Abstract
Durable control of invasive solid tumors necessitates identifying therapeutic resistance mechanisms and effective drug combinations. In this work, we used a network-based mathematical model to identify sensitivity regulators and drug combinations for the PI3Kα inhibitor alpelisib in ER+ PIK3CA mutant breast cancer. The model-predicted efficacious combination of alpelisib and BH3 mimetics, e.g., MCL1 inhibitors, was experimentally validated in ER+ breast cancer cell lines. Consistent with the model, FOXO3 downregulation reduced sensitivity to alpelisib, revealing a novel potential resistance mechanism. Cell line-specific sensitivity to combinations of alpelisib and BH3 mimetics depended on which BCL-2 family members were highly expressed. Based on these results, newly developed cell line-specific network models were able to recapitulate the observed differential response to alpelisib and BH3 mimetics. This approach illustrates how network-based mathematical models can contribute to overcoming the challenge of cancer drug resistance.
Significance
Network-based mathematical models of oncogenic signaling and experimental validation of its predictions can identify resistance mechanisms for targeted therapies, as this study demonstrates for PI3Kα-specific inhibitors in breast cancer.
Introduction
Overcoming intrinsic or acquired resistance to cancer drug therapies is one of the biggest challenges faced today by patients, oncologists, and cancer researchers (1,2). This is particularly the case in the setting of metastatic disease, which accounts for the majority of cancer deaths (3,4). The challenge underlying cancer drug resistance is exemplified by metastatic estrogen receptor positive (ER+) breast cancer, the most common type of metastatic breast cancer. Despite the availability of multiple effective drug therapies (5) (endocrine and targeted therapies, CDK4/6 inhibitors, among others), ER+ metastatic breast cancer eventually develops resistance to all available treatments. As a result, long-term patient survival has not greatly improved, with a median prognosis of around 3–4 years after the development of metastatic disease (5,6).
One of the most recently approved drugs to treat ER+ breast cancer is alpelisib (previously known as BYL719), which is a PI3Kα-specific inhibitor approved for use in combination with fulvestrant (a selective estrogen receptor degrader, a type of ER inhibitor) for ER+ metastatic breast cancer with activating PIK3CA mutations (7). Multiple lines of clinical evidence support PTEN loss-of-function as a clinically relevant resistance mechanism to alpelisib and other PI3Kα inhibitors (8,9). There are several other potential PI3Kα inhibitor resistance mechanisms that have been identified through in vitro or in vivo studies (10–18), but it is still an open question if these will drive resistance in patients and if they will be common enough to be clinically relevant. It is also likely that there are currently unidentified PI3Kα resistance mechanisms that will be found as the landscape of intrinsic and acquired resistance mechanisms is revealed, as has happened in the case of endocrine therapies (19–22) and CDK4/6 inhibitors (23,24).
How can we identify these currently unknown PI3Kα resistance mechanisms? Based on what we have learned from other targeted drugs (e.g. BRAF(25) or EGFR inhibitors(26)), resistance mechanisms act by reactivating the targeted pathway or by activating alternative oncogenic pathways that bypass it (1,2,27). Therefore, a promising avenue to identify such resistance mechanisms is to use approaches that combine a deep understanding of the oncogenic pathways driving ER+ PIK3CA mutant breast cancer, their cross-talk, and their integration by cancer cells to make the cellular decisions that result in tumor growth. One such approach is provided by mathematical models of cancer (28,29), particularly dynamic network models, which mechanistically model the dynamics of the molecular network underlying cancer cells (30,31).
Dynamic network models are based on mechanistic knowledge and explicitly model how information flows through signaling networks under distinct genetic conditions (e.g. in a context where known or potential resistance mechanisms are active) and in response to perturbations (e.g. drugs or environmental signals). A particularly attractive property of dynamic network models in the context of cancer is that these models can be customized to become specific to particular cancer cell lines, or even to individual patient tumors, by introducing known genetic alterations and/or by mimicking the known response (or lack thereof) to a perturbation (31). Tested and validated network models can predict the degree of effectiveness of single-agent and combinatorial drug interventions, and predict potential mechanisms of drug resistance, which can then be used to prioritize drug combinations for in vitro and in vivo testing in relevant cancer model systems (30–36).
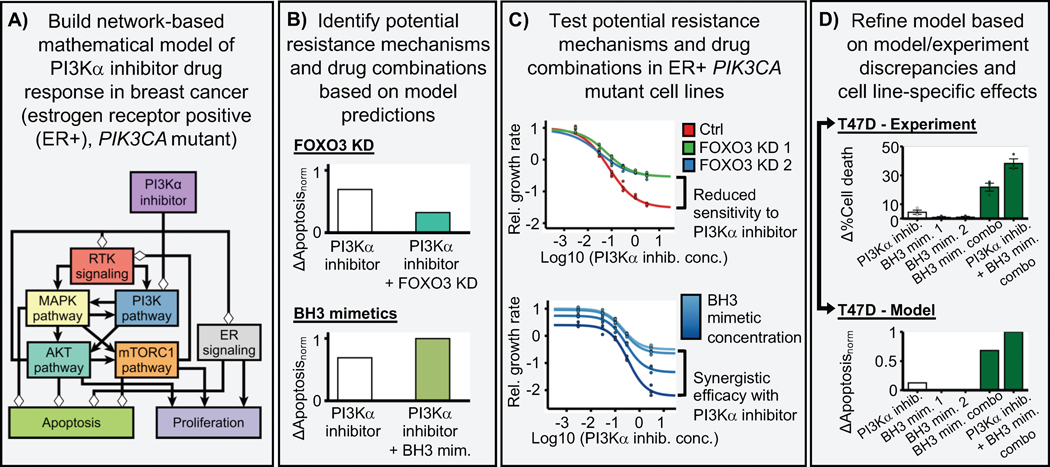
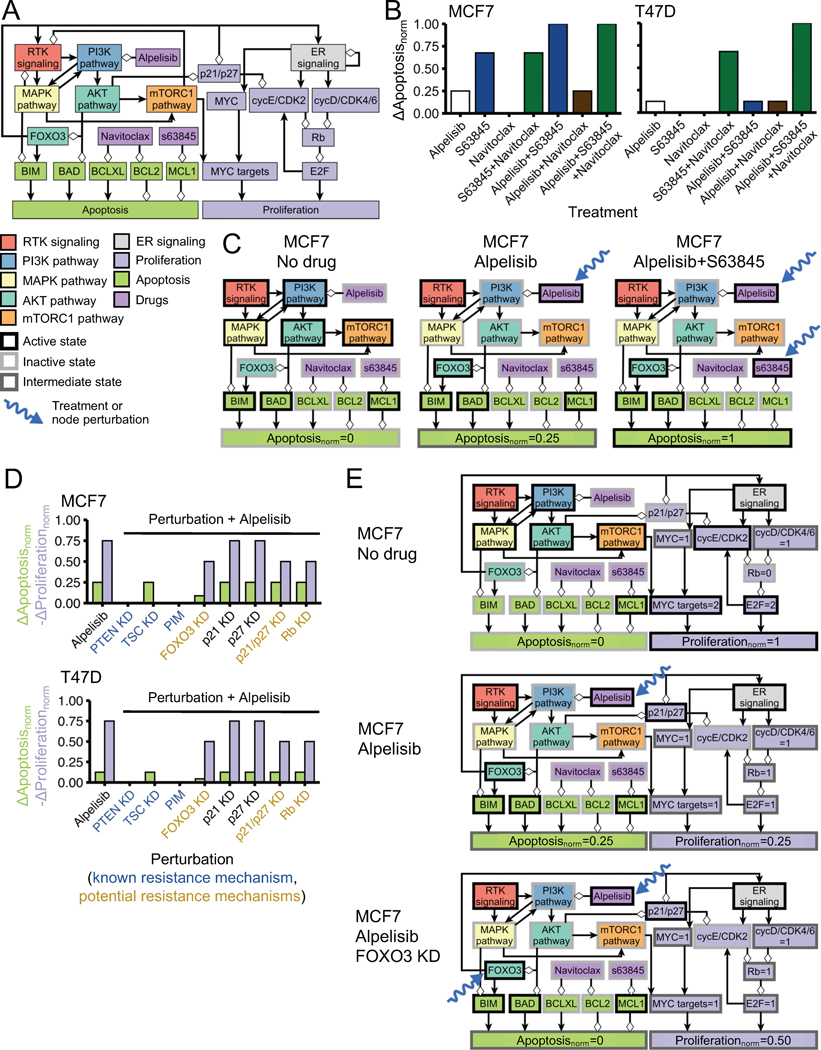
We previously built a network-based mathematical model of oncogenic signal transduction in ER+ PIK3CA mutant breast cancer (Fig. 1A) (32). The model integrated the mechanistic knowledge of the response/resistance to PI3Kα inhibitors and other drugs relevant in the context of ER+ breast cancer (e.g. CDK4/6 inhibitors, ER inhibitors, or mTORC1 inhibitors). The model was based on the combined knowledge of in vitro, in vivo, and clinical literature; it recapitulated the drug resistance phenotype of a variety of resistance mechanisms to PI3Kα inhibitors (e.g. PTEN loss-of-function, mTORC1 activity, and PIM overexpression) and other drugs (e.g. RB1 loss in response to CDK4/6 inhibitors, and ER transcriptional reactivation in response to ER inhibitors). By exhaustively testing the effect of all single- and double-element perturbations, the model predicted potential resistance mechanisms and efficacious drug combinations for PI3Kα inhibitors (Fig. 1B). In particular, it predicted the knockdown of tumor suppressors FOXO3, RB1, or p21 and p27 as potential PI3Kα inhibitor resistance mechanisms, and identified BH3 mimetics targeting anti-apoptotic proteins MCL1, BCL-XL, and BCL2 as efficacious drug combinations. Note that our mathematical model rephrases as a single problem what is traditionally viewed as two separate problems: identifying potential therapeutic drug combinations or identifying potential resistance mechanisms. From the model’s perspective, both are part of the same problem, that is, which perturbations/alterations increase (potential therapeutic drug combinations) or decrease (potential resistance mechanisms) sensitivity to a drug.
Fig. 1. Outline of the research strategy followed in this work.
(A) We use a network-based mathematical model of oncogenic signal transduction in estrogen receptor positive (ER+), PIK3CA mutant breast cancer. This model integrates the mechanistic knowledge of the response/resistance to PI3Kα inhibitors and recapitulates the drug resistance phenotype of a variety of known resistance mechanisms. (B) Based on this model, we predict potential resistance mechanisms (e.g. FOXO3 knockdown) and drug combinations (BH3 mimetics) for PI3Kα inhibitors. (C) We experimentally test these predicted resistance mechanisms and drug combinations in ER+, PIK3CA mutant breast cancer cell lines. Based on these experiments, we can verify the model’s predictions and pinpoint any discrepancies between the model and the experimental results. (D) We use the experimental results and the discrepancies between the model and the experiments to refine the network-based mathematical model and incorporate the observed cell line-specific effects into a new version of the model.
