Abstract
Aspergillus flavus is second to A. fumigatus as a cause of invasive aspergillosis, but no standard method exists for molecular typing of strains from human sources. A repetitive DNA sequence cloned from A. flavus and subcloned into a pUC19 vector, pAF28, was used to type 18 isolates from diverse clinical, environmental, and geographic sources. The restriction fragment length polymorphisms generated with EcoRI- or PstI-digested genomic DNA and probed with digoxigenin-labeled pAF28 revealed complete concordance between patterns. Eighteen distinct fingerprints were observed. The probe was used to investigate two cases of cutaneous A. flavus infection in low-birth-weight infants in a neonatal intensive care unit (NICU). Both infants were transported by the same ambulance and crew to the NICU on the same day. A. flavus strains of the same genotype were isolated from both infants, from a roll of tape used to fasten their umbilical catheters, from a canvas bag used to store the tape in the ambulance, and from the tape tray in the ambulance isolette. These cases highlight the need to consider exposures in critically ill neonates that might occur during their transport to the NICU and for stringent infection control practices. The hybridization profiles of strains from a second cluster of invasive A. flavus infections in two pediatric hematology-oncology patients revealed a genotype common to strains from a definite case patient and a health care worker. A probable case patient was infected with a strain with a genotype different from that of the strain from the definite case patient but highly related to that of an environmental isolate. The high degree of discrimination and reproducibility obtained with the pAF28 probe underscores its utility for typing clinical and environmental isolates of A. flavus.
Members of the genus Aspergillus are among the most widespread fungi in the environment, being found in the soil, on plants, in dust, on food, and in the air. In nonimmunocompromised persons, these molds can cause localized infections of the lungs, sinuses and other sites. In immunocompromised individuals, inhalation of conidia gives rise to invasive infection of the lungs or sinuses, often followed by dissemination to other organs. Involvement of the skin is uncommon, but two forms of cutaneous aspergillosis have been described in compromised patients (26; T. J. Walsh, Editorial Response, Clin. Infect. Dis. 27:453–457, 1998). In primary cutaneous aspergillosis, lesions arise at or near intravenous catheter insertion sites, at surgical wound sites, and at sites associated with occlusive dressings or burns. In secondary cutaneous aspergillosis, the lesions result either from extension to the skin from underlying infected structures or from hematogenous dissemination. Most reported cases of cutaneous aspergillosis have been caused by Aspergillus fumigatus, but a number have been associated with A. flavus, the second most common etiologic agent of human aspergillosis.
The evidence incriminating different environmental sources of A. flavus infection has always been circumstantial because only recently have molecular typing methods been developed to trace the spread of particular subspecific strains. Among the typing methods that have been applied to A. flavus are restriction endonuclease analysis and the detection of restriction fragment length polymorphisms (RFLPs) by Southern hybridization and probing with ribosomal or other repetitive sequences (19, 20). The application of randomly amplified polymorphic DNA (RAPD) analysis as a genotyping method has also been reported (14).
Restriction endonuclease analysis of total cellular DNA has not proved to be a suitable method for discrimination of strains of A. flavus (2). Moody and Tyler (19) identified mitochondrial DNA RFLPs and used them to propose the occurrence of species within the A. flavus group: A. flavus, A. parasiticus, and A. nomius. RFLP analysis of A. flavus nuclear DNA probed with recombinant DNA clones from A. nidulans and Neurospora crassa supported the results obtained with mitochondrial DNA but revealed limited geographic correlations among A. flavus strains (20). RAPD analysis has been used to type strains of A. flavus, but the profiles may suffer from a lack of reproducibility (14). Moreover, RAPD analysis may not be appropriate as a tool for epidemiologic tracking of isolates or for surveying the genetic variation in natural populations because of its inability to discriminate artifactual variation from actual polymorphism (24, 25). Buffington et al. (2) combined the products from RAPD analysis-RFLP analysis of a tester strain of A. flavus to produce a DNA probe for Southern blot analysis. Although a high degree of discrimination among strain types was achieved, the probe and target sequences are anonymous and the probe is available in only a single laboratory.
In an effort to develop an improved method for molecular typing of clinical and environmental isolates of A. flavus, we evaluated a DNA fingerprinting procedure that uses a repetitive DNA sequence cloned from A. flavus var. flavus to probe RFLPs of genomic DNA (16). The method was evaluated with a panel of clinical and environmental isolates of A. flavus from different sources and then was applied to strains from two small clusters of pediatric cases of invasive aspergillosis: cutaneous A. flavus infection in low-birth-weight infants from a neonatal intensive care unit (NICU) and invasive pulmonary aspergillosis in a hematology-oncology unit (2).
MATERIALS AND METHODS
Isolates.
A total of 31 isolates of A. flavus were studied (Table 1). These included six clinical and environmental isolates (isolates AFL1 to AFL6) obtained from a cluster of cutaneous aspergillosis cases in the NICU at Cook Children's Medical Center, Fort Worth, Tex. A single control isolate of A. flavus unrelated to this cluster (isolate AFL7) was provided by the Texas Department of Health. The remainder of the collection consisted of seven A. flavus isolates (isolates AFL8 to AFL14) collected during an outbreak of invasive aspergillosis in the hematology-oncology and bone marrow transplant (BMT) units at Los Angeles Children's Hospital, Los Angeles, Calif. (2); four nasal sinus isolates (isolates AFL15 to AFL18) obtained from patients with allergic fungal sinusitis at the Memorial Medical Center in Savannah, Ga.; four isolates (isolates AFL19 to AFL22) obtained from the American Type Culture Collection; six isolates from human sources (isolates AFL23 to AFL28) obtained from the culture collections of the Mycotic Diseases Branch, Centers for Disease Control and Prevention, Atlanta, Ga., or the National Institutes of Health, Bethesda, Md.; and two aflatoxin-producing isolates (isolates AFL29 and AFL30) and one non-aflatoxin-producing isolate (isolate AFL31) obtained from the National Peanut Research Laboratory, Dawson, Ga.
