Abstract
Some bacteria are difficult to identify with phenotypic identification schemes commonly used outside reference laboratories. 16S ribosomal DNA (rDNA)-based identification of bacteria potentially offers a useful alternative when phenotypic characterization methods fail. However, as yet, the usefulness of 16S rDNA sequence analysis in the identification of conventionally unidentifiable isolates has not been evaluated with a large collection of isolates. In this study, we evaluated the utility of 16S rDNA sequencing as a means to identify a collection of 177 such isolates obtained from environmental, veterinary, and clinical sources. For 159 isolates (89.8%) there was at least one sequence in GenBank that yielded a similarity score of ≥97%, and for 139 isolates (78.5%) there was at least one sequence in GenBank that yielded a similarity score of ≥99%. These similarity score values were used to defined identification at the genus and species levels, respectively. For isolates identified to the species level, conventional identification failed to produce accurate results because of inappropriate biochemical profile determination in 76 isolates (58.7%), Gram staining in 16 isolates (11.6%), oxidase and catalase activity determination in 5 isolates (3.6%) and growth requirement determination in 2 isolates (1.5%). Eighteen isolates (10.2%) remained unidentifiable by 16S rDNA sequence analysis but were probably prototype isolates of new species. These isolates originated mainly from environmental sources (P = 0.07). The 16S rDNA approach failed to identify Enterobacter and Pantoea isolates to the species level (P = 0.04; odds ratio = 0.32 [95% confidence interval, 0.10 to 1.14]). Elsewhere, the usefulness of 16S rDNA sequencing was compromised by the presence of 16S rDNA sequences with >1% undetermined positions in the databases. Unlike phenotypic identification, which can be modified by the variability of expression of characters, 16S rDNA sequencing provides unambiguous data even for rare isolates, which are reproducible in and between laboratories. The increase in accurate new 16S rDNA sequences and the development of alternative genes for molecular identification of certain taxa should further improve the usefulness of molecular identification of bacteria.
Accurate identification of bacterial isolates is an essential task for clinical microbiology laboratories. For slow-growing and fastidious organisms, traditional phenotypic identification is difficult and time-consuming, and when phenotypic methods are used to identify bacteria, interpretation of test results can involve a substantial amount of subjective judgement (20). Phenotypic variability among strains belonging to the same species also results in some bacterial isolates presenting characteristics that are atypical for a candidate identification. Reference laboratories, including the Centers for Disease Control and Prevention in the United States and the Collection de l'Institut Pasteur and BioMérieux laboratories in France, collected unidentified microorganisms isolated from environmental, veterinary, and clinical specimens from various geographic origins and developed extensive flow charts for their accurate phenotypic identification. However, numerous isolates remained unidentifiable after the application of all available phenotypic tests. In these situations, 16S ribosomal DNA (rDNA)-based molecular identification could achieve identification, for reasons including its universal distribution among bacteria (30) and the presence of species-specific variable regions. This molecular approach has been extensively used for bacterial phylogeny (32), leading to the establishment of large public-domain databases (13, 27) and its application to bacterial identification, including that of environmental and clinical uncultured microorganisms (17, 22), unique or unusual isolates (7), and collections of phenotypically identified isolates (23, 24). In this situation, 16S rDNA-based identification has been favorably compared to computer-assisted cell wall fatty acid analysis and computer-assisted biochemical profile analysis of a collection of 72 aerobic gram-negative bacilli (23) and a collection of 52 coryneform isolates (24). However, its reliability and performance have never before been evaluated with a collection of unidentifiable isolates. We have now evaluated 16S rDNA sequence analysis as a tool for molecular identification of unidentifiable isolates by application of this molecular tool to the BioMérieux collection of 177 unidentifiable isolates.
MATERIALS AND METHODS
Bacterial isolates and conventional identification methods.
As part of its commitment to diagnosis in microbiology, BioMérieux offers microbiologists the opportunity to submit for study isolates that remain unidentified when tested using its commercial identification strips. After the organisms are tested for purity, they are subjected to an extensive phenotypic investigation, including the study of respiratory type and temperature of growth, cell morphology after Gram staining, spore-forming ability, oxidase and catalase activities, and biochemical profile. Gram-positive bacilli were tested using the APICoryne, catalase-negative gram-positive cocci were tested using the APIStrep, catalase-positive gram-positive cocci were tested using the APIStaph and the ID32Staph, oxidase-negative gram-negative bacilli were tested using the API20E, oxidase-positive gram-negative bacilli were tested using the API NE, and Bacillus spp. were tested using the API20E and the API50CH. Every questionable test was repeated twice. Once these tests were completed, phenotypic identifications were achieved by reference to published descriptions of bacterial species (15, 31). On average, 300 strains are received every year for analysis, and about 20% of these strains remain unidentified. A collection of 177 such unidentifiable bacterial isolates collected over 3 years was tested in this study. These isolates had been taken from environmental sources (79 of 177; 44.6%), veterinary clinical samples (17 of 177; 9.7%), and medical clinical samples (81 of 177; 45.7%). Phenotypic data were reassessed after molecular analysis allowed for identification (see below) in order to determine what caused the conventional identification to fail. These faults were classified as growth requirement determination failures, morphology and Gram stain determination failures, oxidase and catalase activity determination failures, and biochemical determination failures.
