Abstract
Helicobacter pylori infects up to 50% of the human population worldwide. The infection occurs predominantly in childhood and persists for decades or a lifetime. H. pylori is believed to be transmitted from person to person. However, tremendous genetic diversity has been reported for these bacteria. In order to gain insight into the epidemiological basis of this phenomenon, we performed molecular typing of H. pylori isolates from different families. Fifty-nine H. pylori isolates from 27 members of nine families were characterized by using restriction fragment length polymorphism analysis of five PCR-amplified genes, by pulsed-field gel electrophoresis (PFGE) of chromosomal DNA, and by vacA and cagA genotyping. The 16S rRNA gene exhibited little allelic variation, as expected for a unique bacterial species. In contrast, the vacA, flaA, ureAB, and lspA-glmM genes were highly polymorphic, with a mean genetic diversity of 0.83, which exceeds the levels recorded for all other bacterial species. In conjunction with PFGE, 59 H. pylori isolates could be differentiated into 21 clonal types. Each individual harbored only one clone, occasionally with a clonal variant. Identical strains were always found either between siblings or between a mother and her children. Statistical analysis revealed clonality of population structure in all isolates. The results of this study suggest the possible coexistence of a large array of clonal lineages that are evolving in each individual in isolation from one another. Transmission appears to occur primarily from mother to child and perhaps between siblings.
Helicobacter pylori-associated gastritis is today recognized as the major cause of duodenal and gastric ulcers, gastric adenocarcinoma and mucosa-associated lymphoid tissue lymphoma (9). H. pylori infects up to 50% of the human population worldwide. The infection occurs predominantly in childhood. Once the stomach is colonized, the organism persists for decades, if not for a lifetime. Many lines of evidence suggest person-to-person transmission of H. pylori, possibly by an oral-oral, fecal-oral, or gastric-oral route (4, 8, 9, 32–35, 44, 48). However, tremendous genetic diversity has been reported for these bacteria (23, 26). The molecular and epidemiological bases for this phenomenon are not understood.
Limited data suggest that the H. pylori population exhibits a panmictic or recombinational structure (1, 19, 24, 37, 39), and horizontal gene transfer followed by frequent DNA recombination has been considered to be an important source of the immense genetic diversity. However, clonal groupings of H. pylori have also been detected from different ethnic groups in New Zealand, China, The Netherlands, and other countries (1, 7, 25, 43, 45, 46). Moreoveor, comparison of the complete genomic sequences of two unrelated H. pylori isolates (2, 42) reveals that the overall genomic organizations, gene orders, and predicted proteomes are quite similar, with the majority of nucleotide changes representing synonymous substitutions. Based on these observations, Wang et al. (47) pointed out that mutation must be of major importance in generating the genetic variation of H. pylori.
In the present study, 59 H. pylori isolates from 27 different family members of nine families were characterized in order to define the H. pylori population structure. The results are most compatible with the concept that a large array of clonal lineages coexist and are evolving in isolation from one another. The evidence also indicates that transmission of H. pylori occurs predominantly and perhaps even exclusively within families. If this is correct, H. pylori would represent the first human pathogen recognized to display this remarkably restricted mode of transmission.
(Part of this work was performed in fulfillment of the requirements for a doctoral thesis by H.-C. E. Zschausch.)
MATERIALS AND METHODS
Subjects.
Nine index children from different families, designated families A through I, were referred to the Department of Pediatrics, Johannes Gutenberg University, Mainz, Germany, due to recurrent abdominal pain. One or more gastric (antral and corpus) and duodenal biopsy specimens were obtained from each of these patients and from at least one additional family member (see Fig. 2). In total, 27 members from nine families were investigated, represented by 2 to 6 members in each family. These patients were 23 children (11 boys and 12 girls with an age range of 2 to 18 years) and 4 adults from three families (2 women and 2 men from 29 to 44 years of age). The other parents were not sampled. They lived either within or in close proximity to Mainz.
FIG. 2.
Genetic relationships among 59 H. pylori isolates from nine different families. The dendrogram was constructed from PCR-RFLP typing data by the unweighted pair group cluster method with arithmetic means. Each distinct combination of 10 individual gene and enzyme patterns was designated a PCR-RFLP type, corresponding to a clonal type. Ten major lineages, termed I through X, separating at a genetic distance of 0.3 are indicated to the left. The columns to the right indicate the designation of H. pylori isolates, the origin of the isolates, and the molecular typing results. n.d., not determined; n.t., not typeable.
Culture and PCR.