TABLE 1.
Origins of A. flavus isolates collected from patients with aspergillosis and the hospital environment and strains of diverse geographic and clinical originsa
| Isolate no. | Geographic origin | Source | Culture no. | Genotype by Southern blot analysis |
|---|---|---|---|---|
| AFL1 | Fort Worth, Tex. | Abdominal skin lesion, patient A | 98-019777 | A |
| AFL2 | Fort Worth, Tex. | Abdominal skin lesion, patient B | 98-019778 | A |
| AFL3 | Fort Worth, Tex. | Adhesive tape | 98-019779 | A |
| AFL4 | Fort Worth, Tex. | Blood culture, patient C | 98-019781 | A |
| AFL5 | Fort Worth, Tex. | Transport isolette | 98-019782 | A |
| AFL6 | Fort Worth, Tex. | Canvas tape bag | 98-019783 | A |
| AFL7 | Fort Worth, Tex. | TDOH | 98-019784 | B |
| AFL8 | Los Angeles, Calif. | Ward 4 West vent | B5105 | C |
| AFL9 | Los Angeles, Calif. | BMT nursing station vent | B5106 | D |
| AFL10 | Los Angeles, Calif. | Nasal sinus of HCW | B5107 | E |
| AFL11 | Los Angeles, Calif. | Probable IA, primary blood culture, patient 2 | B5108 | D |
| AFL12 | Los Angeles, Calif. | Probable IA, secondary blood culture, patient 2 | B5109 | F |
| AFL13 | Los Angeles, Calif. | Definite IA, blood culture, patient 1 | B5110 | E |
| AFL14 | Los Angeles, Calif. | Ward 3 West vent (adjacent NICU) | B5117 | D |
| AFL15 | Savannah, Ga. | Nasal sinus | AFS1 | G |
| AFL16 | Savannah, Ga. | Nasal sinus | AFS2 | H |
| AFL17 | Savannah, Ga. | Nasal sinus | AFS3 | I |
| AFL18 | Savannah, Ga. | Nasal sinus | AFS4 | J |
| AFL19 | Ohio | Human sputum | ATCC 64025 | K |
| AFL20 | New Jerseyb | Proteinase production | ATCC 11497 | D |
| AFL21 | Thailand | Soy sauce, Koji | ATCC 44310 | L |
| AFL22 | Virginia | Prosthetic mitral valve | ATCC 34896 | M |
| AFL23 | Unknown | Unknown clinical isolate | 96001 | N |
| AFL24 | Maryland | Amputation stump, 1959 | NIH 5239 | O |
| AFL25 | Maryland | Unknown clinical isolate, 1959 | NIH 5251 | P |
| AFL26 | Maryland | Unknown clinical isolate, 1960 | NIH 5250 | Q |
| AFL27 | Brazil | Lung tissue | B4955 | R |
| AFL28 | Pennsylvania | Prosthetic heart valve | B5096 | S |
| AFL29 | Dawson, Ga. | Peanut | NPRL1 (high aflatoxin) | T |
| AFL30 | Dawson, Ga. | Peanut | NPRL2 (low aflatoxin) | U |
| AFL31 | Dawson, Ga. | Peanut | NPRL3 (no aflatoxin) | V |
Abbreviations; HCW, health care worker; TDOH, Texas State Department of Health; IA, invasive aspergillosis; NPRL, National Peanut Research Laboratory, Dawson, Ga.; ATCC, American Type Culture Collection, Rockville, Md.; NIH, National Institutes of Health, Bethesda, Md.
Culture was originally deposited in 1929 by Selman Waksman, New Brunswick, N.J.
Isolates were confirmed as A. flavus on the basis of their macroscopic and microscopic characteristics in culture (12). Isolates were maintained on 2.5% malt extract agar slants and were stored at 4°C. In preparation for DNA extraction and molecular typing, monoconidial isolates of each A. flavus culture were produced by the limiting dilution method. Conidial suspensions were harvested from a 7-day growth of a single conidium on potato dextrose agar and were stored in 50% (vol/vol) sterile glycerol–phosphate-buffered saline at −20°C.
Epidemiologic investigation.
Two preterm infants delivered by emergency cesarean section at separate outlying hospitals were transported to the NICU, Cook Children's Medical Center in an isolette by the same ambulance and crew on March 10, 1998. Baby A (patient A in Table 1) weighed 792 g at birth. On day 3 of life, the infant developed black abdominal skin lesions under adhesive tape fastening umbilical catheters. Culture of the lesions resulted in the isolation of A. flavus, and biopsy of the site revealed scattered septate, branching fungal hyphae consistent with an Aspergillus species. The baby lived for 12 days, and the immediate cause of death was respiratory arrest. No autopsy was performed.
Baby B (patient B in Table 1) weighed 420 g at birth. On day 7 of hospitalization, the infant developed black abdominal skin lesions at the site of umbilical catheters secured with tape from the same roll used for baby A. A. flavus grew from cultures of these lesions. The infant was treated with intravenous amphotericin B for 7 days prior to his death on day 17. No autopsy was performed, but cutaneous aspergillosis was considered to be a contributing cause of death.
