ABSTRACT
The gastrointestinal tract is continuously exposed to trillions of commensal microbes, collectively termed the microbiota, which are environmental stimuli that can direct health and disease within the host. In addition to well-established bacterial sensing pathways, microbial signals are also integrated through epigenetic modifications that calibrate the transcriptional program of host cells without altering the underlying genetic code. Microbiota-sensitive epigenetic changes include modifications to the DNA or histones, as well as regulation of non-coding RNAs. While microbiota-sensitive epigenetic mechanisms have been described in both local intestinal cells and as well in peripheral tissues, further research is required to fully decipher the complex relationship between the host and microbiota. This Review highlights current understandings of epigenetic regulation by gut microbiota and important implications of these findings in guiding therapeutic approaches to prevent or combat diseases driven by impaired microbiota-host interactions.
KEYWORDS: Microbiota, microbiome, epigenetics, epigenome, chromatin, intestine, histone modification, SCFAs, HDAC
Introduction
The mammalian gastrointestinal tract is a complex ecosystem that is densely populated by trillions of commensal bacteria, fungi, archaea, and viruses, collectively termed the microbiota. Research from recent decades has highlighted the importance of the microbiota in the development and maintenance of normal intestinal development and physiology, including digestion and nutrient uptake, metabolism, tissue development, and immunity.1–3 Importantly, though, changes in the composition or abundance of the microbiota have been associated with chronic human conditions such as inflammatory bowel disease (IBD), metabolic disorders and cancer.1,4,5 Commensal microbes interact closely with intestinal epithelial cells (IECs) which exist at the direct interface between the microbiota and underlying host cells, and are therefore optimally poised to respond to resident microbes and their metabolites.6 Microbial signals are equally important in the development and maintenance of innate and adaptive immune cells. In addition to regulating local intestinal cells, the microbiota can also impact peripheral tissues through systemic trafficking of microbial components and microbiota-derived metabolites.7–9 The combined abundance and proximity of commensal microbes to mammalian cells emphasize the microbiota as a large source of environmental stimuli that can shape host cellular function. In support of this notion, epigenetic regulation by the microbiota has emerged as a fundamental interface by which commensal microbes dynamically influence intestinal biology.
Epigenetic regulation linking the microbiota and mammalian cells
Epigenetic modifications enable mammalian cells to alter gene expression without modifying the genetic code, and, therefore, represent a fundamental mechanism by which mammalian cells can adapt their transcriptional program to environmental cues.10 In eukaryotic cells, genetic information encoded within the DNA is packaged around histone proteins and organized into a compact structure called chromatin. In general, epigenetic modifications that promote relaxation of the chromatin (euchromatin) are associated with active gene transcription, whereas condensation of the histone-DNA complexes (heterochromatin) denote inaccessible and silenced regions.11 Epigenetic modifications that dynamically regulate chromatin accessibility include post-translational modification of histones and DNA methylation, which are controlled by the activity of opposing epigenetic modifying enzymes. Furthermore, non-covalent epigenetic mechanisms that control gene expression, such as micro-RNAs (miRNAs) and long-noncoding RNAs (lncRNA), have also been implicated in initiating and sustaining epigenetic change.12 Together, these epigenetic mechanisms influence the cellular transcriptional program by directing accessibility of histone modifiers, transcription factors and transcriptional/translational machinery to the DNA or RNA transcripts. Epigenetic regulation is being increasingly recognized as a potent mechanism through which the microbiota influence host physiology and can occur through multiple potential mechanisms: 1) microbial biosynthesis or metabolism that influence the availability of chemical donors for DNA or histone modifications; 2) regulation of epigenetic modifying enzyme expression and/or activity; or 3) activation of host-cell intrinsic processes that direct epigenetic pathways.
Microbiota-derived metabolites as epigenetic substrates and enzymatic regulators
Epigenetic-modifying enzymes require appropriate substrates to catalyze changes to the chromatin. For instance, DNA/histone methyltransferases (DNMTs/HMTs) and histone acetyltransferases (HATs) generally rely on methyl and acetyl donors, respectively, for their catalytic activity. While many of these donor substrates can be generated from host-intrinsic pathways, the microbiota is being increasingly appreciated as an additional source of these molecules. Indeed, the microbiota can synthesize a plethora of biological compounds including several that serve as epigenetic substrates, co-factors or regulators of epigenetic enzyme activity.13,14 In particular, the intestinal microbiota is known to generate folate and other B vitamins (B2, B12) that donate methyl groups for DNA or histone methylation (Figure 1). Folate is an essential nutrient produced by numerous commensal microbes including probiotic Bifidobacterium and Lactobacillus species that participate in one-carbon metabolism to generate S-adenosylmethionine (SAM), the primary substrate for DNA and histone methylation.15 Commensal microbes can also metabolize dietary methionine into SAM.16 Thus, changes in bacterial composition can influence SAM availability and correspondingly shift the DNA or histone methylation status in the host.
Figure 1.
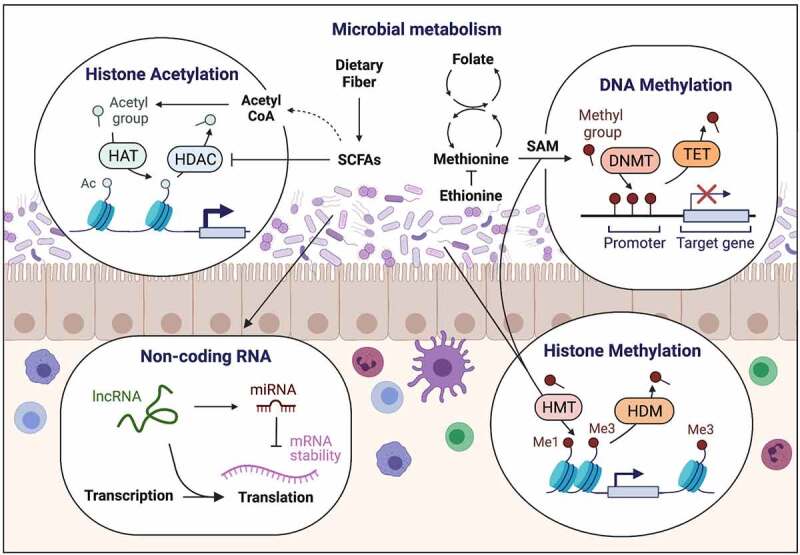
Intestinal microbiota interact with the mammalian epigenome through production of epigenetic substrates or regulators of chromatin-modifying enzymes. This figure was created using BioRender.com.
