Abstract
Imaging neuronal activity in awake, behaving animals has become a groundbreaking method in neuroscience that has rapidly enhanced our understanding of how the brain works. In vivo microendoscopic imaging has enabled researchers to see inside the brains of experimental animals and thus has emerged as a technology fit to answer many experimental questions. By combining microendoscopy with cutting edge targeting strategies and sophisticated analysis tools, neuronal activity patterns that underlie changes in behavior and physiology can be identified. However, new users may find it challenging to understand the techniques and to leverage this technology to best suit their needs. Here we present a background and overview of the necessary components for performing in vivo optical calcium imaging and offer some detailed guidance for current recommended approaches.
Keywords: functional imaging, microendoscopy, neuronal circuits, GRIN lens, GCaMP, GECI
1. Introduction
Optical monitoring of neuronal activity in freely moving animals has begun to crack the circuitry underlying interaction with the environment, physiology, and behavior. Recent functional imaging studies have significantly advanced our understanding of how specific neuronal circuits deep in the brain encode processes essential to survival such as feeding (Betley et al., 2015; Jennings et al., 2015; Douglass et al., 2017; Rossi et al., 2019), sleep (Cox et al., 2016; Weber and Dan, 2016), and nociception (Corder et al., 2019). Moreover, the use of these techniques has shed light on neuronal mechanisms involved in complex processes like place coding (Ziv et al., 2013; Kinsky et al., 2018), substance abuse (Siciliano et al., 2019), and learning and memory (Grewe et al., 2017; Kamigaki and Dan, 2017; Kitamura et al., 2017). The shift from electrical to optical recording methods has brought several key improvements including (a) the ability to target specific cell subpopulations with fluorescent, genetically-encoded activity sensors (e.g., calcium sensors), (b) the ability to record from the same exact population of cells over multiple sessions, (c) a vast increase in the numbers of simultaneously recorded cells, (d) the ability to map the relative spatial locations of groups of cells, and (e) increased accessibility to deep structures in the brain. Additionally, miniaturization of the microendoscope has allowed for interrogation of the neuronal circuits involved in the complex, waking behavioral states listed above, freeing researchers from the constraints of anesthetized or head-fixed configurations (Table 1). The decreasing costs of microendoscopy systems combined with increasingly user-friendly and elegant data analysis pipelines are also increasing the appeal of these approaches.
Table 1.
Microendoscopy system features
| Feature | Fiber Photometry | Single-photon Imaging | Two-photon Imaging |
|---|---|---|---|
| Resolution | Population dynamics, no single-cell resolution | Cellular resolution, overlapping cells in Z-space | Subcellular resolution, optical sectioning |
| Restraint | Freely moving | Freely moving | Typically head-fixed; freely moving not widely available |
| Motion Correction | Second channel used to remove motion artifacts | Required | Required |
| Skill required | + | ++ | +++ |
| Footprint | Small | Moderate, dual-site imaging possible | Large |
| Cost | $ | $$ − $$$ | $$$$ |
However, despite these advantages, the multi-faceted nature of in vivo functional imaging approaches can be overwhelming to inexperienced users. Obtaining satisfactory end results heavily depends on many factors including viral strategy, surgical technique, microscopy system, experimental design, programming experience, data preprocessing, and postprocessing analysis (Kamigaki and Dan, 2017). While excellent detailed methodological protocols have been described (Resendez et al., 2016; Yang and Yuste, 2017; Martianova et al., 2019; Lee and Han, 2020), the field is progressing rapidly, and additional recent advances have been made. Thus, here we present a background and overview of the key components for performing in vivo optical calcium imaging and offer some detailed guidance for current recommended approaches in microendoscopy.