A third child, a 4-year-old male (patient C in Table 1), was admitted to the same hospital and was treated for brucellosis during the same week that the two low-birth-weight infants were treated. The hospital laboratory recovered A. flavus from a culture of a single blood sample from this child. This patient received no antifungal therapy and recovered, and he was discharged from the hospital.
Construction was in progress to replace double doors at the entrance to the NICU during the time that the infected infants were being treated in the unit. Environmental samples were obtained from various sources, including the NICU itself, the ambulance, and materials from the ambulance. The environmental samples culture positive for A. flavus included a roll of adhesive tape, a canvas bag used to store rolls of tape, the transport isolette, and a roll of clear plastic film stored in a closet in the NICU. The roll of tape kept in the ambulance was used to fasten umbilical intravascular catheters to each infant.
Isolation of A. flavus genomic DNA.
A 10-μl loopful of conidia from each monoconidial isolate was used to seed 50 ml of yeast-peptone-dextrose (YPD) broth (Difco Laboratories, Detroit, Mich.) supplemented with 50 μg of ampicillin (FisherBiotech, Fair Lawn, N.J.) per ml in a 250-ml Erlenmeyer flask. Cultures were incubated in a rotary shaker at 175 rpm and 35°C for 36 h. The mycelia were harvested by filtration through a Whatman no. 1 filter cup (Whatman International, Ltd., Maidstone, United Kingdom) and rinsed with sterile distilled water. The mycelial mat was then transferred to a precooled porcelain mortar containing 0.5-mm-diameter sterile glass beads, frozen with liquid N2, and ground to a fine powder in a biological safety cabinet. A total of 10 ml of the following lysis buffer equilibrated at 65°C was added: 20 mM EDTA, 10 mM Tris, 1% Triton X-100, 500 mM guanidine thiocyanate, and 250 mM NaCl (pH 8.0). The suspension was incubated at 65°C for 10 min to inactivate nuclease activity and to induce protein denaturation. Proteinase K was added to a concentration of 1.0 mg/ml, and the suspension was incubated at 55°C for 3 h with gentle agitation. Potassium acetate was added to a final concentration of 1 M, the mixture was incubated in an ice bath for 30 min, and the suspension was centrifuged for 15 min at 4°C to remove precipitated materials. The cleared lysate was transferred to a 30-ml Oak Ridge tube, and nucleic acids were precipitated with 1 volume of isopropanol and centrifuged at 7,500 × g for 15 min. The nucleic acid pellet was dried and dissolved in TE buffer (0.01 M Tris, 1 mM EDTA [pH 8.0]) and diluted with 5 to 10 volumes of QBT buffer (Qiagen Inc., Chatsworth, Calif.) for purification on a Qiagen tip of the appropriate capacity. A. flavus genomic DNA was purified according to the manufacturer's recommendations (3; Qiagen Genomic DNA Handbook, Qiagen GmbH, Qiagen Inc., Hilden, Germany, 1995). Purified DNA was washed with 70% ethanol, dried, resuspended in an appropriate volume of TE buffer, and stored at −20°C. DNA concentrations were determined with Hoechst 33258 fluorescent dye (Sigma Chemical Co., St. Louis, Mo.) in a fluorometer (Fluorescence Concentration Analyzer; IDEXX Laboratory, Westbrook, Maine).
Restriction endonuclease digestion and Southern hybridization.
Restriction endonuclease digestion of 3 μg of A. flavus genomic DNA with EcoRI or PstI (Roche Molecular Systems, Indianapolis, Ind.) was performed according to the manufacturer's specifications. After digestion, restriction endonuclease fragments were resolved by electrophoresis through a 0.9% agarose (I.D.na agarose; FMC Bioproducts, Rockland, Maine) gel immersed in TBE buffer (100 mM Tris, 90 mM borate, 1 mM EDTA [pH 8.3]) for 18 h at 1.2 V/cm. DNA was transferred to positively charged nylon membranes (Roche Molecular Systems) by standard procedures (15). The probe, plasmid pAF28, contains a 6.2-kb EcoRI fragment containing a repetitive DNA sequence isolated from A. flavus var. flavus NRRL 6541 (16). The nucleotide sequence contained in pAF28 is unknown, so at present, any homology to transposons remains to be determined. Plasmid pAF28 was labeled with digoxigenin by a random priming method according to the manufacturer's directions (DIG DNA Labeling and Detection kit; Roche Molecular Systems). Prehybridization and hybridization were carried out at 65°C in DIG Easy Hyb buffer (Roche Molecular Systems). Digoxigenin-labeled molecular size markers consisting of EcoRI- and HindIII-digested bacteriophage λ DNA were used (Roche Molecular Systems). After posthybridization washing and blocking, the membranes were developed chromogenically and photographed. The Southern blot patterns of different A. flavus strains were compared visually for the presence or absence of bands. Two isolates were considered identical when individual DNA fingerprints could be superimposed.
Analysis of data.