16S rDNA sequencing.
Each isolate was plated onto either Trypticase soy agar, 5% sheep blood agar, or chocolate agar (BioMérieux). Bacteria were lysed either by boiling for 15 min (gram-negative bacilli) or by boiling for 20 min in a 20% Chelex suspension (gram-positive cocci) (21). Alternatively, gram-positive bacilli were lysed using a 1-h incubation at 37°C in 100 μl of Tris-EDTA buffer (10 mM Tris, 1 mM EDTA, 0.1 M NaCl, pH 8.0) followed by a 1-h incubation at 55°C in a solution of 25 mg of proteinase K per ml and 10% sodium dodecyl sulfate. Next, 200 μl of 4 M guanidine thiocyanate was added to each tube, left for an hour at room temperature, and then heated at 100°C for 10 min with 50 μl of 0.5 M NaOH. Extraction of nucleic acids was carried out using a QIAamp kit (Qiagen, Hilden, Germany). Extracted DNA was amplified by using PCR technology and the universal 16S rDNA primers fD1 and rp2 (30) (Eurogentec, Seraing, Belgium). Amplifications and sequencing of amplified products were done as previously described (6). 16S rDNA sequences were compared with those available in the GenBank, EMBL, and DJB databases using the gapped BLASTN 2.0.5 program through the National Center for Biotechnology Information server (1). Comparisons were performed using the BLOSUM 62 matrix with default parameters including a gap existence cost of 11, a cost-per-residue gap of 1, and a lambda ratio of 0.85. Every sequence was aligned with the first 10 database sequences giving the highest scores of sequence similarity, and the quality of the database sequences was assessed. Only 16S rDNA database sequences containing <1% undetermined positions were retained for analysis; unknown positions (N), purine positions (R), and pyrimidine positions (Y) were considered undetermined bases. In case a database sequence exhibited >1% undetermined positions, the 16S rDNA gene sequence was determined for the type strain.
Criteria for identification.
Identification to the species level was defined as a 16S rDNA sequence similarity of ≥99% with that of the prototype strain sequence in GenBank; identification at the genus level was defined as a 16S rDNA sequence similarity of ≥97% with that of the prototype strain sequence in GenBank. A failure to identify was defined as a 16S rDNA sequence similarity score of lower than 97% with those deposited in GenBank at the time of analysis (May 1999).
Phylogenetic analysis of unidentified isolates.
For those isolates which were not identified by 16S rDNA sequence analysis, taxonomic relationships were inferred from 16S rDNA sequence comparison. Sequences were obtained from the GenBank database and aligned by using the multisequence alignment program ClustalW (26) in the BISANCE software package (5). Phylogenetic relationships were inferred from this alignment by using programs in version 3.4 of the PHYLIP software package (8, 9). A distance matrix was generated using DNADIST under the assumptions of Jukes and Cantor (11) and Kimura (12). Phylogenetic trees were derived from these matrices using neighbor joining. Isolates were assigned to the taxonomic group of the two bacterial strains forming the taxonomic frame of the unidentified isolate.
Analysis of discrepancies.
In the case of a low similarity score resulting from 16S rDNA sequences containing >1% undetermined positions in GenBank (as defined above), the 16S rDNA sequences of type strains obtained from the Collection de l'Institut Pasteur (Institut Pasteur, Paris, France) and the American Type Culture Collection (Manassas, Va.) were determined in order to refine molecular analysis.
Statistics.
Comparisons of identification ratios were performed with Epiinfo, version 6 (Centers for Disease Control and Prevention).
RESULTS
16S rDNA sequence analysis and bacterial identification.
