Abstract
MUTYH-associated polyposis (MAP) is an autosomal recessive disorder characterized by the development of multiple adenomatous colonic polyps and an increased lifetime risk of colorectal cancer. Germline biallelic pathogenic variants in MUTYH are responsible for MAP. The MUTYH c.934-2A > G (NM_001128425.1) variant, which is also known as c.850-2A > G for NM_001048174.2, has been identified in our laboratory in more than 800 patients, including homozygous and compound heterozygote carriers. The variant was initially classified as a variant of uncertain significance (VUS) because of lack of a MAP phenotype in biallelic carriers. In two unrelated female patients who were heterozygous carriers of this variant, further testing by RNA sequencing identified an aberrant transcript with a deletion of 9 nt at the start of exon 11 (MUTYH r.934_942del9). This event is predicted to lead to an in-frame loss of three amino acids in a noncritical domain of the protein. This was the only splice defect identified in these patients that was not present in the controls, and the aberrant transcript is derived exclusively from the variant allele, strongly supporting the cause of this splice defect as being the intronic variant, MUTYH c.934-2A > G. The splicing analysis demonstrating a small in-frame skipping of three amino acids in a noncritical domain, along with the absence of a MAP phenotype in our internal cohort of biallelic carriers, provides evidence that the variant is likely benign and not of clinical significance.
Keywords: hamartomatous polyps, neoplasm of the gastrointestinal tract
CASE PRESENTATION
The MUTYH gene (NM_001128425.1) encodes an adenine DNA glycosylase that is involved in base excision repair necessary for the repair of oxidative damage and prevention of tumorigenesis (Pezzi et al. 2009). Biallelic pathogenic mutations in MUTYH cause the autosomal recessive disorder MUTYH-associated polyposis (MAP; OMIM #604933), which is typically characterized by the presence of 20–99 colonic adenomatous polyps (although other types of polyps may occur) and an 80% risk of colorectal cancer by age 70 (Syngal et al. 2015; Kantor et al. 2017). Molecular genetic testing is recommended for patients suspected of having MAP, in which early diagnosis can reduce morbidity and mortality through available surveillance and prophylaxis. National Comprehensive Cancer Network (NCCN) Guidelines recommend colonoscopy and polypectomy every 1–2 yr starting as early as age 25 for biallelic MUTYH carriers. The NCCN further recommends prophylactic colectomy and ileorectal anastomosis when adenomas cannot be treated endoscopically (NCCN 2019).
Rare, canonical splice site alterations are often classified as likely pathogenic by clinical diagnostic laboratories based on the American College of Medical Genetics and Genomics/Association for Molecular Pathology (ACMG/AMP) guidelines (Richards et al. 2015; Pesaran et al. 2016). However, MUTYH c.934-2A > G occurs at high frequency (∼1.5%–2.6%) in East Asian general populations and is reported as homozygous in some of these individuals (Lek et al. 2016). Taken together, these conflicting data led to an initial classification MUTYH c.934-2A > G as a variant of uncertain significance (VUS). Furthermore, this variant has not been identified in a patient with MAP in a homozygous state or in conjunction with a second pathogenic mutation in MUTYH. In two cases in which this alteration was identified in unrelated Asian females by multigene panel testing (MGPT), further investigation was performed by RNA sequencing (Table 1).
Table 1.
Clinical characteristics and results of RNA sequencing analysis for the two cases presented in this study
| Case | Sex | Age at diagnosis | Cancer history | Colorectal polyps | Family history | MUTYH NM_001128425.1 variants identified | Zygosity | Results of RNA analysis | Other variants identified |
|---|---|---|---|---|---|---|---|---|---|
| 1 | F | Early 30s | Colon cancer | No report of polyps | Pancreatic, head and neck, gastric, and breast cancers | c.934-2A > G | Heterozygous | r.934_942del9 (in-frame deletion of 9 nt in exon 11) | None |
| 2 | F | No data | No personal history of cancer | 7–8 polyps | Breast, pancreatic, prostate cancers, and lymphoma; possible colon and stomach cancers | c.934-2A > G | Heterozygous | r.934_942del9 (in-frame deletion of 9 nt in exon 11) | VUS in RAD50 |
(VUS) Variant of uncertain significance.
