Abstract
The Mediator complex is central to transcription by RNA polymerase II (Pol II) in eukaryotes. In budding yeast (Saccharomyces cerevisiae), Mediator is recruited by activators and associates with core promoter regions, where it facilitates preinitiation complex (PIC) assembly, only transiently before Pol II escape. Interruption of the transcription cycle by inactivation or depletion of Kin28 inhibits Pol II escape and stabilizes this association. However, Mediator occupancy and dynamics have not been examined on a genome-wide scale in yeast grown in nonstandard conditions. Here we investigate Mediator occupancy following heat shock or CdCl2 exposure, with and without depletion of Kin28. We find that Pol II occupancy shows similar dependence on Mediator under normal and heat shock conditions. However, although Mediator association increases at many genes upon Kin28 depletion under standard growth conditions, little or no increase is observed at most genes upon heat shock, indicating a more stable association of Mediator after heat shock. Unexpectedly, Mediator remains associated upstream of the core promoter at genes repressed by heat shock or CdCl2 exposure whether or not Kin28 is depleted, suggesting that Mediator is recruited by activators but is unable to engage PIC components at these repressed targets. This persistent association is strongest at promoters that bind the HMGB family member Hmo1, and is reduced but not eliminated in hmo1Δ yeast. Finally, we show a reduced dependence on PIC components for Mediator occupancy at promoters after heat shock, further supporting altered dynamics or stronger engagement with activators under these conditions.
Transcription of protein-coding genes in eukaryotes is a complex process involving recruitment of coactivators, assembly of the preinitiation complex (PIC), including RNA polymerase II (Pol II), and transition from promoter melting and initiation to productive elongation (Schier and Taatjes 2020). The Mediator complex, a large, multisubunit complex present in plants, animals, and single-celled eukaryotes, is central to this process (Jeronimo and Robert 2017; Soutourina 2018). In yeast, Mediator has been divided into head, middle, tail, and cyclin-CDK modules on the basis of structural and genetic data (Plaschka et al. 2016). Functionally, yeast Mediator is first recruited to upstream activation sites (UASs) that are 200–400 bp upstream of the transcription start sites (TSSs) of most genes by sequence-specific DNA-binding activator proteins via the Mediator tail module (Jeronimo et al. 2016; Jeronimo and Robert 2017; Knoll et al. 2018; Soutourina 2018). Subunits in the middle and head module then engage PIC components, including Pol II, to facilitate PIC assembly (Plaschka et al. 2016; Robinson et al. 2016; Jeronimo and Robert 2017; Soutourina 2018). This process entails bridging between the UAS and the core promoter, where the PIC is assembled, by a single Mediator complex (Jeronimo et al. 2016; Petrenko et al. 2016). Association of Mediator with the core promoter is normally brief (estimated as being <1 sec for a single transcription initiation event) but is stabilized by inhibiting Pol II escape, for example, by depletion or inactivation of Kin28, which facilitates Pol II escape by phosphorylating the C-terminal domain of Rpb1 (Jeronimo and Robert 2014; Wong et al. 2014). Transit of Mediator from UASs to the core promoter depends on Spt15 (also known as TBP, as it will be referred to hereafter), and its association with the core promoter in the absence of Kin28 is destabilized by depletion of Taf1 or Pol II (Knoll et al. 2018).
Mediator occupancy measured by ChIP during the normal transcription cycle (i.e., with Kin28 active) is much higher at induced genes that are controlled by strong activators, such as those activated by heat shock or growth in galactose (Fan et al. 2006; Fan and Struhl 2009; Kim and Gross 2013). This could reflect stronger interactions between Mediator and activators, and hence less dependence on PIC components or other factors for stabilizing Mediator occupancy, at strongly induced genes compared with constitutively active genes during growth in rich medium (YPD). Increased Mediator occupancy has been observed at a few individual promoters induced by heat shock following Kin28 depletion (Petrenko et al. 2016); however, no genome-wide analysis has been reported examining whether promoter occupancy by Mediator is stabilized by Kin28 depletion at strongly induced genes or depends on PIC components as it does in yeast grown in rich medium (YPD). In this work, we examine genome-wide Mediator occupancy in Saccharomyces cerevisiae following two perturbations, heat shock and CdCl2 administration, that induce large and rapid genome-wide transcriptional responses.
Results
Pol II recruitment upon heat shock depends on Mediator
To identify genes most active and most highly induced after heat shock, we performed ChIP-seq to determine Pol II occupancy in the commonly used laboratory strain BY4741 and in YFR1321, the parent strain (originally derived from W303) (Haruki et al. 2008) to that used for anchor-away experiments described in later sections, before and after 15 and 30 min of heat shock. Heat shock resulted in increased Pol II occupancy at targets of Hsf1, the primary transcription factor responding to heat shock, and the stress-responsive activators Msn2 and Msn4, whereas Pol II occupancy was nearly completely lost from RP genes, consistent with previous determinations of mRNA levels and nascent RNA production following heat shock (Supplemental Fig. S1A; Warner 1999; Gasch et al. 2000; Causton et al. 2001; Pincus et al. 2018). K-means clustering revealed two groups of genes strongly up-regulated by heat shock; these groups were both enriched for association with Hsf1 and Msn2 and Msn4 (Supplemental Fig. S1B). Transcription induced by Hsf1 has been reported to occur independently of Mediator (Lee and Lis 1998; McNeil et al. 1998), but we found a similar two- to threefold reduction in Pol II occupancy at Hsf1 and Msn2/4 targets after heat shock upon depletion of the essential Mediator head module subunit Med17, using the anchor-away method (see next section), as at other genes in the absence of heat shock, in agreement with recent results from the Struhl laboratory (Supplemental Fig. S1C; Petrenko et al. 2017).
Effect of Kin28 depletion on Mediator recruitment with and without heat shock
To examine Mediator dynamics at genes induced upon heat shock, we performed ChIP-seq against Gal11 (also known as Med15) (Bourbon et al. 2004), from the Mediator tail module, in yeast engineered to allow depletion of Kin28 from the cell nucleus using the anchor-away method, and in the parent strain, YFR1321 (Jeronimo and Robert 2014; Wong et al. 2014; Knoll et al. 2018). The kin28AA yeast strain expresses Kin28 with a C-terminal FRB tag and the ribosomal subunit Rpl13A C-terminally tagged with the FKBP12 fragment; upon administration of rapamycin, the FRB and FKBP12 moieties are tightly coupled, and Kin28 is evicted from the nucleus following nuclear processing of Rpl13A. The parent strain is identical except for lacking the FRB tag, and both strains harbor tor1-1 and fpr1Δ mutations that abrogate the stress response and toxicity otherwise caused by exposure to rapamycin (Haruki et al. 2008). Yeast cultures were grown to early log phase (A600 ∼ 0.6–0.8), treated with rapamycin for 1 h to allow Kin28 depletion, and then subjected to heat shock by a rapid temperature shift for 15 min before cross-linking for ChIP-seq analysis. Previous work has shown that 1 h of rapamycin treatment results in efficient depletion of the target protein and of Kin28 in particular, and robust recruitment of Mediator has been observed after 5–30 min of heat shock (Haruki et al. 2008; Kim and Gross 2013; Wong et al. 2014; Knoll et al. 2018).
