. used single-molecule imaging to visualize an NMD reporter and precisely model the overall efficiency of NMD activation. The observed low efficiency of activation poses a challenge for host cells in limiting virus accumulation. This study also conclusively demonstrated that NMD is not restricted to the pioneer round of translation.
- 72.Park E, Maquat LE. Staufen-mediated mRNA decay. Wiley interdisciplinary reviews RNA 2013;4(4):423–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gleghorn ML, Gong C, Kielkopf CL, Maquat LE. Staufen1 dimerizes through a conserved motif and a degenerate dsRNA-binding domain to promote mRNA decay. Nature structural & molecular biology. 2013;20(4):515–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3’UTRs so as to elicit mRNA decay. Cell. 2005;120(2):195–208. [DOI] [PubMed] [Google Scholar]
- 75.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature. 2011;470(7333):284–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim YK, Furic L, Parisien M, Major F, DesGroseillers L, Maquat LE. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J 2007;26(11):2670–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev 2009;23(1):54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maquat LE, Gong C. Gene expression networks: competing mRNA decay pathways in mammalian cells. Biochemical Society transactions. 2009;37(Pt 6):1287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fang J, Pietzsch C, Ramanathan P, Santos RI, Ilinykh PA, Garcia-Blanco MA, et al. Staufen1 Interacts with Multiple Components of the Ebola Virus Ribonucleoprotein and Enhances Viral RNA Synthesis. mBio 2018;9(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen YM, Ou BT, Chen CY, Chan HH, Chen CJ, Wang RY. Staufen1 Protein Participates Positively in the Viral RNA Replication of Enterovirus 71. Viruses. 2019;11(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rao S, Hassine S, Monette A, Amorim R, DesGroseillers L, Mouland AJ. HIV-1 requires Staufen1 to dissociate stress granules and to produce infectious viral particles. RNA 2019;25(6):727–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ye C, Yu Z, Xiong Y, Wang Y, Ruan Y, Guo Y, et al. STAU1 binds to IBDV genomic double-stranded RNA and promotes viral replication via attenuation of MDA5-dependent β interferon induction. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2019;33(1):286–300.29979632 *This study highlights the pro-viral roles of Stau1 in the IBDV lifecycle by limiting the host interferon response and supporting replication during the late stage of virus infection.
- 83.Rao S, Amorim R, Niu M, Breton Y, Tremblay MJ, Mouland AJ. Host mRNA decay proteins influence HIV-1 replication and viral gene expression in primary monocyte-derived macrophages. Retrovirology. 2019;16(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Abrahamyan LG, Chatel-Chaix L, Ajamian L, Milev MP, Monette A, Clément JF, et al. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. Journal of cell science. 2010;123(Pt 3):369–83. [DOI] [PubMed] [Google Scholar]
- 85.Chatel-Chaix L, Abrahamyan L, Fréchina C, Mouland AJ, DesGroseillers L. The host protein Staufen1 participates in human immunodeficiency virus type 1 assembly in live cells by influencing pr55Gag multimerization. Journal of virology. 2007;81(12):6216–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chatel-Chaix L, Boulay K, Mouland AJ, Desgroseillers L. The host protein Staufen1 interacts with the Pr55Gag zinc fingers and regulates HIV-1 assembly via its N-terminus. Retrovirology. 2008;5:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chatel-Chaix L, Clément JF, Martel C, Bériault V, Gatignol A, DesGroseillers L, et al. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol Cell Biol 2004;24(7):2637–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dixit U, Pandey AK, Mishra P, Sengupta A, Pandey VN. Staufen1 promotes HCV replication by inhibiting protein kinase R and transporting viral RNA to the site of translation and replication in the cells. Nucleic Acids Res 2016;44(11):5271–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Falcón AM, Fortes P, Marión RM, Beloso A, Ortín J. Interaction of influenza virus NS1 protein and the human homologue of Staufen in vivo and in vitro. Nucleic Acids Res 1999;27(11):2241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.de Lucas S, Peredo J, Marión RM, Sánchez C, Ortín J. Human Staufen1 protein interacts with influenza virus ribonucleoproteins and is required for efficient virus multiplication. Journal of virology. 2010;84(15):7603–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee JH, Oh JY, Pascua PN, Kim EG, Choi YK, Kim HK. Impairment of the Staufen1-NS1 interaction reduces influenza viral replication. Biochemical and biophysical research communications. 2011;414(1):153–8. [DOI] [PubMed] [Google Scholar]