Case 1: This proband had a history of colon cancer diagnosed in her late 30s and the tumor was microsatellite stable with normal expression of the MLH1, MSH2, and MSH6 mismatch repair proteins by immunohistochemical (IHC) staining. No history of polyps was noted. The IHC staining result for PMS2 was not provided. There was a family history of pancreatic cancer, head and neck cancer, gastric cancer, and breast cancer in several first- and second-degree relatives. The results of MGPT in this individual did not identify any other alterations of potential clinical relevance in MUTYH nor in any other gene on the panel.
Case 2: This proband was reported to have seven to eight colon polyps of unspecified histology, but no personal history of cancer. There was a family history of breast cancer, pancreatic cancer, prostate cancer, lymphoma, and possibly colon and stomach cancer in several first- and second-degree relatives. The results of MGPT in this individual did not identify any other alterations of potential clinical relevance in MUTYH; however, there was a missense VUS in RAD50, c.3902A > G (p.K1301R).
VARIANT INTERPRETATION
MUTYH c.934-2A > G is reported in the ClinVar public database (access date 7/19/2021: VCV000041766.23) with conflicting interpretations of pathogenicity: pathogenic, likely pathogenic, variant of uncertain significance, and likely benign. In addition, there is conflicting evidence of pathogenicity in the literature (Tao et al. 2004; Taki et al. 2016; Thibodeau et al. 2019). Within the framework of the ACMG/AMP guidelines, canonical splice site alterations are considered as inherently having very strong prior probability of being pathogenic (Richards et al. 2015; Pesaran et al. 2016). However, this weight may be reduced when the predicted splice impact results in an in-frame transcript not subject to nonsense-mediated decay (NMD) and/or when there is a lack of supporting clinical phenotype. In support of this variant being interpreted carefully, thorough review of the literature identified no homozygous or compound heterozygous patients with polyposis despite a high general population frequency of >1% in several Asian populations (Tao et al. 2004, 2008; Miyaki et al. 2005; Kim et al. 2007; Johnston et al. 2012; Bodian et al. 2014; Kurian et al. 2014; Jang et al. 2015; Olfson et al. 2015; Jamuar et al. 2016; Kline et al. 2016; Lin et al. 2016; Taki et al. 2016; Cheng et al. 2017; DeRycke et al. 2017; Hansen et al. 2017; Kobayashi et al. 2017; Zhang et al. 2017; Reuter et al. 2018; Takao et al. 2018). Furthermore, in the general population database, gnomAD, this variant has been observed in the Asian general population five times in a homozygous state (gnomAD, dbSNP: rs77542170). Although the phenotypes of these individuals are unknown, they represent the general population as opposed to a cancer cohort. The frequency of this variant combined with the lack of homozygotes and compound heterozygotes in MAP patients is evidence that this variant is benign.