Cluster analysis was conducted with Diversity Database fingerprinting software (Bio-Rad Laboratories, Hercules, Calif.). Similarity values were computed between A. flavus strains by using the Jaccard coefficient formula. Most of the Southern hybridization patterns are shown in Fig. 1, 3, and 4. The similarity matrix values were converted into a Euclidean squared distance matrix by unweighted pair group analysis with arithmetic mean (UPGMA) and were displayed as a dendrogram. The tolerance limit of ±2% was used to determine if two bands were shared or unshared between two different A. flavus strains. A numerical index of discrimination, D, based on the probability that two unrelated strains sampled from a test population will be characterized as different molecular types, was used to assess the method described in this report. Discriminatory power was calculated as described by Hunter and Gaston (11) by using Simpson's index of diversity as given in the following equation:
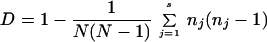 |
where N is the total number of strains in the test population, s is the number of strain types observed, and nj is the number of strains in the population that belong to the jth strain type.
FIG. 1.
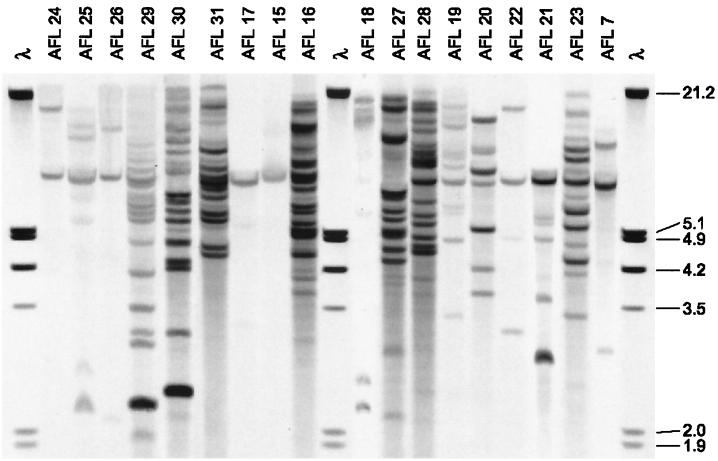
Southern blot patterns of EcoRI-digested DNAs of 18 unrelated isolates of A. flavus hybridized with pAF28. Molecular size markers of EcoRI- and HindIII-digested bacteriophage λ DNA (in kilobases) are shown on the right. Strains AFL29, AFL30, AFL31, and AFL20 were from environmental sources.
FIG. 3.
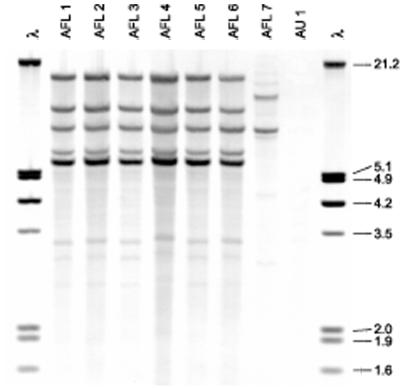
Southern blot patterns of EcoRI-digested DNAs of six isolates of A. flavus collected at Cook Children's Medical Center. See Table 1 for descriptions of individual isolates. Molecular size markers (in kilobases) are shown in lanes 1 and 9 and contain EcoRI- and HindIII-digested DNA of bacteriophage λ.
FIG. 4.
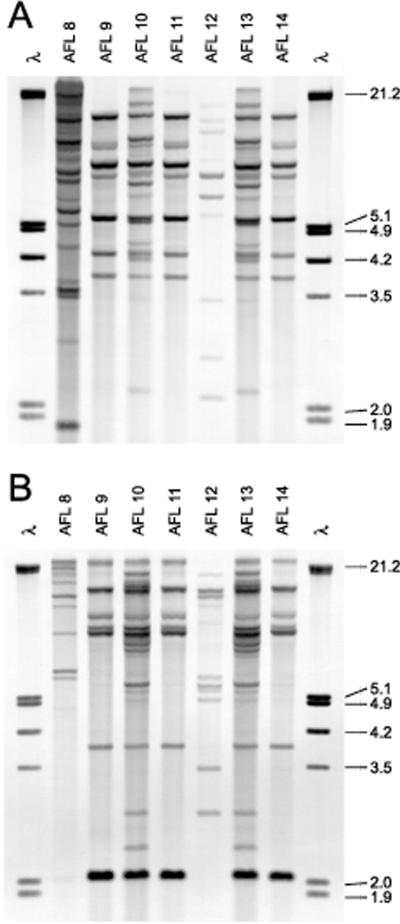
Southern blot patterns of EcoRI-digested (A) and PstI-digested (B) DNAs of clinical and environmental strains of A. flavus from the hematology-oncology and BMT units of the Los Angeles Children's Hospital. See Table 1 for descriptions of individual isolates. Molecular size markers (in kilobases) are shown in lanes 1 and 9 of each panel and contain EcoRI- and HindIII-digested DNA of bacteriophage λ.
RESULTS
DNA fingerprinting.
In order to establish whether the repetitive DNA sequence probe pAF28 might be suitable for the subspecific discrimination of A. flavus, 18 isolates from diverse clinical, environmental, and geographic sources (isolates AFL7 and AFL15 to AFL31) were hybridized with the probe. Eighteen distinct DNA fingerprints were observed (Fig. 1). The size markers did not hybridize with pAF28; the strong signal in the hybridizations is because in the final round of experiments digoxigenin-labeled size markers were used which were stained in the immunoperoxidase reaction. Cluster analysis of the fingerprints resulted in the dendrogram shown in Fig. 2, which suggests that the isolates can be divided into two distinct clusters or groups, each of which comprises nine isolates composed of both clinical and environmental isolates (isolates AFL20 and AFL29 to AFL31). With EcoRI digestion, 2 to 17 fragments were produced that bound to the probe in the molecular mass range of 2 to 23 kb. PstI digestion produced a greater number of fragments, up to 21, ranging in mass from 1 to 24 kb (data not shown).
