ABSTRACT
The ongoing SARS-CoV-2 coronavirus pandemic of 2020–2021 underscores the need for manufacturing platforms that can rapidly produce monoclonal antibody (mAb) therapies. As reported here, a platform based on Nicotiana benthamiana produced mAb therapeutics with high batch-to-batch reproducibility and flexibility, enabling production of 19 different mAbs of sufficient purity and safety for clinical application(s). With a single manufacturing run, impurities were effectively removed for a representative mAb product (the ZMapp component c4G7). Our results show for the first time the reproducibility of the platform for production of multiple batches of clinical-grade mAb, manufactured under current Good Manufacturing Practices, from Nicotiana benthamiana. The flexibility of the system was confirmed by the results of release testing of 19 different mAbs generated with the platform. The process from plant infection to product can be completed within 10 days. Therefore, with a constant supply of plants, response to the outbreak of an infectious disease could be initiated within a matter of weeks. Thus, these data demonstrated that this platform represents a reproducible, flexible system for rapid production of mAb therapeutics to support clinical development.
KEYWORDS: Plant made pharmaceuticals, monoclonal antibody, Nicotiana benthamiana, biologic
Introduction
Protein-based biologic therapeutics represent a substantial proportion of new drugs in development and newly approved treatments. In particular, monoclonal antibodies (mAbs) or antibody-derived drugs are now available for many diseases, including autoimmune disorders, macular degeneration, prevention of transplant rejection, and cancer. Based on the current commercial therapeutic market, this sector is predicted to be a $300 billion industry by 2025.1,2 Additionally, mAbs represent an important therapeutic strategy for dealing with emerging infectious agents,3 including SARS-CoV-24,5 and Ebola virus.6,7
Plant-made pharmaceuticals (PMPs), including therapeutic antibodies, are a relatively inexpensive alternative to similar products generated in mammalian, bacterial, or yeast systems. For mAbs, individuals exposed to the infectious agent or administered a vaccine are also a source of therapeutic antibodies. Indeed, isolation of neutralizing antibodies from individuals often provides the necessary starting point for developing therapeutic mAbs. Plant-produced mAbs formulated in various ways have recently reached clinical trials, including a cocktail of three mAbs (ZMapp) used for treating Ebola infection;8 a mAb conjugated to keyhole limpet hemocyanin for use as a personalized lymphoma vaccine;9 a human immunodeficiency virus (HIV)-neutralizing mAb;10 two anti-viral mAbs (one against HIV and another against herpes simplex virus (HSV)) incorporated into a vaginal film for prevention of sexually transmitted diseases;11 and a human contraceptive mAb incorporated into a vaginal film for prevention of pregnancy.12 Several of these investigational products have completed a phase of clinical investigation and published outcomes have been made available. Both the lymphoma vaccine13 and anti-viral vaginal film11 have completed Phase 1 safety trials; the human contraceptive antibody is currently in Phase 1; the ZMapp antibody cocktail for Ebola was evaluated under Phase 1 and Phase 2 clinical testing concurrently to meet the needs of an active outbreak and based on compassionate use experience.6,7 Of the products mentioned, Kentucky BioProcessing (KBP) produced the ZMapp cocktail, the anti-viral mAb-containing vaginal films, and the contraceptive mAb films using their previously described Good Manufacturing Practice (GMP) platform process.14,15
Although individual products generated with plant-based platforms have been described, few data regarding the flexibility and reproducibility of these systems for clinical GMP production are available. Here, we present data showing that a GMP platform based on Nicotiana benthamiana14 provides a reproducible, flexible system for rapid production of mAb therapeutics. Through the analysis of samples for a representative mAb product, c4G7 of the ZMapp cocktail16 over a single manufacturing batch, we showed the effectiveness of removal of impurities at each step. By comparing the final characteristics of 14 batches of c4G7, we demonstrated the reproducibility of producing a plant-made GMP clinical-grade mAb. To illustrate the flexibility of generating mAbs in N. benthamiana, we showed the final quality certification analysis for 19 different mAbs.
An effective mAb manufacturing platform for production of clinical material must show consistency from batch to batch of the final product and have the ability to meet GMP. Ideally, the system should also have flexibility for changing between products, scalability, and rapid production, especially for use in case of emerging infectious diseases. Our data with the KBP GMP platform process for generating therapeutic mAbs suggest that it is an effective mAb manufacturing platform.
Results
Production of a single lot of c4G7
KBP developed a scalable GMP manufacturing process for producing mAbs in N. benthamiana (Figure 1).14 After infiltration of the plants with the Agrobacterium tumefaciens carrying plasmids for the expression of heavy and light chains of c4G7, we harvested the aerial parts of the plants, and extracted, clarified, and purified c4G7 through a series of chromatography steps: Protein A affinity chromatography (Figure 2a), anion exchange chromatography (Figure 2b), and multimodal chromatography (Figure 2c).
Figure 1.
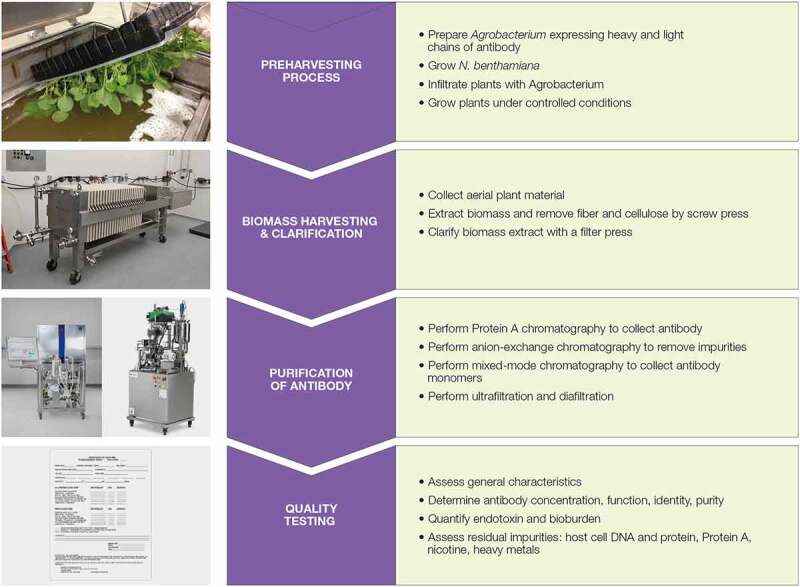
Overview of the workflow for manufacture of mAbs in N. benthamiana. Flow chart outlining a general overview of the manufacturing process including Agrobacterium infiltration prior to harvest, harvesting of biomass, filter press clarification, chromatography purification, formulation, and quality testing.
Figure 2.