Short-chain fatty acids (SCFAs) (discussed in more detail below) represent another important group of epigenetically relevant molecules that are exclusively produced by commensal microbes through fermentation of complex non-digestible carbohydrates and fiber. Importantly, SCFAs inhibit deacetylase activity of histone deacetylases (HDACs), resulting in chromatin changes generally associated with increased expression of target genes (Figure 1). Germ-free (GF) mice possess minimal levels of both intestinal and peripheral SCFAs,17–19 and supplementation of SCFAs in GF mice can largely restore transcriptional and epigenetic changes instilled by the microbiota.20 Concurrent with HDAC inhibition, SCFAs also contribute to cellular acetyl coenzyme A levels (acetyl donor), which subsequently affects other TCA intermediates such as a-ketoglutarate, fumarate, and succinate that modulate enzymatic activity of ten-eleven translocation (TET) methylcytosine dioxygenases that are involved in DNA methylation.21
Microbiota direct DNA methylation patterns during homeostasis and inflammation
DNA methylation is an epigenetic modification whereby methyl groups (-CH3) are covalently added to cytosine or adenine bases by DNA methyltransferases (DNMTs) using the donor metabolite SAM. Methylated DNA residues are often located within CpG Islands and are generally associated with gene repression by physically restricting access of regulatory mediators to the DNA, especially when located near gene promoters. Changes in DNA methylation were initial evidence that the microbiota influences the host epigenome. Specifically, DNA methylation of the 5ʹ CpG Island in the toll-like receptor 4 (Tlr4) gene was decreased in large intestinal IECs from GF mice compared to conventionally housed (CNV) mice, resulting in reduced Tlr4 expression and responsiveness to the pathogen-associated molecular pattern lipopolysaccharide (LPS).22 Demethylation was also detected at the chemokine ligand Cxcl16 gene in the colon and lungs of GF mice compared to CNV mice, which is consistent with decreased mucosal invariant natural killer T cell accumulation in absence of the microbiota.23 Microbiota also directed DNA methylation in regulatory T cells (Tregs) by increasing expression of Uhrf1, a DNA methylation adaptor protein that complexes with Dnmt1 and HDAC1.24 T-cell specific deletion of Uhrf1 impaired normal cell cycle gene expression in Tregs and consequently led to spontaneous colitis.
Subsequent studies have since discovered that the microbiota more broadly impacts the mammalian DNA methylome in multiple cell types and tissues during homeostasis and disease. Transcriptome and global DNA methylation analyses on colonic IECs from GF and CNV mice revealed that microbiota-exposure induced DNA hypomethylation and increased expression of anti-bacterial and anti-inflammatory genes.25 This microbiota-dependent demethylation corresponded with increased expression and activity of the DNA demethylase enzymes Tet3 and Dnmt1 in CNV IECs,25,26 and IEC-specific Tet3 deletion led to an increase in global DNA methylation including microbiota-sensitive regions.26 During chemically induced dextran sulfate sodium (DSS) colitis, CNV IECs undergo further hypomethylation, whereas GF mice remain relatively unchanged.25 In humans, DNA methylation in colonic biopsies correlated with microbiota composition, inflammation status, and disease classification of ulcerative colitis (UC) and Crohn’s disease patients.27 UC has been shown to precede the development of colorectal cancer in certain patients, and disease progression has been associated with microbial dysbiosis.28,29 Notably, high Fusobacterium levels were associated with increased DNA methylation at colorectal-cancer related genes in UC patients.30 Colorectal cancer-associated microbiota further correlated with DNA methylation in a large patient cohort and, when colonized into GF mice, promoted epithelial hyperplasia and DNA methylation in the intestine.31 The link between intestinal microbial composition and DNA methylation was also observed in obese human subjects, especially at genes involved in energy regulation in the blood and adipose tissue.32 Collectively, these findings highlight the relationship between microbiota and DNA methylation signatures under homeostatic and pathologic conditions, and that microbiota-associated DNA methylation changes may predict inflammation-driven carcinogenesis in the intestine.
In addition to microbiota-dependent DNA methylation in mammalian cells in adults, recent studies have emphasized the importance of DNA methylation induced by commensal microbes during early life. The neonatal intestine is uniquely sensitive to initial microbial colonization after birth, with the complexity and density of the microbiota continuously expanding into adulthood. This critical early window is important for post-natal tissue development and establishing normal immune responses.33–36 Microbiota-sensitive differential DNA methylation and gene expression were detected in mouse small intestinal IECs during postnatal development and correlated with shifts in transcript levels of DNA methylation/demethylation enzymes Dnmt3a and Tet3.37 Early microbial colonization was also associated with altered DNA methylation in genes associated with immunity, metabolism, and vascular regulation in ileal IECs from CNV and antibiotic-treated preterm piglets.38 Similar changes were demonstrated in intestinal stem cells in the colon, where developmentally established DNA methylation patterns, particularly at the 3′ CpG Islands of glycosylation genes involved in cellular maturation, were substantially blunted in the absence of gut microbiota.39 Further, this was not due to impaired Dnmt1 activity, contrary to findings in mature IECs.25,26 Neonatal colonization also protected against pulmonary and enteric disease by preventing accumulation of invariant natural killer T cells in lungs and intestines of mice by decreasing Cxcl16 DNA methylation and gene expression in those cells.23 In vitro, differential DNA methylation was observed in human fetal and mature intestinal epithelial cell lines exposed to commensal or pathogenic bacteria, and fetal cells were more sensitive to pathogen-induced modifications compared to adult.40 These findings emphasize the importance of microbiota-epigenome crosstalk during early life stages, and that timing, density, and diversity of microbial colonization likely influence this interaction into adulthood.
Host–microbiota interactions through histone modifications
Post-translational modifications of histone proteins represent another central mechanism for microbial regulation of host chromatin. Instead of targeting DNA directly, these types of modifications are typically covalently added to lysine [K] residues on histone tails to influence chromatin conformation and gene expression. While over 20 unique classes of histone modifications have been identified,41 histone acetylation and methylation have been most commonly studied in context of microbial regulation. These modifications are balanced by the activity of opposing classes of epigenetic enzymes (i.e. HATs versus HDACs; histone methyltransferases (HMTs) versus demethylases (HDMs)). Importantly, though, new studies are carving out potential roles for additional histone modifications, such as crotonylation and ethylation, in facilitating crosstalk between the microbiota and host.42,43
Changes in chromatin accessibility as a result of microbiota-induced histone modifications has been described in multiple tissues and cell populations. For instance, gut microbiota was found to globally alter histone H3 and H4 acetylation and methylation in various tissues (colon, liver, and adipose tissue) in a diet-dependent manner, and supplementation of SCFAs to GF mice partially restored microbiota-sensitive histone modifications and gene expression.20 A recent study also showed that cecal microbiota transfer from dams to pups exposed to antibiotics early in life restored histone modifications in the ileum and liver.44 Specifically, this microbiota-transfer led to lysine 27 (K27) deacetylation and trimethylation, potentially through enhanced Jumanji family-related lysine demethylase D3 (Jmjd3/Kdm6b) activity in response to Tlr2 induction, indicating that microbiota prevents expression of antibiotic-induced genes. In the intestinal epithelium, differential H3K4me3, a histone methylation associated with active transcription, was also identified at genes in ileal IECs from GF compared to CNV mice, a subset of which overlapped with H3K4me3 profiles unique to newly diagnosed IBD patients compared to healthy controls.45 This modification was also found to be regulated specifically at Wnt-responsive gene promoters by microbiota-derived gallic acid in a colorectal cancer mouse model,46 supporting that the microbiota not only interact with host chromatin through histone modifications at steady-state, but also in context of inflammation and disease.