2. Genetically encoded calcium indicators
The premise of in vivo functional imaging is to use changes in fluorescence of a genetically encoded calcium indicator (GECI) as a proxy for neuronal activity. Although indicators for other molecules such as GABA (Marvin et al., 2019), glutamate (Marvin et al., 2013), and dopamine (Patriarchi et al., 2018) are now available, this review will focus on GECIs as they are the most widespread in imaging studies. During an action potential, intracellular calcium levels increase, leading to increased fluorescence in neurons expressing GECIs, and this can be interpreted as an increase in cellular activity. By far the most widely used GECIs in optical imaging are the green fluorescent protein (GFP)-based calcium sensors of the GCaMP family. Initially developed in 2001 (Nakai et al., 2001), GCaMP was formulated as a fusion protein of GFP, the calcium-binding protein calmodulin (CaM), and the CaM-interacting M13 peptide. A conformational change in the structure of GCaMP when calcium is bound allows GFP to fluoresce, and hence indicates the presence of the cation. Since then, seven main iterations of GCaMPs have occurred, with the vast majority of literature over the past decade coming from the use of GCaMP6 (Chen et al., 2013), which was designed in three kinetic variants: fast (GCaMP6f), medium (GCaMP6m), and slow (GCaMP6s). The latest round of structure-guided design has yielded the family of jGCaMP7 sensors, which offer improved sensitivity and signal-to-noise ratios (SNR) for the jGCaMP7s and jGCaMP7f variants, as well as optimization for neuropil imaging with jGCaMP7b (“high baseline”), and improved contrast with jGCaMP7c (“high contrast”) (Dana et al., 2019).
While calcium imaging with single-wavelength GCaMPs has yielded many novel insights toward understanding the activity of neuronal circuits underlying behavior and pathology, there also was a need for calcium sensors that fluoresce at longer wavelengths. Longer wavelength light penetrates tissue more effectively (Nikolaos et al., 2008), as green light emission is absorbed by blood (Svoboda and Block, 1994), and thus red-wavelength imaging results in comparable image quality at lower illumination levels to reduce photobleaching and to enhance the quality of imaging in deep brain structures. Longer-wavelength light emission also facilitates “all-optical” approaches of circuit characterization, permitting the use of optogenetics combined with calcium imaging through independent spectral channels for photostimulation and recording (Akerboom et al., 2013). With these aims in mind, multiple red-shifted GECIs were developed from circularly permutated mApple and mRuby using structure-guided mutagenesis by testing variants with automated neuronal assays. These GECIs, including R-GECO and RCaMPs, are similar to GCaMP in that they incorporate calmodulin to bind calcium, as well as a binding peptide such as M13 or ckkap (Akerboom et al., 2013; Wu et al., 2014; Dana et al., 2016). These red-shifted GECIs have different advantages depending on the experimental application. For example, mApple-based GECIs are sensitive to calcium transients but exhibit photoswitching when illuminated with blue light, preventing simultaneous use with optogenetic photostimulation of the light-activated cation channel channelrhodopsin (Akerboom et al., 2013; Dana et al., 2016). On the other hand, RCaMP1, developed from mRuby, has lower sensitivity to calcium transients but is compatible with the blue light used for stimulation of channelrhodopsin, as it is not stimulated by blue light itself (Akerboom et al., 2013; Dana et al., 2016). The recent development of red-shifted opsins (e.g., Chrimson, bReaChES) allows latest-generation GCaMP indicators to be used in all-optical configurations (Klapoetke et al., 2014; Jennings et al., 2019). Another powerful approach offered by indicators accessible in different spectral channels is the simultaneous, parallel imaging of different genetically defined populations (Inoue et al., 2015; Dana et al., 2016). Continuing advances in cell targeting strategies will allow increasing specificity and control over opsin and calcium indicator targeting to facilitate our understanding of how the activity of neuronal circuits relates to behavior.
Importantly, there has been extensive use of ratio-metric (i.e, Fura-2 and Indo-1) and single-wavelength (i.e., Oregon Green BAPTA-1 and Fluo-4) dyes to study intracellular calcium dynamics in cultured cells and brain slices (Aponte et al., 2008; Paredes et al., 2008; Barreto-Chang and Dolmetsch, 2009). To use these dyes in vivo, multicell bolus loading (MCBL) was designed to microinject calcium sensitive acetoxymethyl (AM) ester dyes into brain regions of interest (Stosiek et al., 2003). However, similar to the challenges encountered when using most dyes, this approach does not achieve high enough signal-to-noise ratio for in vivo imaging of subcellular structures (Garaschuk et al., 2006). The dye Cal-520 has been used due to its improved sensitivity over other dyes, but it is still only suitable in vivo for very signal-rich regions such as neocortical neurons or cerebellar Purkinje cells (Tada et al., 2014). Though other dyes such as Fluo-4 are preferred for detecting action potential events, they fade rather quickly and are not conducive for staining large populations (Golshani et al., 2009).