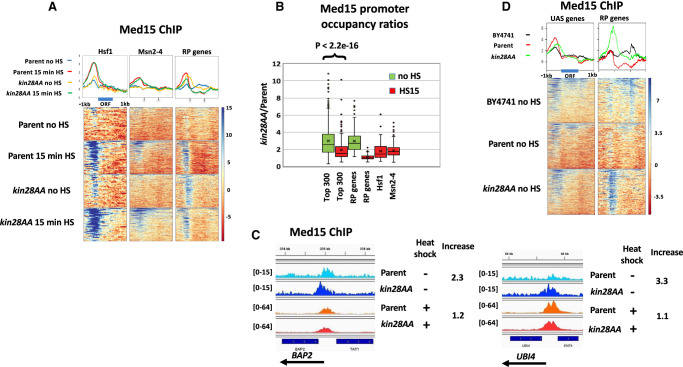
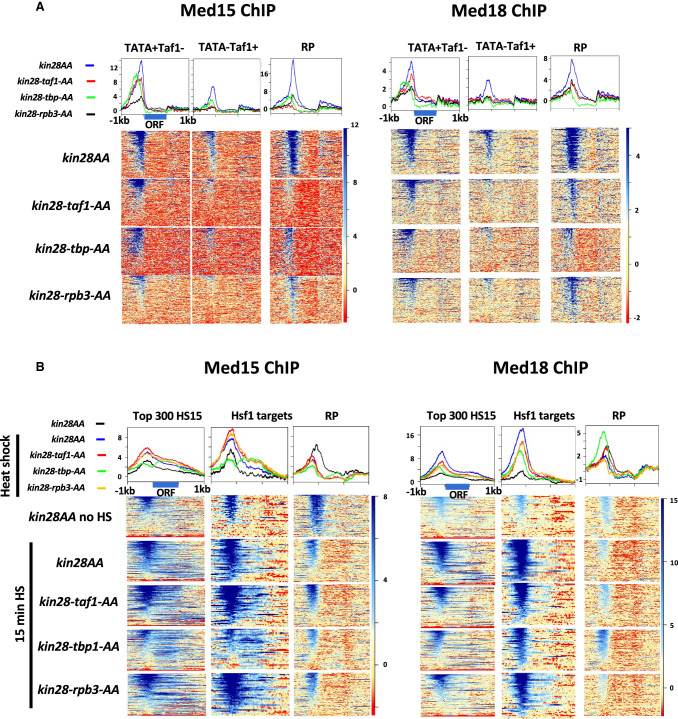
In the absence of heat shock, depletion of Kin28 resulted in an increase in Med15 ChIP signal at promoters of Hsf1 targets and RP genes, consistent with prior work (Fig. 1A, cf. “Parent no HS” and “kin28AA no HS”; for biological replicate experiment, see also Supplemental Fig. S2A; Jeronimo and Robert 2014; Wong et al. 2014). Many Hsf1 target genes are active even in the absence of heat shock (Gross et al. 1990; Solís et al. 2016; de Jonge et al. 2017; Pincus et al. 2018), so the association of Mediator with promoters of Hsf1 targets in the absence of heat shock seen upon depletion of Kin28 is not unexpected. Heat shock resulted in an increased Med15 ChIP signal at Hsf1 and Msn2/4 targets with or without Kin28 depletion (Fig. 1A). Quantitation of Med15 ChiP signal in non-heat-shocked cells at RP genes and at the approximately 300 non-RP gene promoters having highest Med15 occupancy after Kin28 depletion (also in non-heat-shocked cells) revealed an increase in signal in kin28AA compared with wild-type yeast, as expected (Fig. 1B; Supplemental Fig. S2B). In contrast, Med15 occupancy showed little or no increase upon Kin28 depletion in heat-shocked cells, either at Hsf1 targets or the 300 genes most highly occupied by Med15 in kin28AA yeast after heat shock (Fig. 1A,B; Supplemental Fig. S2A,B). Analysis of the 300 non-RP gene promoters with highest Pol II occupancy (in non-heat-shocked and heat-shocked YFR1321 yeast) yielded similar results (Supplemental Fig. S2C), as did comparison of genes having similar Pol II occupancy in the absence and presence of heat shock (Supplemental Fig. S2D).
Figure 1.
Effect of heat shock on Mediator association. (A) Heat maps and line graphs depicting normalized occupancy of the Mediator tail module subunit, Med15, in kin28AA yeast treated with rapamycin and the parent strain YFR1321, also treated with rapamycin, before and after 15 min of heat shock, at 42 Hsf1 targets and 213 Msn2-4 targets and 137 RP genes (see Methods; Supplemental Table S2). (B) Box and whisker plots showing the ratios of Med15 occupancy with and without Kin28 depletion for the approximately 300 genes showing the highest Med15 occupancy in Kin28-depleted cells without or with heat shock; ratios are also shown for RP genes, Hsf1 targets, and Msn2-4 targets in heat-shocked cells. (C) Browser scans showing Med15 occupancy upstream of BAP2 and UBI4 in kin28AA yeast and the parent strain YFR1321, both treated with rapamycin, with and without heat shock. UBI4 is a target of Hsf1, whereas BAP2 is not a target of Hsf1 or Msn2-4. Scale, in reads per million mapped reads, is indicated for each scan. (D) Heat maps and line graphs depicting occupancy of the Mediator tail module subunit, Med15, in BY4741 yeast, kin28AA yeast treated with rapamycin, and the parent strain YFR1321, also treated with rapamycin, in the absence of heat shock, at “UAS genes” (see text and Supplemental Table S2) and RP genes.
Consistent with the trends evident from Figure 1, A and B, stronger induction of Med15 occupancy upon Kin28 depletion under non–heat shock than under heat shock conditions was observed upon inspection of browser scans at many genes (Fig. 1C; Supplemental Fig. S3, YDJ1). At the same time, some promoters behave counter to this trend, reflecting the range of altered Med15 occupancy observed upon Kin28 depletion (Fig. 1B; Supplemental Fig. S3, TPS1). Variable effects on the increase in Med15 occupancy upon Kin28 depletion were observed for Hsf1 and Msn2/4 targets as well as for genes induced by heat shock that are not known to be targets of either of these stress-responsive activators (Supplemental Fig. S3; Supplemental Table S1). The gene-specific increase in Mediator ChIP signal upon Kin28 depletion or inactivation is similarly evident in previous reports, but the cause of this remains unexplained (Jeronimo and Robert 2014; Knoll et al. 2018).
Under normal growth conditions in rich medium, many active gene promoters, most conspicuously those of many RP genes, show little or no association with Mediator as measured by ChIP (Jeronimo and Robert 2014; Wong et al. 2014; Paul et al. 2015). Some active genes, however, do show Mediator ChIP signal at UAS regions, but not at core promoter regions, under normal growth conditions; depletion or inactivation of Kin28 results in a shift in Mediator occupancy at these genes from UAS to core promoter, with the shift being greatest for head and middle module subunits that contact the PIC and least for tail module subunits (Jeronimo et al. 2016; Petrenko et al. 2016; Knoll et al. 2018). Correspondingly, we observed a shift toward the core promoter with little change in intensity of the Med15 ChIP signal upon Kin28 depletion in the absence of heat shock at a set of 498 genes, which we refer to as “UAS genes,” that show Mediator ChIP signal at UAS regions under normal growth conditions (Fig. 1D; Supplemental Table S2; Jeronimo et al. 2016). In contrast, RP genes show increased Mediator occupancy upon Kin28 depletion, along with a shift in the peak toward the TSS (Fig. 1D).
Taken together, these results indicate that Hsf1 targets that are active under non-heat-shocked conditions behave similarly to the large cohort of genes, exemplified by RP genes, that show little or no Mediator signal unless Kin28 is depleted, whereas under heat shock conditions, Hsf1 and Msn2/4 targets behave more like the “UAS genes” at which Mediator ChIP signal is seen under normal growth conditions and shows modest increase upon depletion of Kin28.
Mediator remains associated at genes repressed by heat shock
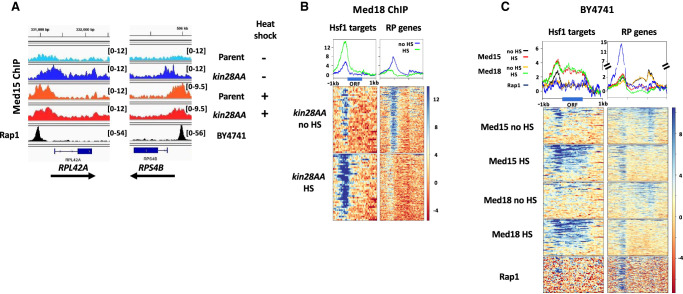
We were surprised to note a prominent Med15 peak upstream of RP genes following heat shock (Fig. 1A), despite the near absence of Pol II occupancy (Supplemental Fig. S1A). The position of this peak was not affected by Kin28 depletion in heat-shocked cells and was shifted upstream relative to the Med15 peak observed following Kin28 depletion in the absence of heat shock (Fig. 1A, RP genes, and Fig. 2A). Thus, upon heat shock, Mediator association with RP genes, under conditions of depleted Kin28, shifts from a core promoter location close to the TSS to an upstream position that closely coincides with that of the RP gene activator Rap1 (see below; Fig. 2A). ChIP-seq against the Mediator head module subunit Srb5 (also known as Med18) (Bourbon et al. 2004) in kin28AA yeast treated with rapamycin also showed persistent association with the repressed RP genes following heat shock, albeit at reduced intensity relative to that seen with Med15, and again accompanied by an upstream shift of the peak (Fig. 2B; Supplemental Fig. S4).
Figure 2.
Mediator association persists at RP genes after heat shock. (A) Browser scans showing normalized occupancy of Med15 upstream of RPL42A and RPS4B in kin28AA yeast and the parent strain YFR1321, both treated with rapamycin, with and without heat shock (top four scans) or Rap1 in strain BY4741 (bottom scan). Scale, in reads per million mapped reads, is indicated for each scan. (B) Heat maps and line graphs depicting normalized occupancy of the Mediator head module subunit, Med18, in kin28AA yeast treated with rapamycin, with and without heat shock, at Hsf1 targets and RP genes. (C) Heat maps and line graphs depicting normalized occupancy of Mediator subunits Med15 (tail) and Med18 (head), and Rap1, at Hsf1 target genes and RP genes in BY4741. The signal observed at transcribed ORF regions (seen at Hsf1 targets under heat shock conditions, and at RP genes under non-heat-shocked conditions) is a ChIP artifact frequently observed at highly transcribed ORFs (Eyboulet et al. 2013; Park et al. 2013; Teytelman et al. 2013; Jeronimo and Robert 2014; Knoll et al. 2020).