Consistent with global changes in histone acetylation driven by the microbiota, studies have demonstrated that HDAC enzymes are involved in maintaining microbiota-dependent intestinal homeostasis. In particular, HDAC3, a class I histone deacetylase that is highly expressed in the intestinal epithelium, is sensitive to microbial signals. IEC-specific deletion of HDAC3 (HDAC3∆IEC) increased IEC-H3K9 acetylation and gene expression, altered Paneth cell homeostasis and intestinal barrier function, and worsened response to DSS-induced intestinal inflammation.47 Importantly, abnormalities in HDAC3∆IEC mice were only evident in CNV mice, and not in GF mice, indicating that HDAC3 is essential to maintain normal commensal-host relationships. In context of a high-fat diet, alterations in microbiota-derived signals and disruption of intestinal epithelial HDAC3 prevented the development of obesity.48,49 Consistent with these studies, increased enrichment of H3K27Ac, a marker of active enhancers, was described at metabolic genes in colonic IECs in response to obesity-associated microbiota transfer compared to control microbiota recipients.50 Microbiota also improved defense against the bacterial pathogen Citrobacter rodentium through microbiota-dependent regulation of IEC-intrinsic HDAC3.51,52
The most abundant SCFAs in the mammalian intestinal lumen include acetate, propionate, and butyrate, which are found at molar ratios of 60:20:20 in the intestinal tract and can reach peak concentrations of up to 100 mM.53 SCFAs inhibit HDACs, often resulting in increased histone acetylation at direct targets (Figure 1). Despite the abundance of HDAC-inhibiting SCFAs in the gut, recent work found that HDAC3 activity was elevated in IECs from CNV mice compared to GF mice due to production of an HDAC-activating metabolite, inositol triphosphate (IP3), by intestinal commensal bacteria.54 Consistent with increased HDAC activity, IECs from mice mono-colonized with IP3-producing bacteria exhibited decreased H3K9Ac at HDAC3 target genes relative to mice with lower IP3 in the intestine. In intestinal epithelial stem cells, which reside at the base of the crypts and are anatomically protected from SCFA exposure, administration of butyrate directly to these cells inhibited HDAC activity and increased H3K9 and K27 acetylation, which coincided with impaired proliferation and response to injury.55 Thus, HDAC3 regulation by the microbiota occurs in multiple epithelial cell populations and reflects more than SCFA-mediated inhibition alone, and instead relies on integration of multiple microbiota-derived signals. Additional studies are needed to determine whether other histone modifiers known to control intestinal physiology, such as HDAC1 and HDAC2,56–58 are similarly regulated by the microbiota.
Lysine crotonylation is a relatively less studied histone modification that is often enriched at active promotors and enhancers in mammalian cells.59 While the addition or removal of crotonyl motifs can be catalyzed by specialized histone crotonyltransferases and decrotonylases, HATs and HDACs have also been reported to exhibit histone crotonyl-modifying activity.60 Consistent with this finding, Histone H3 and H4 crotonylation levels (H3K18Cr, H3K8Cr, and H4K8Cr) were significantly decreased in the colon of mice treated with antibiotics and administering the HDAC inhibitor butyrate promoted histone crotonylation.42 Taken together, these studies demonstrate that the microbiota stimulate multiple types of histone modifications and regulate activity of histone-modifying enzymes to calibrate local and extra-intestinal chromatin landscapes.
Immune cells similarly undergo broad histone modifications in response to microbial colonization. Non-mucosal mononuclear phagocytes (macrophages and dendritic cells) require microbial signals to epigenetically and transcriptionally activate genes involved in interferon (IFN) signaling and initiating normal T cell responses.61,62 Specifically, GF or antibiotic-treated mice have decreased H3K4me3 (activating) at genes involved in pro-inflammatory responses including type I IFNs and increased H3K27me3 (repressive) at metabolic pathways compared to microbiota-exposed cells. The microbiota further maintain this anti-inflammatory phenotype in macrophages through HDAC3-mediated histone deacetylation of the pro-inflammatory cytokine IL-12β.63 Similar changes in chromatin accessibility defined by differential H3K4me2 enrichment were shown at lineage-defining regulatory elements in innate lymphoid cells (ILCs) to favor an ILC3 phenotype over ILC1 or ILC2 in response to the microbiota.64 Ethionine was recently discovered as a novel microbiota-derived metabolite produced by the commensal bacterium Lactobacillus reuteri through a 2-carbon folate cycle.43 In agreement with previous reports that ethionine inhibits histone methylation,65 mass spectrometric analyses revealed that human monocytic THP-1 cells treated with ethionine preferentially incorporated ethyl groups into lysine residues of histone H3 (K9/K10/K26) instead of methyl groups. Moreover, ethionine-treated monocytes failed to activate NF-kB signaling or TNF-α expression in response to LPS treatment.43
Similar to IECs, distinct bacterial populations have also been shown to modulate histone modifications in immune cells. Colonization with the commensal bacteria segmented filamentous bacteria (SFB) or administration of SFB-induced serum amyloid A (SAA) increased expression of the demethylase Jmjd3 and increased global H3K27me3 in bone marrow-derived cells, resulting in improved protection against Entamoeba histolytica infection.66 Clostridium scindens colonization in mice promoted expression of the same H3K27-targeting demethylase in bone marrow-derived granulocyte-monocyte progenitors (GMPs) and increased H3K4me3 (activating) and decreased H3K27Ac (repressive) in the promotor regions of granulopoiesis genes C/EBPA and C/EBPB, respectively, leading to increased neutrophils and enhanced protection against E. histolytica infection.67 C. scindens was also demonstrated to metabolize the bile salt cholate into deoxycholate, and administration of deoxycholate increased Jmjd3 expression in GMPs.67 Collectively, these studies demonstrate the intestinal microbiota or individual commensal bacterial populations alter the chromatin landscape in multiple immune cell types by directing covalent modifications of histones or regulating the activity of histone-modifying enzymes.
In addition to global changes in the histone landscape at steady state, recent insights reveal that the microbiota exhibits a circadian rhythmicity that programs the host transcriptome and epigenome at the histone modification level.49,68 Diurnal cycles in microbiota composition and abundance that occurred throughout the day in mice corresponded to similar rhythmicity in histone modifications marking active promoters (H3K27Ac and H3K4me3), enhancer activity (H3K27Ac and H3K4me2), and gene expression of circadian genes in colonic IECs. Oscillations in chromatin landscape and transcriptional responses were largely lost upon microbiota-depletion using antibiotics or comparing GF mice, indicating that normal oscillations of the microbiota and microbiota-mediated control of the circadian transcriptome is necessary to maintain normal circadian cycles.68 Similar cycling patterns in H3K27 and K9 acetylation were detected in small intestines of CNV, but not GF mice that was attributed to a lack of oscillating expression of the histone deacetylase HDAC3 in absence of commensal microbes.49 Interestingly, mono-colonization of GF mice with Bacteroides thetaiotamicron was sufficient to increase HDAC3 expression,49 re-iterating that specific commensal species can have potent effects on the host epigenome, and that the natural circadian rhythmicity of the microbiota adds an additional layer of complexity in this regulation.