In this work we experimentally test the above predictions of the 2017 version of our mathematical model (Fig. 1C) (32). We find that some of the model’s predictions are verified experimentally, particularly the efficacy of the drug combination of alpelisib and BH3 mimetics, and the reduced sensitivity to PI3Kα inhibitors caused by FOXO3 knockdown. We also find some discrepancies between our experimental findings and the model, some of which are cell line-specific. In particular we find a cell line-specific response to BH3 mimetics in MCF7 and T47D, two of the most commonly used cell line models for ER+ PIK3CA mutant breast cancer. Finally, these experimental results as well as the most recent work on PI3Kα inhibitors (17,18) are used to refine the network-based mathematical model and incorporate the observed cell line-specific effects into a new version (the 2020 version) of the model (Fig. 1D).
Materials and Methods
Cell culture
293T, T47D and MCF7 cells were purchased from American Type Culture Collection (ATCC). 293T cells were cultured in DMEM (GIBCO #11995–065) with 10% fetal bovine serum (Gemini #100–119). T47D cells were cultured in Phenol Red-free RPMI-1640 (GIBCO #11835–030) with 10% FBS. MCF7 cells were cultured in Phenol Red-free MEM-α (GIBCO #41061–029) with 10% FBS. We used cell lines of passage number ranging from 4 to 15 in the described experiments. All cell lines tested negative for Mycoplasma contamination by ATCC Universal Mycoplasma Detection Kit. The last test was performed in November 2019.
Kill curves and CellTiter-Glo viability assay
Cells were plated in 96-well tissue culture ViewPlate (Perkin Elmer # 6005181) on Day 1 and treated with drug on Day 2. Media with or without drugs was refreshed on Day 5. On Day 8, cells were equilibrated to room temperature, media was removed, and cells were lysed in a mixture of 50 μL media and 50 μL CellTiter-Glo 2.0 reagent (Promega # G9243) per well. Plates were then incubated on an orbital shaker for 2 mins. Following another 10 mins of incubation at room temperature to stabilize the signal, luminescence was recorded to measure cell viability on an Infinite M200 Pro microplate reader (Tecan).
Relative Growth Rate
The relative growth rate () for a final time for our experiments) and drug concentration , is , where . In all cases, the growth rates are calculated separately for each cell line condition (MCF7 parental, MCF7 sgRNA NT, T47D parental, etc.). For the single-dose experiments optimized for the highest PI3K inhibitor concentrations, we used a larger number of initial cells, which overflew the plate for the DMSO condition and did not allow us to calculate the DMSO growth rate. For these experiments, we instead used the DMSO growth rate from the drug response curves in each cell line condition.
Colony formation assay
1,000–10,000 cells were plated in 6-well plates on Day 1 and treated on Day 2. Media was refreshed every 3–4 days until crystal violet staining. On the day of staining, cells were fixed with ice-cold 100% methanol for 10 minutes and then incubated with 0.5% crystal violet solution (Sigma Aldrich #C6158) in 25% methanol at room temperature for 10 minutes.
Synergy scores
We quantify drug combination synergy using MuSyC (37). MuSyC calculates the synergy score from drug combination kill curves and considers separately the effect of efficacy and potency. In this work, we focus on the synergy score of efficacy ( measures the percentage change in drug response of the drug combination when compared to that of each drug alone (at the highest concentration used for each drug alone and their combination). The synergy score of efficacy is such that means the drugs are synergistic, that they are antagonistic, and means the drugs show no synergistic or antagonistic effect.
Analysis of loss-of-function genetic screens, gene expression, and protein expression in cell line datasets
Gene dependency data from CRISPR knockout (Avana dataset), RNAi knockdown (combined Achilles, DRIVE, and Marcotte datasets), gene expression (38) (RNA-seq from the Cancer Cell Line Encyclopedia (CCLE)), relative protein expression (39) (quantitative mass-spectroscopy proteomics from CCLE), and associated cell line annotations were taken from the Cancer Dependency Data (DepMap) 21Q2 Public data release (40) (https://depmap.org/portal/download). Gene dependency is measured using the gene effect, which measures the impact of CRISPR knockout or RNAi knockdown of a gene on cell viability. The gene effect is normalized for each cell line so that a gene effect of 0 is the median dependency score of a negative control gene and a gene effect of −1 is the median dependency score of pan-essential genes (40).
Dynamic network modeling
Our dynamic network modeling approach follows the methodology in (32), which uses a type of dynamic network model known as discrete dynamic network model. In a discrete dynamic network model, each node in the directed network that underlies the model has an associated variable that can only take a discrete number of states. The state of a variable is determined using a regulatory function , which depends on the variable of a node only if there is a directed interaction from to ).
The state of the system , where is the state of variable at time , changes in time following a general asynchronous updating scheme. In general asynchronous updating, the state of the system is updated in discrete time units by (i) choosing one randomly selected node at each time step (with update probability ) and updating its state based on the state of the system at the previous time step () and (ii) transferring the node state from the previous time step of the rest of the nodes (, ).
Model simulations
Model simulations were performed using similar methodology as in (32). The updating probabilities are chosen by categorizing nodes into either a fast or slow node, according to whether activation of the node denotes a (fast) signaling event or a (slow) transcriptional or translational event. The regulatory functions of all the nodes are indicated and explained in the Supplemental Text and Supplemental File 1. We perform 10,000 simulations in each modeled scenario because this was found to be sufficient to estimate the average value of a node state at the end of a simulation with a <1%. The of each node state at the end of all model simulations is included in the GitHub repository of the model. The number of time units is 25 for all simulations, where a time unit is equal to the average number of time steps needed to update a slow node. More details on the network model and the model simulations can be found in the Supplemental Text and Supplemental File 1.
The code used to simulate the model is available on GitHub (https://github.com/jgtz/BreastCancerModelv2), and includes a Jupyter notebook that generates all the model results. To allow easy reuse of the model, we use bioLQM (version 0.6.1, https://github.com/colomoto/bioLQM) to make the model available in Systems Biology Markup Language (SBML) format on the GitHub repository.
Results
PI3Kα inhibitors and BH3 mimetics are a synergistically efficacious drug combination and induce apoptosis in ER+ PIK3CA-mutant breast cancer cell lines
In the 2017 network model (32), PI3Kα inhibition has a cytotoxic effect mediated by the increased activity of the pro-apoptotic BCL-2 family members BIM and BAD (through upregulation of FOXO3-mediated transcription (41) and through a decrease of an inactivating AKT-mediated phosphorylation (42), respectively). The cytotoxic effect of PI3Kα inhibition is countered by the presence of anti-apoptotic BCL-2 family members MCL1 and BCL2 (which in the model represents BCL-2 and other anti-apoptotic family members such as BCL-XL). As a result, the combined inhibition of PI3Kα and anti-apoptotic family members MCL1, BCL-2, or BCL-XL, each of which can be targeted using BH3 mimetics, is predicted to be an effective and synergistic drug combination (32). Specifically, the model predicts that inhibition of the anti-apoptotic family members would be more effective to induce apoptosis than the combinations used with alpelisib in clinical practice (fulvestrant) and in clincal trials (palbociclib, everolimus), all of which predominantly inhibit cell proliferation.
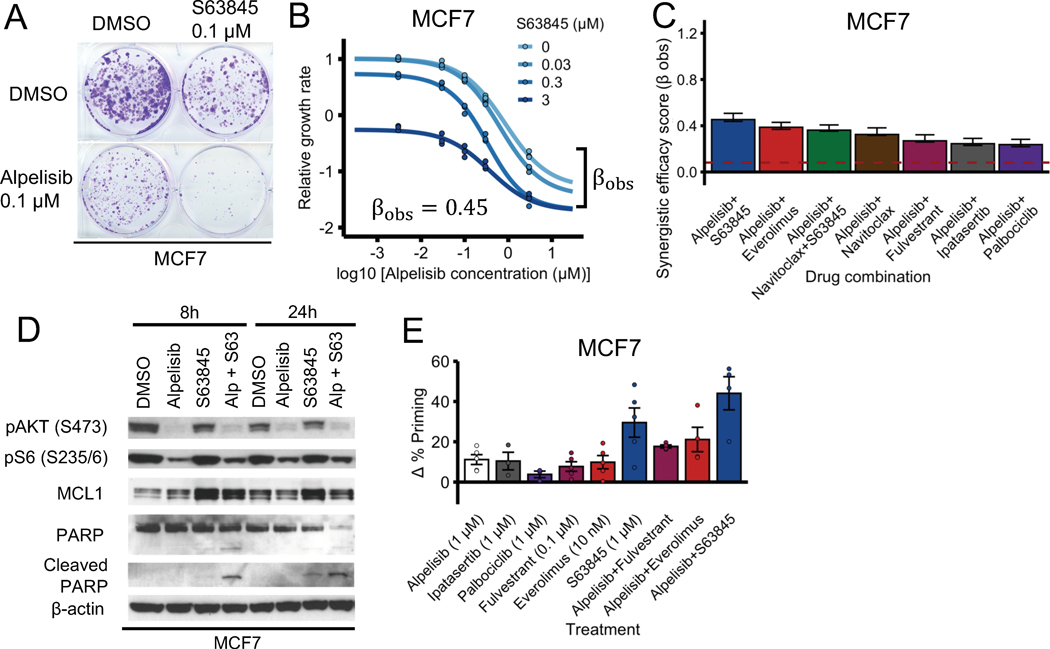
To test the effect of combining PI3Kα inhibitors and BH3 mimetics of anti-apoptotic proteins, we first treated MCF7 cells (ER+, PIK3CA-mutant) with PI3Kα inhibitor alpelisib and MCL1 inhibitor s63845 (a BH3 mimetic (43)) separately and in combination using both a long-term colony formation assay (2 weeks, Fig. 2A) and a short-term cell viability assay (6 days, Fig. 2B). In both cases the combination of alpelisib and s63485 was more efficacious than each drug on its own. We calculated the synergy score of the combination using MuSyC (37) and found that it was synergistically efficacious (, where means the combination is synergistically efficacious and is the null-hypothesis value expected from doubling the alpelisib concentration). To compare the synergistic efficacy of the alpelisib + s63845 combination with other proposed alpelisib drug combinations, we performed drug response curves of the combination of alpelisib with fulvestrant (the FDA-approved combination for the treatment for ER+ PIK3CA-mutant metastatic breast cancer (7,15)), palbociclib (44), or everolimus (which had previously been found to increase alpelisib-induced cell death (10)), and calculated their synergy scores (Fig. 2C, Supplemental Fig. S1). We found that all the tested drug combinations were synergistically efficacious (i.e. , with their range being 0.24 – 0.45; Fig. 2C), but the alpelisib + s63845 combination had the highest synergy score among all the combinations ().
Fig. 2. PI3Kα inhibitors and MCL1 inhibitors are a synergistically efficacious combination and induce apoptosis in MCF7.