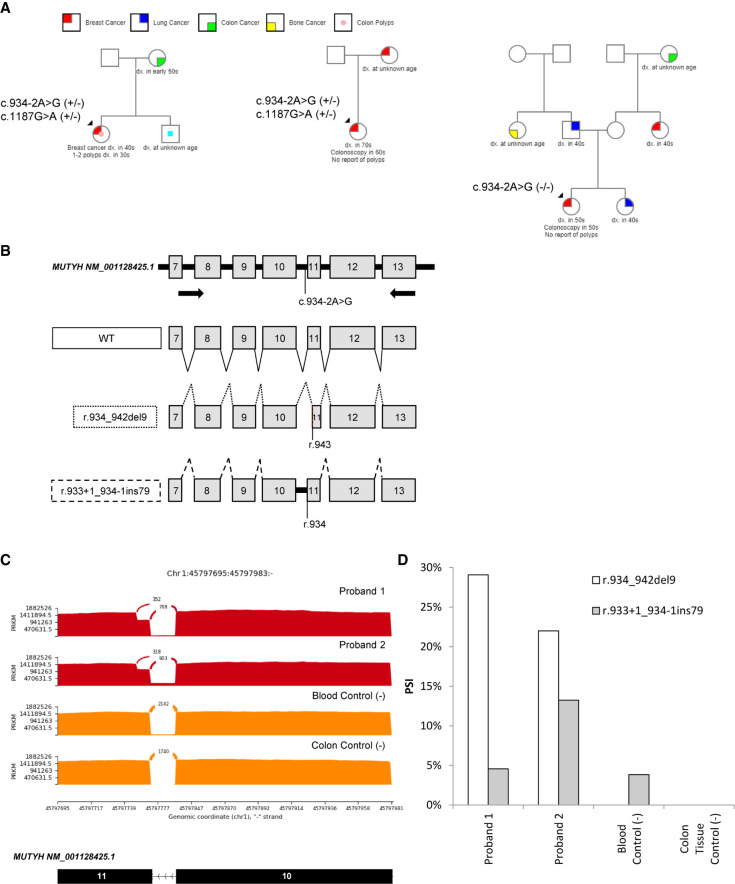
A retrospective review of our laboratory's clinical testing database was performed to identify all probands with this variant in a homozygous state or a compound heterozygous state with another pathogenic MUTYH alteration. Of 418 probands with this variant, one was a confirmed homozygous case, whereas the other two had co-occurrence of this variant with the pathogenic founder mutation MUTYH c.1187G > A (p.G396D), but phase was not determined (Fig. 1A). Only one of these reported a history of colon polyps (one to two colon polyps in her 30s). The other two probands reported having colonoscopies, but no polyp history was reported.
Figure 1.
Family histories and massively parallel RNA sequencing (RNA-seq) provide evidence that MUTYH NM_001128425.1 c.934-2A > G is likely benign. (A) Pedigrees from retrospective analysis of cohort representing a homozygous proband (right) and two compound heterozygote probands (center and left). (B) Schematic representation of reverse transcription polymerase chain reaction (RT-PCR) primer design (top), the wild type (WT; solid lines) transcript, and the two splicing events observed: r.934_942del9 (dotted lines) and r.933 + 1_934-1ins79 (dashed lines). (C) Sashimi plot of the two heterozygous probands (red) and two controls (orange) showing RNA-seq reads supporting the partial exon deletion (top arch), wild type (middle arch), and intron inclusion (bottom line). (D) Quantitation of percent spliced in (PSI) for r.933 + 1_934-1ins79 (gray) and r.934_942del9 (white) observed in massively parallel RNA-seq.
For this study, two probands were available and willing to provide an additional blood sample for quantitative RNA sequencing (RNA-seq) analysis of MUTYH c.934-2A > G, as described previously (Schafer et al. 2015; Farber-Katz et al. 2018; Karam et al. 2019; Landrith et al. 2020). RNA-seq reflected that the majority of abnormal transcript present in both probands was r.934_942del9 (del9; Fig. 1C,D). Of note, one patient carries the polymorphism MUTYH c.1014G > C. Analysis of this polymorphism revealed that all the del9 transcripts were derived exclusively from the allele harboring MUTYH c.934-2A > G. There was no allele bias in the amplification of this transcript as the polymorphism was present in equal amounts in the pool of transcripts (data not shown).