Biopsy specimens were cultured on Columbia agar with 7% human erythrocytes and an H. pylori-selective supplement at 37°C under microaerobic conditions. H. pylori was identified and bacterial genomic DNA was prepared as previously described (23). PCR amplification of the vacA, flaA, ureAB, lspA-glmM (formerly ureCD), or cagA gene fragments was performed using the primer pairs described previously (23). The cagA gene status was determined by the presence or absence of cagA amplicons. The vacA genotypes, including signal(s) sequences and midregion (m) types, were characterized by a one-step PCR method (22). For ribotyping, a 1.5-kb fragment was amplified with broad-specificity 16S ribosomal DNA (rDNA) primer pairs pA-f (5′-AGAGTTTGATCCTGGCTCAG-3′) and pH-b (5′-AAGGAGGTGATCCAGCCGCA-3′) (15), using an initial denaturation step at 94°C for 2 min, followed by 35 cycles of denaturation at 94°C, annealing at 55°C, and extension at 72°C (each for 1 min), and a final extension at 72°C for 10 min. In order to eliminate contaminating DNA within PCR reagents, UV light and 8-methoxypsoralen (Sigma, Munich, Germany) were used (31). Genomic DNA was added after irradiation of the PCR mixture with UV light (366 nm) at room temperature for 20 min.
PCR-RFLP analysis.
PCR-amplified vacA, flaA, ureAB, lspA-glmM, or 16S rDNA fragments were digested with HaeIII (for ureAB or 16S rDNA), HhaI (for vacA, flaA, or lspA-glmM), HinfI (for 16S rDNA), HphI (for vacA), or Sau3AI (for flaA, ureAB, or lspA-glmM) for 3 h at 37°C in the appropriate buffer recommended by the supplier (New England Biolabs, Schwalbach/Taunus, Germany), and the DNA digests were analyzed on ethidium bromide-stained 2% agarose gels. Each isolate was thus characterized by 10 individual gene-enzyme patterns. A combination of these 10 patterns was designated a PCR-based locus-specific restriction fragment length polymorphism (PCR-RFLP) type, which defined the clonal type of an H. pylori isolate. Additionally, a combination of two restriction patterns of 16S rDNA was defined as a ribotype. The cagA gene fragments were not analyzed by PCR-RFLP, since the cagA gene could be detected in only 34 of 59 H. pylori isolates.
PFGE.
The methods previously described by Taylor et al. (41) were modified. Fresh 2-day cultures of H. pylori were harvested and washed three times using normal saline. Alternatively, in order to inhibit DNase activity, 0.9-ml portions of the cell suspensions were incubated with 0.1 ml of 37% formaldehyde solution for 1 h at room temperature, and then washed (18). Approximately 1 ml of cell suspension with an optical density at 600 nm of 0.9 was needed for the preparation of two agarose plugs. The required volume of cell suspension was centrifuged, and bacteria were resuspended in 10 mM Tris-HCl–10 mM EDTA (pH 8.0) (0.1 ml for 2 plugs). This suspension was mixed with an equal volume of 1.2% agarose equilibrated to 56°C to prepare 0.6% agarose plugs. The solidified plugs were incubated overnight in ESP lysis buffer (0.25 M EDTA [pH 9.0], 1% lauroyl sarcosine, 1 mg of proteinase K per ml) (Sigma, St. Louis, Mo.) at 56°C. The plugs were washed two times in 15 ml of the Tris-EDTA buffer described above for 2 h each at 4°C. For the subsequent enzyme reactions, the plugs were washed once in 15 ml of 10 mM Tris-HCl–0.5 mM EDTA (pH 8.0) for 30 min at 4°C. The plugs could be stored in 10 mM Tris-HCl–50 mM EDTA (pH 8.0) at 4°C for 3 months.
For restriction enzyme digestion, plugs were incubated with 1 ml of the appropriate 1× enzyme buffer for 30 to 60 min at room temperature. The enzyme buffer was replaced with 100 μl of fresh enzyme buffer containing 25 U of NotI, NruI, or SmaI (New England Biolabs) and incubated overnight at 37°C (25°C for SmaI). The plugs were chilled on ice for 15 min, and the enzyme buffer was replaced with 10 mM Tris-HCl–10 mM EDTA (pH 8.0), followed by loading of the plugs into the 1% running gel (Pulsed field certified agarose; Bio-Rad Laboratories, Hercules, Calif.). A lambda ladder PFG marker (New England Biolabs) served as a size standard. For pulsed-field gel electrophoresis (PFGE), CHEF-DR III PFGE systems (Bio-Rad Laboratories) were used. An agarose gel prepared in 0.5× Tris-borate-EDTA running buffer (Bio-Rad Laboratories) was subjected to electrophoresis for 20.3 h at 200 V and a 0.5- to 60.4-s switch time at a constant temperature of 14°C. Agarose gels were stained with ethidium bromide and photographed. A combination of three restriction patterns in an isolate was designated a PFGE type.