3. Cell targeting strategies
The molecular markers present in individual cells are indicators of the presence of structural machinery that could be related to function. Distinct brain regions are comprised of a diverse array of cells including neurons, oligodendrocytes, microglia, and astrocytes that can be categorized by their molecular markers, physical boundaries, or functional importance. Due to such diversity, the dissection of cellular populations into individual groups of neurons has been very challenging. For example, a given marker can indicate neurons that have a broad array of functions across brain regions and thus genetic strategies to specifically manipulate such neurons could not be achieved simply through cross-breeding of mouse strains. Initially, there was hope that microinjection of promoter-driven viruses would be sufficient to target molecularly defined populations in a region-specific manner (Klein et al., 2002). While these strategies can be effective at transducing a target population under highly transcribed promoters (Saito et al., 2013), it leaves the chance of ectopic expression in cells also exposed to the virus (one form of “leaky” expression).
Because of these challenges, a new standard was adopted for the induction of expression of a desired transgene in a cellular population of interest. By combining breeding and viral microinjection strategies, regionally restricted groups of neurons can be further subdivided by molecularly identified populations. The implementation of the Cre-Lox Flip Excision (FLEX) switch (Schnutgen et al., 2003), also known as DIO (Double-floxed Inverse Open reading frame), enabled a level of specificity that was previously unattainable by ensuring that expression occurs only in cells that express Cre recombinase. The FLEX switch strategy resolves ectopic expression by incorporating the key genetic construct for the transgene of interest in a non-transcriptionally functional, reverse orientation (Schnutgen et al., 2003; Atasoy et al., 2008; Sohal et al., 2009). By flanking the transgene of interest with a pair of heterotypic lox sites, such as loxP and lox2272, in opposing orientations, the inversion of the transgene proceeds in a Cre recombinase-dependent manner. Thus, cells expressing Cre recombinase will express functional transgenes, whereas cells lacking Cre recombinase will not. Alternatively, the system can be used for Cre-OFF transgene control by packaging the key construct in the forward direction whereby the FLEX switch reverses the transgene to the non-functional orientation. Variations of this system have been designed to allow simultaneous expression of Cre-ON and Cre-OFF constructs (Saunders et al., 2012). Recombinant viruses (typically adeno-associated viruses) expressing GECIs can be injected into transgenic Cre mouse lines or Cre recombinase can be introduced in a region-specific manner by viral injection (Gong et al., 2007). Constraining GECI expression to relevant cell types is critical to limit functional heterogeneity and to improve the ability to detect a relationship between neuronal activity of a specific population of neurons and behavior.
4. Surgical techniques for deep-brain imaging
Although cranial window preparations may be sufficient for shallow (1 mm) depth imaging of calcium sensors using optics with appropriate working distances (Svoboda et al., 1997), deeper structures (> 1 mm) can only be accessed by relay lenses that extend the length of the imaging system to permit imaging in deep brain regions. Gradient-index (GRIN) lenses are used for this purpose. GRIN lenses are rod-shaped and are available in small diameters (< 1 mm) and multiple lengths. Their miniature profile makes them ideal for implantation into rodent brains and their optical properties make them ideal for use as relay lenses within microendoscopy systems (Jung and Schnitzer, 2003; Jung et al., 2004; Levene et al., 2004; Bocarsly et al., 2015).