The c.934-2A > G alteration results in the in-frame deletion of 9 nt from the 5′ end of MUTYH exon 11, which is predicted by in silico splice site analysis (Jaganathan et al. 2019). This transcript was absent in normal blood and tissue controls, further confirming the observed splice defect is caused by the variant (Fig. 1C,D). This small deletion is predicted to lead to an in-frame loss of three amino acids (p.V312_Q314del). Another transcript (r.933 + 1_934-1ins79), which results in out-of-frame inclusion of intron 10, was identified in both the probands and control samples. This transcript is known to be a low-level, naturally occurring, alternative splicing event (Fig. 1C; Tao et al. 2004; Taki et al. 2016). RNA data from other published studies of MUTYH c.934-2A > G are derived from reverse transcription polymerase chain reaction (RT-PCR) followed by gel electrophoresis, which would be unable to resolve a small-nucleotide change such as del9. In Tao et al. (2004), the authors followed up with quantitative RT-PCR of patient material, but primers designed to detect the variant-type and wild-type transcripts would not detect the del9 transcript. The forward primer for the variant-type transcript was designed to anneal to intron 10, which is absent in the del9 transcript. In addition, the reverse primer for the wild-type transcript spans exons 10/11 and loss of 9 nt from the 5′ end of exon 11 in the del9 transcript would prevent annealing of the primer (Taki et al. 2016). The del9 transcript was detected in the Taki et al. (2016) study after sequencing of the wild-type RT-PCR product from patient samples. A third study identified both the wild-type and del9 transcripts using RNA-seq, but results were not quantitative and a comparison with control samples was not performed (Thibodeau et al. 2019).
The significance of the amino acids predicted to be lost by the del9 transcript (p.V312_Q314del) was investigated by searching for evidence of pathogenicity of missense changes at these three amino acids in public databases (ClinVar, HGMD) and in the literature, although there was limited evidence available. A computational-based structural analysis of this portion of the protein was also limited by the fact that the residues constitute part of a partially flexible linker between the domains and do not engage in any apparent specific interactions (Wang et al. 2015). Flexible linkers are often more tolerant of sequence variation, with any effects often being too subtle to estimate by computational methods.
The cumulative evidence described in this work led to a reclassification of this variant from a variant of uncertain significance to a likely benign variant. The following ACMG criteria were applied: BS1 (subpopulation frequency of 1.539% in East Asian population in gnomAD), BS2 (greater than four to nine homozygotes, presumably without MAP features in gnomAD), and BP2 (co-occurrence with the founder mutation, MUTYH c.1187G > A, p.G396D, in two internal cases without MAP features). These data are now available in ClinVar (SCV000183748.8).
SUMMARY
Quantitative RNA sequencing analysis demonstrated that MUTYH c.934-2A > G leads to a single aberrant transcript with deletion of 9 nt from the 5′ end of exon 11. This transcript is predicted to lead to an in-frame loss of three amino acids (p.V312_Q314del), which is unlikely to have a significant effect on protein function. Previous RNA studies reported in the literature either failed to detect the del9 transcript in which gel electrophoresis was applied because of its small size difference from wild type, or it was detected as a very minor proportion of aberrant transcripts because of limitations in the study design and methods (Tao et al. 2004; Taki et al. 2016; Thibodeau et al. 2019). This is also the first time the intron 10 retention event was reported in both MUTYH c.934-2A > G carriers and wild-type controls; it is apparently the result of routine alternative splicing. MUTYH c.934-2A > G was classified as likely benign based on these data combined with high population frequency and homozygous observation of this variant in Eastern Asian general populations, which was further supported by the observation in our laboratory in a homozygous and compound heterozygous state in patients without MAP. Last, it is important to reiterate that caution should be used when applying a very strong prior probability of being pathogenic for canonical splice site alterations if the predicted or demonstrated splice impact results in an in-frame transcript not subject to NMD, especially if there is a lack of clinical phenotype in highly penetrant conditions.
ADDITIONAL INFORMATION
Database Deposition and Access
These data are available in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) under accession number SCV000183748.8.