(A, B) PI3Kα inhibitor alpelisib and MCL1 inhibitor s63845 is an effective and synergistically efficacious drug combination. MCF7 cells were treated with alpelisib, s63845, and their combination. For panel A, cells a colony formation assay was performed after 18 days of drug treatment. For panel B, CellTiter-Glo assay was performed before drug treatment and after 6 days of drug treatment to measure cell viability. The relative growth rate is calculated with respect to the DMSO control treatment using cell viability. Efficacy scores are such that means synergistic behavior and means antagonistic behavior. (C) The combination of alpelisib and s63845 is equally synergistically efficacious (or more) than clinically relevant drugs like fulvestrant, everolimus, and palbociclib. The dotted red line () is the null-hypothesis synergy score value expected from doubling the alpelisib combination (i.e., a combination of alpelisib with itself). Experiments were done and synergy scores were calculated as in panel B. (D) Combined alpelisib (0.1 μM) and s63845 (0.1 μM) result in an increase in apoptosis (as measured by cleaved PARP levels) compared to each separate drug. (E) The combination of alpelisib with s63845 results in an increased apoptosis priming compared to the combination with clinically relevant drugs like fulvestrant and everolimus. Δ% apoptosis priming for a treatment was measured using dynamic BH3 profiling by comparing treated vs untreated cells after 16 h under increased concentration of BIM peptide.
To determine if the effect of the alpelisib + s63845 combination is mediated by apoptosis or other mechanisms, we treated MCF7 with each drug separately and in combination, and measured the levels of cleaved PARP and total PARP (Fig. 2D). We found that alpelisib + s63845 induced increased levels of cleaved PARP compared to each drug alone at both time points (8 and 24 h), with the effect being more evident at the early (8 h) time point (Fig. 2D). To contextualize the apoptotic effect of the alpelisib + s63845 combination, we tested the effect of single agent and drug combinations and measured the resulting apoptotic priming using dynamic BH3 profiling (45) (Fig. 2E). The alpelisib + s63845 combination showed the highest increase in Δ% priming (which measures apoptotic sensitivity) of all combinations, and this effect was larger than each drug on its own. In particular, the alpelisib + s63845 combination showed a higher Δ% priming than alpelisib + fulvestrant and alpelisib + everolimus.
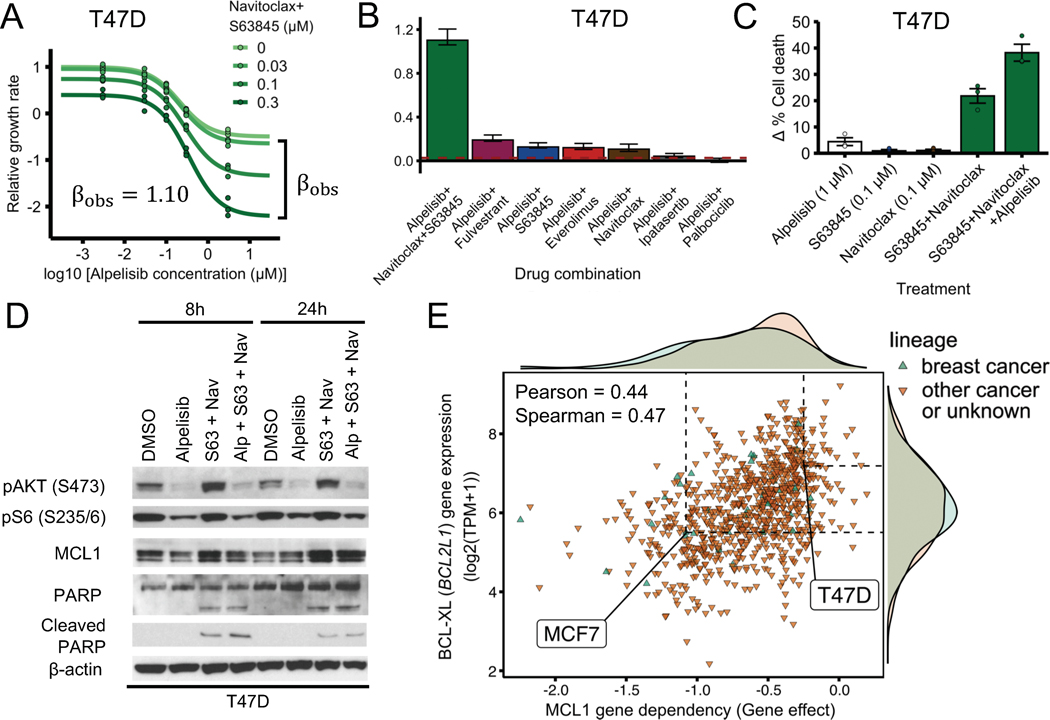
We also performed similar experiments in T47D cells, another ER+ PIK3CA-mutant breast cancer cell line (Fig. 3A–D). In contrast to MCF7, T47D cells did not show a strong synergistic effect under the combination of alpelisib + s63845 (Fig. 3B, Supplemental Fig. S2A). However, the addition of navitoclax (a BH3 mimetic targeting BCL-XL and BCL-2) to the combination of alpelisib and s63845 recapitulated the effects seen in the MCF7 cells (Fig. 3A–B). The alpelisib + s63845 + navitoclax combination had a strong synergistically efficacious effect (; Fig. 3A) and had a significantly higher synergy score than other combinations tested (Fig. 3B). The combination of alpelisib + s63845 + navitoclax in T47D also showed increased apoptosis, measured by cleaved PARP and PARP, which is consistent with the results in MCF7 cells (Fig. 3D). The increased apoptosis was further confirmed by measuring cell death through Annexin V staining (Fig. 3C).
Fig. 3. PI3Kα inhibitors and BH3 mimetics are a synergistically efficacious combination and induce apoptosis in T47D.
(A) PI3Kα inhibitor alpelisib, MCL1 inhibitor s63845, and BCL-2/BCL-XL inhibitor navitoclax is an effective and synergistically efficacious drug combination. T47D cells were treated with alpelisib, s63845 and navitoclax (1 μM), and their combination. CellTiter-Glo assay was performed before drug treatment and after 6 days of drug treatment to measure cell viability. The relative growth rate is calculated with respect to the DMSO control treatment using cell viability. Efficacy scores are such that means synergistic behavior and means antagonistic behavior. (B) The combination of alpelisib, s63845, and navitoclax is more synergistically efficacious than clinically relevant drugs like fulvestrant, everolimus, and palbociclib. The dotted red line () is the null-hypothesis synergy score value expected from doubling the alpelisib combination (i.e., a combination of alpelisib with itself). Experiments were done and synergy scores were calculated as in Fig. 2. (C, D) Combined alpelisib, s63845, and navitoclax result in an increase in apoptosis compared to each separate drug. Apoptosis was measured using annexin V (panel C) and cleaved PARP levels (panel D). Δ% cell death for a treatment was measured using annexin V by comparing treated vs DMSO control cells after 72 h (panel C). Drug concentrations used were: 0.1 μM for alpelisib, 0.3 μM for s63845, and 0.3 μM for navitoclax (panel D). (E) Sensitivity to MCL1 knockout in CRISPR/Cas-9 loss-of-function screens (DepMap) is strongly correlated with BCL-XL gene expression. Sensitivity to gene knockout is measured using gene effect, which is such that gene effect=0 means no effect and gene effect=−1 is the median effect of known common essential genes.
Given that the addition of BCL-XL/BCL-2 inhibitor navitoclax sensitized T47D cells to the combination of alpelisib + s63845, we reasoned that this was because T47D cells had a higher expression of BCL-XL and/or BCL-2 compared to MCF7 cells. To test this hypothesis, we looked at the gene and protein expression of MCF7 and T47D cells in the Cancer Cell Line Encyclopedia (CCLE) (38,39). Gene and protein expression of BCL-XL was higher in T47D compared to MCF7, consistent with our hypothesis (Supplemental Fig. S2B;, difference robust z-score ; , difference robust z-score ). For the case of BCL-2, there was little difference in protein expression and the difference in gene expression was in the opposite direction of the effect we are trying to explain (higher gene expression in MCF7 compared to T47D vs increased sensitivity in T47D when adding navitoclax) (Supplemental Fig. S2B;, difference robust z-score ; , difference robust z-score ).
To further test whether BCL-XL expression can serve as a marker for cells that require inhibition of BCL-XL to become sensitive to MCL1 inhibitors, we looked at how the effect of MCL1 gene knockout correlated with BCL-XL gene expression in various cell lines using the CRISPR/Cas9 loss-of-function genetic screens of the Cancer Dependency Map (DepMap) (40,46). Consistent with the hypothesis of BCL-XL as a marker, sensitivity to MCL1 gene knockout (MCL1 gene dependency) was strongly correlated with reduced BCL-XL expression (Fig. 3E; ) and this was the strongest gene expression correlate (Pearson) among all genes. There was no strong correlation between sensitivity to MCL1 gene knockout and MCL1 expression () or BCL-2 expression (). Consistent with our experimental findings, MCF7 was in the low end of BCL-XL expression (Fig. 3E; , robust z-score ) and was sensitive to MCL1 gene knockout (Fig. 3E; , where a of 0 means no effect, while a of −1 means the same effect as an essential gene). T47D was in the high end of BCL-XL expression (Fig. 3E; , robust z-score ) and was not sensitive to MCL1 gene knockout (Fig. 3E; , 4 times smaller than the gene effect of MCF7). We also found consistent results for the effect of MCL1 gene knockout and BCL-XL expression when looking at a dataset that combines multiple publicly available loss-of-function RNAi screen datasets (Supplemental Fig. S2C), although the dynamic range of gene effects for MCL1 was much smaller for this combined RNAi screen dataset.
In summary, these results support the predictions of the 2017 version of the model that (i) the combined inhibition of PI3Kα and the relevant anti-apoptotic family members (MCL1 for MCF7; MCL1 and BCL-XL for T47D) is an effective and synergistic drug combination, and (ii) combined alpelisib and BH3 mimetics have a larger cell death effect than each drug alone and other effective drug combinations (alpelisib + everolimus, alpelisib + fulvestrant).
FOXO3 knockdown is a potential resistance mechanism to PI3Kα inhibitors
In the 2017 network model (32), PI3Kα inhibition increases FOXO3 nuclear localization and transcriptional activity through AKT-mediated dephosphorylation of FOXO3 (12,47,48). Increased FOXO3 nuclear activity upregulates the expression of downstream targets that result in the suppression of cell growth and induction of cell death (12,49,50). As a result, the network model predicts that knockdown of FOXO3 is a potential resistance mechanism to PI3Kα inhibitors and that this reduced sensitivity is a result of blocking FOXO3 transcriptional activity (32).