- 92.Cho H, Ahn SH, Kim KM, Kim YK. Non-structural protein 1 of influenza viruses inhibits rapid mRNA degradation mediated by double-stranded RNA-binding protein, staufen1. FEBS letters. 2013;587(14):2118–24. [DOI] [PubMed] [Google Scholar]
- 93.Ricci EP, Kucukural A, Cenik C, Mercier BC, Singh G, Heyer EE, et al. Staufen1 senses overall transcript secondary structure to regulate translation. Nature structural & molecular biology. 2014;21(1):26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Laver JD, Li X, Ancevicius K, Westwood JT, Smibert CA, Morris QD, et al. Genome-wide analysis of Staufen-associated mRNAs identifies secondary structures that confer target specificity. Nucleic Acids Res 2013;41(20):9438–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, D’Ambrogio A, et al. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature. 2015;519(7544):491–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Fischer JW, Busa VF, Shao Y, Leung AKL. Structure-Mediated RNA Decay by UPF1 and G3BP1. Mol Cell. 2020;78(1):70–84.e6.32017897 **Fischer et. al. demonstrated structure-mediated RNA decay (SRD) of mRNAs and circRNAs was mediated by Upf1 and G3BP1. This novel pathway could potentially target diverse viruses since the 3’ UTRs of viral RNAs are often highly structured. Also, since non-coding circRNAs are targeted, it is conceivable that non-coding viral RNAs could also be susceptible to SRD.
- 97.Matsuki H, Takahashi M, Higuchi M, Makokha GN, Oie M, Fujii M. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes to cells : devoted to molecular & cellular mechanisms. 2013;18(2):135–46. [DOI] [PubMed] [Google Scholar]
- 98.Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R. Distinct stages in stress granule assembly and disassembly. Elife 2016;5:e18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ivanov P, Kedersha N, Anderson P. Stress Granules and Processing Bodies in Translational Control. Cold Spring Harb Perspect Biol 2019;11(5):a032813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Krapp S, Greiner E, Amin B, Sonnewald U, Krenz B. The stress granule component G3BP is a novel interaction partner for the nuclear shuttle proteins of the nanovirus pea necrotic yellow dwarf virus and geminivirus abutilon mosaic virus. Virus research. 2017;227:6–14. [DOI] [PubMed] [Google Scholar]
- 101.Yang W, Ru Y, Ren J, Bai J, Wei J, Fu S, et al. G3BP1 inhibits RNA virus replication by positively regulating RIG-I-mediated cellular antiviral response. Cell death & disease. 2019;10(12):946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim SS, Sze L, Liu C, Lam KP. The stress granule protein G3BP1 binds viral dsRNA and RIG-I to enhance interferon-β response. The Journal of biological chemistry. 2019;294(16):6430–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cobos Jiménez V, Martinez FO, Booiman T, van Dort KA, van de Klundert MA, Gordon S, et al. G3BP1 restricts HIV-1 replication in macrophages and T-cells by sequestering viral RNA. Virology. 2015;486:94–104. [DOI] [PubMed] [Google Scholar]
- 104.Visser LJ, Medina GN, Rabouw HH, de Groot RJ, Langereis MA, de Los Santos T, et al. Foot-and-Mouth Disease Virus Leader Protease Cleaves G3BP1 and G3BP2 and Inhibits Stress Granule Formation. Journal of virology. 2019;93(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.White JP, Cardenas AM, Marissen WE, Lloyd RE. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2007;2(5):295–305. [DOI] [PubMed] [Google Scholar]
- 106.Ng CS, Jogi M, Yoo J-S, Onomoto K, Koike S, Iwasaki T, et al. Encephalomyocarditis Virus Disrupts Stress Granules, the Critical Platform for Triggering Antiviral Innate Immune Responses. Journal of virology. 2013;87(17):9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cristea IM, Rozjabek H, Molloy KR, Karki S, White LL, Rice CM, et al. Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: role for G3BP1 and G3BP2 in virus replication. Journal of virology. 2010;84(13):6720–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Götte B, Panas MD, Hellström K, Liu L, Samreen B, Larsson O, et al. Separate domains of G3BP promote efficient clustering of alphavirus replication complexes and recruitment of the translation initiation machinery. PLoS Pathog 2019;15(6):e1007842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hosmillo M, Lu J, McAllaster MR, Eaglesham JB, Wang X, Emmott E, et al. Noroviruses subvert the core stress granule component G3BP1 to promote viral VPg-dependent translation. Elife 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]