Ethics Statement
This study was approved by the Western Institutional Review Board. All patients provided written consent to genetic testing and research-based RNA studies at Ambry Genetics.
Acknowledgments
The authors of this study acknowledge the patients who participated in this study.
Author Contributions
F.H., B.R.C., M.E.R., H.L., E.C., T.P., and R.K. participated in manuscript preparation and editing. B.R.C. and F.H. participated in data acquisition and analysis. E.C., T.P., and R.K. were responsible for study design.
Funding
This study was funded by Ambry Genetics. All authors were employees of Ambry Genetics for the duration of this study.
Competing Interest Statement
All authors were employees of Ambry Genetics for the duration of this study.
REFERENCES
- Bodian DL, McCutcheon JN, Kothiyal P, Huddleston KC, Iyer RK, Vockley JG, Niederhuber JE. 2014. Germline variation in cancer-susceptibility genes in a healthy, ancestrally diverse cohort: implications for individual genome sequencing. PLoS ONE 9: e94554. 10.1371/journal.pone.0094554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng DT, Prasad M, Chekaluk Y, Benayed R, Sadowska J, Zehir A, Syed A, Wang YE, Somar J, Li Y, et al. 2017. Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med Genomics 10: 33. 10.1186/s12920-017-0271-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRycke MS, Gunawardena S, Balcom JR, Pickart AM, Waltman LA, French AJ, McDonnell S, Riska SM, Fogarty ZC, Larson MC, et al. 2017. Targeted sequencing of 36 known or putative colorectal cancer susceptibility genes. Mol Genet Genomic Med 5: 553–569. 10.1002/mgg3.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber-Katz S, Hsuan V, Wu S, Landrith T, Vuong H, Xu D, Li B, Hoo J, Lam S, Nashed S, et al. 2018. Quantitative analysis of BRCA1 and BRCA2 germline splicing variants using a novel RNA-massively parallel sequencing assay. Front Oncol 8: 286. 10.3389/fonc.2018.00286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen MF, Johansen J, Sylvander AE, Bjørnevoll I, Talseth-Palmer BA, Lavik LAS, Xavier A, Engebretsen LF, Scott RJ, Drabløs F, et al. 2017. Use of multigene-panel identifies pathogenic variants in several CRC-predisposing genes in patients previously tested for Lynch syndrome. Clin Genet 92: 405–414. 10.1111/cge.12994 [DOI] [PubMed] [Google Scholar]
- Jaganathan K, Panagiotopoulou KS, McRae JF, Darbandi SF, Knowles D, Li YI, Kosmicki JA, Arbelaez J, Cui W, Schwartz GB, et al. 2019. Predicting splicing from primary sequence with deep learning. Cell 176: 535–548.e24. 10.1016/j.cell.2018.12.015 [DOI] [PubMed] [Google Scholar]
- Jamuar SS, Kuan JL, Brett M, Tiang Z, Tan WL, Lim JY, Liew WK, Javed A, Liew WK, Law HY, et al. 2016. Incidentalome from genomic sequencing: a barrier to personalized medicine? EBioMedicine 5: 211–216. 10.1016/j.ebiom.2016.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MA, Lee SH, Kim N, Ki CS. 2015. Frequency and spectrum of actionable pathogenic secondary findings in 196 Korean exomes. Genet Med 17: 1007–1011. 10.1038/gim.2015.26 [DOI] [PubMed] [Google Scholar]