Statistical analysis.
For the RFLP data, different patterns of each of the analyzed genes were treated as different alleles. A distinct combination of 10 individual patterns in an H. pylori isolate was designated an RFLP type (clonal type) and equated with an allelic combination. Genetic diversity (h) for a locus j is calculated as hj = 1 − Σχi2[n/(n − 1)], where χi is the frequency of the ith allele at the locus, n is the number of isolates in the sample, and n/(n − 1) is a correction for bias in small samples (38). Genotypic diversity was calculated by the same formula, in which χi is the frequency of the ith RFLP type and n is the number of RFLP types. Genetic diversity and genotypic diversity have a range of between 0 and 1. The mean diversity per locus is the arithmetic average of h over all loci examined. The genetic distance between pairs of isolates was calculated as the proportion of loci at which dissimilar alleles occur, i.e., the proportion of mismatches (38). The dendrogram was constructed from PCR-RFLP typing data by the unweighted pair group cluster method with arithmetic means in the computer program MEGA (28).
The index of association between loci (IA) was calculated as described previously (30). The IA value is a measure of the degree of association between loci and has an expected value of zero for a large random-mating (panmictic) bacterial population. In contrast, for bacterial species that are clonal at all levels as a result of geographic isolation or infrequent genetic recombination, the expected value of IA should deviate significantly from zero. To determine whether IA is significantly greater than zero, a Monte Carlo procedure can be performed. Computer programs written by J. G. Lorén (17) were used to calculate locus and genotypic diversity, to calculate IA, and to perform the Monte Carlo simulations.
RESULTS
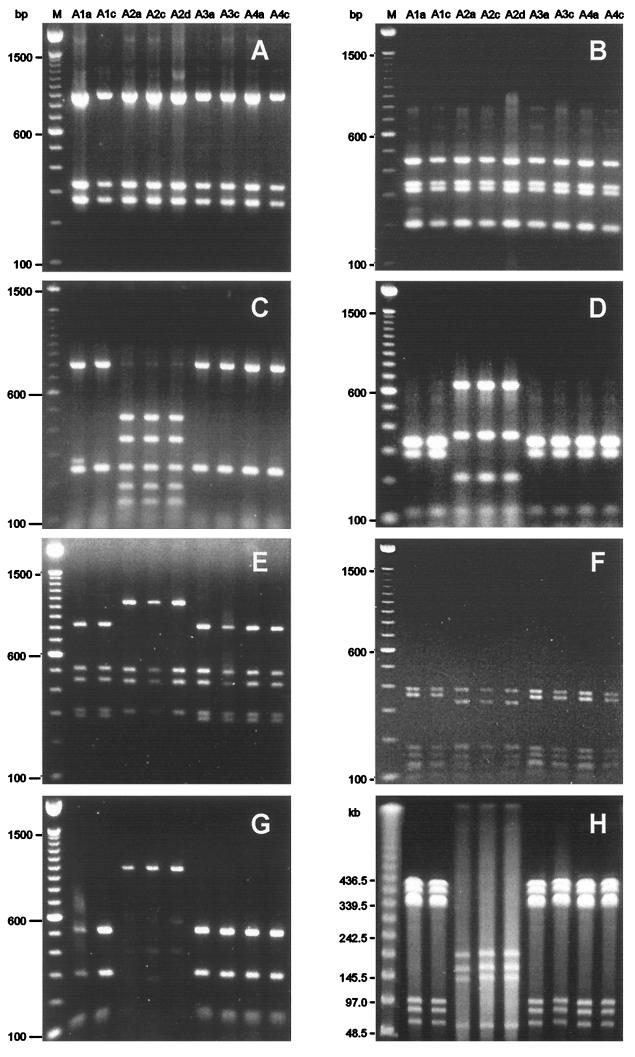
Figure 1 illustrates the RFLP and PFGE patterns obtained with nine isolates from the mother, the father, and two children of family A. These results are representative. Whenever possible, analyses were performed on isolates obtained from the antrum (a), corpus (c), and duodenum (d). Most patients harbored only a single H. pylori isolate. If a second isolate was detected, it differed only in a single restriction pattern, except for one isolate (H4a). An example is the isolate from the antrum of the mother (A1a), which varied from the corpus isolate A1c exclusively in the flaA-HhaI patterns. We considered isolate A1a to be a mutational variant of the parent isolate A1c.