Regulation of immune cells by SCFAs
SCFAs have become well-appreciated intermediary between the microbiota and mucosal immune cell populations. Importantly, regulation by SCFAs extends to innate and adaptive immune compartments locally in the intestine as well as systemically. This has been described to occur through a combination of HDAC inhibition and G-protein coupled receptor (GPR) activation.69,70 Treatment of intestinal or bone marrow-derived macrophages with butyrate inhibited HDAC activity and increased histone H3 acetylation at secondary response genes including IL-6, IL-12, and mediators of STAT6 signaling.19,71 However, In contrast to the predicted inhibitory effect of SCFAs on HDACs, expression of these same genes were instead decreased in response to butyrate, reflecting an anti-inflammatory M2 macrophage state that attenuated intestinal inflammation in response to DSS-induced colitis.19,71 Furthermore, butyrate, and to a lesser degree propionate, improved defense against enteric pathogens by promoting monocyte to macrophage differentiation and activating an antimicrobial transcriptional program through HDAC3 inhibition.72 SCFAs also promoted IL-22 histone acetylation and production by ILCs and CD4+ T cells through a combination of GPR41 activation and HDAC inhibition, resulting in enhanced protection from intestinal inflammation during bacterial infection.73 Butyrate was further shown to inhibit HDACs and activate GPR109A in colonic macrophages and dendritic cells, enabling them to promote anti-inflammatory regulatory T cells (Tregs).18,74,75 Thus, SCFAs control epigenetic mechanisms and transcriptional responses in immune cells to support an overall anti-inflammatory phenotype during chemically induced colitis and pathogenic infection.
Tregs are defined by their expression of the transcription factor Foxp3 and act to suppress aberrant inflammatory and effector responses, thereby playing a critical role in immune homeostasis and maintaining tolerance to the microbiota. Colonization with Clostridial species, or treatment with Clostridia-derived butyrate, increased the frequency of circulating Tregs by inducing acetylation on histone H3 within regulatory regions of the Foxp3 locus.17 Post-transcriptional acetylation of Foxp3 proteins also occurred in peripheral CD4+ T cells, thereby enhancing expression and protein stability.18 In agreement with these findings, treatment with the SCFA propionate, or deletion of HDAC3, HDAC6, or HDAC9 in mice resulted in both increased H3K9 acetylation at the Foxp3 locus and enhanced Foxp3 protein stability.76–78 Acetate, another microbiota-derived SCFA, also inhibited HDAC activity and increased overall acetylation of histones H3 (K9) and H4 (K5/8/16) in CD3/CD28 activated primary human CD4+ T cells,79 although HDAC inhibition by acetate is somewhat controversial.18,80,81
In contrast to physiological concentrations of butyrate, elevated butyrate levels induced expression and H3 acetylation of Th1-associated genes including IFNγ and Tbx21 (encodes T-bet) in T cells under Treg-inducing conditions, while expression of Th17- and Th2-promoting transcription factors RORγt and Gata3, respectively, were not induced.82 Moreover, higher concentrations of butyrate administered to GF mice resulted in elevated IFNγ production and exacerbated disease during DSS-induced colitis. Another study similarly found that butyrate promoted Th1, but suppressed Th17 polarization from naïve T cells in culture and in vivo by inhibiting HDAC activity independently from GPR43 activation to induce expression of T-bet and the anti-inflammatory cytokine IL-10.83 Congruent with enhanced IL-10 in response to butyrate treatment of bulk CD4+ T cells, microbiota-specific T cells isolated from butyrate-treated CBir1 transgenic mice caused less severe colitis in Rag−/− mice compared to control T cells.83,84 Faecalibacterium prausnitzii-derived butyrate similarly suppressed Th17 differentiation and simultaneously promoted Tregs by inhibiting HDAC3 and HDAC1 deacetylase activity in vitro and in vivo.85,86 This is similar to HDAC3 inhibition in IECs in response to F. prausnitzii colonization.54 Besides butyrate, Pentanoate, a 5-carbon SCFA, also reduced IL-17A production by CD4+ T cells through HDACs, and induced IL-10 gene histone acetylation and expression in CD4+ T cells and regulatory B cells, thereby dampening T-cell mediated autoimmune responses.87 Thus, SCFAs exert context-and concentration-dependent effects that generally promote a low Th17/high Treg phenotype that is protective against intestinal inflammation.
CD8+ cytotoxic T lymphocytes (CTLs) defend against intracellular pathogens and possess cancer-targeting abilities. Recent findings demonstrate that butyrate, and to some degree propionate, increased effector molecule expression including CD25, IFN-γ and TNF-α in splenic and mesenteric CD8+ T cells independently from GPR41 or GPR43 activation, resulting in enhanced anti-tumor activity.88 Instead, this regulation was driven by HDAC inhibition with corresponding increases in histone H4 acetylation directly at the promotors of IFNγ and Tbx21, a similar result described for CD4+ T cells exposed to butyrate under Treg, Th1- or Th2-polarizing conditions.82,89 It was subsequently also discovered that butyrate, and Megasphaera massiliensis-derived pentanoate, improved the anti-tumor reactivity of antigen-specific CD8+ T cells by increasing their production of effector cytokines IFNγ and TNF-α through HDAC inhibition.90 Additionally, studies showed that increased SCFAs from high-fiber feeding in mice encouraged CD8+ T cell activation and later transition into memory cells in context of viral infections.91,92
Microbiota-derived SCFAs also supported intestinal B cell differentiation and antibody production by inhibiting HDAC activity leading to increased histone acetylation (H3K9Ac) and gene expression of multiple genes associated with B cell function.93 Similar effects were observed in human and mouse B cells treated specifically with butyrate and propionate that dose-dependently counteracted HDAC-mediated H3K9 deacetylation and transcriptional repression of miRNAs that target genes involved in B cell differentiation.94 Furthermore, butyrate and propionate restricted normal B cell intrinsic functions including immunoglobulin class switching and somatic hypermutation. Thus, microbiota-derived SCFAs have potent immunomodulatory effects on both innate and adaptive immune cells in the host that actively maintain homeostasis and dampen inflammation in the intestine.
Non-covalent modifications
Epigenetic regulation by non-coding RNAs (ncRNAs), which are RNA molecules that do not encode a functional protein, has recently been linked to microbiota–host interactions. ncRNAs are functionally categorized based on nucleotide length, and recent studies have shown that ncRNAs can regulate gene expression at the gene or chromosome level. Long non-coding RNAs (lncRNAs) typically contain over 200 nucleotides and regulate gene expression by acting directly as DNA scaffolding to modify chromatin interactions or by complexing with chromatin modifiers and/or transcriptional machinery.95 Initial investigation into commensal-regulated lncRNAs focused on characterizing these transcripts in intestinal epithelial tissues from GF, CNV, or GF mice mono-colonized with E. coli or E. coli expressing a bile salt hydrolase.96 These analyses identified hundreds of lncRNA differentially regulated between IECs from GF and CNV mice. Interestingly, expression of only six lncRNAs overlapped between the different colonizations, and lncRNA signatures were able to distinguish mice with unique microbial compositions, demonstrating a degree of commensal species-specificity in this regulation. Microbiota-induced lncRNAs and were also found to be highly expressed in the spleen and thymus, suggesting that microbiota-dependent lncRNAs may be involved in host immune regulation.96 Global transcriptional profiling of lncRNAs in GF and CNV mice further revealed tissue-specific modulation of lncRNAs in the intestine and peripheral tissues in response to the microbiota.97 Furthermore, predicted interactions between lncRNAs and target protein-coding RNAs were functionally enriched for genes related to nutrient absorption and metabolic pathways across different tissues. Thus, the microbiota not only regulates expression of lncRNAs in the intestine and immune organs, but also remotely in metabolic tissues. Additional studies are needed to further elucidate the tissue-specific and functional relevance of microbiota-sensitive lncRNAs and mechanisms underlying their regulation.