- Johnston JJ, Rubinstein WS, Facio FM, Ng D, Singh LN, Teer JK, Mullikin JC, Biesecker LG. 2012. Secondary variants in individuals undergoing exome sequencing: screening of 572 individuals identifies high-penetrance mutations in cancer-susceptibility genes. Am J Hum Genet 91: 97–108. 10.1016/j.ajhg.2012.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor M, Sobrado J, Patel S, Eiseler S, Ochner C. 2017. Hereditary colorectal tumors: a literature review on MUTYH-associated polyposis. Gastroenterol Res Pract 2017: 8693182. 10.1155/2017/8693182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam R, Conner B, LaDuca H, McGoldrick K, Krempely K, Richardson ME, Zimmermann H, Gutierrez S, Reineke P, Hoang L, et al. 2019. Assessment of diagnostic outcomes of RNA genetic testing for hereditary cancer. JAMA Netw Open 2: e1913900. 10.1001/jamanetworkopen.2019.13900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, DW., Kim, IJ., Kang, HC., Jang, SG., Kim, K., Yoon, HJ., Ahn, SA., Han, SY., Hong, SH., Hwang, JA., et al. Germline mutations of the MYH gene in Korean patients with multiple colorectal adenomas. Int J Colorectal Dis 2007; 22: 1173–1178. 10.1007/s00384-007-0289-8 [DOI] [PubMed] [Google Scholar]
- Kline CN, Joseph NM, Grenert JP, van Ziffle J, Yeh I, Bastian BC, Mueller S, Solomon DA. 2016. Inactivating MUTYH germline mutations in pediatric patients with high-grade midline gliomas. Neuro Oncol 18: 752–753. 10.1093/neuonc/now013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Yang S, Nykamp K, Garcia J, Lincoln SE, Topper SE. 2017. Pathogenic variant burden in the ExAC database: an empirical approach to evaluating population data for clinical variant interpretation. Genome Med 9: 13. 10.1186/s13073-017-0403-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian AW, Hare EE, Mills MA, Kingham KE, McPherson L, Whittemore AS, McGuire V, Ladabaum U, Kobayashi Y, Lincoln SE, et al. 2014. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol 32: 2001–2009. 10.1200/JCO.2013.53.6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrith L, Li B, Cass AA, Conner BR, LaDuca H, McKenna DB, Maxwell KN, Domchek S, Morman NA, Heinlen C, et al. 2020. Splicing profile by capture RNA-seq identifies pathogenic germline variants in tumor suppressor genes. NPJ Precis Oncol 4: 4. 10.1038/s41698-020-0109-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Exome Aggregation Consortium. http://exac.broadinstitute.org/.
- Lin PH, Kuo WH, Huang AC, Lu YS, Lin CH, Kuo SH, Wang MY, Liu CY, Cheng FT, Yeh MH, et al. 2016. Multiple gene sequencing for risk assessment in patients with early-onset or familial breast cancer. Oncotarget 7: 8310–8320. 10.18632/oncotarget.7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaki M, Iijima T, Yamaguchi T, Hishima T, Tamura K, Utsunomiya J, Mori T. 2005. Germline mutations of the MYH gene in Japanese patients with multiple colorectal adenomas. Mutat Res 578: 430–433. 10.1016/j.mrfmmm.2005.01.017 [DOI] [PubMed] [Google Scholar]
- NCCN Clinical Practice Guidelines in Oncology. 2019. Genetic/Familial High-Risk Assessment: Colorectal. V.3.2017. Available at nccn.org.