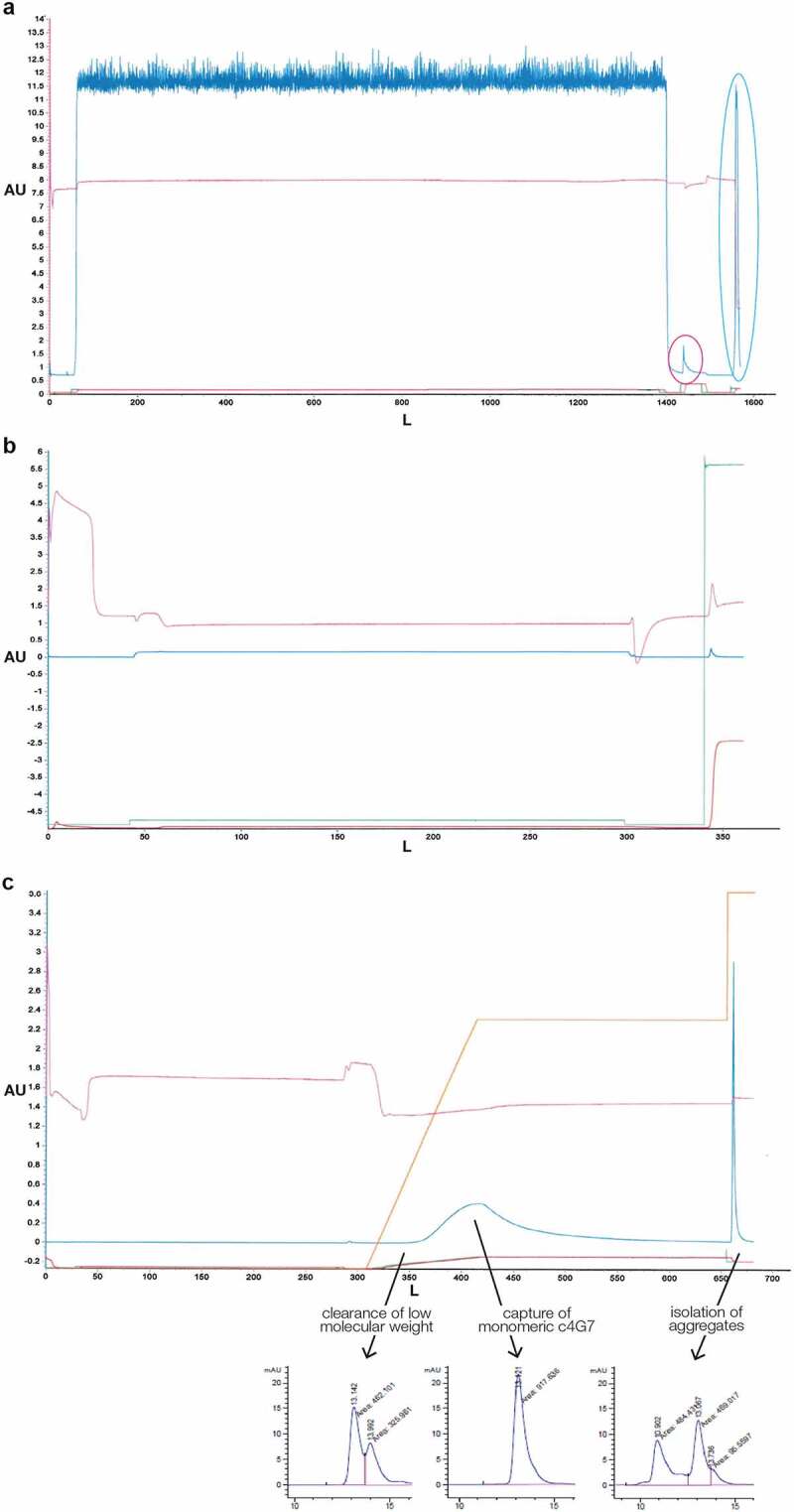
Representative chromatograms trace of three-step chromatography purification process for c4G7 (a) Protein A affinity chromatography. The large peak in the dark blue line before 1600 L on the x axis, circled in light blue, is the product elution. The small peak circled in red before the elution represents nonspecifically bound proteins released during washes. Dark blue line, UV A280; pink line, pH; green line, pre-column conductivity; red line, post column conductivity. (b) Anion-exchange chromatography. Antibody product is present in the flow through
. Blue line, UV A280; pink line, pH; green line, pre-column conductivity; red line, post column conductivity. The large strip peak at is seen at the end of the chromatogram in Figure 2b. The strip fraction contains host cell DNA, HCP, endotoxin, and color. (c) Mixed-mode chromatography.
Blue line, UV A280; pink line, pH; green line, pre-column conductivity; red line, post column conductivity; Orange line, program gradient. In the top chromatogram, the product is in the blue peak at ~400 L on the x axis. The CHT purification step allows separation of product species of different sizes. LMW fractions elute first, monomer elutes second, and HMW aggregates are bound until the stripping step as represented by the lower chromatogram. A singular UV peak during the elution indicates all species are relatively close in size and structure. The large strip peak by is shown at the end of the chromatogram. The strip fraction contains host cell DNA, HCP, endotoxin, and HMW aggregates. 2aTypical Protein A affinity purification chromatogram. Multiple washes after product load remove nonspecific impurities as indicated by a small peak circled. The product is eluted after washing as indicated by a large peak circled. 2b. Typical Capto Q anion exchange chromatogram. Product is captured in the flow through fraction as indicated by a slight but consistent increase at UV A280. Negatively charged impurities are bound to the resin and removed post loading via a high salt wash. Impurities are observed by a large peak at UV A280. 2 c. Typical CHT multimodal chromatogram. Product is bound to the resin and then eluted over a 30 CV gradient. The gradient is held at UV A280 peak max to isolate monomeric mAb.
We collected and evaluated 11 samples at 5 different unit operations throughout the extraction, clarification, and purification process. SDS-PAGE analysis was used to evaluate protein purity and integrity of the 11 in-process samples, representing both the product stream and selected washes or solutions from stripping the chromatography columns (Figure 3a). Additionally, size-exclusion high performance liquid chromatography (SE-HPLC) analysis was performed on the material recovered from anion-exchange chromatography and mixed-mode chromatography (Figure 3b, c). These samples showed that, following mixed-mode chromatography, the purified c4G7 in the eluate was predominately a single monomeric species.
Figure 3.