The kin28AA strain and its parent, YFR1321, harbor tor1-1 and fpr1Δ mutations, which prevent the normal stress response accompanying rapamycin administration (Haruki et al. 2008). The TOR pathway is intimately connected with ribosome function (Mayer and Grummt 2006), and Fpr1 associates with the UAS of RP gene promoters and promotes binding of Fhl1/Ifh1 (Kasahara et al. 2020), raising the possibility that the unexpected association of Mediator with RP genes repressed by heat shock was a consequence of the tor1-1 or fpr1Δ mutation. We therefore performed ChIP-seq against Med15 and Med18 in the wild-type strain BY4741 before and after 15 min of heat shock. Again, we observed peaks at RP genes for both Med15 and Med18 following heat shock (Fig. 2C; Supplemental Fig. S4). Furthermore, as with the anchor-away parent strain YFR1321, Mediator ChIP-seq signal at RP genes was stronger after heat shock. Thus, persistent Mediator association with RP genes repressed by heat shock is independent of the TOR pathway mutations present in the anchor-away strain background.
Mediator peaks upstream of repressed RP genes in heat-shocked yeast were nearly coincident with the peak observed for Rap1, which binds upstream of the large majority of RP genes and is essential for recruitment of the PIC to most RP genes (Fig. 2C; Lieb et al. 2001; Mencı´a et al. 2002; Ansari et al. 2009; Zeevi et al. 2011; Knight et al. 2014; Reja et al. 2015). This observation suggests that Mediator is recruited to these genes in heat-shocked cells but does not transit to core promoters as it normally does in non-heat-shocked cells. This notion is also consistent with the lower signal observed at RP genes repressed by heat shock for Med18 (from the head module) than for Med15 (from the tail module), as Med15 is expected to have more direct contact with activators bound to UASs and thus be more efficiently cross-linked than Med18 (Jeronimo et al. 2016; Petrenko et al. 2016).
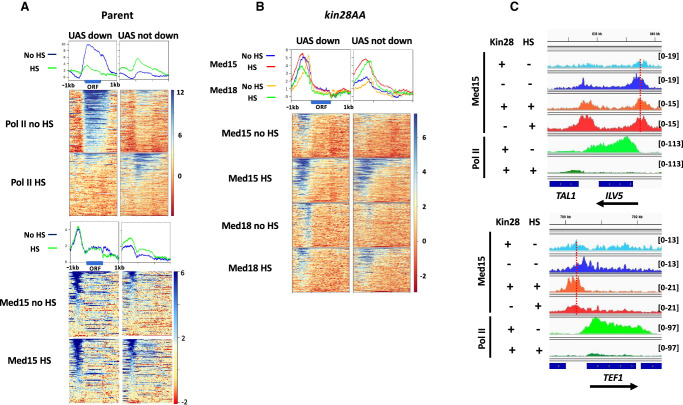
Many non-RP genes are also repressed by heat shock (Supplemental Fig. S1; Gasch et al. 2000; Causton et al. 2001). To assess whether Mediator occupies promoters of such genes after heat shock, we examined ChIP-seq signal at the UAS genes defined earlier (Supplemental Table S2; Jeronimo et al. 2016). We divided these genes into those showing decreased Pol II occupancy after heat shock by at least twofold (116 “UAS down” genes after removing RP genes) and the 231 “UAS not down” genes having a ratio of Pol II occupancy in heat-shocked compared with non-heat-shocked cells ≥1, and compared the Med15 signal at these two sets of genes before and after heat shock (Fig. 3A). We observed Med15 signal for both the “UAS down” and “UAS not down” cohorts in YFR1321 yeast (parent strain for kin28AA) grown at 30°C; upon heat shock, Med15 signal was unchanged for “UAS down” genes and showed a slight increase for “UAS not down” genes (Fig. 3A). Thus, despite the divergent behavior of these two cohorts with regard to Pol II occupancy, both show continued Mediator association with their UAS regions after heat shock.
Figure 3.
Mediator association persists at non-RP genes repressed by heat shock. (A) Heat maps and line graphs showing normalized occupancy of Pol II and Med15 at non-RP UAS genes (see text) having Pol II occupancy decreased by at least twofold upon heat shock (“UAS down”) (Supplemental Table S2) or having Pol II occupancy unchanged or increased upon heat shock (“UAS not down”) (Supplemental Table S2) in rapamycin-treated YFR1321, the parent strain to kin28AA yeast, with and without heat shock. (B) Heat maps and line graphs showing normalized occupancy of Med15 (tail) and Med18 (head) at “UAS down” and “UAS not down” genes in kin28AA yeast treated with rapamycin, with and without heat shock. (C) Browser scans showing Med15 and Pol II occupancy at ILV1 and TEF1 genes in kin28AA yeast (“−Kin28”) or the parent strain YFR1321 (“+Kin28”), both treated with rapamycin, with or without heat shock. Scale, in reads per million mapped reads, is indicated for each scan. Note that Pol II occupancy is reduced at both genes upon heat shock; the vertical dashed lines emphasize the shift of the Med15 peak toward the promoters only when Kin28 is depleted in the absence of heat shock.
We then examined association of Med15 and Med18 at these same cohorts under conditions of Kin28 depletion to allow assessment of Mediator association with core promoter regions (Fig. 3B). In the absence of heat shock, “UAS down” genes showed stronger Mediator peaks than “UAS not down” genes, consistent with the higher expression of the former (Fig. 3A). Med18, from the head module, showed a relatively narrow peak close to the TSS, whereas Med15, from the tail module, showed a broader peak extending from near the TSS to farther upstream at both cohorts. This is consistent with previous observations and concordant with Mediator being recruited to UASs via the tail module and then transiting to the core promoter upon engagement of the head module with PIC components (Jeronimo et al. 2016; Petrenko et al. 2016; Knoll et al. 2018).
Upon heat shock, the two cohorts behaved differently with regard to Mediator association. At “UAS not down” genes, both Med15 and Med18 peaks increased in amplitude while not changing much in position, consistent with the overall increased expression of this cohort. In contrast, “UAS down” genes behaved similarly to the repressed RP genes, with both Med15 and Med18 showing peaks that were shifted upstream from their positions near the TSS, at the core promoter, with little change in amplitude (Fig. 3B, left panel, C).
Altogether, these results reveal persistent Mediator association with genes repressed by heat shock. This association was localized to UASs even under conditions of Kin28 depletion, suggesting that Mediator is recruited by activators at these repressed loci but is unable to facilitate PIC assembly and transcription. We next sought to gain insight into the mechanism underlying the persistent association of Mediator with genes repressed by heat shock.
Hmo1 contributes to persistent Mediator association at genes repressed by heat shock but is not the sole determinant
To test whether the nonproductive retention of Mediator at genes repressed by heat shock could be associated with specific activators or promoter configurations, we first examined the RP genes in more detail. Based on ChIP-seq experiments, RP genes have been divided into three categories. The large majority binds Rap1, which recruits Fhl1 and Ifh1, with Ifh1 being critical for transcriptional activation (Martin et al. 2004; Schawalder et al. 2004; Wade et al. 2004; Rudra et al. 2005). These 129 RP genes fall into two distinct classes, one of which binds the high mobility group family protein, Hmo1, downstream from Rap1 and is characterized by smaller Rap1-TSS and Rap1-Fhl1 separation than the other class, which lacks Hmo1 binding (Hall et al. 2006; Kasahara et al. 2007; Knight et al. 2014; Reja et al. 2015; Zencir et al. 2020). The remaining RP genes do not bind Rap1, Ifh1, or Fhl1 and are instead under control of the general regulatory factor Abf1.