- Olfson E, Cottrell CE, Davidson NO, Gurnett CA, Heusel JW, Stitziel NO, Chen LS, Hartz S, Nagarajan R, Saccone NL, et al. 2015. Identification of medically actionable secondary findings in the 1000 genomes. PLoS ONE 10: e0135193. 10.1371/journal.pone.0135193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaran T, Karam R, Huether R, Li S, Farber-Katz S, Chamberlin A, Chong H, LaDuca H, Elliott A. 2016. Beyond DNA: an integrated and functional approach for classifying germline variants in breast cancer genes. Int J Breast Cancer 2016: 2469523. 10.1155/2016/2469523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzi A, Roncucci L, Benatti P, Sassatelli R, Varesco L, Di Gregorio C, Venesio T, Pedroni M, Maffei S, Reggiani Bonetti L, et al. 2009. Relative role of APC and MUTYH mutations in the pathogenesis of familial adenomatous polyposis. Scand J Gastroenterol 44: 1092–1100. 10.1080/00365520903100481 [DOI] [PubMed] [Google Scholar]
- Reuter MS, Walker S, Thiruvahindrapuram B, Whitney J, Cohn I, Sondheimer N, Yuen RKC, Trost B, Paton TA, Pereira SL, et al. 2018. The Personal Genome Project Canada: findings from whole genome sequences of the inaugural 56 participants. Can Med Assoc J 190: E126–E136. 10.1503/cmaj.171151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–424. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer S, Miao K, Benson CC, Heinig M, Cook SA, Hubner N. 2015. Alternative splicing signature in RNA-seq data: percent spliced in (PSI). Curr Protoc Hum Genet 87: 11.16.1–11.16.14. 10.1002/0471142905.hg1116s87 [DOI] [PubMed] [Google Scholar]
- Syngal S, Brand RE, Church JM, Giardiello FM, Hampel HL, Burt RW. 2015. American College of Gastroenterology. ACG clinical guideline: genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol 110: 223–262. 10.1038/ajg.2014.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao M, Yamaguchi T, Eguchi H, Tada Y, Kohda M, Koizumi K, Horiguchi S, Okazaki Y, Ishida H. 2018. Characteristics of MUTYH variants in Japanese colorectal polyposis patients. Int J Clin Oncol 23: 497–503. 10.1007/s10147-017-1234-7 [DOI] [PubMed] [Google Scholar]
- Taki K, Sato Y, Nomura S, Ashihara Y, Kita M, Tajima I, Sugano K. 2016. Arai M Mutation analysis of MUTYH in Japanese colorectal adenomatous polyposis patients. Fam Cancer 15: 261–265. 10.1007/s10689-015-9857-1 [DOI] [PubMed] [Google Scholar]
- Tao H, Shinmura K, Hanaoka T, Natsukawa S, Shaura K, Koizumi Y, Kasuga Y, Ozawa T, Tsujinaka T, Li Z, et al. 2004. A novel splice-site variant of the base excision repair gene MYH is associated with production of an aberrant mRNA transcript encoding a truncated MYH protein not localized in the nucleus. Carcinogenesis 25: 1859–1866. 10.1093/carcin/bgh206 [DOI] [PubMed] [Google Scholar]
- Tao H, Shinmura K, Suzuki M, Kono S, Mibu R, Tanaka M, Kakeji Y, Maehara Y, Okamura T, Ikejiri K, et al. 2008. Association between genetic polymorphisms of the base excision repair gene MUTYH and increased colorectal cancer risk in a Japanese population. Cancer Science 99: 355–360. 10.1111/j.1349-7006.2007.00694.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau ML, Zhao EY, Reisle C, Ch'ng C, Wong HL, Shen Y, Jones MR, Lim HJ, Young S, Cremin C, et al. 2019. Base excision repair deficiency signatures implicate germline and somatic MUTYH aberrations in pancreatic ductal adenocarcinoma and breast cancer oncogenesis. Cold Spring Harb Mol Case Stud 5: a003681. 10.1101/mcs.a003681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lee SJ, Verdine GL. 2015. Structural basis for avoidance of promutagenic DNA repair by MutY adenine DNA glycosylase. J Biol Chem 290: 17096–17105. 10.1074/jbc.M115.657866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wang X, de Voer RM, Hehir-Kwa JY, Kamping EJ, Weren RDA, Nelen M, Hoischen A, Ligtenberg MJL, Hoogerbrugge N, et al. 2017. A molecular inversion probe-based next-generation sequencing panel to detect germline mutations in Chinese early-onset colorectal cancer patients. Oncotarget 8: 24533–24547. 10.18632/oncotarget.15593 [DOI] [PMC free article] [PubMed] [Google Scholar]