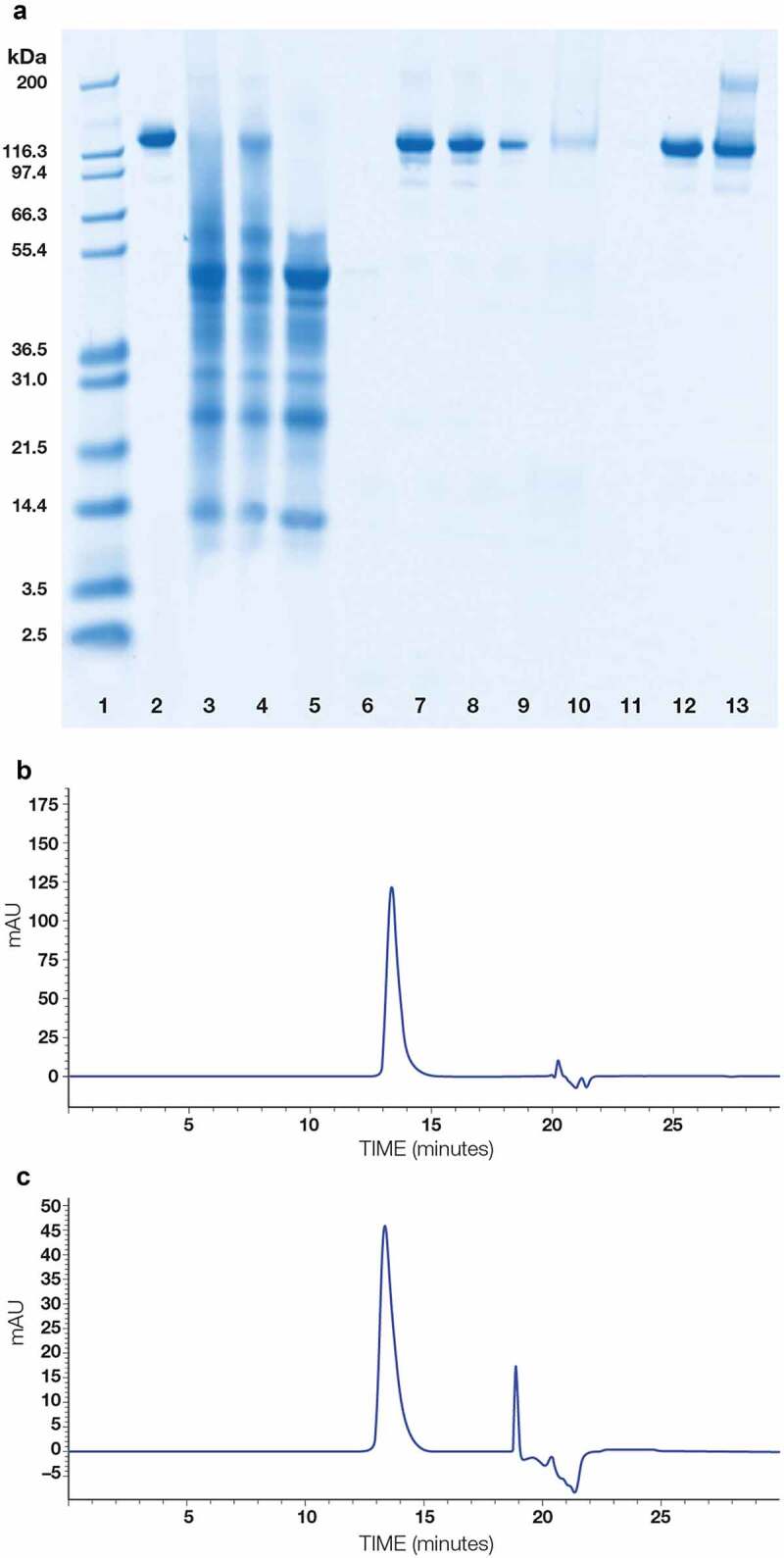
Purity of c4G7 and evaluation of protein components in various steps of the purification process. (a) Non-reducing SDS-PAGE analysis of 11 samples during purification process. Lane 1: molecular weight standards; lane 2: reference antibody; lane 3: green juice; lane 4: filter-press filtrate and wash; lane 5: Protein A flow through and washes 1 and 3; lane 6: Protein A wash 2; lane 7: Protein A eluate; lane 8: anion-exchange flow through; lane 9: anion-exchange wash; lane 10: anion-exchange strip; lane 11: mixed-mode (CHT) flow through and wash; lane 12: mixed-mode eluate; lane 13: mixed-mode strip. (b) SE-HPLC analysis of anion-exchange flow through. Main peak is the monomeric mAb c4G7. (c) SE-HPLC analysis of mixed-mode chromatography eluate. Main peak is the monomeric mAb c4G7. Non-reduced SDS Page analysis of all in-process samples demonstrates the removal of impurities at each step resulting in a final pure product of a single species post CHT purification. 3b. SEC-HPLC analysis of Capto Q anion exchange flow through. Main peak indicates primarily monomeric mAb. Other peaks observed are sample buffer related. 3 c. SEC-HPLC analysis of CHT multimodal elution. Main peak indicates primarily monomeric mAb. Other peaks observed are sample buffer related.
We evaluated samples at multiple steps in the purification process for a single sub-lot of one harvest of plants expressing c4G7 (Table 1). We calculated protein concentration using UV absorbance at 280 nm (A280) and the molecule specific extinction coefficient. For samples of the product stream at the protein A chromatography and anion-exchange chromatography steps, we subtracted A320 from A280 to account for background color, reflective of residual plant host-derived pigments. We determined the yield on the basis of product per amount of biomass processed (mg/kg). The amount of c4G7 monomer, as well as high molecular weight (HMW) and low molecular weight (LMW) protein contaminants, were determined by calculating the percent area under the curve from SE-HPLC. These in-process results showed that most c4G7 produced by the plants was recovered in the Protein A affinity purification step and that the anion-exchange and mixed-mode chromatography steps removed HMW aggregates of the antibody as well as other process-related impurities.
Table 1.
In-process testing for a single lot of c4G7
|
Product stream from step in process |
Concentration (mg/ml) |
Background-adjusted concentration (mg/mL) |
Total amount (g) |
Recovery (%) |
Yield (mg/kg) |
Proportion of protein components based on area under the curve from SE-HPLC |
Endotoxin(EU/mg) |
Bioburden(CFU/ml) |
||
| HMW% |
Monomer % |
LMW % |
||||||||
| Protein A chromatography | 2.74 | 2.60* | 42.63 | N/A | 47.13 | 7.60 | 92.40 | 0.00 | 103.80 | 0 |
| Anion-exchange chromatography | 0.17 | 0.16* | 36.32 | 85% | 40.15 | N.D. | N.D. | N.D. | < 0.625 | N.D. |
| Mixed-mode chromatography | 0.20 | N.D. | 29.42 | 81% | 32.52 | 0.00 | 100.00 | 0.00 | < 0.025 | N.D. |
| Ultrafiltration and diafiltration | 25.39 | N.D. | 60.94** | 100%** | 34.35 | 0.00 | 100.00 | 0.00 | 0.01 | N.D. |
Table notes: Data for the product stream at the ultrafiltration and diafiltration step represent the combination of two sub-lots that comprised the final lot. Data for all other in-process samples are for one sub-lot. * To account for background color in sample, protein concentration was calculated after subtracting the absorbance at 320 nm from the absorbance at 280 nm. ** The increase in product after ultrafiltration and diafiltration resulted from the combination of two sub-lots to make the final lot. Abbreviations: N/A, not applicable; N.D., not determined.
Process-related impurities include host cell protein (HCP), HMW aggregates of the antibody, endotoxin, bioburden, and nicotine. We calculated the percent reduction in each of these impurities at multiple steps in the purification process (Table 2). After the Protein A chromatography step, no detectable nicotine was present and HCP was also reduced below the limit of detection (LOD). Product-specific impurity requirements dictated bioburden of ≤ 1 colony forming unit (CFU) per mL and endotoxin units (EU) of <0.3 EU/mg. After the Protein A chromatography step, the sample was free of bioburden, but still retained an unacceptable endotoxin content (Table 1). We attributed this high initial endotoxin content to the Agrobacterium used for producing the antibody heavy and light chains. However, the two subsequent chromatography steps eliminated most of the remaining endotoxin, resulting in drug substance that met the pre-specified requirements.
Table 2.
Percent impurity reduction of in-process samples from a single sub lot of c4G7 from a single harvest
| Process Step | Endotoxin | Nicotine | HCP | Residual Protein A | LMW protein | HMW aggregates |
|---|---|---|---|---|---|---|
| Raw plant extract (green juice) | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| Filter press filtrate + wash | 43.70% | 29.70% | N.D. | N.D. | N.D. | N.D. |
| Protein A elution | 99.90% | 100.00% | 100.00% | N.D. | N.D. | N.D. |
| Anion-exchange flow through | 99.80% | N.D. | N.D. | 66.00% | 70.60% | 43.20% |
| Mixed-mode chromatography elution | 83.40% | N.D. | N.D. | 100.00% | 100.00% | 100.00% |
Table notes: Data are for the same sub-lot described in Table 1. Nicotine Assay LOD = 10 µg/mL; HCP Assay LOD = 31 ng/mL. Abbreviations: HCP, host cell protein; N.D., not determined.
Reproducibility of multiple batches of c4G7
Multiple analyses were performed on the final c4G7 product for clinical batch release, and these were compared across 14 manufacturing batches to determine reproducibility of the production platform. A manufacturing batch was comprised of c4G7 formulated from up to three individual harvests of c4G7. The quality and functionality of the mAb, as well as the removal of biological, chemical, or metal impurities, were documented on a certificate of analysis (Table 3).