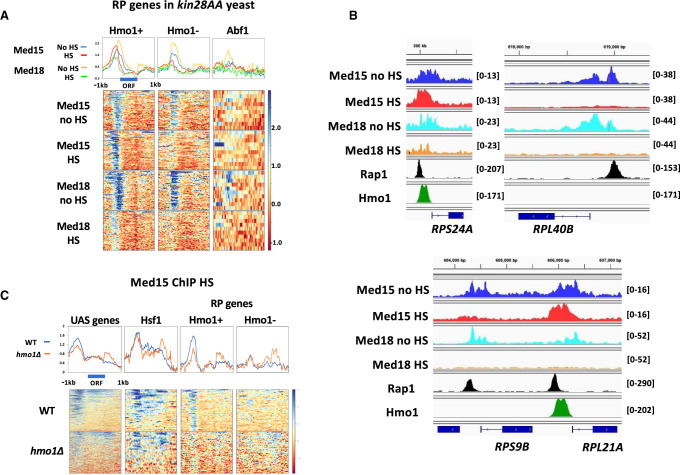
All three classes of RP genes are repressed by heat shock (Supplemental Fig. S5A; Reja et al. 2015). However, these cohorts behave differently with respect to Mediator association: Mediator association persists at Hmo1-binding RP genes after heat shock in both kin28AA and parent yeast strains, whereas association is much lower in non-Hmo1-binding RP genes (Fig. 4A,B; Supplemental Fig. S5B). Abf1-bound RP genes show weak Med18 association only in non-HS kin28AA yeast and show no discernible Med15 peak. Med15 peaks after heat shock approximately coincide with Rap1 peaks (Supplemental Fig. S5C); browser scans of individual loci indicate Med15 occupancy slightly downstream from Rap1 binding sites and overlapping Hmo1 binding sites (Fig. 4B). These results suggest that Hmo1 could be important for persistent Mediator association at RP genes repressed by heat shock.
Figure 4.
Persistent Mediator association preferentially occurs at Hmo1-binding RP genes repressed by heat shock. (A) Heat maps and line graphs showing normalized occupancy of Med15 and Med18 in kin28AA yeast treated with rapamycin, with and without heat shock, at RP genes divided into Hmo1-binding, non-Hmo1-binding, and Abf1-binding genes (Supplemental Table S2). (B) Browser scans showing occupancy of Med15 and Med18 in kin28AA yeast treated with rapamycin, with and without heat shock, and Rap1 and Hmo1 in non-heat-shocked yeast, at RPS24A and RPL21A (Hmo1-binding) and RPL40B and RPS9B (non-Hmo1-binding). Scale, in reads per million mapped reads, is indicated for each scan. (C) Heat maps and line graphs showing normalized occupancy of Med15 and Med18 at UAS genes, Hsf1 targets, and Rap1-binding RP genes that do or do not bind Hmo1, in wild-type (BY4741) and hmo1Δ yeast after 15 min of heat shock.
To determine whether Hmo1 itself or some other feature common to Hmo1-binding RP gene promoters determines persistent Mediator association after heat shock, we sought to examine Mediator association with RP genes following heat shock in hmo1Δ yeast. RP gene repression by heat shock is not affected in hmo1Δ yeast (Hall et al. 2006). However, the parent anchor-away strain, YFR1321, is fpr1Δ, and hmo1Δ fpr1Δ yeast are severely slow growing or inviable (Dolinski and Heitman 1999; Kasahara et al. 2020). We therefore compared Mediator association following heat shock in hmo1Δ and wild-type BY4741 yeast. We found that Med15 association was still evident upstream of repressed, Hmo1-binding RP genes in hmo1Δ yeast but was reduced by approximately twofold relative to wild-type cells (Fig. 4C). Med15 peaks upstream of Hsf1 targets, and non-Hmo1-binding RP genes showed a similar intensity in wild-type and hmo1Δ yeast, providing internal controls for the reduced intensity observed at Hmo1-binding RP genes repressed by heat shock in hmo1Δ yeast.
We also noted a decrease in intensity of the Med15 peak upstream of UAS genes in heat-shocked hmo1Δ yeast (Fig. 4C). Some RP genes are included among the set of UAS genes (Supplemental Table S2), but many non-RP genes also show Hmo1 peaks at upstream promoter regions, albeit of lower intensity (Supplemental Fig. S6A; Hall et al. 2006; Kasahara et al. 2007; Knight et al. 2014; Reja et al. 2015). To examine whether persistent Mediator binding in non-RP genes repressed by heat shock was influenced by Hmo1, we identified, among UAS genes repressed by heat shock, five genes having high-intensity Hmo1 ChIP-seq peaks and 105 genes having low-intensity Hmo1 peaks (Supplemental Fig. S6A; Supplemental Table S2). Med15 signal was observed at both sets of genes after heat shock both in kin28AA yeast treated with rapamycin and in the parent strain and, similarly to the RP genes, showed a greater increase upon heat shock at the small set of high Hmo1-binding than at those genes binding Hmo1 at lower levels (Supplemental Fig. S6B). In addition, Med15 association decreased about twofold at high Hmo1-binding UAS genes in hmo1Δ yeast while remaining nearly unchanged at low Hmo1-binding UAS genes repressed by heat shock (Supplemental Fig. S6C).
From these results, we conclude that Hmo1 evidently contributes to persistent Mediator association with both RP genes and UAS genes down-regulated by heat shock, although conclusions regarding the latter are limited by the small size of the high Hmo1-binding cohort. At the same time, some additional feature common to Hmo1-binding promoters also apparently promotes persistent Mediator association with genes down-regulated by heat shock, as association is observed even in hmo1Δ yeast.
As mentioned above, the large majority of RP gene promoters bind Rap1. Rap1 also binds to promoters of many non-RP genes, including several that are highly transcribed. All five of the non-RP, Rap1-bound genes having the highest Pol II occupancy in our ChIP data are strongly repressed by heat shock. Four of these show increased (PMA1, ZEO1, and PMP1) or unchanged (TEF1) Med15 association that coincides with Rap1 binding sites; only at PDC1 was repression accompanied by decreased Med15 ChIP signal (Supplemental Fig. S7). Thus, persistent Mediator binding also occurs at non-RP, Rap1-bound promoters that are repressed upon heat shock.
Many additional non-RP genes that do not bind Rap1 and that are repressed upon heat shock also show retention of Mediator at UAS regions, whether or not Kin28 is depleted (Fig. 3). To ascertain whether Hmo1 or other TFs might specify Mediator retention at these genes, we identified the 500 genes showing the highest Med15 occupancy under conditions of Kin28 depletion and no heat shock, the rationale being that this cohort would best allow the determination of retention or loss of Mediator signal upon heat shock. After excluding nonverified ORFs and RP genes from this set, we identified 39 genes from the remaining 304 genes that showed decreased Pol II occupancy by at least fourfold after heat shock (Supplemental Table S3; Supplemental Fig. S8). Only one of these 39 genes showed decreased Med15 occupancy by more than twofold after heat shock, whereas 33 showed an occupancy >75% of that seen before heat shock. Only nine of these 39 genes showed Hmo1 binding more than two times above background levels in an early microarray study (Hall et al. 2006). Gene Ontology analysis indicated that among TFs, only LEU3 showed enrichment for binding, although 47 different TFs bind to these 39 gene promoters (Supplemental Table S3). We conclude that no single TF, including Hmo1, is required for persistent Mediator association at genes repressed by heat shock.
Role of PIC components in Mediator association following heat shock
We next addressed the role of PIC components TBP, TFIID, and Pol II in stabilizing Mediator occupancy at gene promoters in heat-shocked cells. Previously, we reported on the effect of depletion of Taf1, TBP, or the Pol II subunit Rpb3 on association of Mediator with gene promoters in yeast growing in rich medium (Knoll et al. 2018). In KIN28+ yeast, depletion of PIC components had little effect on Mediator association with UAS regions. In contrast, when PIC components were depleted together with Kin28, allowing Mediator association with core promoters to be detected, a strong dependence on PIC components for Mediator association was observed. We examined the effects at two categories of gene promoters: TFIID-dominated genes, which lack consensus TATA elements, are relatively enriched for association with the Taf subunits of TFIID, and comprise ∼85% of yeast genes, including most constitutively active genes; and SAGA-dominated genes, which possess consensus TATA elements, show relatively reduced Taf association, and are enriched for inducible genes (Huisinga and Pugh 2004; Tirosh and Barkai 2008; Rhee and Pugh 2012; Donczew et al. 2020; Knoll et al. 2020). Under conditions of Kin28 depletion, Mediator association was affected by the deletion of any of the PIC components examined. Concordant with the distinction between TFIID-dominated and SAGA-dominated genes, depletion of Taf1 reduced association of Med15 and Med18 to a greater degree at TFIID-dominated genes than at SAGA-dominated genes, whereas Rpb3 depletion reduced Mediator association strongly at all genes (Knoll et al. 2018). In contrast, the simultaneous depletion of TBP and Kin28, when compared with depletion of Kin28 alone, resulted in Mediator ChIP-seq peaks shifting from the core promoter region to UAS sites at both SAGA-dominated and TFIID-dominated genes, indicating that TBP is required for transit of Mediator from its initial site of recruitment to its normally transient engagement with the core promoter. Similar effects of PIC depletion are observed in comparing TATA-containing, Taf1-depleted genes (enriched for SAGA-dominated genes) with TATA-less, Taf1-enriched genes (enriched for TFIID-dominated genes) (Rhee and Pugh 2012): Taf1 depletion reduces Mediator association at the latter class and also at RP genes, much more than at TATA-containing, Taf1-depleted genes, whereas TBP depletion results in an upstream shift together with a decrease in intensity in the average ChIP-seq peaks for Med15 and Med18 for both categories (Fig. 5A).