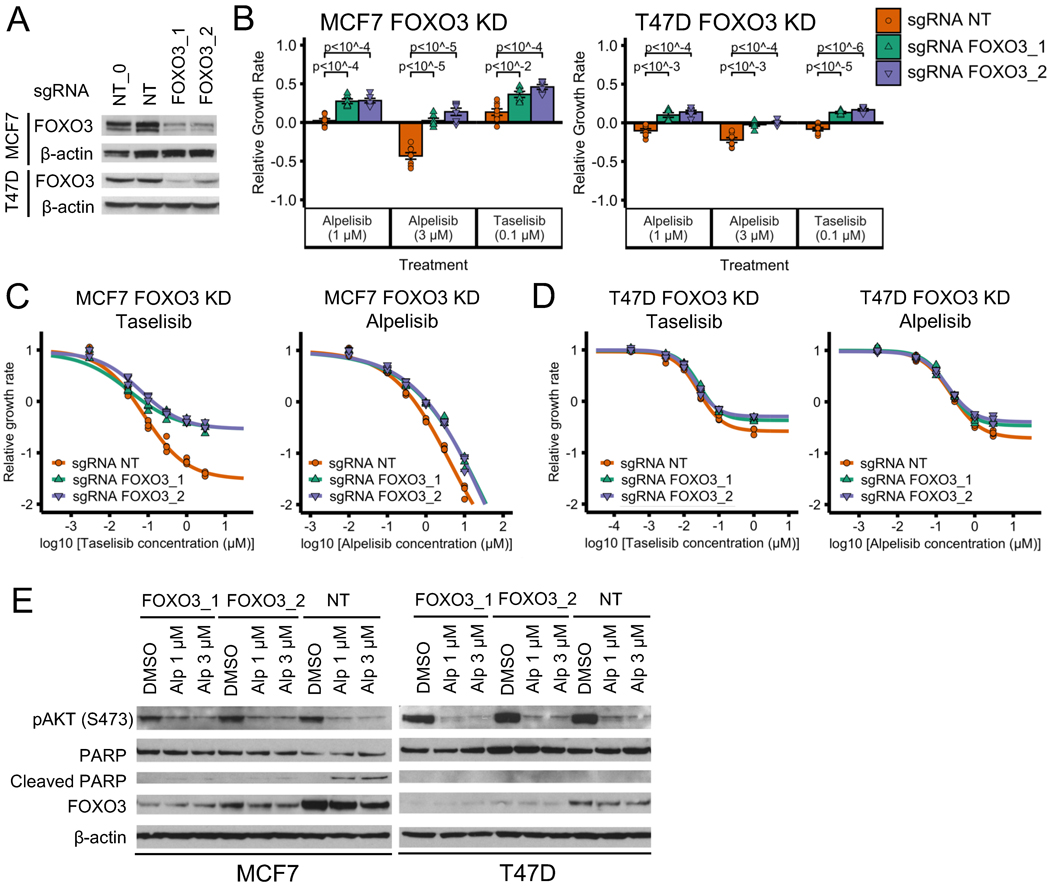
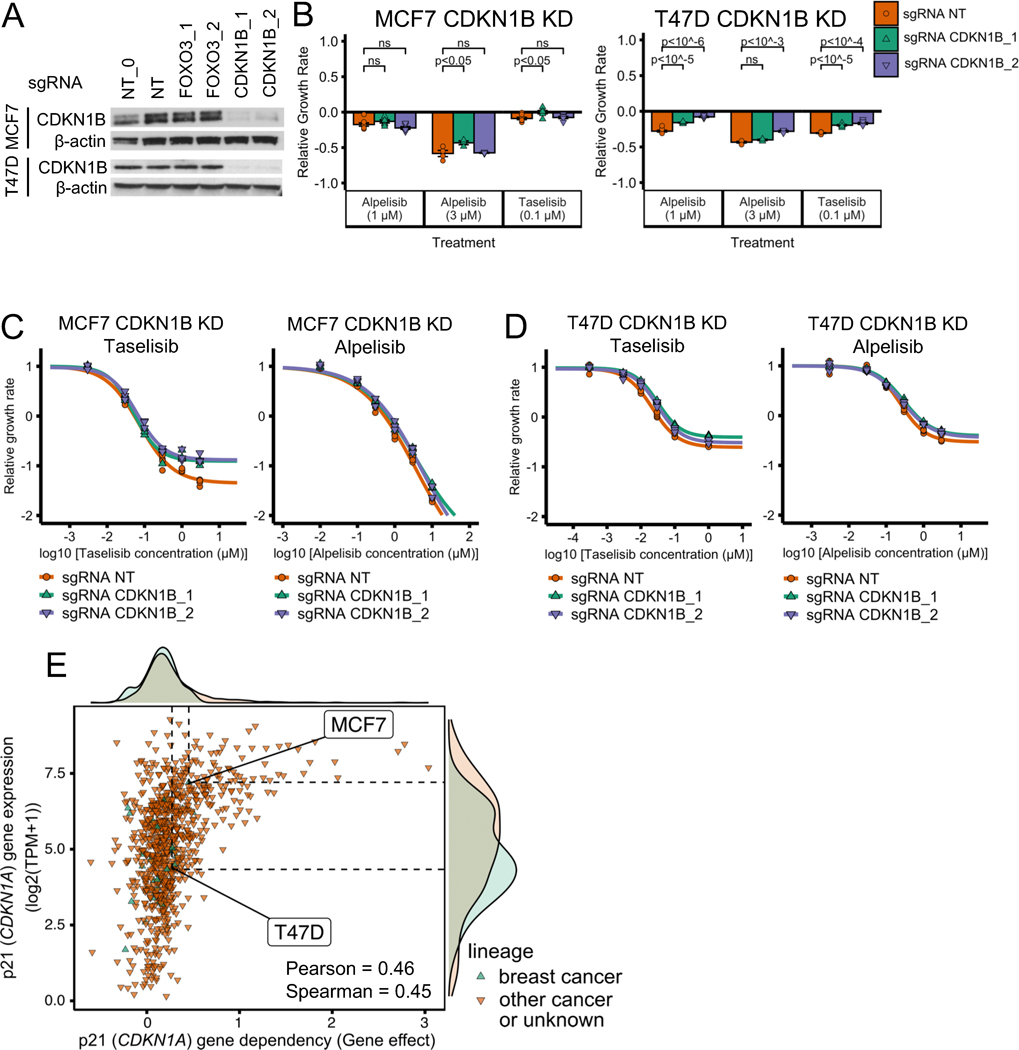
To test the model’s prediction, we generated MCF7 and T47D cell lines with FOXO3 knockdown (KD) using CRISPR-Cas9 technology and verified the knockdown of FOXO3 by western blot (Fig. 4A). We tested the response of FOXO3 KD MCF7 and T47D cells to two PI3Kα inhibitors (alpelisib and taselisib) and found that both cell lines show reduced sensitivity to both PI3Kα inhibitors when compared to control cells with a non-targeting (NT) sgRNA (Fig. 4B; two-sided t-test for each sgRNA vs NT control comparison). To further test this reduced sensitivity to PI3Kα inhibitors, we performed drug response curves to varying concentrations of alpelisib or taselisib (Fig. 4C–D). We found that the drug response curves show a reduced sensitivity to PI3Kα inhibitors in FOXO3 KD MCF7 cells as compared to control cells, (Fig. 4C), consistent with the resistant phenotype in Fig. 4B. For FOXO3 KD T47D cells we did not see significant effect on PI3Kα inhibitor sensitivity from FOXO3 knockdown in the drug response curves (compare Fig. 4D and 3B), a result that may be attributed to the methodology being insufficiently sensitive to measure moderate effects (Supplemental Fig. S3A–B).
Fig. 4. FOXO3 knockdown reduces sensitivity to PI3Kα inhibitors in ER+ breast cancer cell line models.
(A) Stable cell lines generated using CRISPR FOXO3 knockdown have reduced proteins levels of FOXO3. NT and NT_0 are two distinct non-targeting control guides; FOXO3_1 and FOXO3_2 are two distinct guides targeting FOXO3. The experiment and samples from Figs. 4A and 5A are the same, thus, so are the β-actin blots. In Fig. 4A we show part of the blot, while in Fig. 5A we show the full blot. (B-D) FOXO3 knockdown reduces sensitivity to PI3Kα inhibitors alpelisib and taselisib in ER+ breast cancer cell lines MCF7 and T47D. CellTiter-Glo assay was used to measure cell viability before drug treatment and after 6 days of drug treatment. An unpaired, two-sided t-test was used for comparison between groups (panel B). Experiments in panel B single-dose experiments were optimized for the tested PI3Kα inhibitor concentrations: panel B used a larger number of initial cells than panels C and D (5,000 vs 1,000 cells). The relative growth rate is calculated with respect to the growth rate of the DMSO control of each cell line condition. (E) FOXO3 knockdown reduces PI3Kα-inhibitor-induced cell death in MCF7. Cell death was measured using cleaved PARP levels.
Many drug resistance mechanisms for targeted therapies involve reactivation of the signaling pathway inhibited by targeted therapy. However, according to the model, FOXO3 KD does not reactivate the PI3K pathway. We decided to test this experimentally by measuring AKT phosphorylation, which is indicative of PI3K pathway activation. We found that AKT(S473) phosphorylation was similar when comparing FOXO3 KD and control cells at baseline and in response to alpelisib in both MCF7 and T47D (Fig. 4E). We conclude that FOXO3 KD does not reactivate PI3K pathway activity. This is consistent with the current knowledge on FOXO3, which indicates that it mainly acts downstream of the PI3K pathway but might also act upstream of the PI3K pathway due to feedback activation, and suggests that the effect of feedback activation by FOXO3 is weak in this context.
One important prediction of the 2017 model is that FOXO3 KD reduces PI3Kα inhibitor sensitivity by decreasing drug-induced apoptosis. To test this, we determined the effect of alpelisib on cell death by measuring the levels of cleaved PARP and PARP in FOXO3 KD vs control cells (Fig. 4E). We found that the FOXO3 KD MCF7 cells have reduced levels of cleaved PARP compared to control (Fig. 4E, left). We did not see this difference in cleaved PARP for T47D (Fig. 4E, right). The reduced levels of cleaved PARP seen only in MCF7 FOXO3 KD cells is consistent with the reduced cytotoxic effect seen in the alpelisib drug response curves of FOXO3 KD in MCF7 but not in T47D (Fig. 4C–D).
Overall, we found that FOXO3 KD reduces sensitivity to PI3Kα inhibitors in both T47D and MCF7, and that this effect was more pronounced in MCF7 cells, which is mediated by a reduction in drug-induced apoptosis in MCF7. These results are consistent with the predictions of the 2017 version of the model and suggest that downregulation of FOXO3 is a potential resistance mechanism for PI3Kα inhibitors.
Published in vivo experiments and patient tumor data are consistent with a potential role for FOXO transcription factors in mediating resistance to PI3Kα inhibitors
Our experiments showed that FOXO3 KD reduces the sensitivity to PI3Kα inhibitors in ER+ PIK3CA mutant breast cancer cell lines and is a potential resistance mechanism, but there is no guarantee that the same effect will be seen in vivo or in patient tumors. To test our hypothesis that FOXO3 KD is a potential resistance mechanism to PI3Kα inhibitors, we looked at whether the FOXO3 interactions responsible for PI3K inhibitor sensitivity seen in vitro have been observed in vivo and in patient tumor samples. We also looked at whether publicly available breast cancer tumor datasets showed any evidence consistent with our hypothesis. The summary of our findings in the breast cancer tumor datasets are presented in Supplemental Table S1.