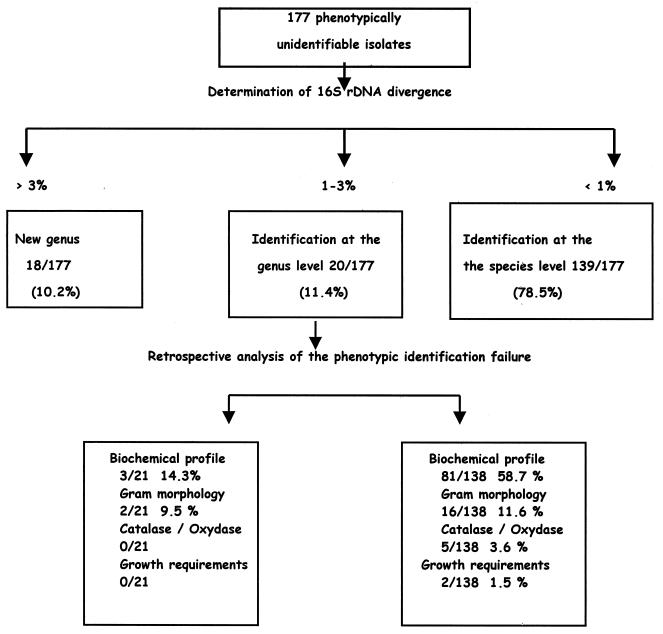
An almost-complete 16S rDNA sequence containing fewer than 1% undetermined positions was obtained for all of the isolates included in the study; thus, 177 query sequences were available for comparison (Table 1). For three isolates (1.7%) belonging to the genus Corynebacterium and an unidentified species, DNA extraction had to be repeated after initial 16S rDNA amplification attempts failed. For two isolates (1.1%), the 16S rDNA sequencing procedure had to be carried out twice after the first analysis demonstrated probable mixed sequences. 16S rDNA-based analysis resulted in the classification of the isolates into three categories (Table 1 and Fig. 1). A total of 139 of 177 isolates (78.5%) possessed a 16S rDNA sequence with ≥99% similarity to that of a previously characterized bacterial species. A total of 159 of 177 (89.8%) possessed a 16S rDNA sequence with ≥97% similarity to that of a genus member. Among these 159 isolates, Enterobacter and Pantoea exhibited a 99% 16S rDNA sequence similarity with GenBank sequences significantly less frequently than isolates belonging to the other genera (P = 0.04; odds ratio = 0.32 [95% confidence interval, 0.10 to 1.14] [Fischer's exact test]). A total of 18 of 177 isolates (10.2%) had a 16S rDNA sequence with <97% similarity with the closest sequence in GenBank. The efficiency in achieving a 99% 16S rDNA similarity level was not significantly different between isolates obtained from clinical or environmental sources. However, 12 of 18 isolates with <97% similarity to other GenBank sequences originated from environmental sources (P = 0.07 by the Mantel-Haenszel test). A total of 41 original 16S rDNA sequences corresponding to new species and new genera have been deposited in public databases (Table 1).
TABLE 1.
16S rDNA-based identification of a collection of 177 phenotypically unidentified bacterial isolates
| 16S rDNA-based identification | No. of isolates
|
GenBank accession no.d | ||
|---|---|---|---|---|
| Ea | Vb | Cc | ||
| Isolates exhibiting >99% 16S rDNA sequence homology with a deposited sequence (n = 139)e | ||||
| Abiotrophia defectiva | 1 | D50541 | ||
| Actinobacillus capsulatus | 1 | 2 | AF145255 | |
| Actinobacillus pleuropneumoniae | 1 | AF146371 | ||
| Actinomyces europaeus | 2 | Y08828 | ||
| Actinomyces neuii | 1 | X71861 | ||
| Actinomyces pyogenes | 2 | X79225 | ||
| Actinomyces turicensis | 1 | X78720 | ||
| Aerococcus urinae | 3 | U63458 | ||
| Aeromonas media | 2 | X74679 | ||
| Aranicola proteolyticus | 1 | U93263 | ||
| Arcobacter skirrowi | 1 | L14625 | ||
| Bacillus agri | 1 | U65892 | ||
| Bacillus alcalophilus | 1 | X76436 | ||
| Bacillus cereus | 1 | 2 | D16266 | |
| Bacillus psychrophilus | 1 | X54969 | ||
| Bacillus pumilus | 1 | AF071856 | ||
| Bacillus sphaericus | 1 | 1 | D16280 | |
| Bacillus sporothermodurans | 1 | U49079 | ||
| Bacillus subtilis | 3 | AB018484 | ||
| Bacillus thuringiensis | 1 | D16281 | ||
| Brevibacillus thermoruber | 1 | Z26921 | ||
| Brevibacterium casei | 1 | X76564 | ||
| Brevundimonas diminuta | 1 | X87274 | ||
| Caulobacter intermedius | 1 | AJ007802 | ||
| Citrobacter freundii | 4 | 1 | AF025365 | |
| Clostridium ramosum | 1 | M23731 | ||
| Corynebacterium bovis | 1 | AJ222965 | ||
| Corynebacterium minutissimum | 1 | X84678 | ||
| Corynebacterium mucifaciens | 1 | Y11200 | ||
| Corynebacterium striatum | 1 | X84442 | ||
| Corynebacterium xerosis | 1 | AF145257 | ||
| Dermabacter hominis | 1 | X76728 | ||
| Enterobacter amnigenus | 3 | AB004749 | ||
| Enterococcus avium | 1 | AF133535 | ||
| Enterococcus cecorum | 2 | 1 | X54290 | |