FIG. 2.
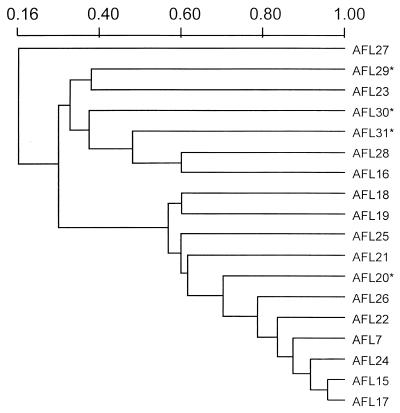
UPGMA-based dendrogram of the 18 unrelated A. flavus isolates based on the Jaccard similarity coefficient of the hybridization patterns generated with the repetitive DNA sequence probe, pAF28. See Table 1 for descriptions of individual isolates. Asterisks indicate strains from environmental sources.
Molecular typing of A. flavus isolates from Cook Children's Medical Center.
Figure 3 shows the Southern blot patterns of EcoRI-digested DNAs from the six A. flavus isolates (isolates AFL1 to AFL6) obtained from Fort Worth, Tex. The hybridization patterns observed with the EcoRI and PstI digests were identical. The isolates obtained from the infants' abdominal lesions were indistinguishable from each other and appeared to be identical to the isolates recovered from the roll of adhesive tape used to anchor their umbilical catheters, the canvas bag used to store the tape in the ambulance, and the tape tray in the ambulance isolette (Fig. 3; Table 1). The DNA fingerprint of an A. flavus isolate (isolate AFL4) recovered in a culture of blood from a 4-year-old child treated during the same week as the two low-birth-weight infants was identical to those of the isolates from the two case infants (isolates AFL1 and AFL2). A seventh A. flavus isolate (isolate AFL7), obtained from the Texas State Department of Health, had a distinctly different fingerprint unrelated to the fingerprints of the isolates in the cluster (Fig. 3).
Molecular typing of A. flavus isolates from the Los Angeles Children's Hospital.
Figure 4 shows the Southern blot patterns of EcoRI- and PstI-digested DNAs from seven clinical and environmental isolates of A. flavus (isolates AFL8 to AFL14) from the hematology-oncology and BMT units of the Los Angeles Children's Hospital. The results are summarized in Table 1. Analysis of this cluster entailed the isolation in culture of monoconidia from the primary stock culture. Consequently, two monoconidial subcultures from the probable invasive aspergillosis case isolate were produced (isolates AFL11 and AFL12). The result indicated the presence of two infecting strains from the same patient. Four distinct strain types were detected, but none of these were identical to the genotype observed in the Cook Children's Medical Center cluster.
Discriminatory power.
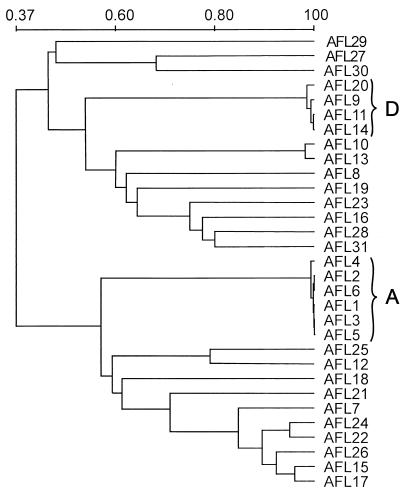
Cluster analysis of the DNA fingerprints of the entire set of 31 A. flavus isolates is illustrated in the dendrogram in Fig. 5. We observed 22 distinct DNA fingerprints among these A. flavus isolates. Of these, 19 strain types were unique, but the remaining three types (types A, D, and E) comprised six, four, and two identical isolates, respectively (Table 1).
FIG. 5.
UPGMA-based dendrogram of the 31 A. flavus isolates based on the Jaccard similarity coefficient of the hybridization patterns generated with the repetitive DNA sequence probe, pAF28. See Table 1 for descriptions of individual strains and letter designations of two clusters of identical strain genotypes (genotypes A and D).
To assess the discriminatory index of the pAF28 repetitive DNA sequence probe, the 31 A. flavus isolates were evaluated on the basis of the method of Hunter and Gaston (11). The discriminatory power was calculated, and the index obtained by this method was shown to be highly discriminatory (D = 0.9526). The reproducibility of the method was assessed in triplicate with different DNA preparations isolated from A. flavus cultures. The reproducibility of our experiments was 100%.
DISCUSSION
Previous studies with the pAF28 repetitive DNA sequence probe distinguished among 29 individual A. flavus strains obtained from agricultural sources representing 22 different vegetative compatibility groups by using PstI to digest A. flavus DNA (16). This study demonstrated that DNA fingerprinting with the pAF28 repetitive probe is a highly reproducible and discriminatory method for tracing the transmission of human cases of A. flavus infection, expanding its utility beyond the agricultural purpose for which it was developed (16). These findings are consistent with the successful findings obtained with the AfutI repetitive DNA sequence probe in investigations of the nosocomial transmission of A. fumigatus infection (7, 8, 21). We compiled an early indication of the degree of strain diversity within A. flavus var. flavus; however, this database needs to be expanded to obtain a more complete appreciation of the population genetics of the group. A better understanding of the nature of the probe is needed after sequence analysis, and its behavior with respect to other species in A. flavus group should be determined. Pending the availability of such information, phylogenetic analysis could proceed with a larger collection of isolates of A. flavus involved in human infections. Information in such a database would be analogous to that from the characterization of clinical and environmental isolates of A. fumigatus (4, 5).