Figure 5.
Effect of depleting PIC components on Mediator association with gene promoters. (A) Heat maps and line graphs showing normalized occupancy of Med15 (tail) and Med18 (head) at TATA-containing, Taf1-depleted promoters from the 1000 genes with highest Pol II occupancy (228 genes); TATA-less, Taf1-enriched genes excluding RP genes from the 1000 genes with highest Pol II occupancy (330 genes); and RP genes after depletion of Kin28 alone or together with Taf1, TBP, or Rpb3, as indicated. (B) Heat maps and line graphs showing normalized occupancy of Med15 (tail) and Med18 (head) at the approximately 300 genes with highest Pol II occupancy after heat shock, Hsf1 targets, and RP genes after depletion of Kin28 alone or together with Taf1, TBP, or Rpb3, as indicated, without or with 15 min of heat shock, as indicated.
To determine whether Mediator association is similarly dependent on PIC components in heat-shocked cells, we performed ChIP-seq against Med15 and Med18 following heat shock in yeast cells depleted for Kin28 alone or in combination with Taf1, TBP, or Rpb3. In contrast to previous observations made in the absence of heat shock (Knoll et al. 2018), Mediator association with the approximately 300 genes showing the highest Pol II occupancy after 15 min of heat shock was relatively insensitive to depletion of either Taf1 or Rpb3. This was also true of Hsf1 targets and for both the TATA-containing (or SAGA-dominated) and TATA-less (or TFIID-dominated) genes in these cohorts (Fig. 5B; Supplemental Figs. S9, S10). Depletion of TBP resulted in a slight upstream shift in both Med15 and Med18 ChIP-seq signals, consistent with the effect seen in the absence of heat shock (Fig. 5). (We note that ChIP-seq signal was seen over transcribed ORF regions in these experiments. This signal appears to be artifactual, although its origin is not completely understood [Eyboulet et al. 2013; Park et al. 2013; Teytelman et al. 2013; Jeronimo and Robert 2014; Paul et al. 2015; Grünberg et al. 2016]). Finally, Mediator signal at RP genes after heat shock appeared mostly insensitive to depletion of Taf1, TBP, or Rpb3, with the exception being an increased Med18 signal upon depletion of Kin28 together with TBP (Fig. 5B). This insensitivity was not surprising, inasmuch as all three of these PIC components are depleted from RP genes under heat shock conditions (Supplemental Fig. S1; Reja et al. 2015; Vinayachandran et al. 2018).
Effect of exposure to cadmium on Pol II and Mediator occupancy
The unexpected observation of persistent Mediator association with genes repressed by heat shock prompted us to examine the effect of another environmental stress on association of Mediator with gene promoters. To this end, we chose to examine the effect of exposure to CdCl2. Exposure of yeast to this toxic metal at submillimolar concentrations induces a rapid transcriptional response in which about 150–500 genes are induced and from 18 to around 400 genes repressed (Momose and Iwahashi 2001; Cormier et al. 2010; Hosiner et al. 2014; Huang et al. 2016). Induced genes are enriched for binding sites for Hsf1, Msn2, and Msn4, and genes involved in ribosomal biogenesis are repressed by CdCl2 exposure, in common with the response to heat shock and other environmental stresses (Gasch et al. 2000; Causton et al. 2001; Hosiner et al. 2014). However, CdCl2 exposure also induces genes involved in sulfur compound metabolism, such as the MET genes, among others, that are not involved in the heat shock response (Momose and Iwahashi 2001; Cormier et al. 2010; Hosiner et al. 2014; Huang et al. 2016), thus providing an environmental perturbation that results in a distinct but overlapping response to that elicited by heat shock.
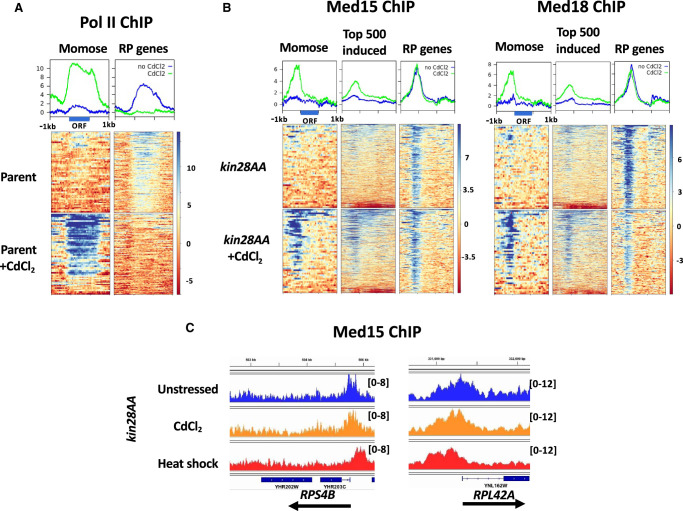
We first tested the effect of CdCl2 exposure on Pol II association by ChIP-seq (Fig. 6A). Marked induction of Pol II association was observed with the 50 genes showing the largest increase in mRNA abundance upon CdCl2 exposure (Momose and Iwahashi 2001), whereas association of Pol II with RP genes decreased to near baseline levels, in agreement with previous studies showing a decrease in RP gene transcript levels after CdCl2 exposure (Momose and Iwahashi 2001; Hosiner et al. 2014; Huang et al. 2016). Promoters showing at least a threefold increase in normalized Pol II occupancy upon CdCl2 administration, and from the top 1000 Pol II-occupied genes after CdCl2 exposure, overlapped strongly with those identified in microarray studies (Supplemental Table S4; Momose and Iwahashi 2001). Gene Ontology analysis revealed enrichment for categories related to sulfur compound metabolism and amino acid biosynthesis, as well as with response to stress, as expected (Supplemental Table S4). Transcription factors enriched for binding to induced genes included Hsf1, Met4, Met32, Msn2, Msn4, and Yap1, all identified previously in a microarray study (Hosiner et al. 2014), as well as Aft2, Cbf1, Met31, and Skn7 (Supplemental Table S4).
Figure 6.
Effect of CdCl2 exposure on Pol II and Mediator occupancy. (A) Heat maps and line graphs showing normalized occupancy of Pol II at 50 strongly induced genes (Supplemental Table S2; Momose and Iwahashi 2001) and RP genes in the anchor-away parent strain, YFR1321. (B) Heat maps and line graphs showing normalized occupancy by Med15 (tail module) and Med18 (head module) at 50 strongly induced genes, the 500 genes having the highest ratio of induced to uninduced Pol II occupancy, and RP genes. (C) Browser scans showing Med15 occupancy in reads per million mapped reads after Kin28 depletion in unstressed cells, cells exposed to CdCl2, and after 15 min of heat shock, as indicated, at the RPS4B and RPL42A loci. Scale, in reads per million mapped reads, is indicated for each scan.
We next examined the effect of CdCl2 exposure on Mediator association. ChIP-seq of Med15 and Med18 under conditions of Kin28 depletion revealed increased association at genes identified as showing increased expression upon CdCl2 administration and at those showing increased Pol II association (Fig. 6B). Similar to our observations of the effect of heat shock, RP genes, despite showing greatly reduced Pol II association upon CdCl2 exposure, showed persistent association of both Med15 and Med18 (Fig. 6B). Also, in accord with the effect of heat shock, Med18 showed relatively lower signal at RP genes after CdCl2 exposure than did Med15. Unlike Mediator association after heat shock, little if any upstream shift in the peaks for Med15 and Med18 at RP genes was observed after CdCl2 exposure (Fig. 6B,C; compare Figs. 1, 2). Also, in contrast to the effect of heat shock on Mediator association with repressed RP genes, persistent Mediator association was observed at both Hmo1-binding and non-Hmo1-binding RP genes, although it appeared slightly lower at the latter (Supplemental Figs. S11, S12). Finally, as with UAS genes following heat shock, Med15 and Med18 association persisted at UAS genes showing decreased Pol II occupancy, while increasing (Med15) or staying constant (Med18) at genes having unchanged or increased Pol II occupancy (Supplemental Fig. S13). However, unlike the case for heat shock and mirroring the results for RP genes, little or no shift of Mediator to more upstream sites was observed after CdCl2 exposure. We conclude that the stresses of heat shock and CdCl2 exposure both allow continued Mediator association with repressed genes while suppressing Pol II association, but they differ in the extent to which they allow Mediator to remain associated with core promoters rather than UAS regions under conditions of Kin28 depletion.
Discussion
Mediator association with strongly induced genes, as assayed by ChIP, appears stronger than at many constitutively active genes, even those expressed at high levels (Fan et al. 2006; Fan and Struhl 2009; Kim and Gross 2013). However, whether this reflects altered dynamics of Mediator has not been closely examined. In this work, we used ChIP-seq in combination with rapid depletion of Kin28 and PIC components to investigate Mediator association with activated and repressed genes following heat shock.