FIG. 1.
Representative molecular typing results for H. pylori isolates from family A. PCR-RFLP analyses of 16S rDNA fragments digested with HaeIII (A), 16S rDNA fragments digested with HinfI (B), flaA fragments digested with HhaI (C), flaA fragments digested with Sau3AI (D), ureAB fragments digested with HaeIII (E), lspA-glmA fragments digested with HhaI (F), and vacA fragments digested with HphI (G) and PFGE of genomic DNA digested with NotI (H) are shown. Lanes: M, size markers; A1a to A4c, H. pylori isolates, designated according to the origins of the isolates (A, family A; 1 to 4, family members; a, c, and d, antrum, corpus, and duodenum, respectively).
Conspicuously, apparently identical isolates were very frequently found within families, where they were identified between siblings or between mother and child. The collective results of molecular typing are summarized in Fig. 2. For 59 H. pylori isolates, analysis of the 16S rRNA gene demonstrated an average of 2.5 RFLP patterns, a mean genetic diversity of 0.08, and a mean genotypic diversity of 0.14. The sparse allelic variation in the 16S rRNA gene was as expected for a unique bacterial species (14). In contrast, the vacA, flaA, ureAB, and lspA-glmM genes were highly polymorphic, with an average of 11.25 RFLP patterns per gene, a mean genetic diversity of 0.83, and a mean genotypic diversity of 0.91 for all of 59 H. pylori isolates. These values exceed the level of diversity recorded for all other bacterial species (19). The results of PFGE typing were concordant with the PCR-RFLP analysis in 46 of 52 cases. Three variants (A1a, D2a, and I2c) were distinct from their parent strains by PCR-RFLP analysis but showed PFGE patterns identical to those of the parent strains. Vice versa, three variants (B1c, H4a, and H4c) were distinguishable from their parent strains by PFGE but not by PCR-RFLP analysis. Hence, detection of strain variants is sometimes possible only by implementing both PCR-RFLP and PFGE analysis.
The genetic relationship among the 59 H. pylori isolates exhibited a tree-like phylogenetic structure with a deep but limited branching pattern (Fig. 2). Twenty-one clonal types clustered into 10 divisions (designated I to X). Isolates from one family tended to cluster into the same division. Isolates from different families were sometimes also found to be related, as evident in divisions II and IV.
Siblings tended to harbor identical isolates (families A, C, D, G, and H), but distinct H. pylori isolates could also be found between siblings (families D, E, F, and I). We were able to analyze H. pylori isolates from only one pair of parents (family A). The mother harbored the same isolate as her children, whereas the father harbored an unrelated strain. Intriguingly, the mother in family B also had the same isolate as her son. In contrast, the H. pylori strain of the father in family F was distinct from the isolates recovered in either of his children. These results indicate that H. pylori can be transmitted within families and possibly from parent (mother) to child or between siblings. However, a common source of exposure for H. pylori infection could not be completely ruled out.
The complete set of H. pylori isolates and subsets of the population were analyzed for multilocus linkage disequilibrium (Table 1). The 59 isolates differed on average at 6.80 of the 10 loci examined, and the IA was 0.43. The Monte Carlo procedure indicated that the IA was significantly different from zero. Moreover, calculation of IA for the 21 PCR-RFLP types resulted in a value of 1.42, which was significantly different from zero (P < 0.001) based on the Monte Carlo procedure. These results, analyzed at the level of all isolates or RFLP types, demonstrated a significant degree of linkage disequilibrium among the H. pylori isolates from different families, consistent with the clonal structure described by Maynard Smith et al. (30).
TABLE 1.