miRNAs are short, single-stranded ncRNAs (~22 nucleotides) that are abundantly expressed in mammalian cells and can also circulate systemically.98 Unlike lncRNAs that target the chromatin, miRNAs primarily interfere with post-transcriptional gene expression by base-pairing with complementary protein-coding mRNA sequences. miRNAs can also occasionally mediate DNA methylation or histone modifications at gene promoters to influence target gene expression,99,100 however this has yet to be explored in context of the microbiota. miR-181a and miR-181b, two miRNAs reported to control inflammation- and obesity-associated pathologies, were recently found to be significantly upregulated in epididymal white adipose tissue from specific pathogen-free (SPF) mice as compared to GF mice.101 It was also recently shown that transfer of maternal cecal contents into antibiotic treated pups restored global ileal epithelial miRNA expression profiles including miRNAs that target microbiota-sensitive genes involved in regulating innate and adaptive immunity such as CD44, Tlr2, and Reg3g.44 Taken together, these studies indicate that further investigation into miRNA regulation by the microbiota has the potential to expand our understanding of microbiota-induced control of host physiology and disease beyond epigenetic regulation of histones and the DNA.
Conclusions and future perspectives
Recent research has highlighted that epigenetic modification of the host by commensal microbes represents a broad and fundamental level of regulation during early life development, homeostasis, and disease. Indeed, studies have shown that host cells integrate beneficial microbial cues that mediate these processes through epigenetic pathways, and that perturbation of the microbiota–host relationship associated with pathologies, including IBD and colorectal cancer, correlate with altered epigenetic patterning. However, our mechanistic understanding of microbiota–host epigenome interactions underlying this regulation is still in need of deeper investigation.
While much emphasis has been placed on SCFAs, implementation of multi-omics approaches has revealed that the microbiota is responsible for the production of many more biologically active metabolites that may facilitate epigenetic modification. Future work will determine the involvement of more recently identified microbiota-derived molecules on the host epigenome. Furthermore, comparative metagenomic microbial analyses in humans and the discovery of commensal species-specific epigenetic modifications indicate that enrichment for certain microbial species has the potential to modulate unique gene expression signatures. Recent associations between microbiota composition and epigenetic modifications in IBD, cancer, and obesity also indicate that epigenetic patterns could serve as diagnostic tools to link genetic predisposition and microbial dysbiosis with disease pathogenesis. Improved chromatin analytical techniques, including the ability to examine epigenetic marks on single cells and limited cell numbers could reveal epigenetic heterogeneity within a cell population or in rare cell types in response to microbiota. Finally, genetic manipulation of epigenetic-modifying bacteria or controlling microbial-produced epigenetic substrate levels could represent a new avenue for pre- or probiotic approaches to provide a more targeted, localized method to control epigenetic enzymes in the intestine.
Acknowledgments
We thank members of the Alenghat lab for discussions and critical reading of this Review.
Funding Statement
This work is supported by the National Institutes of Health (DK114123, DK116868 to T.A), Cardell Fellowship to V.W., and a Kenneth Rainin Foundation award to T.A. T.A. holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Fan Y, Pedersen O.. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2020;19(1):55–14. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- 2.Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30(6):492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268. doi: 10.1126/SCIENCE.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat Med. 2019;25(3):377–388. doi: 10.1038/s41591-019-0377-7. [DOI] [PubMed] [Google Scholar]
- 5.Caruso R, Lo BC, Núñez G. Host–microbiota interactions in inflammatory bowel disease. Nat Rev Immunol. 2020;20(7):411–426. doi: 10.1038/s41577-019-0268-7. [DOI] [PubMed] [Google Scholar]
- 6.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14(3):141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 7.Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17(4):223–237. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 8.Matsumoto M, Kunisawa A, Hattori T, Kawana S, Kitada Y, Tamada H, Kawano S, Hayakawa Y, Iida J, Fukusaki E, et al. Free D-amino acids produced by commensal bacteria in the colonic lumen. Sci Rep. 2018;8(1):17915. doi: 10.1038/s41598-018-36244-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106(10):3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17(8):487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 11.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11(5):384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 12.Cech TR, Steitz JA. The noncoding RNA revolution—trashing old rules to forge new Ones. Cell. 2014;157(1):77–94. doi: 10.1016/J.CELL.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Miro-Blanch J, Yanes O. Epigenetic regulation at the interplay between gut microbiota and host metabolism. Front Genet. 2019;10(JUL). doi: 10.3389/fgene.2019.00638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shock T, Badang L, Ferguson B, Martinez-Guryn K. The interplay between diet, gut microbes, and host epigenetics in health and disease. J Nutr Biochem. 2021:95. doi: 10.1016/J.JNUTBIO.2021.108631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossi M, Amaretti A, Raimondi S. Folate production by probiotic bacteria. Nutrients. 2011;3(1):118–134. doi: 10.3390/nu3010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Wang Z, Cai H, Zhou C . Progress in the microbial production of S-adenosyl-L-methionine. World J Microbiol Biotechnol. 2016. doi: 10.1007/s11274-016-2102-8. [DOI] [PubMed] [Google Scholar]
- 17.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 18.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014;111(6):2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-Wilt GA, Rabaglia ME, Keller MP, Attie AD, Rey FE, Denu JM, et al. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell. 2016;64(5):982–992. doi: 10.1016/j.molcel.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Etchegaray J-P, Mostoslavsky R. Interplay between metabolism and epigenetics: a nuclear adaptation to environmental changes. Mol Cell. 2016;62(5):695. doi: 10.1016/J.MOLCEL.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi K, Sugi Y, Hosono A, Kaminogawa S. Epigenetic regulation of TLR4 gene expression in intestinal epithelial cells for the maintenance of intestinal homeostasis. J Immunol. 2009;183(10):6522–6529. doi: 10.4049/jimmunol.0901271. [DOI] [PubMed] [Google Scholar]