Table 3.
Release testing for 14 manufacturing lots of c4G7
| Tested parameter | Test method and reported units | Average result |
|---|---|---|
| General characteristics | Visible appearance | Clear, colorless |
| pH | 5.9 ± 0.07 | |
| Osmolality (mOsm/kg) | 365.4 ± 4.12 | |
| Protein concentration | UV absorbance (A280 converted to mg/mL) | 20.1 ± 1.12 |
| Identity | Ion exchange-HPLC | Conforms to standard |
| Ebola glycoprotein ELISA | Binding occurs | |
| Potency | Ebola glycoprotein ELISA (% of A280) | 106.80% ± 17.9 |
| Purity | Size Exclusion – HPLC (% area under the curve) | 99.5% ± 0.69 monomer mAb |
| 0.3% ± 0.30 HMW aggregates | ||
| Reduced and Coomassie-stained SDS-PAGE (% main band) | 98.6% ± 0.89 | |
| Safety | Endotoxin assay (EU/mg) | 0.2 ± 0.1 |
| Bioburden assay (CFU/mL) | 0 ± 0 | |
| Impurities | Host cell DNA detected by qPCR (pg/mg) | < 3.1 ± 0 |
| ELISA for Protein A (ng/mg) | 4.4 ± 3.6 | |
| ELISA for HCP (ng/mg) | < LOD | |
| Gas chromatography for nicotine (µg/mL) | < LOD | |
| Inductively coupled plasma mass spectrometry for heavy metals | Meets USP <232 > |
Abbreviations: A280, absorption at 280 nm wavelength; ELISA, enzyme-linked immunosorbent assay; HCP, host cell protein; HMW, high molecular weight; HPLC, high performance liquid chromatography; LOD, limit of detection; mAb, monoclonal antibody; N/A, not applicable; qPCR, quantitative polymerase chain reaction; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; USP, United States Pharmacopoeia
Analyses included standard chemical properties, such as pH and osmolality. Product-specific qualities were also determined. In particular, protein concentration was evaluated by ultraviolet absorbance and converted to mg/mL, the identity of the protein as c4G7 was confirmed by ion-exchange (IEX) HPLC, and functionality was evaluated by enzyme-linked immunosorbent assay (ELISA). The purity of c4G7 in each batch was assessed by SE-HPLC (Figure 4a) to determine the percent of the product that is LMW, monomeric product, and HMW aggregates (Table 3). Reducing and non-reducing SDS-PAGE also confirmed product purity (Figure 4b). Overall, the average and standard deviations of the release results indicated a highly consistent product was obtained for each batch, indicating that N. benthamiana is an effective source for production of mAbs using this platform.
Figure 4.
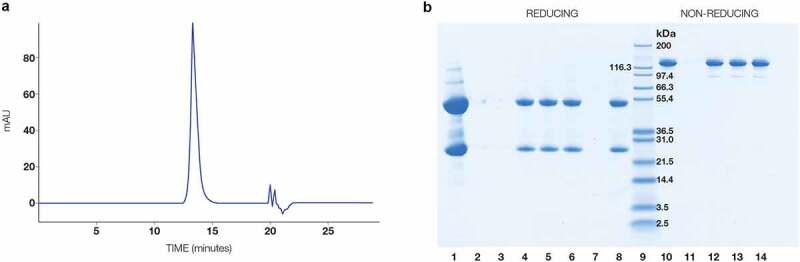
Purity analysis by SE-HPLC and SDS-PAGE of c4G7. (a) SE-HPLC chromatogram of final product. (b) Reducing and non-reducing SDS-PAGE for final product from 3 batches of c4G7. Lane 1: reduced mAb standard, lane 2–3: blank, lane 4: reduced c4G7 batch 1, lane 5: reduced c4G7 batch 2, lane 6: reduced c4G7 batch 3, lane 7: blank, lane 8: reduced mAb standard, lane 9: molecular weight standards, lane 10: non-reduced mAb standard, lane 11: blank, lane 12: non-reduced c4G7 batch 1, lane 13: non-reduced c4G7 batch 2, lane 14: non-reduced c4G7 batch 3. SEC-HPLC analysis of c4G7 final product. Main peak indicates monomeric product. Other peaks observed are sample buffer related. 4b. Reduced and Non-Reduced SDS-Page analysis of three different batches of c4G7. Each sample matches the mAb standard indicating conformity.
We analyzed impurities and recovery of c4G7 across 20 harvests (Table 4) (note: one lot may include multiple harvests). There was a wide range of endotoxin, HCP, and nicotine at early steps in the manufacturing process. After the Protein A chromatography step, we detected residual protein A in the eluate. However, in the eluate from mixed-mode chromatography, these analytes were all either below the LOD or within the acceptable levels for all harvests (Table 4).
Table 4.
Summary of percent impurity reduction and product purity and recovery across 20 lots of c4G7
| Process Step | Product recovery (%) | Endotoxin (EU/mg) | Nicotine (µg/mL) | HCP (ng/mg) | Protein A (ng/mg) | LMW protein (% of total protein) | HMW aggregates (% of total protein) | Monomer mAb product (% of total protein) |
|---|---|---|---|---|---|---|---|---|
| Raw plant extract (green juice) | N/A | 685,392 ± 212,353 | 9.9 ± 4.8 | N/A | N/A | N/A | N/A | N/A |
| Filter press filtrate + wash | 79 ± 11 | 385,448 ± 101,754 | 7.0 ± 4.0 | 137,713 ± 75,117 | N/A | N/A | N/A | N/A |
| Protein A elution | 98 ± 1 | 80 ± 35 | < LOD | < LOD | 177 ± 217 | 1.42 ± 0.53 | 5.21 ± 0.83 | 93.4 ± 0.97 |
| Anion-exchange flow through | 85 ± 7 | 0.16 ± 0.09 | < LOD | < LOD | 60 ± 46 | 0.42 ± 0.88 | 2.96 ± 0.48 | 96.6 ± 0.92 |
| Mixed-mode chromatography elution | 88 ± 5 | 0.026 ± 0.002 | < LOD | < LOD | < LOD | < LOD | < LOD | 100 ± 0.0 |
Across the 20 manufacturing runs of c4G7, 100% of the mAb was monomeric by the end of chromatographic purification (Table 4). In the final batches for clinical use, the mAb was concentrated and formulated by tangential flow filtration (TFF). A small amount of residual protein A was detectable by ELISA in the concentrated product, but was below the acceptance criteria (Table 3).
Abbreviations: N/A, not applicable; LOD, limit of detection; HCP, host cell protein.
Flexibility of the plant-based mAb manufacturing platform
To demonstrate the flexibility of the plant-based manufacturing platform, we evaluated the protein concentration, antibody purity, chemical properties, and safety (endotoxin and bioburden) of individual manufacturing lots for 19 different IgG1 mAbs (Table 5). Each of the mAb products met the pre-specified requirements for concentration, osmolality, and pH. For the 19 antibodies, the target concentrations were 10 mg/mL or 20 mg/mL. Only two antibodies were below 10 mg/ml and all exhibited high purity, with product representing a minimum of 95.8% of monomeric protein. Additionally, bioburden was undetectable in each of the 19 mAb products. Endotoxin evaluation depended on the LOD of the assay, which implied more variability than was likely present. However, endotoxin was consistently reduced to ≤1 EU/mg.
Table 5.