Analysis of association between loci in 59 H. pylori isolates from family membersa
| Isolate group | Isolates
|
PCR-RFLP types only
|
||||||
|---|---|---|---|---|---|---|---|---|
| No. | X | IA | P | No. | X | IA | P | |
| vacA s1 type | 44 | 6.83 | 0.61 | <0.001 | 16 | 7.67 | 1.64 | <0.001 |
| vacA s2 type | 10 | 4.02 | 0.78 | >0.05 | 3 | 6.00 | 0 | >0.05 |
| vacA m1 type | 34 | 6.05 | 0.99 | <0.001 | 11 | 7.58 | 2.52 | <0.001 |
| vacA m2 type | 25 | 6.51 | 0.24 | >0.05 | 10 | 7.00 | 1.36 | 0.001 |
| cagA positive | 34 | 6.78 | 0.36 | >0.05 | 13 | 7.89 | 1.66 | <0.001 |
| cagA negative | 25 | 5.88 | 1.59 | <0.001 | 9 | 6.64 | 2.92 | <0.001 |
| From antrum | 27 | 7.13 | 0.74 | <0.001 | 19 | 7.60 | 1.14 | <0.001 |
| From corpus | 26 | 6.90 | 0.22 | >0.05 | 16 | 7.44 | 0.50 | >0.05 |
| All | 59 | 6.80 | 0.43 | <0.01 | 21 | 7.58 | 1.42 | <0.001 |
X, mean number of allelic mismatches between pairs of H. pylori isolates or between PCR-RFLP types; IA, index of association between loci; P, probability based on the Monte Carlo procedure. Values of IA that differ significantly from zero indicate a clonal structure in bacterial populations, with rare or absent recombination. In contrast, values of IA that do not differ significantly from zero indicate a panmictic structure, with frequent recombination.
DISCUSSION
Previous studies have suggested that H. pylori is panmictic and that the immense genetic diversity among H. pylori strains arises by frequent horizontal DNA transfer and recombination (1, 19, 24, 37, 39). Indeed, direct evidence for recombination between different H. pylori strains has been observed in humans (27) and in mice (10). However, several lines of evidence indicate that mutations may also play an important role in generating the observed diversity in H. pylori (47). We show in this report that the bacterium H. pylori does in fact exhibit a clonal structure. According to these data and previous studies (23), each individual will harbor very few H. pylori strains (usually only one). This hypothesis was substantiated by our data (Fig. 2) that a large array of clonal lineages coexist, which evolve in isolation, i.e., in each individual. This would account for the finding that separate lineages of H. pylori have been detected from different ethnic groups in New Zealand (Polynesians and Europeans), in China, in The Netherlands, and in other countries (1, 7, 25, 43, 45, 46). Identical flaA, flaB, or vacA alleles were also found in unrelated H. pylori strains from a population in Capetown, South Africa (39). These results are consistent with the existence of divergent H. pylori clonal lineages at the level of different ethnic groups or different geographic regions.
The results presented in this report strongly suggest that mutations which occur within the individual host are capable of generating variant H. pylori strains. Recombination, although possible, will be a rather rare event in the absence of multiple infections. In fact, infection with multiple H. pylori strains appears to be infrequent in the developed world. The “different” strains recovered from the same patient often seemed more closely related by fingerprinting than other strains recovered from different patients. In experimentally infected rhesus monkeys, a mixed infection tends to resolve quickly, with just one strain generally predominating within a year (8). If the working hypothesis is correct, the clonal population structure of H. pylori would only slowly be eroded by recombination. The fact that the mode of H. pylori transmission is probably very restricted gains particular importance in this context. Although the number of subjects included here was still relatively small, all of the results would be in line with the contention that H. pylori transmission occurs mainly, if not exclusively, within families. Thereby, transmission from parent (mother) to child and perhaps from child to child predominates, occurring conceivably via the gastric-oral, oral-oral, or fecal-oral route. This contention would be consistent with many studies that have utilized epidemic (5, 11–13, 16, 35) or molecular typing (4, 8, 32, 34, 44, 48) methods. A small number of epidemic studies have concluded otherwise (3, 6, 20, 21), but this may have been due to factors affecting H. pylori transmission, e.g., the general prevalence of H. pylori in different age groups, the timing of acquisition, and the socioeconomy and ethnic composition of the studied population. An investigation by Goodman and Correa (21) with rural Colombian children appeared to reveal transmission of H. pylori from older to younger siblings. However, Rowland (36) has commented on the problems surrounding the interpretation of those data. The hypothesis that H. pylori transmission occurs predominantly from parents to child, and not vice versa, is supported by the observations that (i) H. pylori acquisition occurs predominantly in childhood and (ii) the prevalence of H. pylori infection in parents is much higher than that in children. Additionally, interspousal transmission of H. pylori occurs rarely (29, 40). Our molecular analysis of H. pylori suggests the vertical spread from parent (mother) to child, perhaps combined with the horizontal transmission between siblings, as the pathway for H. pylori transmission. If this proves to be correct, H. pylori would represent the first human pathogen recognized to display this remarkably restricted mode of transmission.