Geiser et al. (6) conducted PCR-RFLP analysis of 11 nuclear genes and concluded that a collection of A. flavus, A. parasiticus, and A. oryzae strains could be divided into two reproductively isolated clades, groups I and II, with group I comprising isolates of A. oryzae. Cluster analysis of 18 unrelated A. flavus isolates in this study also revealed two clades (Fig. 2). The clade distribution observed in this study may be a reflection of the probe, which results in two general hybridization profiles: strains with several pAF28-binding fragments and other strains with few such fragments. The A. flavus strains from agricultural sources, strains AFL20 and AFL29 to AFL31, were distributed in both clades. The occurrence of pAF28 sequences and the number of copies per strain was confirmed with a small number of strains representative of closely related species or varieties of the A. flavus group (16): A. oryzae (strong hybridizer, several copies), A. parasiticus (lower hybridization signal, several copies), A. sojae (low signal, small copy number), and A. nomius (single intense copy). A larger number of strains representing these species should be analyzed by Southern blotting with pAF28 and phylogenetic analysis to confirm the occurrence of these clades.
The extent to which other members of the A. flavus group may occur in human infections is unknown, and a clarification of the taxonomic status of a larger collection of A. flavus clinical isolates may contribute to a better understanding of whether there is a dichotomy in the pathogenic potential of A. flavus. Molecular typing of an expanded collection of clinical and environmental isolates is required to address this question and to determine if there is segregation of clinical from environmental isolates within phylogenetic groups I and II of A. flavus reported by Geiser et al. (6). Similar approaches involving A. fumigatus have already yielded important information about strain diversity in that species (5).
As a fungus which primarily propagates asexually, A. flavus is limited in its ability to exchange genetic material between compatible strains. Barriers to vegetative fusions are widespread among individual strains of A. flavus (16). Limited parasexual recombination may occur, however, within isolated populations. This interpretation, if correct, is promising for the application of genomic typing to trace epidemiologically significant clonal transmission. The contribution of the airborne distribution of A. flavus conidia to strain diversity in a particular geographic region or hospital environment has been suggested but not confirmed by molecular typing (4).
Heterogeneity within the A. flavus group is under investigation (6). In this study we found that some strains weakly hybridized to the repetitive DNA sequence, consistent with earlier observations that some varieties within the A. flavus group possess smaller numbers of copies of the repetitive sequence. A previous study conducted to determine the percent relatedness of varieties within the A. flavus group on the basis of DNA-DNA reassociation showed that A. oryzae exhibited 100% DNA homology with A. flavus var. flavus, whereas the DNA-DNA homology of A. sojae was 74% and that of A. parasiticus was 70% (13).
Molecular typing of clinical and environmental isolates of A. flavus from Cook Children's Medical Center indicated that a roll of adhesive tape was the probable source of the infecting strain of A. flavus recovered from both low-birth-weight infants. The isolate of A. flavus from a culture of blood from a third child, who did not develop aspergillosis and who could not be epidemiologically linked to the NICU infants, had an DNA fingerprint identical to those of the isolates from the NICU infants (Fig. 3; Table 1). This relationship was confirmed by cluster analysis (Fig. 5). Possibly, this genotype may have been introduced from the contaminated tape into the hospital ventilation system. Alternatively, this A. flavus genotype could have been an endemic strain in the hospital ventilation system antedating the tape problem. Both of these possibilities allow an isolate of this genotype to be a laboratory contaminant in the culture of blood from the third patient. A more thorough environmental survey of this hospital would have been required in order to determine the wider prevalence of the A. flavus genotype implicated in these infections.
In 1994 Buffington et al. (2) reported one definite and one probable case of invasive infection with A. flavus among immunosuppressed children with underlying hematologic diseases in the Los Angeles Children's Hospital. Environmental isolates of A. flavus were obtained from air exhaust vents in three areas: the BMT unit nursing station, an area adjacent to Ward 4 West, and Ward 3 West adjacent to the NICU on the floor below. A. flavus was also isolated from the nasal sinus of a health care worker who visited patients in all units. Buffington et al. (2), using hybridization profiles obtained by RAPD analysis-RFLP analysis, concluded that the isolates recovered from the health care worker, the probable case patient, and the BMT unit air vent were identical. The A. flavus isolate from the definite case patient was shown to be different from all the other clinical and environmental isolates. In the present study, the pAF28 hybridization profiles of A. flavus isolates from the definite case patient and from the health care worker were highly related (Table 1; Fig. 4). The probable case patient was infected with a strain with a genotype different from that of the strain from the definite case patient, but the pattern was highly related to those observed for the A. flavus isolates from the BMT nursing station and from Ward 3 West. Moreover, the probable case patient appeared to be infected with two different A. flavus genotypes (isolates AFL11 and AFL12).