Release testing results and variability from a single manufacturing lot of 19 different mAbs
|
Tested parameter |
Antibody A |
Antibody B |
c4G7 |
Antibody D |
Antibody E |
Antibody F |
Antibody G |
Antibody H |
Antibody I |
Antibody J |
Antibody K |
Antibody L |
Antibody M |
Antibody N |
Antibody O |
Antibody P |
Antibody Q |
Antibody R |
Antibody S |
Minimum across all 19 mAbs |
Maximum across all mAbs |
| Protein concentration (mg/mL) | 19.6 | 20 | 18.9 | 20.4 | 21.2 | 11.3 | 10.9 | 20 | 20.5 | 15.5 | 23.5 | 17.1 | 20.9 | 17.5 | 18.2 | 20 | 22 | 9.1 | 7.6 | 7.60 | 23.50 |
| pH | 5.9 | 5.9 | 5.9 | 6.4 | 6.6 | 5.6 | 5.57 | 5.8 | 5.8 | 5.6 | 5.6 | N/A | 5.57 | 5.64 | 5.58 | 5.58 | 6.5 | 5.6 | 5.65 | 5.57 | 6.60 |
| Osmolality (mOsm/kg) | 373 | 367 | 369 | 27 | 28 | 316 | 311 | 356 | 441 | 381 | 361 | N/A | 317 | 310 | 358 | 313 | 25 | 328 | 315 | 25.00 | 441.00 |
| Purity based on SDS-PAGE (%) | > 99 | > 99 | > 99 | > 99 | 95 | > 99 | > 99 | > 99 | > 99 | > 99 | > 99 | > 99 | > 99 | > 99 | > 99 | > 99 | > 99 | > 99 | > 99 | N/A | N/A |
| HMW protein based on SE-HPLC (%) | 1.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.2 | 0.8 | 1.1 | 0 | 0 | 2 | 1 | 0 | 2 | 0 | 0.00 | 2.20 |
| Monomeric mAb based on SE-HPLC (%) | 98.5 | 98.7 | 100 | 95.8 | 100 | 100 | 100 | 100 | 100 | 97.8 | 99.2 | 98.9 | 100 | 100 | 98 | 99 | 100 | 97 | 98 | 95.80 | 100.00 |
| LMW protein based on SE-HPLC (%) | 0 | 1.3 | 0 | 4.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.1 | 2 | 0.00 | 4.20 |
| Endotoxin (EU/mg) | 0.02 | 0.01 | 0.02 | 0.64 | 0.41 | 0.04 | 0.05 | <1.00 | 0.07 | 0.22 | 0.08 | 0.09 | 0.01 | <0.02 | <0.25 | <0.05 | 0.01 | 0.03 | 0.03 | 0.01 | 1.00 |
| Bioburden (CFU/ml) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0.00 |
Table notes: Target concentration for mAb F, G, R, and S was 10 mg/mL and for remaining mAbs was 20 mg/ml. Endotoxin assays were used with varying LOD based on the dilution.
Five of the KBP-produced mAb products have been in clinical trials for treatment of disease caused by Ebola virus, HSV, and HIV, and for contraception (Table 6), indicating that the platform can produce a variety of mAb products of sufficient purity and safety for use in patients. The remaining mAb products target viral diseases, such as rabies, Venezuelan equine encephalitis virus, or hemorrhagic fever caused by Marburg virus, Junin virus, or Ebola virus. Some mAbs target bacterial infections, such as Staphylococcal enterotoxin B, and others are intended to neutralize toxins, such as ricin. Thus, antibodies produced with this platform have diverse potential applications.
Table 6.
KBP-manufactured mAbs in clinical trials
| mAb | Purpose | Clinical phase | Trial ID |
|---|---|---|---|
| A, B, c4G7 | Ebola treatment | 1/2 | NCT02363322 |
| D | HSV treatment | 1 | NCT02579083 |
| E | HIV treatment | 1 | NCT02579083 |
| Q | Contraception | 1 | NCT04731818 |
Table note: Antibody nomenclature matches that in Table 5.
Discussion
Rapid production in response to emerging pathogens
The production of the three mAbs evaluated in Phase 1/2 clinical trials for treatment of Ebola virus infection provides an example of the speed with which this platform can be deployed (Figure 5). The initial case for the 2014–2016 Ebola outbreak occurred in December 2013 and a medical alert was issued in January 2014 for the region in Guinea where multiple cases occurred.17 Within one month of identifying the potentially therapeutic antibodies, small quantities of the three antibodies were produced for a pilot study in non-human primates. Based on the positive outcome of that study,18 in August of the same year, GMP production was initiated, and a few patients received the mAb cocktail, named ZMapp, through expanded access/compassionate use.19 By October of 2014, ZMapp was available, and the investigational new drug (IND) application was approved in February 2015, enabling initiation of the clinical trials.
Figure 5.
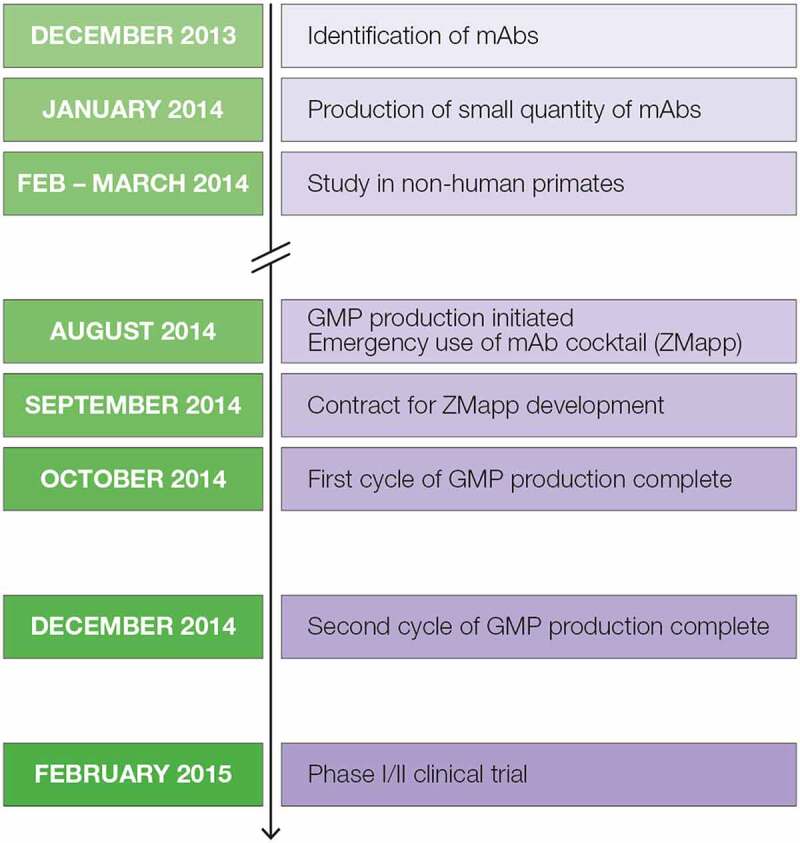
Timeline of development and production of the ZMapp antibody cocktail for emergency use and clinical trials for treating Ebola virus disease during the outbreak of 2014.7 Timeline representing development of the ZMapp antibody therapeutic. In December 2013 mAbs were identified, January 2014 small scale production initiated, February through March 2014 non-human primate studies performed, August 2014 GMP production initiated for emergency use of ZMapp, September 2014 a contract for ZMapp development initiated, October through December 2014 the first and second GMP production cycles completed, and February 2015 Phase I/II clinical trials begin.