| Enterococcus faecalis | 1 | AF039902 | ||
| Enterococcus faecium | 1 | 1 | AF145258 | |
| Erwinia herbicola | 1 | AB004757 | ||
| Escherichia coli | 2 | 2 | 5 | U18997 |
| Fusobacterium russii | 1 | M58681 | ||
| Haemophilus aphrophilus | 1 | M75041 | ||
| Klebsiella oxytoca | 2 | Y17661 | ||
| Klebsiella planticola | 1 | X93216 | ||
| Klebsiella pneumoniae | 1 | AJ233420 | ||
| Klebsiella terrigena | 3 | Y17228 | ||
| Listeria seeligeri | 1 | X98531 | ||
| Methylobacterium mesophilicum | 1 | D25306 | ||
| Microbacterium sp. | 2 | Y17228 | ||
| Micrococcus varians | 1 | X87754 | ||
| Moraxella cuniculi | 1 | AF005188 | ||
| Moraxella osloensis | 1 | X95304 | ||
| Neisseria gonorrhoeae | 1 | AF146369 | ||
| Neisseria polysaccharea | 1 | L06167 | ||
| Oerskovia xanthineolytica | 1 | X79453 | ||
| Oligella urethralis | 1 | AF133538 | ||
| Paenibacillus pabuli | 1 | X60630 | ||
| Paenibacillus polymyxa | 2 | D16276 | ||
| Pantoea agglomerans | 1 | 2 | AB004691 | |
| Pasteurella canis | 1 | M75049 | ||
| Pasteurella multocida | 2 | M35018 | ||
| Pediococcus pentosaceus | 1 | M58834 | ||
| Phyllobacterium rubiacearum | 1 | D12790 | ||
| Propionibacterium acnes | 1 | 1 | AF145256 | |
| Pseudomonas aeruginosa | 1 | 2 | AF157689 | |
| Pseudomonas huttiensis | 1 | AB021366 | ||
| Ralstonia eutropha | 1 | AF027047 | ||
| Ralstonia picketii | 1 | AB004790 | ||
| Rathayibacter rathayi | 1 | U96186 | ||
| Riemerella anatipestifer | 1 | 1 | U60101 | |
| Rhodococcus equi | 1 | X80614 | ||
| Salmonella enterica Dublin | 1 | AF227868 | ||
| Salmonella enterica Montevideo | 1 | AF227867 | ||
| Salmonella enterica Typhi | 1 | AF170176 | ||
| Salmonella enterica Typhimurium | 2 | AF227869 | ||
| Serratia fonticola | 1 | AJ233429 | ||
| Serratia marcescens | 1 | M59160 | ||
| Shigella sonnei | 1 | 1 | X96964 | |
| Staphylococcus aureus | 1 | D83353 | ||
| Staphylococcus epidermidis | 1 | 1 | AF128279 | |
| Staphylococcus haemolyticus | 2 | L37600 | ||
| Staphylococcus saprophyticus | 1 | 1 | L37596 | |
| Staphylococcus schleiferi | 1 | 1 | AB009945 | |
| Streptococcus anginosus | 1 | 1 | AF104678 | |
| Streptococcus mitis | 2 | AJ007426 | ||
| Streptococcus salivarius | 1 | M58839 | ||
| Streptococcus sanguis | 1 | AF003928 | ||
| Streptomyces scabiei | 1 | D63863 | ||
| Uncultured sp. | 2 | AF227866, AF234634 | ||
| Weeksella virosa | 1 | AF133539 | ||
| Isolates exhibiting 97–99% 16S rDNA sequence homology with a deposited sequence (n = 20) | ||||
| Bacillus sp. | 1 | 1 | AF227844, AF227852 | |
| Bifidobacterium sp. | 1 | AF227870 | ||
| Bordetella sp. | 1 | AF227829 | ||
| Brevibacillus sp. | 1 | AF227853 | ||
| Clostridium sp. | 1 | AF227826 | ||
| Corynebacterium sp. | 2 | 1 | AF227854, AF227825, AF227828 | |
| Enterobacter sp. | 1 | AF227845 | ||
| Nocardia | 1 | AF227864 | ||
| Paenibacillus sp. | 1 | AF227827 | ||
| Pantoea sp. | 1 | 1 | 2 | AF227846, AF227851, AF227860, AF227832 |
| Pasteurella sp. | 1 | 1 | AF227861, AF227862 | |
| Pseudomonas sp. | 1 | AF227841 | ||
| Rahnella sp. | 1 | AF227838 | ||
| Isolates exhibiting <97% 16S rDNA sequence homology with a deposited sequence (n = 18) | ||||
| 1 | 1 | AF227843 | ||
| 2 | 1 | AF227847 | ||
| 3 | 1 | AF227849 | ||
| 4 | 1 | AF227863 | ||
| 5 | 1 | AF227865 | ||
| 6 | 1 | AF227858 | ||
| 7 | 1 | AF227857 | ||
| 8 | 1 | AF227855 | ||
| 9 | 1 | AF227833 | ||
| 10 | 1 | AF227834 | ||
| 11 | 1 | AF227835 | ||
| 12 | 1 | AF227836 | ||
| 13 | 1 | AF227859 | ||
| 14 | 1 | AF227831 | ||
| 15 | 1 | AF227830 | ||
| 16 | 1 | AF227837 | ||
| 17 | 1 | AF227840 | ||
| 18 | 1 | AF227839 | ||
E, environmental source.