- 23.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science (80-). 2012;336(6080):489–493. doi: 10.1126/SCIENCE.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obata Y, Furusawa Y, Endo TA, Sharif J, Takahashi D, Atarashi K, Nakayama M, Onawa S, Fujimura Y, Takahashi M, et al. The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nat Immunol. 2014;15(6):571–579. doi: 10.1038/ni.2886. [DOI] [PubMed] [Google Scholar]
- 25.Ansari I, Raddatz G, Gutekunst J, Ridnik M, Cohen D, Abu-Remaileh M, Tuganbaev T, Shapiro H, Pikarsky E, Elinav E, et al. The microbiota programs DNA methylation to control intestinal homeostasis and inflammation. Nat Microbiol. 2020;5(4):610–619. doi: 10.1038/s41564-019-0659-3. [DOI] [PubMed] [Google Scholar]
- 26.Poupeau A, Garde C, Sulek K, Citirikkaya K, Treebak JT, Arumugam M, Simar D, Olofsson LE, Bäckhed F, Barrès R, et al. Genes controlling the activation of natural killer lymphocytes are epigenetically remodeled in intestinal cells from germ-free mice. FASEB J. 2019;33(2):2719–2731. doi: 10.1096/FJ.201800787R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan FJ, Ahern AM, Fitzgerald RS, Laserna-Mendieta EJ, Power EM, Clooney AG, O’Donoghue KW, McMurdie PJ, Iwai S, Crits-Christoph A, et al. Colonic microbiota is associated with inflammation and host epigenomic alterations in inflammatory bowel disease. Nat Commun. 2020;11(1):1–12. doi: 10.1038/s41467-020-15342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eaden JA, Abrams KR, Mayberry JF. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut. 2001;48(4):526–535. doi: 10.1136/gut.48.4.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linson EA, Hanauer SB. Epidemiology of colorectal cancer in inflammatory bowel disease – the evolving landscape. Curr Gastroenterol Rep. 2021;23(9):16. doi: 10.1007/S11894-021-00816-3. [DOI] [PubMed] [Google Scholar]
- 30.Tahara T, Hirata I, Nakano N, Tahara S, Horiguchi N, Kawamura T, Okubo M, Ishizuka T, Yamada H, Yoshida D, et al. Potential link between Fusobacterium enrichment and DNA methylation accumulation in the inflammatory colonic mucosa in ulcerative colitis. Oncotarget. 2017;8(37):61917–61926. doi: 10.18632/ONCOTARGET.18716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sobhani I, Bergsten E, Couffin S, Amiot A, Nebbad B, Barau C, De’angelis N, Rabot S, Canoui-Poitrine F, Mestivier D, et al. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc Natl Acad Sci. 2019;116(48):24285–24295. doi: 10.1073/PNAS.1912129116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramos-Molina B, Sánchez-Alcoholado L, Cabrera-Mulero A, Lopez-Dominguez R, Carmona-Saez P, Garcia-Fuentes E, Moreno-Indias I, Tinahones FJ . Gut microbiota composition is associated with the global DNA methylation pattern in obesity. Front Genet. 2019;(JUL):613. doi: 10.3389/FGENE.2019.00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pennisi E. The right gut microbes help infants grow. Science (80-). 2016;351(6275):802. doi: 10.1126/SCIENCE.351.6275.802. [DOI] [PubMed] [Google Scholar]
- 34.Schwarzer M, Makki K, Storelli G, Machuca-Gayet I, Srutkova D, Hermanova P, Martino ME, Balmand S, Hudcovic T, Heddi A, et al. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science (80-). 2016;351(6275):854–857. doi: 10.1126/SCIENCE.AAD8588. [DOI] [PubMed] [Google Scholar]
- 35.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science (80-). 2016;352(6285):539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gollwitzer ES, Marsland BJ. Impact of early-life exposures on immune maturation and susceptibility to disease. Trends Immunol. 2015;36(11):684–696. doi: 10.1016/j.it.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 37.Pan W-H, Sommer F, Falk-Paulsen M, Ulas T, Best P, Fazio A, Kachroo P, Luzius A, Jentzsch M, Rehman A, et al. Exposure to the gut microbiota drives distinct methylome and transcriptome changes in intestinal epithelial cells during postnatal development. Genome Med. 2018;10(1):1–15. doi: 10.1186/S13073-018-0534-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan X, Gong D, Nguyen DN, Zhang X, Hu Q, Lu H, Fredholm M, Sangild PT, Gao F. Early microbial colonization affects DNA methylation of genes related to intestinal immunity and metabolism in preterm pigs. DNA Res. 2018;25(3):287–296. doi: 10.1093/DNARES/DSY001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu DH, Gadkari M, Zhou Q, Yu S, Gao N, Guan Y, Schady D, Roshan TN, Chen M-H, Laritsky E, et al. Postnatal epigenetic regulation of intestinal stem cells requires DNA methylation and is guided by the microbiome. Genome Biol. 2015;16(1):1–16. doi: 10.1186/s13059-015-0763-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cortese R, Lu L, Yu Y, Ruden D, Claud EC. Epigenome-Microbiome crosstalk: a potential new paradigm influencing neonatal susceptibility to disease. Epigenetics. 2016;11(3):205–215. doi: 10.1080/15592294.2016.1155011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Y, Garcia BA. Comprehensive catalog of currently documented histone modifications. Cold Spring Harb Perspect Biol. 2015;7(9):a025064. doi: 10.1101/CSHPERSPECT.A025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fellows R, Denizot J, Stellato C, Cuomo A, Jain P, Stoyanova E, Balázsi S, Hajnády Z, Liebert A, Kazakevych J, et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat Commun. 2018;9(1):105. doi: 10.1038/s41467-017-02651-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Röth D, Chiang AJ, Hu W, Gugiu GB, Morra CN, Versalovic J, Kalkum M. Two-carbon folate cycle of commensal Lactobacillus reuteri 6475 gives rise to immunomodulatory ethionine, a source for histone ethylation. FASEB J. 2019;33(3):3536. doi: 10.1096/FJ.201801848R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X-S, Yin YS, Wang J, Battaglia T, Krautkramer K, Li WV, Li J, Brown M, Zhang M, Badri MH, et al. Maternal cecal microbiota transfer rescues early-life antibiotic-induced enhancement of type 1 diabetes in mice. Cell Host Microbe. 2021;29(8):1249–1265.e9. doi: 10.1016/J.CHOM.2021.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelly D, Kotliar M, Woo V, Jagannathan S, Whitt J, Moncivaiz J, Aronow BJ, Dubinsky MC, Hyams JS, Markowitz JF, et al. Microbiota-sensitive epigenetic signature predicts inflammation in Crohn’s disease. JCI Insight. 2018;3(18). doi: 10.1172/jci.insight.122104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kadosh E, Snir-Alkalay I, Venkatachalam A, May S, Lasry A, Elyada E, Zinger A, Shaham M, Vaalani G, Mernberger M, et al. The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature. 2020;586(7827):133–138. doi: 10.1038/s41586-020-2541-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alenghat T, Osborne LC, Saenz SA, Kobuley D, Ziegler CGK, Mullican SE, Choi I, Grunberg S, Sinha R, Wynosky-Dolfi M, et al. Histone deacetylase 3 coordinates commensal-bacteria-dependent intestinal homeostasis. Nature. 