Antibody-based drugs have many applications, including as prophylaxes and therapeutics for infectious diseases. The potential rapid growth and spread of infectious pathogens, as demonstrated with Ebola virus,20 Zika virus,21 and SARS-CoV-2,22 requires a system to support quick and scalable responses in the manufacture of candidate antibody therapies. During the Prevail Phase 2 trial for ZMapp, clinical promise was observed with a strong trend toward improved survival, although efficacy metrics were not reached due to patient access as the Ebola outbreak dissipated. PMPs have the potential to meet such a need, yet the use of plant-based platforms for producing biologics, such as mAbs, remains limited. This limited use is likely attributed to concerns around regulatory acceptance of a currently unapproved technology. Scalability of indoor growth systems is another concern. With the increasing popularity and sophistication of indoor farming, plant growth facilities are expected to become more scalable in the near future.23 Using the KBP manufacturing platform as a benchmark, mAb yield averages ~100 mg/kg with an estimated ability to produce over 8 kg of mAb per year. With improvements and expansion of growth and manufacturing space, PMP commercial output could be easily increased to meet market needs by an order of magnitude. Additional benefits to PMPs include low cost of startup, speed to production, and low risk from the use of plant viruses that do not infect humans.
Other methods for manufacturing mAbs, such as production in Chinese hamster ovary cells, typically require 6–12 months for suitable cell line development from mAb identification to IND readiness.24 Using the KBP manufacturing platform as an example for PMP production, KBP has submitted an IND application to the U.S. Food and Drug Administration within 6 months of identifying the protein. In the KBP growth space, plants require 24–26 days to reach the proper growth stage for infiltration. During this plant growing period, Agrobacterium with the antibody-encoding plasmids is cultured. Post Agrobacterium infiltration, plants grow for 7 additional days prior to harvest. The manufacturing process can be completed in 3 days. With plants of the proper growth stage and plasmids encoding the antibodies in Agrobacterium, we have condensed the timeline to produce a batch of mAb to ~10 days from infiltration of the plants to certificate of analysis of the final product. With this platform, to produce a 3 mAb cocktail, we have performed 70 manufacturing harvests that yielded 38 batches of 3 distinct plant-made mAbs in 20 months. Of the 38 batches, 20 were for a single mAb product (c4G7).
Here, we demonstrated that a plant-based platform based on N. benthamiana represents a reproducible system for GMP production of mAbs for clinical use. The plant-based mAbs were highly consistent among the batches with regards to purity, potency, and low levels of impurities. The process was flexible, enabling production of multiple mAbs. Thus, by leveraging existing plant engineering technologies,25 our results showed that plant-based production of mAbs in N. benthamiana provides an advantageous method for manufacturing mAbs. Furthermore, the speed of this platform for production enabled GMP-quality product for emergency use within 1 month, and product for clinical trials within several months. Therefore, our results show the potential of PMPs to meet the urgent need for rapid development of antibody-based therapeutics to treat emerging pandemics, such as the SARS-CoV-2 pandemic. Multiple mAbs have been identified that neutralize the virus SARS-CoV-2,4,5 and a plant-based manufacturing platform could provide an efficient method for making such therapies available quickly.
Materials and methods
The process of mAb production with the KBP manufacturing platform has been published in detail.14 The process is briefly described here.
N. benthamiana infiltration and growth
Plants deficient in xylose and fucose transferases26 were germinated in soilless tobacco mix medium (Speedling, Bushnell, FL) in an indoor biomass production facility with controlled and monitored temperature (68–76°F), light (300–500 µm/m2/s for 16 hours followed by 8 hours of dark), and humidity (60–80%) using Argus Titan software and controls. Prior to infiltration, plants were watered through a subirrigation system on days one through three post sow, and every three days thereafter. Irrigation water included 250 ppm nitrogen (~59 ppm ammoniacal nitrogen, ~191 ppm nitrate nitrogen), 44 ppm phosphate (P2O5), and 250 ppm soluble potash (K2O).
Agrobacteria were transformed with the heavy chain of the mAb cloned into potato virus X (PVX) and with the light chain of the mAb cloned into turnip vein clearing virus (TVCV).27 The transformed Agrobacteria, with or without p21 silencing suppressor,28 were grown in animal product-free (APF) Luria Broth (LB, Miller) liquid medium containing antibiotics (50 mg/L rifampicin and 50 mg/L kanamycin) to an optical density at 600 nm (OD600) of 1.8–2.2. Agrobacteria cultures were diluted 1:1000 in infiltration buffer [10 mM 2-(N-morpholino) ethanesulfonic acid (MES) and 10 mM MgSO4, pH 5.5].
Plants (~250,000, 4–15 inches tall) were infiltrated with Agrobacteria by submerging in infiltration solution and applying a vacuum (24 inches of mercury for 2 minutes). The vacuum was then released over 3–5 seconds. Following release of the vacuum, the plants were drained, inverted for 2–5 minutes to dry, and then returned to the growth facility. Plants were grown after infiltration for 6–8 days at a temperature of 68–76°F, relative humidity of 60–80%, and light schedule of 16/8 hour on/off with an intensity of 100–300 µm/m.2/s Plants were watered without fertilizer on day 0, 1, 3, 4, and 6 after infiltration.
Extraction and clarification of biomass
Aerial portions of plants were harvested (~850 kg biomass) and extracted. The plant cells were ruptured with a double-stack disintegrator (Corenco) in the presence of 0.5 L aqueous extraction buffer per kg biomass (100 mM Tris-Base, 40 mM ascorbic acid, 1 mM EDTA, pH 8.5). Fiber and cellular debris were separated from the liquid extract with a hydraulic screw press (Vincent). After adjusting the liquid extract pH to 8.0 using concentrated sodium hydroxide or concentrated hydrochloric acid, the extract was clarified by depth filtration. The extract (referred to as green juice) was mixed for at least 15 minutes with diatomaceous earth (DE) (33 g/L Celpure 300). The mixture was then filtered through a plate and frame filter press (24 inch) (Ertel Alsop) equipped with 0.3-µm filter sheets (46 kg of biomass per filter frame). The filter press was washed with a 10× quantity of DE (1 kg DE = 10 L wash) in extraction buffer.
Samples for in-process analysis were collected of the extract and the filter-press filtrate with wash.
Chromatographic purification
Clarified extract was filtered through a 1.2-micron glass fiber pre-column filter (Sartorius) and loaded, up to 35 mg of protein per mL of resin, onto a Protein A column [bed height of ≥8 cm and column screen size of 23 µm, (Mab SelectSuRe, Cytiva)] equilibrated with 50 mM Tris-HCl, pH 8.0. The column was washed with 5 column volumes of 50 mM Tris-HCl, pH 8.0. After the first wash, the inline glass fiber filter was removed. The column was then washed with 10 column volumes of 500 mM arginine, pH 8.0, followed by 7–10 column volumes of 50 mM Tris-HCl, pH 8.0. The bound mAb was eluted with 3–5 column volumes of 100 mM acetic acid, 200 mM arginine, pH 3.0. The eluate was immediately neutralized with 1 M Tris-HCl, pH 8.0, keeping the pH below the antibody’s isoelectric point.
Samples were collected for in-process analysis from initial column load, column washes, and neutralized eluate.