We find that the transcriptional upregulation of genes associated with the activation of FOXO3 in response to PI3K inhibitors has been observed in vivo and in tumor samples (15,16,51). The most relevant is the one in (51), who found an increase in FOXO3 activity in their patient tumor-derived cells and in vitro models in response to alpelisib. They also observed the upregulation of BIM in response to alpelisib in early passage patient tumor-derived breast epithelial cells and in allograft models, and attributed this pro-apoptotic effect to FOXO3. These findings are evidence for the activity of FOXO3 in vivo and in tumor samples in response to PI3K inhibitors, and are consistent with FOXO3 KD leading to a decreased sensitivity to PI3Ki in this setting.
PI3Kα inhibitor alpelisib was approved in May 2019, so there is very limited tumor data to test whether FOXO3 loss of function is a resistance mechanism. As a first step, we looked at the prevalence of genomic alteration in FOXO3 and its paralog FOXO1 in multiple publicly available breast cancer tumor datasets (TCGA PanCancer, METABRIC, MSK IMPACT 2018, and The Metastatic Breast Cancer Project; see Supplemental Table S1 and Supplemental File 2). We looked at both FOXO3 and FOXO1 because we expect mutations in these paralog genes to be rare and because we think it’s likely they have a similar effect on sensitivity to PI3Kα inhibitors; their activity as a transcription factor is regulated by PI3K/AKT signaling through direct phosphorylation by AKT (52). Focusing on loss-of-function mutations (frameshift and nonsense mutations, and excluding those in tumors with high mutation burden) and copy number deep deletions, we find that these genes are altered in breast cancer but that this occurs rarely. To be more specific, we find that FOXO3 has deep deletions in 0.4–0.8% of samples and loss-of-function mutations in 0–0.5% of samples in these datasets (FOXO3 is not profiled in MSK IMPACT 2018). For FOXO1, we find deep deletions in 0.1–1.4% of samples and loss-of-function mutations in 0–0.1% of samples in these datasets.
To check whether there is evidence for FOXO3 or FOXO1 to be associated with resistance to alpelisib, we looked at the most comprehensive clinical study of alpelisib to date with publicly available genomic data (9). This study used a gene panel that did not include FOXO3 and only a small number of samples (20/141) used a gene panel with FOXO1. There was one sample with a FOXO1 alteration; this sample had a loss-of-function mutation (FOXO1 Q529*). This sample was a primary tumor sample; the same patient’s pre-treatment and post-treatment cell-free DNA samples (which shared truncal mutations with the primary sample) were sequenced using a gene panel that did not include FOXO1. The patient had no clinical benefit from the treatment received (alpelisib + exemestane), and did not have genomic alterations in the genes identified by the study to drive resistance to therapy (PTEN alterations for alpelisib, ESR1 mutations for aromatase inhibitors). The other annotated mutations in the patient, apart from the hotspot PIK3CA mutation that is required for eligibility in the study (PIK3CA E524K for this patient), are TP53 K320*, and the hotspot mutations ERBB3 E928G and ERCC2 N238S (ERBB3 and ERCC2 were not included in the panel used for the cell-free DNA samples). Only ERBB3 is known to be related to resistance. The lack of clinical benefit in this patient, despite the absence of PTEN or ESR1 mutations, is suggestive of FOXO1 playing a role in the observed intrinsic resistance to alpelisib + exemestane.
Overall, the available in vivo and tumor data are consistent with a potential role for the downregulation of FOXO transcription factors in mediating resistance to PI3Kα inhibitors in breast cancer.
CDKN1B (p27) knockdown does not have a strong effect on PI3Kα inhibitor sensitivity
The 2017 network model contains a node, , that merges the tumor suppressor CDKN1B (p27) and its paralogue CDKN1A (p21). In the model, PI3Kα inhibition results in an increased activity of this node, mediated by the upregulation of FOXO3-mediated CDKN1B transcription (50) and a decrease in AKT-mediated p27 and p21 phosphorylation (53,54). p27 and p21 exert their tumor suppressor function by binding and inactivating the cyclin E-CDK2 complex (55,56), which upon release from p27 or p21 promotes the G1-S transition and progression into the cell cycle. As a consequence, the model predicts that inactivation of the merged node will interfere with the cytostatic effect of PI3Kα inhibition, and result in a reduced sensitivity to PI3Kα inhibitors (32). As the model does not specify the contribution of p27 and p21 to this merged node, inactivation of this merged node might require simultaneous knockdown of CDKN1B and CDKN1A, or, if the contribution of p21 is minor, might be reproduced by knockdown of CDKN1B only. Given that p27 but not p21 has been shown to be transcriptionally upregulated by AKT-mediated FOXO3 activation (50,57), we chose to investigate the contribution of p27 by knocking down CDKN1B only.
We generated MCF7 and T47D cell lines with CDK1NB knockdown, which we verified through western blotting (Fig. 5A). We then tested the response of CDKN1B KD cells to the PI3Kα inhibitors alpelisib and taselisib (Fig. 5B–D). We found that the CDKN1B KD cell lines did not show a consistent reduced sensitivity to PI3Kα inhibition compared to control, neither in the drug response curves nor in the further optimized drug treatment experiment at selected doses. (Fig. 4B–D).
Fig. 5. CDKN1B (p27) knockdown does not have a strong effect on PI3Kα inhibitor sensitivity in ER+ breast cancer cell line models.
(A) Stable cell lines were generated using CDKN1B CRISPR knockdown and showed reduced proteins levels of CDKN1B. NT and NT_0 are two distinct non-targeting control guides. CDKN1B_1 and CDKN1B_2 are two distinct guides targeting CDKN1B. NT, NT_0, FOXO3_1, and FOXO3_2 are the same guides as in Fig. 4. The experiment and samples from Figs. 4A and 5A are the same, thus, so are the β-actin blots. In Fig. 4A we show part of the blot, while in Fig. 5A we show the full blot. (B-D) CDKN1B knockdown does not strongly reduce sensitivity to PI3Kα inhibitors alpelisib and taselisib in ER+ breast cancer cell lines MCF7 and T47D. The same experimental procedures and methods as in panels B-D of Fig. 4 were used for panels B-D of this figure. (E) Sensitivity to p21 (CDKN1A) knockout in CRISPR/Cas-9 loss-of-function screens (DepMap) is strongly correlated with p21 gene expression. Sensitivity to gene knockout is measured using gene effect, which is such that gene effect=0 means no effect and gene effect=−1 is the median effect of known common essential genes.
The lack of a strong effect of CDKN1B (p27) knockdown on PI3Kα inhibitor sensitivity can still be consistent with the 2017 model, if we hypothesize that only joint KD of CDKN1A (p21) and CDKN2B (p27) can accomplish what we observe in the model for the inactivation of the node . To test the plausibility of this hypothesis, we looked at the gene expression and gene knockout effect of p21 and p27 using the Cancer Cell Line Encyclopedia (CCLE) and the CRISPR/Cas9 loss-of-function genetic screens of the Cancer Dependency Map (DepMap). Across all cell lines, p21 tended to show a tumor suppressor effect ( for all 851 cell lines) and this effect was correlated with the cell line’s p21 (CDKN1A) gene expression (). This was not the case for p27, which did not show a consistent tumor suppressor effect ( for the for all 851 cell lines) and its effect was not correlated with its gene expression (). The correlation between p21 (CDKN1A) gene knockout effect and gene expression was also found in a dataset that combines multiple publicly available loss-of-function RNAi screen datasets (Supplemental Fig. S3C).
We also used the DepMap and CCLE database to look at whether we expect p21 knockout to show a difference in gene dependency between the MCF7 and T47D cell lines. MCF7 was in the high end of p21 gene expression (Fig. 5E; , robust z-score ) and grew faster in response to CDKN1A gene knockout (Fig. 5E; ). T47D was in the low end of p21 expression (Fig. 5E; , robust z-score ) and the gene effect of p21 gene knockout on T47D was 1.5 times smaller than that for MCF7 (Fig. 5E; ). MCF7 also showed a larger gene effect than T47D in response to p21 gene knockout in a dataset that combines multiple publicly available loss-of-function RNAi screen datasets (Supplemental Fig. S3C).
Overall, our experimental results suggest that CDK1NB (p27) knockdown does not have a strong effect on PI3Kα inhibitor sensitivity, in contrast with what the 2017 network model predicts. Based on computational analysis of gene expression and loss-of-function cell line databases, we hypothesize that the lack of a strong effect seen in CDK1NB knockdown could be explained by the activity of CDKN1A (p21); we expect MCF7 to show a stronger tumor suppressor effect than T47D in response to CDKN1A knockdown.
RB1 knockdown reduces sensitivity to PI3Kα inhibitors in a cell line-specific manner
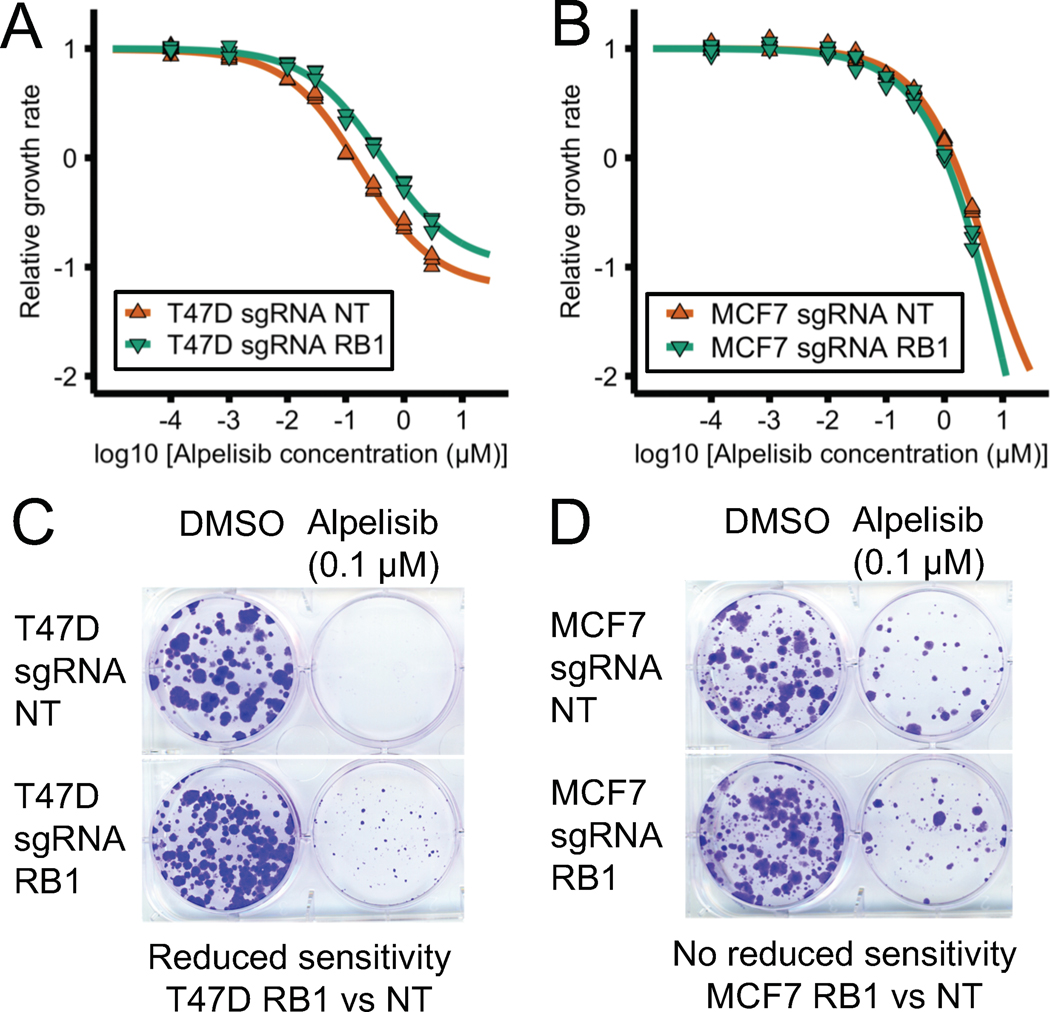
In the 2017 network model, PI3Kα inhibitors result in reduced cell proliferation by two mechanisms: by a decrease in E2F1 activity mediated by Rb or by a decrease in translational machinery mediated by mTORC1. As a result, the model predicts that RB1 knockdown would increase E2F1 activity and reduce the sensitivity to PI3Kα inhibition (32). Consistent with this prediction, there is evidence in ER+ breast cancer that RB1 KD results in the upregulation of E2F transcription factors and that PI3Kα inhibitors cause an increase in Rb activity and a downregulation of E2F transcriptional activity (17,44) (see also Supplemental Text). To test the model’s prediction, we used T47D and MCF7 cell lines with sgRNA CRISPR KD of RB1 we previously generated (23).