Consistent with previous work, we find increased association of Pol II and Mediator with genes induced by heat shock, with Mediator ChIP signal being evident even without Kin28 depletion (Fan et al. 2006; Kim and Gross 2013; Petrenko et al. 2016, 2017). Pol II occupancy was reduced two- to threefold upon depletion of the essential Med17 (head module) subunit of Mediator, under both heat shock and non–heat shock conditions, and the reduction in occupancy did not differ for targets of Hsf1 or Msn2/4 compared with other genes expressed under heat shock conditions (Supplemental Fig. S1C). Reduction in Pol II occupancy or nascent mRNA transcription of two- to eightfold has been reported upon acute depletion or inactivation of essential Mediator subunits in yeast under standard growth conditions (Paul et al. 2015; Plaschka et al. 2015; Petrenko et al. 2017; Warfield et al. 2017; Bruzzone et al. 2018). Thus, genes expressed during heat shock show dependence on Mediator similar to genes expressed during normal growth. Dependence of gene expression on Mediator may show greater variability in mammalian cells: Although degron-induced depletion of the scaffold subunit Med14 in murine B cells resulted in a global, approximately sevenfold reduction in mRNA after 60 h, more acute depletion of core Mediator subunits in “near-haploid” human KBM7 cells strongly affected only a subset of genes, with many genes showing little effect on nascent transcription (El Khattabi et al. 2019; Jaeger et al. 2020).
Depletion of Kin28, which inhibits promoter escape by Pol II (Wong et al. 2014), increases Mediator ChIP signal at proximal promoters in the absence of heat shock (Fig. 1A). Under heat shock conditions, Med15 association is not increased at Hsf1 targets by Kin28 depletion, rather showing only a slight shift toward the promoter (Fig. 1A,B). These observations suggest that Mediator dynamics may be altered during heat shock, such that its residence time and consequent “ChIP-ability” increase relative to non–heat shock conditions. In a possibly related observation, a genetic screen uncovered mutations in several Mediator subunits that reduced the dynamic range in the transcriptional response to heat shock (Singh et al. 2006); whether these mutations might affect dynamics of Mediator is unknown.
An unexpected finding was the persistent association of Mediator with promoters of repressed genes having low or negligible Pol II association following heat shock or exposure to CdCl2 (Figs. 1–3, 6). We first noticed this persistent association at repressed RP genes but additionally found it to occur at many repressed non-RP genes. Mediator association has also been observed by ChEC-seq upstream of genes down-regulated by sulfometuron methyl treatment, which mimics amino acid starvation (Grünberg et al. 2016). At genes repressed by heat shock, ChIP-seq peaks for Mediator were observed upstream of core promoter regions and, at RP genes, were approximately coincident with binding sites for Rap1, which is required for Mediator recruitment to most RP genes (Ansari et al. 2009). This was true whether or not Kin28 was depleted; in contrast, Mediator ChIP-seq peaks at active RP and non-RP genes are observed at the core promoter near the TSS when Kin28 is depleted. These results, together with the stronger signal for Med15 (from the tail module) than for Med18 (from the head module) upstream of repressed genes suggest that Mediator is still recruited by UAS-bound activators under repressed conditions but is unable to transit to the core promoter (Supplemental Fig. S14). This scenario is reminiscent of the effect of depleting TBP together with Kin28, which results in Mediator being stranded at UAS regions (Knoll et al. 2018), and is consistent with the absence of a PIC at RP genes repressed by heat shock (Vinayachandran et al. 2018). The lack of a PIC at repressed RP genes also fits well with depletion of TBP, Taf1, or Rpb3 having little effect on association of Med15 and Med18 with RP genes following heat shock (Fig. 5).
Given the localization of Mediator ChIP-seq peaks to UAS regions of repressed genes, it seems likely that bound activators are still able to recruit Mediator under repressive conditions. Consistent with this idea, Mediator can be recruited to activator binding sites in the absence of PIC formation (Bhoite et al. 2001; Knoll et al. 2018). What then prevents PIC formation at Mediator-bound, repressed genes? Two possibilities seem most likely: (1) a factor or factors actively block assembly of the PIC, with consequent failure of Mediator to transit from the UAS to the core promoter (Knoll et al. 2018), or (2) activators or coactivators that are required for PIC assembly at heat shock–repressed genes are prevented from functioning by heat shock, whereas other factors remain that allow continued association of Mediator. In either case, the mechanism must allow transcription of those genes induced or not repressed by heat shock to be active while preventing PIC assembly at a cohort of genes that includes, but is not limited to, RP genes. Considering only the Rap1-binding RP genes, repression by heat shock results in eviction of the critical activator Ifh1, whereas Rap1 and Fhl1 remain bound (Martin et al. 2004; Schawalder et al. 2004; Wade et al. 2004; Rudra et al. 2005). A plausible mechanism, therefore, is that Rap1, possibly together with Fhl1, is able to recruit Mediator, whereas Ifh1 is needed for PIC assembly. Further studies will be needed to test the mechanism by which Mediator is recruited nonproductively to both RP and non-RP genes repressed by heat shock or CdCl2, and to establish whether some activators may recruit Mediator but nonetheless be insufficient to facilitate PIC assembly and activate transcription.
We also discovered a strong correlation between persistent Mediator association after heat shock and of Hmo1 binding at RP genes. This association was reduced, but not eliminated, in hmo1Δ yeast. Hmo1 at RP genes is reduced upon heat shock to ∼30% of non-heat-shocked levels (Knight et al. 2014; Reja et al. 2015); whether the residual Hmo1 directly facilitates Mediator retention and how it might do so remain topics for future research. Furthermore, the observed association of Mediator with genes repressed by heat shock in hmo1Δ yeast indicates that some other feature also contributes to its retention. RP genes that bind or do not bind Hmo1 also differ in the distance between Rap1 and Fhl1 binding sites, Rap1-TSS distance, G/C content, and chromatin structure, with Hmo1-binding RP genes having a larger nucleosome-depleted region (Knight et al. 2014; Reja et al. 2015; Zencir et al. 2020). Moreover, Fhl1 shows binding over an extended region in Hmo1-binding RP gene promoters, in contrast to the narrow binding region seen in RP gene promoters that do not bind Hmo1, and this difference persists even after heat shock (Reja et al. 2015). The relationship among these distinguishing features of Hmo1-binding and nonbinding RP genes is not clear, and one or a combination of these features may contribute to persistent Mediator association after heat shock.
In contrast to our observations following heat shock, Mediator association is seen at both Hmo1-binding and non-Hmo1-binding RP genes following CdCl2 exposure. In addition, under conditions of Kin28 depletion, Mediator ChIP-seq peaks show less of an upstream shift at repressed genes following CdCl2 exposure than after heat shock, suggesting that Mediator still associates with core promoters of repressed genes to some extent after CdCl2 exposure despite the near absence of Pol II. We currently have no explanation for this difference but note that considerable differences exist in the response of yeast to various stresses in both transcriptome changes and signaling pathways used (Gasch and Werner-Washburne 2002; Jin et al. 2008; Zencir et al. 2020).
The dynamics of Mediator recruitment and participation in PIC assembly have only recently begun to be appreciated, principally in the model organism S. cerevisiae (Jeronimo and Robert 2017). It is clear that in budding yeast, these dynamics vary in a gene-dependent fashion, as Mediator occupancy varies at UAS regions and does not correlate with transcriptional output (Jeronimo and Robert 2014; Paul et al. 2015; Grünberg et al. 2016). The work reported here shows that Mediator dynamics, including its association with repressed genes, also can vary in a condition-dependent fashion. Mediator itself is affected in its post-translational modifications and its composition by alterations in environment such as osmotic shock and the transition to stationary phase (Holstege et al. 1998; Miller et al. 2012); whether and how Mediator dynamics are affected by such alterations are currently obscure. Finally, in metazoan organisms, Mediator associates with enhancers that are sometimes many kilobases removed from the sites of PIC assembly at which Mediator participates via loop formation (Kagey et al. 2010); the dynamics of Mediator association and its participation in loop formation and PIC assembly in metazoans are just beginning to be explored (Sun et al. 2021).