The Protein A column eluate was diluted with water to a conductivity of <5 mS/cm and loaded onto a Capto Q anion-exchange resin-packed column [bed height of ≥9 cm and column screen size of 23 µm (Cytiva)] equilibrated with 50 mM HEPES (at a pH sufficiently below the isoelectric point of the antibody) in passive flow through mode. Column flow through was collected from the point of inflection at an absorbance of 280 nm (A280) until the point all product was fully applied (Figure 2b). The column was then washed with 5 column volumes of 50 mM HEPES (at the appropriate product-specific pH). The column was stripped with 5 column volumes of 50 mM HEPES, 3 M NaCl (at the same pH as the equilibration/wash buffer). Protein concentration in the flow through from the Capto Q anion-exchange chromatography was determined by measuring A280 and using a product-specific extinction coefficient to determine protein concentration in mg/mL.
Samples were collected for in-process analysis from the flow through, the wash, and the strip.
Mixed-mode chromatography was performed with ceramic hydroxyapatite (CHT) type II, 80 µm resin (Biorad) in a column [bed height of 9–20 cm and screen size of 10 µm (Cytiva)] using a protein-to-resin ratio of 10–17 mg protein/ml of resin. The column was conditioned with 1 column volume of 250 mM sodium phosphate, pH 6.8 and then equilibrated with 5 column volumes of 5 mM sodium phosphate, pH 6.8. The flow through from anion-exchange chromatography was loaded and the column was washed with 5 column volumes of 5 mM sodium phosphate, pH 6.8. The bound mAb was eluted using a linear salt gradient over 10–30 column volumes by mixing 5 mM sodium phosphate, pH 6.8 and 5 mM sodium phosphate, 500 mM NaCl, pH 6.8. Gradient length was determined empirically for each product. Eluate collection was performed based on increase in A280 above the wash buffer baseline. Once the elution peak reached maximum absorbance the gradient was held isocratically (Figure 2c). The column was then stripped with 5 column volumes of 250 mM sodium phosphate, pH 6.8.
Samples were collected for in-process analysis from the initial column load with wash, the eluate, and the strip.
Chromatography was performed with an AKTA System with Unicorn Software (Cytiva).
Ultrafiltration and diafiltration
CHT eluate was concentrated and formulated using a TFF system for ultrafiltration and diafiltration (Cytiva Uniflux 10 or Pall CM500). The TFF process used a polyethersulfone (PES) membrane with 30 kDa nominal molecular weight cutoff (NMWC) (Pall Omega Series or Cytiva Kvick) installed to the torque specification of the PES membrane (Pall Omega Centramate, 8 Nm; Pall Omega Centrasette, 40–62 Nm, Cytiva Kvick, 20 Nm). The system was equilibrated with at least 1 vessel volume of 5 mM sodium phosphate, 500 mM NaCl, pH 6.8. Eluate from mixed-mode chromatography was loaded at <400 g/m2 of PES membrane surface area. The eluate was concentrated by maintaining a consistent transmembrane pressure (0.5–0.8 bar) and pressure drop across the membrane (Delta P) (0.4–0.7 bar). The retentate was diafiltered against at least seven volumes of formulation buffer: 20 mM citrate, 10 mM glycine, 7% sucrose, pH 5.5; 20 mM histidine, 100 mM NaCl, 4% sucrose, pH 6.0 [with polysorbate 80 (0.001–0.1%)]; or 10 mM histidine, pH 6.5 [with polysorbate 20 (0.005%)]. Formulation buffer was mAb specific. Final product was collected and filtered through a 0.2-µm sterilizing PES filter.
Samples were collected of the permeate and of product in formulation buffer prior to sterilizing filtration and after sterilizing filtration.
In-process and product testing
The physical and chemical properties of the final product were tested using conventional pH (USP <791>), osmolality (USP<785>), and spectrophotometric assays. Target pH was assessed with a VWR Symphony pH meter affixed with a Beckman Coulter Calomel electrode. Osmolality was analyzed using an Advanced Instruments 3320 Osmometer. Protein concentration of the in-process product and final product was determined using a Spectramax M2 Spectrophotometer based on A280 or A280 – A320 for background correction, as necessary, and the molecule-specific predicted extinction coefficient (calculated using the amino acid sequence).
The presence of process-related impurities in mAbs was evaluated by analyses for nicotine, HCP, host cell DNA, and residual Protein A. Nicotine was quantified with a Hewlett Packard 6890 gas chromatograph system equipped with a flame-ionization detector. Analyses of HCP and residual Protein A were performed using ELISA sandwich assays with custom antibodies (goat anti-N. benthamiana, in-house prepared) or rabbit anti-Protein A antibody (Repligen 9333–1), respectively. Host cell DNA was measured using a custom quantitative polymerase chain reaction assay.
Purity of in-process and final product was assessed using both SDS-PAGE and SE-HPLC. Purity by SDS-PAGE was calculated using a Bio-Rad GS900 densitometer from images of both reduced and non-reduced gels. SE-HPLC was performed using a TSKgel SuperSW3000 size-exclusion column (Tosoh Bioscience) on an Agilent 1260 Infinity HPLC system with UV detection set at 280 nm. Target antigen recognition was evaluated by ELISA using either commercial or custom antibodies, depending on the mAb product. Identity was further confirmed through analysis of charge variants by IEX HPLC.
Safety of in-process and final product was evaluated by measurement of residual endotoxin (USP <85>) and microbial count (USP<61>). Microbial count was determined using a standard bioburden testing protocol. Solutions were filtered using a Millipore Milliflex PLUS system. Plate counts were tabulated at least 5, but no more than 8, days post-inoculation. Endotoxin concentration was calculated using the Charles River PTS Endotoxin system.
Acknowledgments
The initial draft of this manuscript was developed by Nancy R. Gough, Ph.D. (a science writer with BioSerendipity, LLC, Elkridge, MD) based on content provided solely by the authors. The final manuscript submitted was under the sole control of the authors.
Funding Statement
This project was funded in part with Federal funds from the Department of Health and Human Services; Office of the Assistant Secretary for Preparedness and Response; Biomedical Advanced Research and Development Authority , under Contract No. HHSO100201400009C.
Abbreviations
APF- Animal Product Free
CFU- Colony Forming Units
CHT- Ceramic Hydroxyapatite
DE- Diatomaceous Earth
ELISA- Enzyme-Linked Immunosorbent Assay
EU- Endotoxin Unit
GMP- Good Manufacturing Practices
HCP- Host Cell Protein
HIV- Human Immunodeficiency Virus
HMW- High Molecular Weight
HSV- Herpes Simplex Virus
IEX- Ion Exchange
IND- Investigational New Drug
LMW- Low Molecular Weight
LOD- Limit of Detection
mAb- Monoclonal Antibody
N/A- Not Applicable
N.D.- Not Determined
NMWC- Nominal Molecular Weight Cutoff
PES- Polyethersulfone
PMP- Plant Made Pharmaceuticals
PVX- Potato Virus X
TFF- Tangential Flow Filtration
TVCV- Turnip Vein Clearing Virus
Disclosure statement
Authors are employed by KBP, a subsidiary of British America Tobacco, ZabBio, and Mapp Biopharmaceutical. Equipment and processes are covered by patents: US10214747B2, CA2971151C, EP3092002B1.