V, veterinary source.
C, clinical source.
Boldface indicates accession numbers assigned from this study.
From comparison with the GenBank database, May 1999 version.
FIG. 1.
Identification scheme for 177 phenotypically unidentifiable bacterial isolates.
Taxonomic relationships of unidentified isolates.
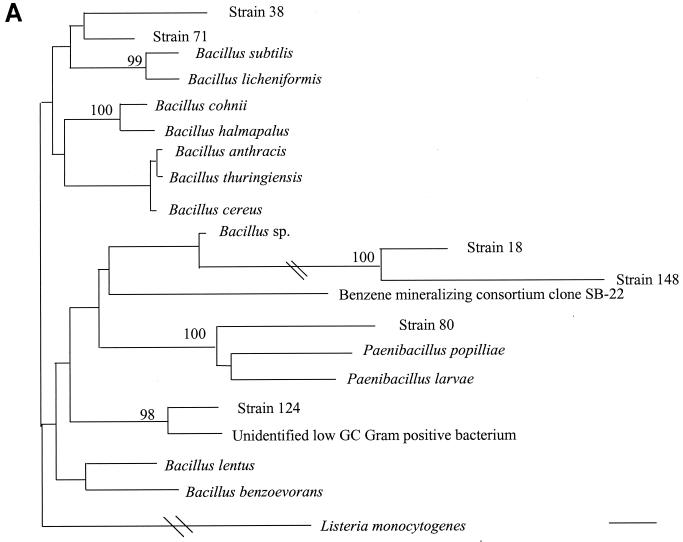
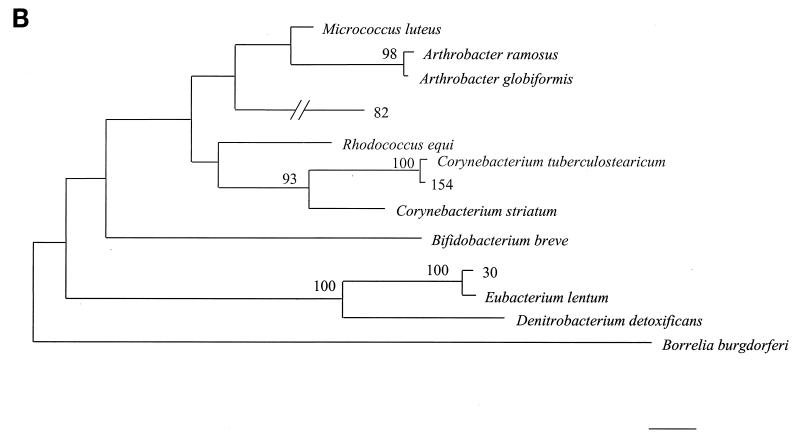
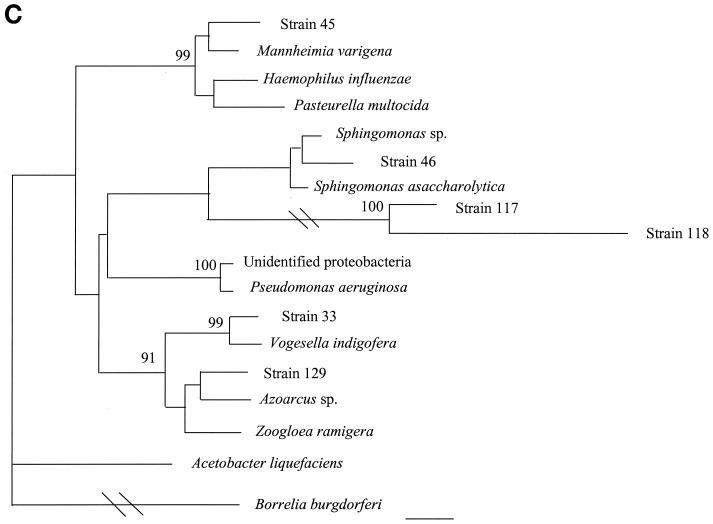
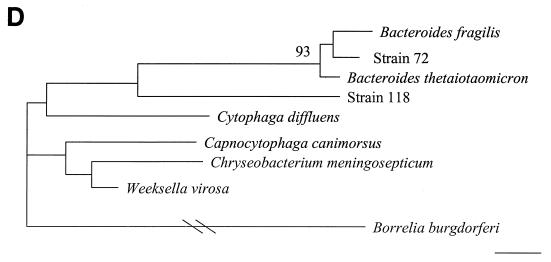
16S rDNA analysis determined that 6 of 18 unidentified isolates belonged to low-percent-G+C-content gram-positive bacteria, 4 of 18 belonged to high-percent-G+C-content gram-positive bacteria, 6 of 18 belonged to gamma subgroup Proteobacteria, and 2 of 18 belonged to the Bacteroides-Cytophaga phylum. The phylogenetic relationships of these isolates as inferred by neighbor-joining analysis are presented in Fig. 2.