Methods
Yeast strains and growth
Additional details can be found in the Supplemental Material as Supplemental Methods. S. cerevisiae strains used in this study are listed in Supplemental Table S5. For simplicity, epitope-tagged strains are referred to by the parent strain names in the text and figures; for example, “BY4741” refers also to TBY100, which harbors the med15-myc allele. Cultures were grown in yeast peptone dextrose (YPD) medium (1% bacto-yeast extract, 2% bacto-peptone extract, 2% glucose). Anchor-away experiments were performed as reported previously (Knoll et al. 2018). In brief, yeasts were grown to 0.6–0.8 OD600 at 30°C and then rapamycin (LC Laboratories) was added from a 1 mg/mL stock solution in ethanol to a final concentration of 1 µg/mL. One hour after rapamycin treatment, one-third volume of prewarmed media (13 mL at 57°C added to 39 mL culture for a final temperature of 37°C), or one-third volume of media at 30°C for non-heat-shocked cells, was added to each flask, and flasks were immediately placed at 37°C (or at 30°C for non-heat-shocked cells) in a shaking incubator for 15 min (30 min for indicated samples in Fig. 1) before cross-linking with formaldehyde. Both kin28AA yeast and the parent strain YFR1321 were treated with rapamycin in all experiments unless indicated otherwise. For CdCl2-treated samples, after 1 h of rapamycin treatment, CdCl2 was added from a 1 M stock solution to a final concentration of 0.5 mM and incubated for another hour before crosslinking.
ChIP-seq experiments and analyses
ChIP experiments, library construction, and analyses were performed as described previously (Knoll et al. 2018). Additional details can be found in the Supplemental Material as Supplemental Methods. The various gene sets used in analyses are listed in Supplemental Table S2. ChIP-seq experiments are summarized, and corresponding deposited files indicated, in Supplemental Table S6. Correlation analysis (determined using the Galaxy server) of ChIP-seq replicate experiments examining Med15 occupancy is shown in Supplemental Figure S15.
Data access
ChIP-seq reads generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA657372.
Supplementary Material
Acknowledgments
We thank Todd Benziger for strain construction, Francois Robert and Kevin Struhl for generously providing yeast strains, and Jason Lieb and Colin Lickwar for help in ChIP-seq of Rap1. We thank the Wadsworth Center Applied Genomics Technology and Tissue Culture and Media Cores for help. This work was supported by the National Science Foundation (MCB1516839 to R.H.M.) and in part by the National Institutes of Health Intramural Research Program at the National Library of Medicine (Z.I.Z. and D.L.).
Author contributions: D.S. conducted most experiments; E.R.K. and R.H.M. also conducted experiments, and E.P. conducted ChIP-seq experiments for Rap1 binding. Z.I.Z., D.L., D.S., and R.H.M. analyzed the data, and Z.I.Z. managed data deposition. D.L. and R.H.M. supervised the project; R.H.M. conceptualized the project and wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275750.121.
Competing interest statement
The authors declare no competing interests.
References
- Ansari SA, He Q, Morse RH. 2009. Mediator complex association with constitutively transcribed genes in yeast. Proc Natl Acad Sci 106: 16734–16739. 10.1073/pnas.0905103106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoite LT, Yu Y, Stillman DJ. 2001. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev 15: 2457–2469. 10.1101/gad.921601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon HM, Aguilera A, Ansari AZ, Asturias FJ, Berk AJ, Bjorklund S, Blackwell TK, Borggrefe T, Carey M, Carlson M, et al. 2004. A unified nomenclature for protein subunits of Mediator complexes linking transcriptional regulators to RNA polymerase II. Mol Cell 14: 553–557. 10.1016/j.molcel.2004.05.011 [DOI] [PubMed] [Google Scholar]
- Bruzzone MJ, Grünberg S, Kubik S, Zentner GE, Shore D. 2018. Distinct patterns of histone acetyltransferase and Mediator deployment at yeast protein-coding genes. Genes Dev 32: 1252–1265. 10.1101/gad.312173.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12: 323–337. 10.1091/mbc.12.2.323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier L, Barbey R, Kuras L. 2010. Transcriptional plasticity through differential assembly of a multiprotein activation complex. Nucleic Acids Res 38: 4998–5014. 10.1093/nar/gkq257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge WJ, O'Duibhir E, Lijnzaad P, van Leenen D, Groot Koerkamp MJ, Kemmeren P, Holstege FC. 2017. Molecular mechanisms that distinguish TFIID housekeeping from regulatable SAGA promoters. EMBO J 36: 274–290. 10.15252/embj.201695621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinski KJ, Heitman J. 1999. Hmo1p, a high mobility group 1/2 homolog, genetically and physically interacts with the yeast FKBP12 prolyl isomerase. Genetics 151: 935–944. 10.1093/genetics/151.3.935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donczew R, Warfield L, Pacheco D, Erijman A, Hahn S. 2020. Two roles for the yeast transcription coactivator SAGA and a set of genes redundantly regulated by TFIID and SAGA. eLife 9: e50109. 10.7554/eLife.50109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Khattabi L, Zhao H, Kalchschmidt J, Young N, Jung S, Van Blerkom P, Kieffer-Kwon P, Kieffer-Kwon KR, Park S, Wang X, et al. 2019. A pliable Mediator acts as a functional rather than an architectural bridge between promoters and enhancers. Cell 178: 1145–1158.e20. 10.1016/j.cell.2019.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyboulet F, Cibot C, Eychenne T, Neil H, Alibert O, Werner M, Soutourina J. 2013. Mediator links transcription and DNA repair by facilitating Rad2/XPG recruitment. Genes Dev 27: 2549–2562. 10.1101/gad.225813.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Struhl K. 2009. Where does Mediator bind in vivo? PLoS One 4: e5029. 10.1371/journal.pone.0005029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Chou DM, Struhl K. 2006. Activator-specific recruitment of Mediator in vivo. Nat Struct Mol Biol 13: 117–120. 10.1038/nsmb1049 [DOI] [PubMed] [Google Scholar]
- Gasch AP, Werner-Washburne M. 2002. The genomics of yeast responses to environmental stress and starvation. Funct Integr Genomics 2: 181–192. 10.1007/s10142-002-0058-2 [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257. 10.1091/mbc.11.12.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DS, English KE, Collins KW, Lee SW. 1990. Genomic footprinting of the yeast HSP82 promoter reveals marked distortion of the DNA helix and constitutive occupancy of heat shock and TATA elements. J Mol Biol 216: 611–631. 10.1016/0022-2836(90)90387-2 [DOI] [PubMed] [Google Scholar]
- Grünberg S, Henikoff S, Hahn S, Zentner GE. 2016. Mediator binding to UASs is broadly uncoupled from transcription and cooperative with TFIID recruitment to promoters. EMBO J 35: 2435–2446. 10.15252/embj.201695020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DB, Wade JT, Struhl K. 2006. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol Cell Biol 26: 3672–3679. 10.1128/MCB.26.9.3672-3679.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell 31: 925–932. 10.1016/j.molcel.2008.07.020 [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728. 10.1016/S0092-8674(00)81641-4 [DOI] [PubMed] [Google Scholar]
- Hosiner D, Gerber S, Lichtenberg-Fraté H, Glaser W, Schüller C, Klipp E. 2014. Impact of acute metal stress in Saccharomyces cerevisiae. PLoS One 9: e83330. 10.1371/journal.pone.0083330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Li Y, Pan J, Li M, Lai Y, Gao J, Li X. 2016. RNA-seq identifies redox balance related gene expression alterations under acute cadmium exposure in yeast. Environ Microbiol Rep 8: 1038–1047. 10.1111/1758-2229.12484 [DOI] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell 13: 573–585. 10.1016/S1097-2765(04)00087-5 [DOI] [PubMed] [Google Scholar]