References
- 1.Lu R-M, Hwang Y-C, Liu I-J, Lee -C-C, Tsai H-Z, Li H-J, Wu H-C.. Development of therapeutic antibodies for the treatment of diseases. J Biomed Sci. 2020;27(1):1. doi: 10.1186/s12929-019-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shukla AA, Wolfe LS, Mostafa SS, Norman C. Evolving trends in mAb production processes. Bioeng Transl Med. 2017;2(1):58–13. doi: 10.1002/btm2.10061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marston HD, Paules CI, Fauci AS. Monoclonal Antibodies for emerging infectious diseases — borrowing from history. N Engl J Med. 2018;378(16):1469–72. doi: 10.1056/NEJMp1802256. [DOI] [PubMed] [Google Scholar]
- 4.Chen P, Nirula A, Heller B, Gottlieb RL, Boscia J, Morris J, Huhn G, Cardona J, Mocherla B, Stosor V, et al. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N Engl J Med [Internet]. 2020. [cited 2021 Jan 28]; Available from 10.1056/NEJMoa2029849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, Musser BJ, Soo Y, Rofail D, Im J, et al. REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N Engl J Med [Internet]. 2020. [cited 2021 Jan 28]; Available from 10.1056/NEJMoa2035002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branswell H. For the first time, clinical trial data show Ebola drugs improve survival rates [Internet]. STAT 2019. [cited 2021 Feb 1]; Available from: https://www.statnews.com/2019/08/12/for-the-first-time-clinical-trial-results-show-ebola-drugs-improve-survival-rates/
- 7.Group TPIWG . A randomized, controlled trial of ZMapp for Ebola virus infection. N Engl J Med. 2016;375(15):1448–56. doi: 10.1056/NEJMoa1604330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mulangu S, Dodd LE, Davey RT. A randomized, controlled trial of Ebola Virus disease therapeutics. N Engl J Med. 2019;381(24):2293–303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tusé D, Ku N, Bendandi M, Becerra C, Collins R, Langford N, Sancho SI, López-díaz de Cerio A, Pastor F, Kandzia R, et al. Clinical safety and immunogenicity of Tumor-Targeted, Plant-made Id-KLH conjugate vaccines for follicular lymphoma. BioMed Res Int 2015.;2015:e648143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma JK-C, Drossard J, Lewis D, Altmann F, Boyle J, Christou P, Cole T, Dale P, van Dolleweerd CJ, Isitt V, et al. Regulatory approval and a first-in-human phase I clinical trial of a monoclonal antibody produced in transgenic tobacco plants. Plant Biotechnol J. 2015;13(8):1106–20. doi: 10.1111/pbi.12416. [DOI] [PubMed] [Google Scholar]
- 11.Politch JA, Cu-Uvin S, Moench TR, Tashima KT, Marathe JG, Guthrie KM, Cabral H, Nyhuis T, Brennan M, Zeitlin L, et al. Safety, acceptability, and pharmacokinetics of a monoclonal antibody-based vaginal multipurpose prevention film (MB66): a Phase I randomized trial. PLOS Med. 2021;18(2):e1003495. doi: 10.1371/journal.pmed.1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson DJ, Politch JA, Cone RA, Zeitlin L, Lai SK, Santangelo PJ, Moench TR, Whaley KJ. Engineering monoclonal antibody-based contraception and multipurpose prevention technologies†. Biol Reprod. 2020;103(2):275–85. doi: 10.1093/biolre/ioaa096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCormick AA, Reddy S, Reinl SJ, Cameron TI, Czerwinkski DK, Vojdani F, Hanley KM, Garger SJ, White EL, Novak J, et al. Plant-produced idiotype vaccines for the treatment of non-Hodgkin’s lymphoma: safety and immunogenicity in a phase I clinical study. Proc Natl Acad Sci U S A. 2008;105(29):10131–36. doi: 10.1073/pnas.0803636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swope K, Morton J, Pogue GP, Hume S, Shepherd J, Simpson C, Pauly M, Bratcher B, Whaley KJ, Zeitlin L, et al. Manufacturing Plant-made monoclonal antibodies for research or therapeutic applications. Methods in Enzymology. Academic Press. 2021. doi: 10.1016/bs.mie.2021.05.011. [DOI] [PubMed] [Google Scholar]
- 15.Pogue GP, Vojdani F, Palmer KE, Hiatt E, Hume S, Phelps J, Long L, Bohorova N, Kim D, Pauly M, et al. Production of pharmaceutical-grade recombinant aprotinin and a monoclonal antibody product using plant-based transient expression systems. Plant Biotechnol J. 2010;8(5):638–54. doi: 10.1111/j.1467-7652.2009.00495.x. [DOI] [PubMed] [Google Scholar]
- 16.Pallesen J, Murin CD, de Val N, Cottrell Ca, Hastie KM, Turner HL, Fusco ML, Flyak AI, Zeitlin L, Crowe JE, et al. Structures of Ebola virus GP and sGP in complex with therapeutic antibodies. Nat Microbiol. 2016;1(9):1–9. doi: 10.1038/nmicrobiol.2016.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaner J, Schaack S. Understanding Ebola: the 2014 epidemic. Glob Health. 2016;12(1):53. doi: 10.1186/s12992-016-0194-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514(7520):47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Largent EA. EBOLA and FDA: reviewing the response to the 2014 outbreak, to find lessons for the future. J Law Biosci. 2016;3(3):489–537. doi: 10.1093/jlb/lsw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Q, Davis KR. The potential of plants as a system for the development and production of human biologics. F1000Research. 2016;5:912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shan C, Xia H, Haller SL, Azar SR, Liu Y, Liu J, Muruato AE, Chen R, Rossi SL, Wakamiya M, et al. A Zika virus envelope mutation preceding the 2015 epidemic enhances virulence and fitness for transmission. Proc Natl Acad Sci. 2020;117(33):20190–97. doi: 10.1073/pnas.2005722117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuccori M, Ferraro S, Convertino I, Cappello E, Valdiserra G, Blandizzi C, Maggi F, Focosi D. Anti-SARS-CoV-2 neutralizing monoclonal antibodies: clinical pipeline. mAbs. 2020;12(1):1854149. doi: 10.1080/19420862.2020.1854149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huebbers JW, Buyel JF. On the verge of the market - Plant factories for the automated and standardized production of biopharmaceuticals. Biotechnol Adv. 2021;46:107681. doi: 10.1016/j.biotechadv.2020.107681. [DOI] [PubMed] [Google Scholar]
- 24.Kelley B. Developing therapeutic monoclonal antibodies at pandemic pace. Nat Biotechnol. 2020;38(5):540–45. doi: 10.1038/s41587-020-0512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeyama N, Kiyono H, Yuki Y. Plant-based vaccines for animals and humans: recent advances in technology and clinical trials. Ther Adv Vaccines. 2015;3(5–6):139–54. doi: 10.1177/2051013615613272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strasser R, Stadlmann J, Schähs M, Stiegler G, Quendler H, Mach L, Glössl J, Weterings K, Pabst M, Steinkellner H, et al. Generation of glyco-engineered Nicotiana benthamiana for the production of monoclonal antibodies with a homogeneous human-like N-glycan structure. Plant Biotechnol J. 2008. May;6(4):392–402. doi: 10.1111/j.1467-7652.2008.00330.x. [DOI] [PubMed] [Google Scholar]
- 27.Giritch A, Marillonnet S, Engler C, van EG, Botterman J, Klimyuk V, Gleba Y. Rapid high-yield expression of full-size IgG antibodies in plants coinfected with noncompeting viral vectors. Proc Natl Acad Sci. 2006;103(40):14701–06. doi: 10.1073/pnas.0606631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamorsky KT, Grooms-Williams TW, Husk AS, Bennett LJ, Palmer KE, Matoba N. Efficient single tobamoviral vector-based bioproduction of broadly neutralizing anti-HIV-1 monoclonal antibody VRC01 in Nicotiana benthamiana plants and utility of VRC01 in combination microbicides. Antimicrob Agents Chemother. 2013;57(5):2076–86. doi: 10.1128/AAC.02588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]