FIG. 2.
Distance-related trees indicating the phylogenetic relationships of 18 unidentified isolates referred as strain numbers. (A) Low-percent-G+C-content gram-positive isolates; (B) high-percent-G+C-content gram-positive isolates; (C) gamma subgroup Proteobacteria isolates; (D) Bacteroides-Cytophaga subgroup isolates. The numbers at nodes are the proportions of 100 bootstrap resamplings that support the topology shown. Only bootstrap values of >90% are indicated. Bars, 1% divergence.
Analysis of conventional-identification failures.
Failures in appropriate conventional identification are presented in Fig. 1. Among 16 Bacillus isolates analyzed, 16S rDNA-based identification confirmed conventional identification for 3 isolates; inaccurate conventional identification was a result of unmatched Gram determination for 7 isolates, unmatched biochemical profile determination for 5 isolates, and unmatched growth requirement determination for 1 isolate. Failure to accurately identify Escherichia coli isolates when using conventional methods was a result of unmatched biochemical profile determination for eight of nine isolates and of inaccurate oxidase activity determination for one of nine isolates. Failure of conventional identification of Staphylococcus spp. resulted from inaccurate biochemical profile determination for all nine isolates examined.
DISCUSSION
In this study, inappropriate DNA extraction prevented 16S rDNA-based identification of 2% of isolates. Various extraction protocols have been published (23), but no optimum approach became widely accepted. Improved, reliable methods are therefore required, particularly with the advent of automation. Mixed cultures led to 1% of 16S rDNA-based identification failures, either because the wrong colony was selected on subculture or because more than one bacterial species was inadvertantly included in the amplification, resulting in ambiguous 16S rDNA data. The frequency of chimeric molecule formation was determined to be as high as 30 to 32% in a model of mixed genomic DNAs from two nearly identical actinomycete 16S rDNA sequences (28, 29). Much attention should therefore be paid to achieving a pure culture prior to 16S rDNA-based identification. Alternatively, mixed 16S rDNA sequences could be separated by, for example, denaturing gradient gel electrophoresis prior to sequencing (18, 25). Likewise, PCR-induced chimeras formed between different rRNA gene copies (29) in bacterial species exhibiting heterogeneous 16S rDNA sequences among multiple ribosomal operons (3) may lead to the description of nonexistent species. Particular attention should be paid to the careful examination of double peaks in the electropherogram. Lastly, unlike protein-encoding genes, the 16S rDNA is not organized into codons, and thus the accuracy of every base position determination cannot be verified before the sequence is compared.
Interpretation of the sequences was hampered by a percentage of position ambiguities higher than 1% (∼15 positions) either in the query sequence or in database sequences, and we recommend routinely obtaining a 16S rDNA sequence with less than 1% ambiguity. For some genera, too few species have been deposited in the databases so that the similarity level for a particular query sequence never exceeds of 97%. When several 16S rDNA sequences are available for the same species, a level of intraspecific 16S rDNA sequence variation exceeding 1% of the sequence has been reported for as many as 48% of the deposited sequences (4). An accurate commercial 16S rDNA database, proposed for the identification of bacteria (23), circumvents this difficulty, but sequence accuracy may be offset by there being limited number of sequences. In assessing the percent similarities between sequences, nongapped programs usually result in higher scores than gapped programs if only a limited portion of the gene is compared (data not shown). Whether only the most variable regions of the gene should be incorporated in 16S rDNA-based identification remains to be determined. This solution has been proposed for the identification of aerobic gram-negative bacilli (23). In this case, nongapped programs should be used. In this study, in order to achieve results using exactly the same method whatever the query sequence, we obtained similarity percentages using a gapped program applied to the entire 16S rDNA sequence. The degree of freedom for gap placement remains to be determined. Indeed, in our experience, when the same sequence was compared against the same database using different programs, different similarity results were obtained, resulting in the assignment of different identities. Similarity scores depend on the lengths of the sequences under analysis and on the number of gaps introduced in the query sequence to optimize the similarity. Unfortunately, there are no guidelines regarding the use of these parameters during the identification process. Based on results of the present study, we recommend that a comparison include at least 1,500 positions, that all of the sequences included in the similarity search have the same length, and that an ungapped program be used. The question of gapped versus ungapped analysis, however, will require more data.