2013;504(7478):153–157. doi: 10.1038/nature12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitt J, Woo V, Lee P, Moncivaiz J, Haberman Y, Denson L, Tso P, Alenghat T. Disruption of epithelial HDAC3 in intestine prevents diet-induced obesity in mice. Gastroenterology. 2018;155(2):501–513. doi: 10.1053/j.gastro.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuang Z, Wang Y, Li Y, Ye C, Ruhn KA, Behrendt CL, Olson EN, Hooper LV. The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science (80-). 2019;365(6460):1428–1434. doi: 10.1126/SCIENCE.AAW3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin Y, Roberts JD, Grimm SA, Lih FB, Deterding LJ, Li R, Chrysovergis K, Wade PA. An obesity-associated gut microbiome reprograms the intestinal epigenome and leads to altered colonic gene expression. Genome Biol. 2018;19(1):1–14. doi: 10.1186/S13059-018-1389-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Navabi N, Whitt J, Wu S, Woo V, Moncivaiz J, Jordan MB, Vallance BA, Way SS, Alenghat T . Epithelial histone deacetylase 3 instructs intestinal immunity by coordinating local lymphocyte activation. Cell Rep. 2017;19(6):1165–1175. doi: 10.1016/j.celrep.2017.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woo V, Eshleman EM, Rice T, Whitt J, Vallance BA, Alenghat T. Microbiota inhibit epithelial pathogen adherence by epigenetically regulating C-type lectin expression. Front Immunol. 2019;10(May):1–10. doi: 10.3389/fimmu.2019.00928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28(10):1221. doi: 10.1136/GUT.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu S, Hashimoto-Hill S, Woo V, Eshleman EM, Whitt J, Engleman L, Karns R, Denson LA, Haslam DB, Alenghat T, et al. Microbiota-derived metabolite promotes HDAC3 activity in the gut. Nature. 2020;586(7827):108–112. doi: 10.1038/s41586-020-2604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaiko GE, Ryu SH, Koues OI, Collins PL, Solnica-Krezel L, Pearce EJ, Pearce EL, Oltz EM, Stappenbeck TS. The colonic Crypt protects stem cells from microbiota-derived metabolites. Cell. 2016;165(7):1708–1720. doi: 10.1016/j.cell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turgeon N, Gagné JM, Blais M, Gendron F-P, Boudreau F, Asselin C. The acetylome regulators Hdac1 and Hdac2 differently modulate intestinal epithelial cell dependent homeostatic responses in experimental colitis. Am J Physiol Liver Physiol. 2014;306(7):G594–G605. doi: 10.1152/ajpgi.00393.2013. [DOI] [PubMed] [Google Scholar]
- 57.Gonneaud A, Turgeon N, Boudreau F, Perreault N, Rivard N, Asselin C. Distinct roles for intestinal epithelial cell-specific Hdac1 and Hdac2 in the regulation of murine intestinal homeostasis. J Cell Physiol. 2016;231(2):436–448. doi: 10.1002/jcp.25090. [DOI] [PubMed] [Google Scholar]
- 58.Zimberlin CD, Lancini C, Sno R, Rosekrans SL, McLean CM, Vlaming H, van den Brink GR, Bots M, Medema JP, Dannenberg JH . HDAC1 and HDAC2 collectively regulate intestinal stem cell homeostasis. FASEB J. 2015;29(5):2070–2080. doi: 10.1096/fj.14-257931. [DOI] [PubMed] [Google Scholar]
- 59.Tan M, Luo H, Lee S, Jin F, Yang J, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wan J, Liu H, Chu J, Zhang H. Functions and mechanisms of lysine crotonylation. J Cell Mol Med. 2019;23(11):7163–7169. doi: 10.1111/JCMM.14650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schaupp L, Muth S, Rogell L, Kofoed-Branzk M, Melchior F, Lienenklaus S, Ganal-Vonarburg SC, Klein M, Guendel F, Hain T, et al. Microbiota-induced type i interferons instruct a poised basal state of dendritic cells. Cell. 2020;181(5):1080–1096.e19. doi: 10.1016/j.cell.2020.04.022. [DOI] [PubMed] [Google Scholar]
- 62.Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, Lienenklaus S, Weiss S, Staeheli P, Aichele P, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37(1):171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 63.Kobayashi T, Matsuoka K, Sheikh SZ, Russo SM, Mishima Y, Collins C, deZoeten EF, Karp CL, Ting JPY, Sartor RB, et al. IL-10 regulates Il12b expression via histone deacetylation: implications for intestinal macrophage homeostasis. J Immunol. 2012;189(4):1792. doi: 10.4049/JIMMUNOL.1200042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gury-benari M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D, Levy M, Salame TM, Weiner A, David E, et al. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell. 2016;166(5):1231–1246.e13. doi: 10.1016/j.cell.2016.07.043. [DOI] [PubMed] [Google Scholar]
- 65.Tuck MT, Cox R. Ethionine inhibits in vivo methylation of nuclear proteins. Carcinogenesis. 1982;3(4):431–434. [Accessed 2021 Aug 6]. http://carcin.oxfordjournals.org/. [DOI] [PubMed] [Google Scholar]
- 66.Burgess SL, Saleh M, Cowardin CA, Buonomo E, Noor Z, Watanabe K, Abhyankar M, Lajoie S, Wills-Karp M, Petri WA, et al. Role of serum amyloid A, granulocyte-macrophage colony-stimulating factor, and bone marrow granulocyte-monocyte precursor expansion in segmented filamentous bacterium-mediated protection from entamoeba histolytica. Infect Immun. 2016;84(10):2824–2832. doi: 10.1128/iai.00316-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burgess SL, Leslie JL, Uddin J, Oakland DN, Gilchrist C, Moreau GB, Watanabe K, Saleh M, Simpson M, Thompson BA, et al. Gut microbiome communication with bone marrow regulates susceptibility to amebiasis. J Clin Invest. 2020;130(8):4019–4024. doi: 10.1172/JCI133605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thaiss C, Levy M, Korem T, Dohnalová L, Shapiro H, Jaitin DA, David E, Winter DR, Gury-benari M, Tatirovsky E. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell. 2016;167(6):1495–1510. doi: 10.1016/j.cell.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 69.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu DI, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, Maruya M, Ian Mckenzie C, Hijikata A, Wong C, et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun. 2015;6(1):1–15. doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- 71.Ji J, Shu D, Zheng M, Wang J, Luo C, Wang Y, Guo F, Zou X, Lv X, Li Y, et al. Microbial metabolite butyrate facilitates M2 macrophage polarization and function. Sci Rep. 2016;6(1):1–10. doi: 10.1038/srep24838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schulthess J, Pandey S, Capitani M, Rue-Albrecht KC, Arnold I, Franchini F, Chomka A, Ilott NE, Johnston DGW, Pires E, et al. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity. 2019;50(2):432–445.e7. doi: 10.1016/j.immuni.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang W, Yu T, Huang X, Bilotta AJ, Xu L, Lu Y, Sun J, Pan F, Zhou J, Zhang W, et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat Commun. 2020;11(1):1–18. doi: 10.1038/s41467-020-18262-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad P, Manicassamy S, Munn D, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40(1):128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaisar MMM, Pelgrom LR, van der Ham AJ, Yazdanbakhsh M, Everts B. Butyrate Conditions Human dendritic cells to prime type 1 regulatory T cells via both histone deacetylase inhibition and G protein-coupled receptor 109A signaling. Front Immunol 8. 