ACKNOWLEDGMENTS
We are indebted to J. G. Lorén, Departament de Microbiologia i Parasitologia Sanitàries, Facultat de Farmàcia, Divisió de Ciències, de la Salut, Universitat de Barcelona, Barcelona, Spain, for providing computer programs of linkage disequilibrium and helpful discussions. We acknowledge G. Rippin, Department of Medical Statistics and Documentation, University of Mainz, for helpful discussions on statistical methods. We thank D. E. Taylor and Q. Jiang, Department of Medical Microbiology and Immunology, University of Alberta, Edmonton, Alberta, Canada, and B. Geilhausen, Department of Medical Microbiology and Hygiene, University of Cologne, Cologne, Germany, for helpful communications concerning the PFGE method.
REFERENCES
- 1.Achtman M, Azuma T, Berg D E, Ito Y, Morelli G, Pan Z-J, Suerbaum S, Thompson S A, van der Ende A, van Doorn L J. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32:459–470. doi: 10.1046/j.1365-2958.1999.01382.x. [DOI] [PubMed] [Google Scholar]
- 2.Alm R A, Ling L-S L, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 3.Ashorn M, Miettinen A, Ruuska T, Laippala P, Maki M. Seroepidemiological study of Helicobacter pylori infection in infancy. Arch Dis Child Fetal Neonatal Ed. 1996;74:F141–F142. doi: 10.1136/fn.74.2.f141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bamford K B, Bickley J, Collins J S A, Johnston B T, Potts S, Boston V, Owen R J, Sloan J M. Helicobacter pylori: comparison of DNA fingerprints provides evidence for intrafamilial infection. Gut. 1993;34:1348–1350. doi: 10.1136/gut.34.10.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassily S, Frenck R W, Mohareb E W, Wierzba T, Savarino S, Hall E, Kotkat A, Naficy A, Hyams K C, Clemens J. Seroprevalence of Helicobacter pylori among Egyptian newborns and their mothers: a preliminary report. Am J Trop Med Hyg. 1999;61:37–40. doi: 10.4269/ajtmh.1999.61.37. [DOI] [PubMed] [Google Scholar]
- 6.Blecker U, Lanciers S, Keppens E, Vandenplas Y. Evolution of Helicobacter pylori positivity in infants born from positive mothers. J Pediatr Gastroenterol Nutr. 1994;19:87–90. doi: 10.1097/00005176-199407000-00014. [DOI] [PubMed] [Google Scholar]
- 7.Campbell S, Fraser A, Holliss B, Schmid J, O'Toole P W. Evidence for ethnic tropism of Helicobacter pylori. Infect Immun. 1997;65:3708–3712. doi: 10.1128/iai.65.9.3708-3712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chalkauskas H, Kersulyte D, Cepuliene I, Urbonas V, Ruzevicene D, Barakauskiene A, Raudonikiene A, Berg D E. Genotypes of Helicobacter pylori in Lithuanian families. Helicobacter. 1998;3:296–302. [PubMed] [Google Scholar]
- 9.Covacci A, Telford J L, Giudice G D, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 10.Danon S J, Luria B J, Mankoski R E, Eaton K A. RFLP and RAPD analysis of in vivo genetic interactions between strains of Helicobacter pylori. Helicobacter. 1998;3:254–259. doi: 10.1046/j.1523-5378.1998.08010.x. [DOI] [PubMed] [Google Scholar]
- 11.Dominici P, Bellentani S, Di Biase A R, Saccoccio G, Le Rose A, Masutti F, Viola L, Balli F, Tiribelli C, Grilli R, Fusillo M, Grossi E. Familial clustering of Helicobacter pylori infection: population based study. Br Med J. 1999;319:537–541. doi: 10.1136/bmj.319.7209.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowsett S A, Archila L, Segreto V A, Gonzalez C R, Silva A, Vastola K A, Bartizek R D, Kowolik M J. Helicobacter pylori infection in indigenous families of Central America: serostatus and oral and fingernail carriage. J Clin Microbiol. 1999;37:2456–2460. doi: 10.1128/jcm.37.8.2456-2460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drumm B, Perez-Perez G I, Blaser M J, Sherman P M. Intrafamilial clustering of H. pylori infection. N Engl J Med. 1990;322:359–363. doi: 10.1056/NEJM199002083220603. [DOI] [PubMed] [Google Scholar]
- 14.Eckloff B W, Podzorski R P, Kline B C, Cockerill F R., III A comparison of 16S ribosomal DNA sequences from five isolates of Helicobacter pylori. Int J Syst Bacteriol. 1994;44:320–323. doi: 10.1099/00207713-44-2-320. [DOI] [PubMed] [Google Scholar]