There are also no accepted guidelines regarding computer-aided comparison of sequence similarity for 16S rDNA-based bacterial identification. A 97% similarity level has been proposed for the bacterial species delineation using the 16S rDNA sequence (19), but this recommendation has been questioned (10). Recently, it has been suggested that a difference rate of >0.5% could be considered indicative of a new species within a known genus (16). In a previous study of 16S rDNA-based bacterial identification, no cutoff values were established (23). In the present study, in the absence of an accepted cutoff value, we retained a 99% similarity as a suitable cutoff for identification at the species level and a 97% similarity as a suitable cutoff for identification at the genus level. While the introduction of these sharp values was necessary to analyze a large collection of unidentified isolates belonging to different genera, further evaluations need to be performed to assess the accuracy of these values. Because bacterial genera do not evolve at the same speed, it may be necessary to use different cutoff values depending on the bacterial genus under investigation.
Evaluation of 16S rDNA-based identification has previously been limited to comparison with phenotypic identification and has found 70 of 72 unusual aerobic gram-negative bacillus isolates (97.2%) identified to the genus level (P = 0.051) and 58 of 65 (89.2%) identified to the species level (P = 0.039; odds ratio = 0.41 [95% confidence interval, 0.15 to 1.01]). We evaluated this molecular tool with a large collection of phenotypically unidentifiable isolates for the first time, and we found this approach efficient in the majority of cases, with 88.7% of isolates being identified to the genus level and 76.3% being identified to the species level. Even those isolates which could not be identified at the genus level could be assigned a phylogenetic position. In contrast to phenotypic identification, which is biased by errors and the variability of character expression, 16S rDNA sequencing provides unambiguous data even for rare isolates, which are reproducible in and between laboratories.
We found that Enterobacter isolates were significantly unidentifiable at the species level. In a previous study (23), six isolates identified as Enterobacter cloacae by the conventional method fell into three different clusters after 16S rDNA sequence analysis, with a 1.52% divergence rate among them. A 16S rDNA-based phylogenetic tree suggests that current Enterobacter taxonomy may not be appropriate, with several species clustering with Escherichia coli. We have previously reported failures in the 16S rDNA-based approach to the identification of enteric bacteria (14). Although not statistically significant, identification of Bacillus isolates to the species level also proved difficult in our study because of low similarity levels, suggesting that too few Bacillus sequences have been deposited in GenBank. In addition, the fact that two distinct Bacillus species may possess identical 16S rDNA sequences has previously been reported (2, 10). In this study, 18 isolates remained unidentified after 16S rDNA sequence analysis, but these were assigned to phylogenetic locations and probably represent new taxa. The majority of these isolates had been collected from environmental sources, suggesting that efforts should be made towards the isolation and culture of fastidious environmental microorganisms and not just towards their 16S rDNA-based detection in environmental samples (22). These isolates may represent prototype strains of new genera or species, which underlines the necessity for a careful description of any unusual bacterial isolate.
The overall performance of 16S rDNA sequence analysis was excellent, since it was able to resolve almost 90% of identifications, when applied to a large collection of phenotypically unidentifiable bacterial isolates. In order to improve this performance, efforts should be made to complete 16S rDNA databases with high-quality sequences and to develop electronic tools for sequence comparison and interpretation. The ongoing progress with DNA microarrays should offer the technological support for its routine application.