2017;(OCT):1429. doi: 10.3389/FIMMU.2017.01429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic T reg cell homeostasis. Science (80-). 2013;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beier UH, Wang L, Han R, Akimova T, Liu Y, Hancock WW. Histone deacetylases 6 and 9 and sirtuin-1 control Foxp3+ regulatory T cell function through shared and isoform-specific mechanisms. Sci Signal. 2012;5(229):ra45–ra45. doi: 10.1126/SCISIGNAL.2002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang L, Liu Y, Han R, Beier UH, Bhatti TR, Akimova T, Greene MI, Hiebert SW, Hancock WW. FOXP3+ regulatory T cell development and function require histone/protein deacetylase 3. J Clin Invest. 2015;125(3):1111–1123. doi: 10.1172/JCI77088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bolduc J-F, Hany L, Barat C, Ouellet M, Tremblay MJ. Epigenetic metabolite acetate inhibits class I/II histone deacetylases, promotes histone acetylation, and increases HIV-1 integration in CD4 + T cells. J Virol. 2017;91(16). doi: 10.1128/jvi.01943-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J Nutr Biochem. 2008;19(9):587–593. doi: 10.1016/J.JNUTBIO.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 81.Jugder BE, Kamareddine L, Watnick PI. Microbiota-derived acetate activates intestinal innate immunity via the Tip60 histone acetyltransferase complex. Immunity. 2021;54(8):1683–1697.e3. doi: 10.1016/j.immuni.2021.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kespohl M, Vachharajani N, Luu M, Harb H, Pautz S, Wolff S, Sillner N, Walker A, Schmitt-Kopplin P, Boettger T, et al. The microbial metabolite butyrate induces expression of Th1-associated factors in CD4+ T cells. Front Immunol 8. 2017;(AUG):1036. doi: 10.3389/FIMMU.2017.01036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen L, Sun M, Wu W, Yang W, Huang X, Xiao Y, Ma C, Xu L, Yao S, Liu Z, et al. Microbiota metabolite butyrate differentially regulates Th1 and Th17 cells’ differentiation and function in induction of colitis. Inflamm Bowel Dis. 2019;25(9):1450–1461. doi: 10.1093/ibd/izz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sun M, Wu W, Chen L, Yang W, Huang X, Ma C, Chen F, Xiao Y, Zhao Y, Ma C, et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat Commun. 2018;9(1):1–15. doi: 10.1038/s41467-018-05901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang M, Zhou L, Wang Y, Dorfman RG, Tang D, Xu L, Pan Y, Zhou Q, Li Y, Yin Y, et al. Faecalibacterium prausnitzii produces butyrate to decrease c-Myc-related metabolism and Th17 differentiation by inhibiting histone deacetylase 3. Int Immunol. 2019;31(8):499–514. doi: 10.1093/intimm/dxz022. [DOI] [PubMed] [Google Scholar]
- 86.Zhou L, Zhang M, Wang Y, Dorfman RG, Liu H, Yu T, Chen X, Tang D, Xu L, Yin Y, et al. Faecalibacterium prausnitzii produces butyrate to maintain Th17/Treg balance and to ameliorate colorectal colitis by inhibiting histone deacetylase 1. Inflamm Bowel Dis. 2018;24(9):1926–1940. doi: 10.1093/ibd/izy182. [DOI] [PubMed] [Google Scholar]
- 87.Luu M, Pautz S, Kohl V, Singh R, Romero R, Lucas S, Hofmann J, Raifer H, Vachharajani N, Carrascosa LC, et al. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat Commun. 2019;10(1):1–12. doi: 10.1038/s41467-019-08711-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Luu M, Weigand K, Wedi F, Breidenbend C, Leister H, Pautz S, Adhikary T, Visekruna A . Regulation of the effector function of CD8+ T cells by gut microbiota-derived metabolite butyrate. Sci Rep. 2018;8(1). doi: 10.1038/S41598-018-32860-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morinobu A, Kanno Y, O’Shea JJ. Discrete roles for histone acetylation in human T helper 1 cell-specific gene expression *. J Biol Chem. 2004;279(39):40640–40646. doi: 10.1074/JBC.M407576200. [DOI] [PubMed] [Google Scholar]
- 90.Luu M, Riester Z, Baldrich A, Reichardt N, Yuille S, Busetti A, Klein M, Wempe A, Leister H, Raifer H, et al. Microbial short-chain fatty acids modulate CD8+ T cell responses and improve adoptive immunotherapy for cancer. Nat Commun. 2021;12(1):1–12. doi: 10.1038/s41467-021-24331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Auré Lien Trompette A, Gollwitzer ES, Pattaroni C, Lopez-Mejia IC, Riva E, Pernot J, Ubags N, Fajas L, Nicod LP, Marsland BJ. Dietary fiber confers protection against flu by shaping Ly6c-patrolling monocyte hematopoiesis and CD8 + T cell metabolism. Immunity. 2018;48:992–1005.e8. doi: 10.1016/j.immuni.2018.04.022. [DOI] [PubMed] [Google Scholar]
- 92.Bachem A, Makhlouf C, Binger KJ, de Souza DP, Tull D, Hochheiser K, Whitney PG, Fernandez-Ruiz D, Dähling S, Kastenmüller W, et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8+ T cells. Immunity. 2019;51(2):285–297.e5. doi: 10.1016/j.immuni.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 93.Kim M, Qie Y, Park J, Kim CH. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe. 2016;20(2):202–214. doi: 10.1016/j.chom.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sanchez HN, Moroney JB, Gan H, Shen T, Im JL, Li T, Taylor JR, Zan H, Casali P. B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids. Nat Commun. 2020;11(1):1–19. doi: 10.1038/s41467-019-13603-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Statello L, Guo C-J, Chen -L-L, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2020;22(2):96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liang L, Ai L, Qian J, Fang J-Y, Xu J. Long noncoding RNA expression profiles in gut tissues constitute molecular signatures that reflect the types of microbes. Sci Reports. 2015;5(1):1–8. doi: 10.1038/srep11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dempsey J, Zhang A, Cui JY. Coordinate regulation of long non-coding RNAs and protein-coding genes in germ-free mice 06 biological sciences 0604 genetics 06 biological sciences 0601 biochemistry and cell biology. BMC Genomics. 2018;19(1). doi: 10.1186/s12864-018-5235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.O’Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne). 2018;9(AUG):402. doi: 10.3389/FENDO.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tan Y, Zhang B, Wu T, Skogerbø G, Zhu X, Guo X, He S, Chen R. Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol Biol. 2009;10(1):1–9. doi: 10.1186/1471-2199-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hawkins P, Morris KV. RNA and transcriptional modulation of gene expression. Cell Cycle. 2008;7(5):602. doi: 10.4161/CC.7.5.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Virtue AT, McCright SJ, Wright JM, Jimenez MT, Mowel WK, Kotzin JJ, Joannas L, Basavappa MG, Spencer SP, Clark ML, et al. The gut microbiota regulates white adipose tissue inflammation and obesity via a family of microRNAs. Sci Transl Med. 2019;11(496):1892. doi: 10.1126/SCITRANSLMED.AAV1892. [DOI] [PMC free article] [PubMed] [Google Scholar]