Fig. 6A–B shows the drug response curve of RB1 KD T47D and MCF7 cell lines in response to PI3Kα inhibitor alpelisib. T47D RB1 KD cells showed a reduced sensitivity to alpelisib compared to control cells (Fig. 6A) and this reduced sensitivity was not seen in MCF7 cells (Fig. 6B). To further verify the reduced sensitivity to PI3Kα inhibitors in T47D RB1 KD cells, we used a long-term colony formation assay (Fig. 6C–D). This assay also indicates that T47D RB1 KD but not MCF7 RB1 KD cells (Fig. 6C) showed reduced sensitivity to alpelisib (Fig. 6D). In summary, we found that RB1 KD can reduce the sensitivity to PI3Kα inhibitor alpelisib, as predicted by the 2017 model, and that this effect was cell line-specific (observed in T47D but not MCF7).
Fig. 6. RB1 knockdown can reduce sensitivity to PI3Kα inhibitors in ER+ breast cancer cell line models.
RB1 knockdown in T47D (panels A and C) but not MCF7 cells (panels B and D) results in a reduced sensitivity to PI3Kα inhibitor alpelisib in short-term and long-term assays. Cell viability was measured using a CellTiter-Glo assay before and after 6 days of drug treatment. Relative growth rate was calculated compared to the DMSO control treatment (panels A and B). Colony formation assay was performed after 30–40 days of drug treatment.
Cell line-specific network models reproduce the efficacy of PI3Kα inhibitor drug combinations and resistance mechanisms
Our experimental results are in agreement with the 2017 model’s predicted efficacy of combining PI3Kα inhibitors with BH3 mimetics and the reduced PI3Kα inhibitor sensitivity that results from FOXO3 knockdown or RB1 knockdown. However, our experimental results also exhibit cell line-specific aspects, which the 2017 model does not fully recapitulate. For example, MCF7 cells were sensitive to combined alpelisib and s63845 (Fig. 2) while T47D cells were only sensitive to combined alpelisib, s63845, and navitoclax (Fig. 3). Because of this, we generated cell line-specific network models for MCF7 and T47D.
The cell line-specific models are based on an updated version of the network model (the 2020 version) that incorporates new knowledge generated on ER+ breast cancer drug resistance since the 2017 model was published (e.g. (17,18,23)) and the discrepancies from the 2017 model found by our experimental results (see Supplemental Text). The 2020 network model separates protein pairs (p21 and p27, BCL-2 and BCL-XL) that were previously denoted by a single combined node (p21/p27 and BCL2). The 2020 model incorporates our hypothesis that joint p21 and p27 KD, but not each of them separately, will result in a reduced sensitivity to PI3Kα inhibitors. The 2020 model includes a new node that denotes the transcriptional status of MYC targets (following the results from (17,58)) and assumes that the mTORC1 and Rb-E2F pathways do not contribute equally to the response to alpelisib (following our findings on RB1 and p27 knockdown). A condensed representation of the 2020 model is shown in Fig. 7A while a more complete representation of the model and the changes introduced is shown in Supplemental Fig. S4A. The 2020 model describes the biological outcomes of proliferation and apoptosis using the multi-state nodes and (each of which has 3 states) and their normalized propensities denoted by and (which take values between 0 and 1).
Fig. 7. Cell line-specific network models reproduce the efficacy of combined PI3Kα inhibitors and BH3 mimetics and the effect of PI3Kα inhibitor resistance mechanisms in MCF7 and T47D.
(A) Condensed representation of the network model in which the cell line-specific models are based. The condensed representation includes nodes denoting pathways, biological outcomes, and drugs or proteins of interest for this work. Positive interactions are drawn with black arrowheads and negative interactions are drawn with white diamonds. A more complete representation of the network model is shown in Supplemental Fig. S4. (B-C) MCF7-specific and T47D-specific network models reproduce the difference in cell death responses to BH3 mimetics, alpelisib, and their combination seen in these cell lines. (D-E) Cell line-specific network models reproduce the drug resistance effect of PI3Kα inhibitor resistance mechanisms. Simulations in which a PI3Kα inhibitor resistance mechanism is active show an increased survivability (reduced and/or increased ) compared to the case of alpelisib alone (panel D). We show the state of the nodes that influence and in several conditions (panels C and E). The models encode the biological outcomes of cell death and proliferation using and , respectively, which are a weighted and normalized (between 0 and 1) measure of the state of the nodes and . Starting from a cancerous state of each model, we perform 10,000 simulations in which the specified treatment or perturbation is maintained throughout the simulation, and obtain the average value of and at the end of the simulations. and of a perturbation denote the difference with respect to the case of no perturbation (), and are such that decreased survivability means an increase in or .
The MCF7-specific and T47D-specific network models differ only in three aspects: (i) the node denoting basal expression of BCL-XL is active in T47D but not MCF7 (in agreement with Fig. 3E), (ii) the node denoting basal expression of p21 is more active in MCF7 than in T47D (in agreement with Fig. 5E), and (iii) the contribution of the lowest active state to is larger in MCF7 than T47D (i.e., results in in MCF7 and in T47D) (in agreement with Figs. 2D and 3D).
The cell line-specific models’ cancerous states (which are steady states) and the time course of their response to alpelisib share many similarities with that of the previous (2017) model. The cancerous states have active RTK, PI3K, MAPK, AKT, mTORC1, and ER signaling, which result in high survivability ( and ). These states can be primed for cell death, a common feature of cancer cells (45,59), by having pro-apoptotic protein BIM active but an anti-apoptotic protein counteracting its effect (BCL-2, BCL-XL, or MCL1 in the updated model). The time course of the MCF7-specific model in response to alpelisib starting from a cancerous state is shown in Supplemental Fig. S4B. In response to alpelisib, there is a quick inactivation of the MAPK, AKT, and mTORC1 pathways in both cell line-specific models, followed by the activation of FOXO3, which transcriptionally upregulates HER3 and ESR1. The inhibition of the mTORC1 pathway downregulates the node , which results in reduced proliferation () in both models. The activation FOXO3 and inactivation of AKT results in the activation of pro-apoptotic proteins BIM and BAD, which result in a small increase in cell death (in MCF7 and in T47D).
The response of the cell line-specific models to multiple drug combinations and resistance mechanisms is shown in Fig. 7B–E and Supplemental Fig. S5A–D. These cell line-specific network models reproduce the observed difference in cell death response to BH3 mimetics and their combination with alpelisib in each cell (see Fig. 7B and Supplemental Fig. S6, where is the difference with respect to the case of no perturbation). In the MCF7 model, alpelisib alone has a small effect on apoptosis (), s63845 alone has larger effect on apoptosis (), and combined alpelisib and s63845 has the largest effect () (Fig. 7B–C). In the T47D model, alpelisib alone has a small effect on apoptosis (), s63845 or navitoclax alone have no effect (), their combination has a larger effect (), and their combination with alpelisib has the largest effect () (Fig. 7B and Supplemental Fig. S5B).
The cell line-specific models recapitulate how FOXO3 knockdown reduces sensitivity to alpelisib in both MCF7 (Fig. 7D–E) and T47D (Fig. 7D and Supplemental Fig. S5D), and the stronger reduction in apoptosis seen in MCF7 (Fig. 7D). For the case of Rb knockdown and p21/p27 knockdown, the models reproduce the observations that their effect on sensitivity to alpelisib is small ( to ) and that p27 knockdown alone does not have a strong effect (no change in ), and also incorporate the hypothesis that the effect of p21 and p27 requires knockdown of both p21 and p27 (Fig. 7D).
The MCF7-specific and T47D-specific network models (2020 version of the model) recapitulate the PI3K inhibitor resistance mechanisms and drug combinations that the 2017 version of the model also did. For example, knockdown of PTEN, knockdown of TSC, or high expression of PIM make the cell insensitive to alpelisib ( with alpelisib changes to with the addition of any of these resistance mechanisms), and the addition of fulvestrant to alpelisib results in a larger effect than any of them alone ( with alpelisib or fulvestrant alone changes to with both) (Fig. 7D and Supplemental Fig. S5A and C). Additional resistance mechanisms to alpelisib or PI3K inhibitors that both 2020 models reproduce include high expression of SGK1/PDK1 (12), the HER3 microenvironment (16), high activity of IGF1R and insulin (18), upregulation of ESR1 expression (15), and high MYC activity (58) (Supplemental Fig. S5C). The 2020 models also recapitulate the resistance mechanisms of other drugs. For example, Rb knockdown is a resistance mechanism to palbociclib ( with palbociclib changes to with added Rb knockdown) but has little effect in the presence of fulvestrant (23) ( with fulvestrant to with the addition of Rb knockdown) (Supplemental Fig. S5C). A comparison of several key experimental and clinical outcomes with the model’s results is included in Supplemental Table S2 and in the Supplemental Text.
Overall, the cell line-specific network models we developed in this work are able to reproduce the current knowledge of PI3Kα inhibitor resistance mechanisms and drug combinations, including the new results from this work on the efficacy of the combination of alpelisib with BH3 mimetics and the reduced sensitivity to PI3Kα inhibitors as a result of FOXO3 knockdown.