- 15.Edwards U, Rogall T, Blöcker H, Emde M, Böttger E C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elitsur Y, Adkins L, Saeed D, Neace C. Helicobacter pylori antibody profile in household members of children with H. pylori infection. J Clin Gastroenterol. 1999;29:178–182. doi: 10.1097/00004836-199909000-00015. [DOI] [PubMed] [Google Scholar]
- 17.Fusté M C, Pineda M A, Palomar J, Viñas M, Lorén J G. Clonality of multidrug-resistant nontypeable strains of Haemophilus influenzae. J Clin Microbiol. 1996;34:2760–2765. doi: 10.1128/jcm.34.11.2760-2765.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson J R, Sutherland K, Owen R J. Inhibition of DNase activity in PFGE analysis of DNA from Campylobacter jejuni. Lett Appl Microbiol. 1994;19:357–358. doi: 10.1111/j.1472-765x.1994.tb00474.x. [DOI] [PubMed] [Google Scholar]
- 19.Go M F, Kapur V, Graham D Y, Musser J M. Population genetic analysis of Helicobacter pylori by multilocus enzyme electrophoresis: extensive allelic diversity and recombinational population structure. J Bacteriol. 1996;178:3934–3938. doi: 10.1128/jb.178.13.3934-3938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gold B D, Khanna B, Huang L M, Lee C-Y, Banatvala N. Helicobacter pylori acquisition in infancy after decline of maternal passive immunity. Pediatr Res. 1997;41:641–646. doi: 10.1203/00006450-199705000-00007. [DOI] [PubMed] [Google Scholar]
- 21.Goodman K J, Correa P. Transmission of Helicobacter pylori among siblings. Lancet. 2000;355:358–362. doi: 10.1016/S0140-6736(99)05273-3. [DOI] [PubMed] [Google Scholar]
- 22.Han S-R, Schneider T, Loos M, Bhakdi S, Maeurer M J. One-step PCR-based typing of Helicobacter pylori vacA gene: association with gastric histopathology. Med Microbiol Immunol. 1999;188:131–138. doi: 10.1007/s004300050115. [DOI] [PubMed] [Google Scholar]
- 23.Han S-R, Schreiber H-J, Bhakdi S, Loos M, Maeurer M J. vacA genotypes and genetic diversity in clinical isolates of Helicobacter pylori. Clin Diagn Lab Immunol. 1998;5:139–145. doi: 10.1128/cdli.5.2.139-145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hazell S L, Andrews R H, Mitchell H M, Daskalopoulous G. Genetic relationship among isolates of Helicobacter pylori: evidence for the existence of a Helicobacter pylori species-complex. FEMS Microbiol Lett. 1997;150:27–32. doi: 10.1111/j.1574-6968.1997.tb10345.x. [DOI] [PubMed] [Google Scholar]
- 25.Ito Y, Azuma T, Ito S, Miyaji H, Hirai M, Yamazaki Y, Sato F, Kato T, Kohli Y, Kuriyama M. Analysis and typing of the vacA gene from cagA-positive strains of Helicobacter pylori isolated in Japan. J Clin Microbiol. 1997;35:1710–1714. doi: 10.1128/jcm.35.7.1710-1714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Q, Hiratsuka K, Taylor D E. Variability of gene order in different Helicobacter pylori strains contributes to genome diversity. Mol Microbiol. 1996;20:833–842. doi: 10.1111/j.1365-2958.1996.tb02521.x. [DOI] [PubMed] [Google Scholar]
- 27.Kersulyte D, Chalkauskas H, Berg D E. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol Microbiol. 1999;31:31–43. doi: 10.1046/j.1365-2958.1999.01140.x. [DOI] [PubMed] [Google Scholar]
- 28.Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis, version 1.01. University Park: The Pennsylvania State University; 1993. [Google Scholar]
- 29.Kuo C-H, Poon S-K, Su Y-C, Su R, Chang C-S, Wang W-C. Heterogeneous Helicobacter pylori isolates from H. pylori-infected couples in Taiwan. J Infect Dis. 1999;180:2064–2068. doi: 10.1086/315130. [DOI] [PubMed] [Google Scholar]