One explanation for the disparity between the two methods of strain typing, RAPD analysis-RFLP analysis and Southern blotting with pAF28, is that the RAPD analysis-RFLP analysis blots from the original published investigation at Los Angeles Children's Hospital were not divided by limiting dilution into cultures of monoconidial strains. In the present investigation with the pAF28 probe, all cultures were derived from each primary culture as monoconidial strains. This explains the recovery of two strains from the primary isolate from the blood of the case patient with probable invasive aspergillosis, who appears to be an example of a patient infected or colonized with two A. flavus genotypes. Another reason is that the two independent probes, pAF28 and the RAPD analysis-RFLP analysis probes (consisting of A. flavus genomic DNA amplified by PCR with two different RAPD analysis primers), may be scanning different regions of the genome or specific chromosomal regions, such that the resulting patterns are not in agreement. Further comparison of the two molecular typing methods would be desirable.
One of the reference isolates, A. flavus AFL20 (a 1946 New Jersey isolate), had a Southern blot genotype identical to those of three of the A. flavus isolates from the Los Angeles Children's Hospital (Table 1; Fig. 5). Identical genotypes were observed in two different situations: (i) a strain from the Los Angeles Children's Hospital was highly related to a culture collection strain from New Jersey and (ii) strains isolated from a patient with cutaneous aspergillosis at Cook Children's Medical Center were highly related or identical to an A. flavus strain isolated as a possible contaminant from a culture of blood from a pediatric patient (patient C) who was in a different ward of the hospital. These examples suggest that the population of A. flavus is not geographically isolated and that typing of a larger number of A. flavus isolates, including sampling at intervals of hospital air and solid surfaces, is warranted.
With one exception, the PstI hybridization patterns observed with the Los Angeles isolates exhibited an intense anonymous DNA fragment with a molecular mass of approximately 2 kb (Fig. 4B) that was also observed with A. flavus AFL20. None of the A. flavus isolates from the Cook Children's Medical Center displayed this particular anonymous fragment or had any distinctive feature in that range of molecular mass.
This investigation has highlighted the risk for cutaneous fungal infection in preterm infants arising from the use of nonsterile adhesive tape to secure umbilical intravascular catheters. The cases reported here emphasize the need to consider exposures in critically ill neonates that might occur during their transport outside the NICU setting and highlight the need for strict adherence to established infection control practices. The use of nonsterile items which may become contaminated with environmental molds poses a special risk to low-birth-weight infants.
We hypothesize that the cases of cutaneous A. flavus infection described in this report arose as a result of direct contact of the skin with contaminated adhesive tape. Contaminated dressings associated with intravascular catheters have previously been described as a cause of cutaneous infection with Aspergillus spp. and Rhizopus spp. in preterm infants, human immunodeficiency virus-infected patients, and other immunocompromised individuals (9, 10, 17, 22). Bryce et al. (1) reported a cluster of four surgical and burn wound aspergillosis cases that were traced to packages of dressings contaminated during construction in the hospital's central supply area. Another report of four preterm infants with cutaneous aspergillosis which subsequently developed into fatal, disseminated infection identified the source of the infection as contaminated latex finger stalls (23). Recently, a cluster of four infants in the NICU of a United Kingdom hospital developed cutaneous mucormycosis due to Rhizopus microsporus. An environmental investigation isolated the fungus from contaminated wooden tongue depressors used as splints to immobilize infants' cannulated limbs (18). Infection control personnel should be aware that critically ill infants and other high-risk patients may be at significant risk for the development of aspergillosis.
The implications of this study underscore the need to avoid the use of such nonsterile items in hospital units housing patients at high risk for the development of invasive fungal infections. In particular, a policy regarding the use of sterile medical supplies by hospital staff, including ambulance transport teams, to avoid this type of exposure should be considered when managing high-risk neonates.
ACKNOWLEDGMENTS
We thank the following investigators who provided A. flavus strains from various origins: J. Noble (Georgia State University, Atlanta), K. J. Kwon-Chung (National Institutes of Health, Bethesda, Md.), and J. Dorner (National Peanut Research Laboratory, Dawson, Ga.).
This research was supported in part by an appointment of M.J.J. to the Emerging Infectious Diseases Fellowship Program administered by the Association of Public Health Laboratories and the Centers for Disease Control and Prevention. Additional support for M.J.J. was provided by the Research Participation Program of the Division of Bacterial and Mycotic Diseases, National Center for Infectious Diseases, Centers for Disease Control and Prevention, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the Centers for Disease Control and Prevention.