Discussion
In this work, we experimentally validated several PI3Kα inhibitor drug resistance and sensitivity factors predicted by a 2017 network-based mathematical model of oncogenic signaling in ER+ PIK3CA mutant breast cancer (Fig. 1). We experimentally verified (i) the efficacy of combining PI3Kα inhibitor alpelisib with BH3 mimetics (MCL1 inhibitor s63845 alone or in combination with BCL-XL/BCL-2 inhibitor navitoclax, in a cell line-specific manner), a promising PI3Kα inhibitor drug combination (Figs. 2 and 3) and (ii) the reduced sensitivity to PI3Kα inhibitors (alpelisib and taselisib) caused by FOXO3 knockdown, a novel candidate resistance mechanism for PI3Kα inhibitors (Fig. 4). We additionally found discrepancies with the 2017 model, in particular, our experiments found that the effect of p27 knockdown and RB1 knockdown did not result in the expected reduced sensitivity or had only a small cell line-specific effect (Figs. 5 and 6). Based on these experiments, we developed updated and cell line-specific network models (2020 version of the model) that can recapitulate our new results and other recent work on PI3Kα inhibitor drug resistance.
Our combined mathematical modeling and experimental approach follows the model/experiment refinement cycle that is a cornerstone of systems biology research (60,61), and applies it to the field of cancer drug resistance. In this approach, cancer biology and clinical knowledge are integrated to form a model, experiments are performed to test and refine the model, new knowledge is subsequently generated, and the cycle then repeats itself. This completion of a model/experiment cycle is a step that is necessary for the development of predictive and validated mathematical models in cancer and systems biology, yet it is rarely accomplished (60–62).
An important result from this work is that the combination of PI3Kα inhibitor alpelisib with BH3 mimetics is synergistically efficacious and that this combination leads to an increase in apoptosis compared to each agent alone (Figs. 2 and 3). Our results are consistent with recent work in (51), which provides strong evidence that the combination of BH3 mimetics and alpelisib is efficacious in ER+ breast cancer in vitro, in vivo, and in early passage patient tumor-derived breast epithelial cells. In our work, we find that the ideal choice of BH3 mimetics in the combination strategy depends on which anti-apoptotic BCL-2 family members (e.g. MCL1, BCL-XL, and BCL-2) are highly expressed in a given cancer cell. For the case of T47D cells, which have high BCL-XL expression, alpelisib was synergistically efficacious only when combined with MCL1 inhibitor s63845 and BCL-XL/BCL-2 inhibitor navitoclax. Meanwhile, for MCF7 cells, which have low BCL-XL expression, alpelisib was synergistically efficacious when combined with s63845 alone.
These cell line-specific results highlight an important lesson for the use of alpelisib and BH3 mimetics in the clinical setting of breast cancer: fully taking advantage of this drug combination will require determining the relevant anti-apoptotic BCL-2 family dependence in the tumor. This could be accomplished by directly quantifying these BCL-2 family members in the tumor and/or by using functional genomic approaches such as dynamic BH3 profiling (45,63,64). In line with this view, recent clinical trials on breast cancer tumors with high BCL-2 expression have shown promising results with the combination of approved breast cancer therapies with the BCL-2 inhibitor venetoclax (65,66). Our results suggest a similar approach using the combination of alpelisib and tumor-specific BH3 mimetics determined through dynamic BH3 profiling.
Another key result from this work is that FOXO3 knockdown reduces the sensitivity to PI3Kα inhibitors, thus revealing a novel candidate resistance mechanism (Fig. 4). Even though increased FOXO3 activity has been associated to response to PI3Kα inhibitors (12), it has never been shown that FOXO3 knockdown (and its resulting effects) could be a resistance mechanism to PI3Kα inhibitors on its own. FOXO3 exemplifies the unique insights that can be obtained from network models and their ability to integrate the context-dependent role of cancer-related genes. FOXO3 is known for its dual role as tumor suppressor and oncogene, acting as one or the other depending on the context. Previous work had highlighted the role of FOXO3 as an oncogene through its PI3K-inhibitor-induced feedback activation and the resulting upregulation of ESR1 and HER3. The upregulation of these receptors leads to increased survival in microenvironments that favor their signaling (15,16). Our network modeling and experimental findings show that the tumor suppressor effect of FOXO3 can dominate.
Overall, we applied a systems biology approach combining a network-based mathematical model and its experimental validation to tackle the problems of PI3Kα inhibitor drug resistance and drug combinations in ER+ PIK3CA mutant breast cancer (Fig. 1). The result was the experimental validation of BH3 mimetics as an efficacious drug combination and of FOXO3 knockdown as a drug sensitivity factor and potential resistance mechanism. These results led to a refinement of the model and the development of cell line-specific network models that recapitulate the observed cell line-specific effects and the current knowledge on drug resistance to PI3Kα inhibitors. The validated drug combinations and sensitivity factors of our approach illustrate how the use of network modeling and systems biology approaches can contribute to overcoming the challenge of cancer drug resistance.
Supplementary Material
Acknowledgements
J.G.T.Z. would like to thank the Wagle lab and the Stand Up To Cancer (SU2C) - National Science Foundation (NSF) Drug Combination Convergence Research Team for many useful discussions. J.G.T.Z. would also like to thank SU2C and The V Foundation (TVF) staff, grant reviewers, and scientific leadership for organizing and making possible multiple meetings, which shaped and enriched this work. J.G.T.Z. acknowledges Christian Meyer and Vito Quaranta for their help in quantifying drug combination synergy using MuSyC. J.M. acknowledges support from the Ramon y Cajal Programme, Ministerio de Economia y Competitividad (RYC-2015-18357). J.M. and C.A. acknowledge the CELLEX foundation. This work was supported by the NSF grants PHY 1545832 (to R.A.), PHY 1545839 (to A.G.L. and N.W.) and 1545853 (to M.S. and G.X.), the SU2C, and SU2C/TVF Convergence Scholar Awards to J.G.T.Z. (D2015-039), J.M. (D2015-037), and P.M..
Footnotes
Competing interests. P.M. is an employee of Bluebird bio. J.B. was an employee of AstraZeneca; was on the Board of Directors of Foghorn; and was a past board member of Varian Medical Systems, Bristol-Myers Squibb, Grail, Aura Biosciences, and Infinity Pharmaceuticals. He had performed consulting and/or advisory work for Grail, PMV Pharma, ApoGen, Juno, Eli Lilly, Seragon, Novartis, and Northern Biologics. He had stock or other ownership interests in PMV Pharma, Grail, Juno, Varian, Foghorn, Aura, Infinity Pharmaceuticals, and ApoGen, as well as Tango and Venthera, of which he was a cofounder. He had previously received honoraria or travel expenses from Roche, Novartis, and Eli Lilly. J.B. was an employee of AstraZeneca, which is currently developing capivasertib, an AKT inhibitor. J.B. was a past board member of Infinity Pharmaceuticals, which markets duvelisib and IPI-549. J.B. had been a paid consultant and/or advisor for Novartis, which markets alpelisib, buparlisib, and everolimus. J.B. had stock or other ownership interests in Venthera (of which he was a cofounder), which is developing topical PI3K inhibitors for dermatological conditions. Companies that have developed or are developing PI3K inhibitors, for which coauthors on this study have a disclosure, include Novartis, AstraZeneca, Eli Lilly, Roche, Infinity Pharmaceuticals, and Venthera. J.B. was an inventor on a patent application (PCT/US2019/047879) submitted by MSKCC that is related to the use of multiple PIK3CA mutations as a biomarker for clinical response to PI3K inhibitors. M.S. is on the scientific advisory board of Menarini Ricerche and the Bioscience Institute; has received research funds from Puma Biotechnology, Daiichi-Sankio, Targimmune, Immunomedics, and Menarini Ricerche; and is a cofounder of Medendi.org. M.S. has received research funds from Menarini Ricerche, which markets MEN1611. A.G.L. reports consulting for AbbVie, sponsored research by Novartis, AbbVie, Astra-Zeneca, and being a co-founder of Flash Therapeutics and Vivid Bioscience. J.M. reports previous consulting for Vivid Biosciences and Oncoheroes Biosciences. N.W. reports the following prior relationships: Foundation Medicine (consultant, stockholder), Novartis (consultant, grant support, advisory board). N.W. reports the following current relationships: Puma Biotechnologies (grant support), Eli Lilly (consultant/advisory board), Section 32 (scientific advisory board), Relay Therapeutics (scientific advisory board, stockholder). No potential conflicts of interests were disclosed by the other authors.