- 30.Maynard Smith J, Smith N H, O'Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meier A, Persing D H, Finken M, Böttger E C. Elimination of contaminating DNA within polymerase chain reaction reagents: implications for a general approach to detection of uncultured pathogens. J Clin Microbiol. 1993;31:646–652. doi: 10.1128/jcm.31.3.646-652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miehlke S, Genta R M, Graham D Y, Go M F. Molecular relationships of Helicobacter pylori strains in a family with gastroduodenal disease. Am J Gastroenterol. 1999;94:364–368. doi: 10.1111/j.1572-0241.1999.859_u.x. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell H M, Hazell S L, Kolesnikow T, Mitchell J, Frommer D. Antigen recognition during progression from acute to chronic infection with a cagA-positive strain of Helicobacter pylori. Infect Immun. 1996;64:1166–1172. doi: 10.1128/iai.64.4.1166-1172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nwokolo C U, Bickley J, Attard A R, Owen R J, Costas M, Fraser I A. Evidence of clonal variants of Helicobacter pylori in three generations of a duodenal ulcer disease family. Gut. 1992;33:1323–1327. doi: 10.1136/gut.33.10.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rothenbacher D, Bode G, Berg G, Knayer U, Gonser T, Adler G, Brenner H. Helicobacter pylori among preschool children and their parents: evidence of parent-child transmission. J Infect Dis. 1999;179:398–402. doi: 10.1086/314595. [DOI] [PubMed] [Google Scholar]
- 36.Rowland M. Transmission of Helicobacter pylori: is it all child's play? Lancet. 2000;355:332–333. doi: 10.1016/S0140-6736(99)00427-4. [DOI] [PubMed] [Google Scholar]
- 37.Salaün L, Audibert C, Lay G L, Burucoa C, Fauchere J-L, Picard B. Panmictic structure of Helicobacter pylori demonstrated by the comparative study of six genetic markers. FEMS Microbiol Lett. 1998;161:231–239. doi: 10.1111/j.1574-6968.1998.tb12953.x. [DOI] [PubMed] [Google Scholar]
- 38.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour M N, Whittam T S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suerbaum S, Maynard Smith J, Bapumia K, Morelli G, Smith N H, Kunstmann E, Dyrek I, Achtman M. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki J, Muraoka H, Kobayasi I, Fujita T, Mine T. Rare incidence of interspousal transmission of Helicobacter pylori in asymptomatic individuals in Japan. J Clin Microbiol. 1999;37:4174–4176. doi: 10.1128/jcm.37.12.4174-4176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor D E, Eaton M, Chang N, Salama S M. Construction of a Helicobacter pylori genome map and demonstration of diversity at the genome level. J Bacteriol. 1992;174:6800–6806. doi: 10.1128/jb.174.21.6800-6806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 43.Van der Ende A, Pan Z-J, Bart A, van der Hulst R W M, Feller M, Xiao S-D, Tytgat G N J, Dankert J. cagA-positive Helicobacter pylori populations in China and The Netherlands are distinct. Infect Immun. 1998;66:1822–1826. doi: 10.1128/iai.66.5.1822-1826.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van der Ende A, Rauws E A J, Feller M, Mulder C J J, Tytgat G N J, Dankert J. Heterogeneous Helicobacter pylori isolates from members of a family with a history of peptic ulcer disease. Gastroenterology. 1996;111:638–647. doi: 10.1053/gast.1996.v111.pm8780568. [DOI] [PubMed] [Google Scholar]
- 45.Van Doorn L-J, Figueiredo C, Megraud F, Pena S, Midolo P, Queiroz D M D M, Carneiro F, Vanderborght B, Pegado M D G F, Sanna R, de Boer W, Schneeberger P M, Correa P, Ng E K W, Atherton J, Blaser M J, Quint W G V. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology. 1999;116:823–830. doi: 10.1016/s0016-5085(99)70065-x. [DOI] [PubMed] [Google Scholar]
- 46.Van Doorn L-J, Figueiredo C, Sanna R, Blaser M J, Quint W G V. Distinct variants of Helicobacter pylori cagA are associated with vacA subtypes. J Clin Microbiol. 1999;37:2306–2311. doi: 10.1128/jcm.37.7.2306-2311.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang G, Humayun M Z, Taylor D E. Mutation as an origin of genetic variability in Helicobacter pylori. Trends Microbiol. 1999;7:488–493. doi: 10.1016/s0966-842x(99)01632-7. [DOI] [PubMed] [Google Scholar]
- 48.Wang J-T, Sheu J-C, Lin J-T, Wang T-H, Wu M-S. Direct DNA amplification and restriction pattern analysis of Helicobacter pylori in patients with duodenal ulcer and their families. J Infect Dis. 1993;168:1544–1548. doi: 10.1093/infdis/168.6.1544. [DOI] [PubMed] [Google Scholar]