- Jaeger MG, Schwalb B, Mackowiak SD, Velychko T, Hanzl A, Imrichova H, Brand M, Agerer B, Chorn S, Nabet B, et al. 2020. Selective Mediator dependence of cell-type-specifying transcription. Nat Genet 52: 719–727. 10.1038/s41588-020-0635-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Robert F. 2014. Kin28 regulates the transient association of Mediator with core promoters. Nat Struct Mol Biol 21: 449–455. 10.1038/nsmb.2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Robert F. 2017. The Mediator complex: at the nexus of RNA polymerase II transcription. Trends Cell Biol 27: 765–783. 10.1016/j.tcb.2017.07.001 [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Langelier MF, Bataille AR, Pascal JM, Pugh BF, Robert F. 2016. Tail and kinase modules differently regulate core Mediator recruitment and function in vivo. Mol Cell 64: 455–466. 10.1016/j.molcel.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YH, Dunlap PE, McBride SJ, Al-Refai H, Bushel PR, Freedman JH. 2008. Global transcriptome and deletome profiles of yeast exposed to transition metals. PLoS Genet 4: e1000053. 10.1371/journal.pgen.1000053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435. 10.1038/nature09380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara K, Ohtsuki K, Ki S, Aoyama K, Takahashi H, Kobayashi T, Shirahige K, Kokubo T. 2007. Assembly of regulatory factors on rRNA and ribosomal protein genes in Saccharomyces cerevisiae. Mol Cell Biol 27: 6686–6705. 10.1128/MCB.00876-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara K, Nakayama R, Shiwa Y, Kanesaki Y, Ishige T, Yoshikawa H, Kokubo T. 2020. Fpr1, a primary target of rapamycin, functions as a transcription factor for ribosomal protein genes cooperatively with Hmo1 in Saccharomyces cerevisiae. PLoS Genet 16: e1008865. 10.1371/journal.pgen.1008865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Gross DS. 2013. Mediator recruitment to heat shock genes requires dual Hsf1 activation domains and Mediator tail subunits Med15 and Med16. J Biol Chem 288: 12197–12213. 10.1074/jbc.M112.449553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight B, Kubik S, Ghosh B, Bruzzone MJ, Geertz M, Martin V, Dénervaud N, Jacquet P, Ozkan B, Rougemont J, et al. 2014. Two distinct promoter architectures centered on dynamic nucleosomes control ribosomal protein gene transcription. Genes Dev 28: 1695–1709. 10.1101/gad.244434.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll ER, Zhu ZI, Sarkar D, Landsman D, Morse RH. 2018. Role of the pre-initiation complex in Mediator recruitment and dynamics. eLife 7: e39633. 10.7554/eLife.39633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll ER, Zhu ZI, Sarkar D, Landsman D, Morse RH. 2020. Kin28 depletion increases association of TFIID subunits Taf1 and Taf4 with promoters in Saccharomyces cerevisiae. Nucleic Acids Res 48: 4244–4255. 10.1093/nar/gkaa165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Lis JT. 1998. Transcriptional activation independent of TFIIH kinase and the RNA polymerase II Mediator in vivo. Nature 393: 389–392. 10.1038/30770 [DOI] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein–DNA association. Nat Genet 28: 327–334. 10.1038/ng569 [DOI] [PubMed] [Google Scholar]
- Martin DE, Soulard A, Hall MN. 2004. TOR regulates ribosomal protein gene expression via PKA and the forkhead transcription factor FHL1. Cell 119: 969–979. 10.1016/j.cell.2004.11.047 [DOI] [PubMed] [Google Scholar]
- Mayer C, Grummt I. 2006. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25: 6384–6391. 10.1038/sj.onc.1209883 [DOI] [PubMed] [Google Scholar]
- McNeil JB, Agah H, Bentley D. 1998. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes Dev 12: 2510–2521. 10.1101/gad.12.16.2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencı´a M, Moqtaderi Z, Geisberg JV, Kuras L, Struhl K. 2002. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol Cell 9: 823–833. 10.1016/S1097-2765(02)00490-2 [DOI] [PubMed] [Google Scholar]
- Miller C, Matic I, Maier KC, Schwalb B, Roether S, Strässer K, Tresch A, Mann M, Cramer P. 2012. Mediator phosphorylation prevents stress response transcription during non-stress conditions. J Biol Chem 287: 44017–44026. 10.1074/jbc.M112.430140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose Y, Iwahashi H. 2001. Bioassay of cadmium using a DNA microarray: genome-wide expression patterns of Saccharomyces cerevisiae response to cadmium. Environ Toxicol Chem 20: 2353–2360. 10.1002/etc.5620201030 [DOI] [PubMed] [Google Scholar]
- Park D, Lee Y, Bhupindersingh G, Iyer VR. 2013. Widespread misinterpretable ChIP-seq bias in yeast. PLoS One 8: e83506. 10.1371/journal.pone.0083506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul E, Zhu ZI, Landsman D, Morse RH. 2015. Genome-wide association of Mediator and RNA polymerase II in wild-type and Mediator mutant yeast. Mol Cell Biol 35: 331–342. 10.1128/MCB.00991-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrenko N, Jin Y, Wong KH, Struhl K. 2016. Mediator undergoes a compositional change during transcriptional activation. Mol Cell 64: 443–454. 10.1016/j.molcel.2016.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrenko N, Jin Y, Wong KH, Struhl K. 2017. Evidence that Mediator is essential for Pol II transcription, but is not a required component of the preinitiation complex in vivo. eLife 6: e28447. 10.7554/eLife.28447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus D, Anandhakumar J, Thiru P, Guertin MJ, Erkine AM, Gross DS. 2018. Genetic and epigenetic determinants establish a continuum of Hsf1 occupancy and activity across the yeast genome. Mol Biol Cell 29: 3168–3182. 10.1091/mbc.E18-06-0353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaschka C, Larivière L, Wenzeck L, Seizl M, Hemann M, Tegunov D, Petrotchenko EV, Borchers CH, Baumeister W, Herzog F, et al. 2015. Architecture of the RNA polymerase II–Mediator core initiation complex. Nature 518: 376–380. 10.1038/nature14229 [DOI] [PubMed] [Google Scholar]
- Plaschka C, Nozawa K, Cramer P. 2016. Mediator architecture and RNA polymerase II interaction. J Mol Biol 428: 2569–2574. 10.1016/j.jmb.2016.01.028 [DOI] [PubMed] [Google Scholar]
- Reja R, Vinayachandran V, Ghosh S, Pugh BF. 2015. Molecular mechanisms of ribosomal protein gene coregulation. Genes Dev 29: 1942–1954. 10.1101/gad.268896.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. 2012. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483: 295–301. 10.1038/nature10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, Trnka MJ, Bushnell DA, Davis RE, Mattei PJ, Burlingame AL, Kornberg RD. 2016. Structure of a complete mediator-RNA polymerase II pre-initiation complex. Cell 166: 1411–1422.e16. 10.1016/j.cell.2016.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D, Zhao Y, Warner JR. 2005. Central role of Ifh1p–Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J 24: 533–542. 10.1038/sj.emboj.7600553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schawalder SB, Kabani M, Howald I, Choudhury U, Werner M, Shore D. 2004. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 432: 1058–1061. 10.1038/nature03200 [DOI] [PubMed] [Google Scholar]
- Schier AC, Taatjes DJ. 2020. Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev 34: 465–488. 10.1101/gad.335679.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Erkine AM, Kremer SB, Duttweiler HM, Davis DA, Iqbal J, Gross RR, Gross DS. 2006. A functional module of yeast Mediator that governs the dynamic range of heat-shock gene expression. Genetics 172: 2169–2184. 10.1534/genetics.105.052738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solís EJ, Pandey JP, Zheng X, Jin DX, Gupta PB, Airoldi EM, Pincus D, Denic V. 2016. Defining the essential function of yeast Hsf1 reveals a compact transcriptional program for maintaining eukaryotic proteostasis. Mol Cell 63: 60–71. 10.1016/j.molcel.2016.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina J. 2018. Transcription regulation by the Mediator complex. Nat Rev Mol Cell Biol 19: 262–274. 10.1038/nrm.2017.115 [DOI] [PubMed] [Google Scholar]
- Sun F, Sun T, Kronenberg M, Tan X, Huang C, Carey MF. 2021. The Pol II preinitiation complex (PIC) influences Mediator binding but not promoter–enhancer looping. Genes Dev 35: 1175–1189. 10.1101/gad.348471.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teytelman L, Thurtle DM, Rine J, van Oudenaarden A. 2013. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci 110: 18602–18607. 10.1073/pnas.1316064110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. 2008. Two strategies for gene regulation by promoter nucleosomes. Genome Res 18: 1084–1091. 10.1101/gr.076059.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayachandran V, Reja R, Rossi MJ, Park B, Rieber L, Mittal C, Mahony S, Pugh BF. 2018. Widespread and precise reprogramming of yeast protein–genome interactions in response to heat shock. Genome Res 28: 357–366. 10.1101/gr.226761.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JT, Hall DB, Struhl K. 2004. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature 432: 1054–1058. 10.1038/nature03175 [DOI] [PubMed] [Google Scholar]
- Warfield L, Ramachandran S, Baptista T, Devys D, Tora L, Hahn S. 2017. Transcription of nearly all yeast RNA polymerase II-transcribed genes is dependent on transcription factor TFIID. Mol Cell 68: 118–129.e5. 10.1016/j.molcel.2017.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440. 10.1016/S0968-0004(99)01460-7 [DOI] [PubMed] [Google Scholar]
- Wong KH, Jin Y, Struhl K. 2014. TFIIH phosphorylation of the Pol II CTD stimulates Mediator dissociation from the preinitiation complex and promoter escape. Mol Cell 54: 601–612. 10.1016/j.molcel.2014.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi D, Sharon E, Lotan-Pompan M, Lubling Y, Shipony Z, Raveh-Sadka T, Keren L, Levo M, Weinberger A, Segal E. 2011. Compensation for differences in gene copy number among yeast ribosomal proteins is encoded within their promoters. Genome Res 21: 2114–2128. 10.1101/gr.119669.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zencir S, Dilg D, Rueda MP, Shore D, Albert B. 2020. Mechanisms coordinating ribosomal protein gene transcription in response to stress. Nucleic Acids Res 48: 11408–11420. 10.1093/nar/gkaa852 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.