References
- 1.Vasan N, Baselga J, Hyman DM. A view on drug resistance in cancer. Nature. 2019;575:299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Konieczkowski DJ, Johannessen CM, Garraway LA. A Convergence-Based Framework for Cancer Drug Resistance. Cancer Cell. 2018;33:801–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steeg PS. Targeting metastasis. Nat Rev Cancer. 2016;16:201–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weigelt B, Peterse JL, van ‘t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591–602. [DOI] [PubMed] [Google Scholar]
- 5.Waks AG, Winer EP. Breast Cancer Treatment: A Review. JAMA. 2019;321:288–300. [DOI] [PubMed] [Google Scholar]
- 6.Caswell-Jin JL, Plevritis SK, Tian L, Cadham CJ, Xu C, Stout NK, et al. Change in Survival in Metastatic Breast Cancer with Treatment Advances: Meta-Analysis and Systematic Review. JNCI Cancer Spectr. 2018;2:ky062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.André F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor–Positive Advanced Breast Cancer. N Engl J Med. Massachusetts Medical Society; 2019;380:1929–40. [DOI] [PubMed] [Google Scholar]
- 8.Juric D, Castel P, Griffith M, Griffith OL, Won HH, Ellis H, et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kα inhibitor. Nature. 2015;518:240–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Razavi P, Dickler MN, Shah PD, Toy W, Brown DN, Won HH, et al. Alterations in PTEN and ESR1 promote clinical resistance to alpelisib plus aromatase inhibitors. Nature Cancer. Nature Publishing Group; 2020;1:382–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elkabets M, Vora S, Juric D, Morse N, Mino-Kenudson M, Muranen T, et al. mTORC1 inhibition is required for sensitivity to PI3K p110α inhibitors in PIK3CA-mutant breast cancer. Sci Transl Med. 2013;5:196ra99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le X, Antony R, Razavi P, Treacy DJ, Luo F, Ghandi M, et al. Systematic Functional Characterization of Resistance to PI3K Inhibition in Breast Cancer. Cancer Discov. 2016;6:1134–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castel P, Ellis H, Bago R, Toska E, Razavi P, Carmona FJ, et al. PDK1-SGK1 Signaling Sustains AKT-Independent mTORC1 Activation and Confers Resistance to PI3Kα Inhibition. Cancer Cell. 2016;30:229–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa C, Ebi H, Martini M, Beausoleil SA, Faber AC, Jakubik CT, et al. Measurement of PIP3 levels reveals an unexpected role for p110β in early adaptive responses to p110α-specific inhibitors in luminal breast cancer. Cancer Cell. 2015;27:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz S, Wongvipat J, Trigwell CB, Hancox U, Carver BS, Rodrik-Outmezguine V, et al. Feedback suppression of PI3Kα signaling in PTEN-mutated tumors is relieved by selective inhibition of PI3Kβ. Cancer Cell. 2015;27:109–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosch A, Li Z, Bergamaschi A, Ellis H, Toska E, Prat A, et al. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor–positive breast cancer. Sci Transl Med. 2015;7:283ra51–283ra51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kodack DP, Askoxylakis V, Ferraro GB, Sheng Q, Badeaux M, Goel S, et al. The brain microenvironment mediates resistance in luminal breast cancer to PI3K inhibition through HER3 activation. Sci Transl Med. 2017;9:eaal4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donnella HJ, Webber JT, Levin RS, Camarda R, Momcilovic O, Bayani N, et al. Kinome rewiring reveals AURKA limits PI3K-pathway inhibitor efficacy in breast cancer. Nat Chem Biol. 2018;14:768–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hopkins BD, Pauli C, Du X, Wang DG, Li X, Wu D, et al. Suppression of insulin feedback enhances the efficacy of PI3K inhibitors. Nature. 2018;560:499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, et al. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell. 2018;34:427–438.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertucci F, Ng CKY, Patsouris A, Droin N, Piscuoglio S, Carbuccia N, et al. Genomic characterization of metastatic breast cancers. Nature. 2019;569:560–4. [DOI] [PubMed] [Google Scholar]
- 21.Nayar U, Cohen O, Kapstad C, Cuoco MS, Waks AG, Wander SA, et al. Acquired HER2 mutations in ER+ metastatic breast cancer confer resistance to estrogen receptor–directed therapies. Nat Genet. 2019;51:207–16. [DOI] [PubMed] [Google Scholar]
- 22.Mao P, Cohen O, Kowalski KJ, Kusiel JG, Buendia-Buendia JE, Cuoco MS, et al. Acquired FGFR and FGF Alterations Confer Resistance to Estrogen Receptor (ER) Targeted Therapy in ER+ Metastatic Breast Cancer. Clin Cancer Res. 2020;26:5974–89. [DOI] [PubMed] [Google Scholar]
- 23.Wander SA, Cohen O, Gong X, Johnson GN, Buendia-Buendia JE, Lloyd MR, et al. The Genomic Landscape of Intrinsic and Acquired Resistance to Cyclin-Dependent Kinase 4/6 Inhibitors in Patients with Hormone Receptor-Positive Metastatic Breast Cancer. Cancer Discov. 2020;10:1174–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, Razavi P, Li Q, Toy W, Liu B, Ping C, et al. Loss of the FAT1 Tumor Suppressor Promotes Resistance to CDK4/6 Inhibitors via the Hippo Pathway. Cancer Cell. 2018;34:893–905.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nat Med. 2013;19:1401–9. [DOI] [PubMed] [Google Scholar]
- 26.Sun C, Bernards R. Feedback and redundancy in receptor tyrosine kinase signaling: relevance to cancer therapies. Trends Biochem Sci. 2014;39:465–74. [DOI] [PubMed] [Google Scholar]
- 27.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarke MA, Fisher J. Executable cancer models: successes and challenges. Nat Rev Cancer. 2020;20:343–54. [DOI] [PubMed] [Google Scholar]
- 29.Rockne RC, Hawkins-Daarud A, Swanson KR, Sluka JP, Glazier JA, Macklin P, et al. The 2019 mathematical oncology roadmap. Phys Biol. 2019;16:041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zañudo J GT, Steinway SN, Albert R. Discrete dynamic network modeling of oncogenic signaling: Mechanistic insights for personalized treatment of cancer. Current Opinion in Systems Biology. 2018;9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saez-Rodriguez J, Blüthgen N. Personalized signaling models for personalized treatments. Mol Syst Biol. 2020;16:e9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gómez Tejeda Zañudo J, Scaltriti M, Albert R. A network modeling approach to elucidate drug resistance mechanisms and predict combinatorial drug treatments in breast cancer. Cancer Converg. 2017;1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steinway SN, Zañudo JGT, Michel PJ, Feith DJ, Loughran TP, Albert R. Combinatorial interventions inhibit TGFβ-driven epithelial-to-mesenchymal transition and support hybrid cellular phenotypes. NPJ Syst Biol Appl. 2015;1:15014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Béal J, Montagud A, Traynard P, Barillot E, Calzone L. Personalization of Logical Models With Multi-Omics Data Allows Clinical Stratification of Patients. Front Physiol. 2018;9:1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fröhlich F, Kessler T, Weindl D, Shadrin A, Schmiester L, Hache H, et al. Efficient Parameter Estimation Enables the Prediction of Drug Response Using a Mechanistic Pan-Cancer Pathway Model. Cell Syst. 2018;7:567–579.e6. [DOI] [PubMed] [Google Scholar]
- 36.Fey D, Halasz M, Dreidax D, Kennedy SP, Hastings JF, Rauch N, et al. Signaling pathway models as biomarkers: Patient-specific simulations of JNK activity predict the survival of neuroblastoma patients. Sci Signal. 2015;8:ra130. [DOI] [PubMed] [Google Scholar]
- 37.Meyer CT, Wooten DJ, Paudel BB, Bauer J, Hardeman KN, Westover D, et al. Quantifying Drug Combination Synergy along Potency and Efficacy Axes. Cell Syst. 2019;8:97–108.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER 3rd, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nusinow DP, Szpyt J, Ghandi M, Rose CM, McDonald ER 3rd, Kalocsay M, et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell. 2020;180:387–402.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dempster JM, Rossen J, Kazachkova M, Pan J, Kugener G, Root DE, et al. Extracting Biological Insights from the Project Achilles Genome-Scale CRISPR Screens in Cancer Cell Lines. bioRxiv. 2019;720243. [Google Scholar]
- 41.Sunters A, Fernández de Mattos S, Stahl M, Brosens JJ, Zoumpoulidou G, Saunders CA, et al. FoxO3a transcriptional regulation of Bim controls apoptosis in paclitaxel-treated breast cancer cell lines. J Biol Chem. 2003;278:49795–805. [DOI] [PubMed] [Google Scholar]
- 42.She Q-B, Solit DB, Ye Q, O’Reilly KE, Lobo J, Rosen N. The BAD protein integrates survival signaling by EGFR/MAPK and PI3K/Akt kinase pathways in PTEN-deficient tumor cells. Cancer Cell. 2005;8:287–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kotschy A, Szlavik Z, Murray J, Davidson J, Maragno AL, Le Toumelin-Braizat G, et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature. 2016;538:477–82. [DOI] [PubMed] [Google Scholar]
- 44.Vora SR, Juric D, Kim N, Mino-Kenudson M, Huynh T, Costa C, et al. CDK 4/6 inhibitors sensitize PIK3CA mutant breast cancer to PI3K inhibitors. Cancer Cell. 2014;26:136–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montero J, Sarosiek KA, DeAngelo JD, Maertens O, Ryan J, Ercan D, et al. Drug-induced death signaling strategy rapidly predicts cancer response to chemotherapy. Cell. 2015;160:977–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49:1779–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muranen T, Selfors LM, Worster DT, Iwanicki MP, Song L, Morales FC, et al. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell. 2012;21:227–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu MC- T, Lee D-F, Xia W, Golfman LS, Ou-Yang F, Yang J-Y, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–37. [DOI] [PubMed] [Google Scholar]
- 50.Medema RH, Kops GJ, Bos JL, Burgering BM. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–7. [DOI] [PubMed] [Google Scholar]
- 51.Stratikopoulos EE, Kiess N, Szabolcs M, Pegno S, Kakit C, Wu X, et al. Mouse ER+/PIK3CAH1047R breast cancers caused by exogenous estrogen are heterogeneously dependent on estrogen and undergo BIM-dependent apoptosis with BH3 and PI3K agents. Oncogene. 2019;38:47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tzivion G, Dobson M, Ramakrishnan G. FoxO transcription factors; Regulation by AKT and 14–3-3 proteins. Biochim Biophys Acta. 2011;1813:1938–45. [DOI] [PubMed] [Google Scholar]
- 53.Viglietto G, Motti ML, Bruni P, Melillo RM, D’Alessio A, Califano D, et al. Cytoplasmic relocalization and inhibition of the cyclin-dependent kinase inhibitor p27(Kip1) by PKB/Akt-mediated phosphorylation in breast cancer. Nat Med. 2002;8:1136–44. [DOI] [PubMed] [Google Scholar]
- 54.Zhou BP, Liao Y, Xia W, Spohn B, Lee MH, Hung MC. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat Cell Biol. 2001;3:245–52. [DOI] [PubMed] [Google Scholar]
- 55.Polyak K, Lee MH, Erdjument-Bromage H, Koff A, Roberts JM, Tempst P, et al. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78:59–66. [DOI] [PubMed] [Google Scholar]
- 56.Planas-Silva MD, Weinberg RA. Estrogen-dependent cyclin E-cdk2 activation through p21 redistribution. Mol Cell Biol. 1997;17:4059–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang X, Tang N, Hadden TJ, Rishi AK. Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta. 2011;1813:1978–86. [DOI] [PubMed] [Google Scholar]
- 58.Ilic N, Utermark T, Widlund HR, Roberts TM. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc Natl Acad Sci U S A. 2011;108:E699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ni Chonghaile T, Sarosiek KA, Vo T-T, Ryan JA, Tammareddi A, Moore VDG, et al. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science. 2011;334:1129–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coutant A, Roper K, Trejo-Banos D, Bouthinon D, Carpenter M, Grzebyta J, et al. Closed-loop cycles of experiment design, execution, and learning accelerate systems biology model development in yeast. Proc Natl Acad Sci U S A. 2019;116:18142–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alberghina L, Westerhoff HV. Systems Biology: Definitions and Perspectives. Springer Science & Business Media; 2007. [Google Scholar]
- 62.Brady R, Enderling H. Mathematical Models of Cancer: When to Predict Novel Therapies, and When Not to. Bull Math Biol. 2019;81:3722–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Montero J, Gstalder C, Kim DJ, Sadowicz D, Miles W, Manos M, et al. Destabilization of NOXA mRNA as a common resistance mechanism to targeted therapies. Nat Commun. 2019;10:5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bhola PD, Ahmed E, Guerriero JL, Sicinska E, Su E, Lavrova E, et al. High-throughput dynamic BH3 profiling may quickly and accurately predict effective therapies in solid tumors. Sci Signal. 2020;13:eaay1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whittle JR, Vaillant F, Surgenor E, Policheni AN, Giner G, Capaldo BD, et al. Dual Targeting of CDK4/6 and BCL2 Pathways Augments Tumor Response in Estrogen Receptor-Positive Breast Cancer. Clin Cancer Res. 2020;26:4120–34. [DOI] [PubMed] [Google Scholar]
- 66.Lok SW, Whittle JR, Vaillant F, Teh CE, Lo LL. A phase Ib dose-escalation and expansion study of the BCL2 inhibitor venetoclax combined with tamoxifen in ER and BCL2–positive metastatic breast cancer. Cancer Discov. 2019;9:354–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.