REFERENCES
- 1.Altschul X, Stephen F, Thomas L, Madden X, Alejandro A, Schäffer X, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ash C, Farrow J A E, Dorsch M, Stackebrandt E, Collins M D. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species based on the basis of reverse transcriptase sequencing of 16S rRNA. Int J Syst Bacteriol. 1991;41:343–346. doi: 10.1099/00207713-41-3-343. [DOI] [PubMed] [Google Scholar]
- 3.Cilia V, Lafay B, Christen R. Sequence heterogeneities exist among the 16S ribosomal RNA sequences of the seven operons in Escherichia coli strain PK3 that can affect phylogenetic analyses at the species level. Mol Biol Evol. 1996;13:451–461. doi: 10.1093/oxfordjournals.molbev.a025606. [DOI] [PubMed] [Google Scholar]
- 4.Clayton R A, Sutton G, Hinkle P S, Jr, Bult C, Fields C. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int J Syst Bacteriol. 1995;45:595–599. doi: 10.1099/00207713-45-3-595. [DOI] [PubMed] [Google Scholar]
- 5.Dessen P, Fondrat C, Valencien C, Munier G. BISANCE: a French service for access to biomolecular sequences databases. CABIOS. 1990;6:355–356. doi: 10.1093/bioinformatics/6.4.355. [DOI] [PubMed] [Google Scholar]
- 6.Drancourt M, Bollet C, Raoult D. Stenotrophomonas africana sp. nov., an opportunistic human pathogen in Africa. Int J Syst Bacteriol. 1997;47:160–163. doi: 10.1099/00207713-47-1-160. [DOI] [PubMed] [Google Scholar]
- 7.Drancourt M, Mainardi J L, Brouqui P, Vandenesch F, Carta A, Lehnert F, Viguier E, Goldstein F, Acar J, Raoult D. Bartonella (Rochalimaea) quintana endocarditis in homeless patients: report of three cases. N Engl J Med. 1995;332:419–423. doi: 10.1056/NEJM199502163320702. [DOI] [PubMed] [Google Scholar]
- 8.Farris J S. PHYLIP—Phylogeny Inference Package version 3.2. Cladistics. 1989;5:164–166. [Google Scholar]
- 9.Felsenstein J. PHYLIP: Phylogeny Inference Package, version 3.5c. Seattle: University of Washington; 1993. [Google Scholar]
- 10.Fox G E, Wisotzkey J D, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 11.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. Vol. 3. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 12.Kimura M. A simple method for estimating evolutionnary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 13.Maidack B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The ribosomal data base project (RDP) Nucleic Acids Res. 1996;24:82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mollet C, Drancourt M, Raoult D. rpoB sequence analysis as a novel basis for bacterial identification. Mol Microbiol. 1997;26:1005–1011. doi: 10.1046/j.1365-2958.1997.6382009.x. [DOI] [PubMed] [Google Scholar]
- 15.Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 16.Palys T, Nakamura L K, Cohan F M. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int J Syst Bacteriol. 1997;47:1145–1156. doi: 10.1099/00207713-47-4-1145. [DOI] [PubMed] [Google Scholar]
- 17.Relman D A, Schmidt T M, MacDermott R P, Falkow S. Identification of the uncultured bacillus of Whipple's disease. N Engl J Med. 1992;327:293–301. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 18.Rolleke S, Muyzer G, Wawer C, Wawer G, Lubitz W. Identification of bacteria in a biodegraded wall painting by denaturating gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1996;62:2059–2065. doi: 10.1128/aem.62.6.2059-2065.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stackebrandt E, Goebel B M. A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 20.Stager C E, Davis J R. Automated systems for identification of microorganisms. Clin Microbiol Rev. 1992;5:302–327. doi: 10.1128/cmr.5.3.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein A, Raoult D. A simple method for amplification of DNA from paraffin-embedded tissues. Nucleic Acids Res. 1992;20:5237–5238. doi: 10.1093/nar/20.19.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strous M, Fuerst J A, Kramer E H, Logemann S, Muyer G, van de Pas-Schoonen K T, Webb R, Kuenen J G, Jetten M S. Missing lithotroph identified as new planctomycete. Nature. 1999;400:446–449. doi: 10.1038/22749. [DOI] [PubMed] [Google Scholar]
- 23.Tang Y-W, Ellis N M, Hopkins M K, Smith D H, Dodge D E, Persing D H. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli. J Clin Microbiol. 1998;36:3674–3679. doi: 10.1128/jcm.36.12.3674-3679.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Y-W, Von Graevenitz A, Waddington M G, Hopkins M K, Smith D H, Li H, Kolbert C P, Montgomery S O, Persing D H. Identification of coryneform bacterial isolates by ribosomal DNA sequence analysis. J Clin Microbiol. 2000;38:1679–1678. doi: 10.1128/jcm.38.4.1676-1678.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teske A, Sigalevich P, Cohen Y, Muyzer G. Molecular identification of bacteria from a coculture by denaturing gradient gel electrophoresis of 16S ribosomal DNA fragments as a tool for isolation in pure culture. Appl Environ Microbiol. 1996;62:4210–4215. doi: 10.1128/aem.62.11.4210-4215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sentivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalities and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van de Peer Y, Nicoläi S, De Rijk P, De Wachter R. Database on the structure of small subunit RNA. Nucleic Acids Res. 1996;24:86–91. doi: 10.1093/nar/24.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang G C, Wang Y. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology. 1996;142:1107–1114. doi: 10.1099/13500872-142-5-1107. [DOI] [PubMed] [Google Scholar]
- 29.Wang G C, Wang Y. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl Environ Microbiol. 1997;63:4645–4650. doi: 10.1128/aem.63.12.4645-4650.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weyant R S, Moss C W, Weaver R E, Hollis D G, Jordan J G, Cook E C, Daneshvar M I. Identification of unusual pathogenic Gram-negative aerobic and facultatively anaerobic bacteria. Baltimore, Md: Williams & Wilkins; 1996. [Google Scholar]
- 32.Woese C R, Kandler O, Wheelis M L. Towards a natural system of organisms: proposal for the domains Archae, Bacteria, and Eukarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]