REFERENCES
- 1.Bryce E A, Walker M, Scharf S, Lim A T, Walsh A, Sharp N, Smith J A. An outbreak of cutaneous aspergillosis in a tertiary-care hospital. Infect Control Hosp Epidemiol. 1996;17:70–72. doi: 10.1086/647266. [DOI] [PubMed] [Google Scholar]
- 2.Buffington J, Reporter R, Lasker B A, McNeil M M, Lanson J M, Ross L A, Mascola L, Jarvis W R. Investigation of an epidemic of invasive aspergillosis: utility of molecular typing with the use of random amplified polymorphic DNA probes. Pediatr Infect Dis J. 1994;13:386–393. [PubMed] [Google Scholar]
- 3.Challen M P, Moore A J, Martínez-Carrera D. Facile extraction and purification of filamentous fungal DNA. BioTechniques. 1995;18:975–976. [PubMed] [Google Scholar]
- 4.Chazalet V, Debeaupuis J P, Sarfati J, Lortholary J, Ribaud P, Shah P, Cornet M, Thien H V, Gluckman E, Brüker G, Latgé J P. Molecular typing of environmental and patient isolates of Aspergillus fumigatus from various hospital settings. J Clin Microbiol. 1998;36:1494–1500. doi: 10.1128/jcm.36.6.1494-1500.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Debeaupuis J P, Sarfati J, Chazalet V, Latgé J P. Genetic diversity among clinical and environmental isolates of Aspergillus fumigatus. Infect Immun. 1997;65:3080–3085. doi: 10.1128/iai.65.8.3080-3085.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geiser D M, Pitt J I, Taylor J W. Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proc Natl Acad Sci USA. 1998;95:388–393. doi: 10.1073/pnas.95.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girardin H, Latgé J P, Srikantha T, Morrow B, Soll D R. Development of DNA probes for fingerprinting Aspergillus fumigatus. J Clin Microbiol. 1993;31:1547–1554. doi: 10.1128/jcm.31.6.1547-1554.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girardin H, Sarfati J, Kobayashi H, Bouchara J P, Latgé J P. Use of DNA moderately repetitive sequence to type Aspergillus fumigatus isolates from aspergilloma patients. J Infect Dis. 1994;169:683–685. doi: 10.1093/infdis/169.3.683. [DOI] [PubMed] [Google Scholar]
- 9.Groll A H, Jaeger G, Allendorf A, Herrmann G, Schloesser R, von Loewenich V. Invasive pulmonary aspergillosis in a critically ill neonate: case report and review of invasive aspergillosis during the first 3 months of life. Clin Infect Dis. 1998;27:437–452. doi: 10.1086/514717. [DOI] [PubMed] [Google Scholar]
- 10.Hunt S J, Nagi C, Gross K G, Wong D S, Mathews W C. Primary cutaneous aspergillosis near central venous catheters in patients with the acquired immunodeficiency syndrome. Arch Dermatol. 1992;128:1229–1232. [PubMed] [Google Scholar]
- 11.Hunter P R, Gaston M A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klich M A, Pitt J I. A laboratory guide to common Aspergillus species and their teleomorphs. North Ryde, New South Wales, Australia: Division of Food Processing, Commonwealth Scientific and Industrial Research Organisation; 1988. [Google Scholar]
- 13.Kurtzman C P, Smiley M J, Robnett C J, Wicklow D T. DNA relatedness among wild and domesticated species in the A. flavus group. Mycologia. 1986;78:955–959. [Google Scholar]
- 14.Leenders A, van Belkum A, Janssen S, DeMarie S, Kluytmans J, Wielenga J, Löwenberg B, Verbrugh H. Molecular epidemiology of apparent outbreak of invasive aspergillosis in a hematology ward. J Clin Microbiol. 1996;34:345–351. doi: 10.1128/jcm.34.2.345-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 16.McAlpin C E, Mannarelli B. Construction and characterization of a DNA probe for distinguishing strains of Aspergillus flavus. Appl Environ Microbiol. 1995;61:1068–1072. doi: 10.1128/aem.61.3.1068-1072.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mead J H, Lupton G P, Dillavou C L, Odoni I W. Cutaneous Rhizopus infection. Occurrence as a postoperative complication associated with an elasticized dressing. JAMA. 1979;242:272–274. doi: 10.1001/jama.242.3.272. [DOI] [PubMed] [Google Scholar]
- 18.Mitchell S J, Gray J, Morgan M E I, Hocking M D, Durbin G M. Nosocomial infection with Rhizopus microsporus in preterm infants: association with wooden tongue depressors. Lancet. 1996;348:441–443. doi: 10.1016/s0140-6736(96)05059-3. [DOI] [PubMed] [Google Scholar]
- 19.Moody S F, Tyler B M. Restriction enzyme analysis of mitochondrial DNA of the Aspergillus flavus group: A. flavus, A. parasiticus, and A. nomius. Appl Environ Microbiol. 1990;56:2441–2452. doi: 10.1128/aem.56.8.2441-2452.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moody S F, Tyler B M. Use of nuclear DNA restriction fragment length polymorphisms to analyze the diversity of the Aspergillus flavus group: A. flavus, A. parasiticus, and A. nomius. Appl Environ Microbiol. 1990;56:2453–2461. doi: 10.1128/aem.56.8.2453-2461.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neuveglise C, Sarfati J, Latgé J P, Paris S. Afut1, a retrotransposon-like element from Aspergillus fumigatus. Nucleic Acids Res. 1996;24:1428–1434. doi: 10.1093/nar/24.8.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papouli M, Roilides E, Bibashi E, Andreou A. Primary cutaneous aspergillosis in neonates: case report and review. Clin Infect Dis. 1996;22:1102–1104. doi: 10.1093/clinids/22.6.1102. [DOI] [PubMed] [Google Scholar]
- 23.Singer S, Singer D, Rüchel R, Mergeryan H, Schmidt U, Harms K. Outbreak of systemic aspergillosis in a neonatal intensive care unit. Mycoses. 1998;41:223–227. doi: 10.1111/j.1439-0507.1998.tb00328.x. [DOI] [PubMed] [Google Scholar]
- 24.Taylor J W, Geiser D M, Burt A, Koufopanou V. The evolutionary biology and population genetics underlying fungal strain typing. Clin Microbiol Rev. 1999;12:126–146. doi: 10.1128/cmr.12.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyler K D, Wang G, Tyler S D, Johnson W M. Factors affecting reliability and reproducibility of amplification based DNA fingerprinting of representative bacterial pathogens. J Clin Microbiol. 1997;35:339–346. doi: 10.1128/jcm.35.2.339-346.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Burik J H, Colven R, Spach D. Cutaneous aspergillosis. J Clin Microbiol. 1998;36:3115–3121. doi: 10.1128/jcm.36.11.3115-3121.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]