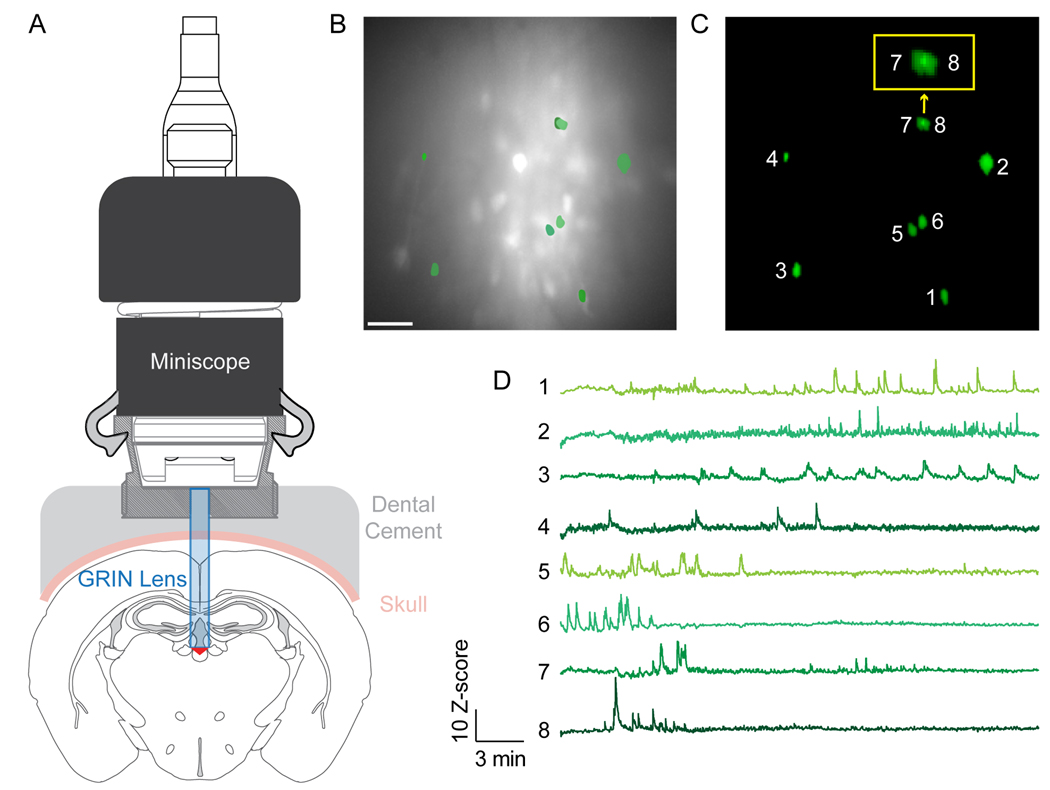
First, a viral-GECI construct is injected, and in the same surgery, a GRIN lens can be implanted (Fig. 1A). Alternatively, the experimenter can perform two surgeries, the first for viral construct injection and the second for lens implantation. Whether one surgery or two, it is advisable to offset the injection (medial, lateral, anterior, or posterior) by at least 100 μm from the center of the desired imaging target to prevent the injection syringe from damaging tissue within the lens field of view (FOV). For GRIN lens implantation a path through the brain tissue must first be created, because implantation of a GRIN lens or GRIN lens cannula displaces tissue along the path (Bocarsly et al., 2015), resulting in tissue compression between the working distance of the lens and the focal plane. To avoid obscuring visualization with this compressed tissue, a number of strategies have emerged. One strategy is to slowly lower into the brain a sterile, beveled-tip syringe with the same diameter as the lens to clear a path for the implant. The sterile syringe should not be lowered all the way to the target site to prevent tissue damage in the FOV (Resendez et al., 2016). Extremely lateral regions may require microprisms, which are right-angled prisms attached to relay lenses allowing placement of the lens lateral to, instead of superior to, the region of interest. For implantation, a straight edge dissecting knife can be used to make a vertical and a lateral incision within the brain to clear the path (Andermann et al., 2013). Alternatively, for some deep brain implants the tissue 1 – 3 mm above the target site can be aspirated with a blunt-tip syringe manually or robotically (Liang et al., 2019). If using aspiration, continue until bleeding has ceased, because blood on the bottom of the surface of the implant will increase the inflammatory response and can obscure the lens (Liang et al., 2019). Finally, the GRIN lens or the GRIN lens cannula can be positioned in the brain at the appropriate working distance from the target. The clearing of excess tissue and blood during lens or cannula insertion is a critical step towards acquisition of high-quality in vivo calcium images, especially for deep brain implants. It is worth noting that after implantation of a GRIN lens, reactive glia form sheaths around the implant. This inflammatory reaction persists for at least two weeks after implantation, but largely subsides by week four (Bocarsly et al., 2015). For this reason, virally induced cellular expression of calcium indicators tends to precede clearing of the inflammatory response—highlighting that clearing of this response is a key contributor to improved visualization of cellular boundaries in the weeks that follow.
Figure 1.
Functional imaging of neuronal activity from a deep-brain region in vivo using a single-photon miniscope.
A) Schematic cross-sectional representation of a 500-μm GRIN lens inserted above the paraventricular thalamus (PVT) and miniscope attachment for imaging in freely moving mice. An adeno-associated virus with jGCaMP7f (AAV9-syn-FLEX-jGCaMP7f-WPRE; Addgene:104492-AAV9) was injected into the PVT of Drd2-Cre transgenic mice (MGI:3836635). B) Representative image depicting the maximum intensity per pixel from a motion-corrected video. Scale bar = 100 μm. C) Selected sample ROIs from (B) extracted by CNMF-E. (Inset outlined in yellow is a zoomed-in view of neurons 7 and 8, demonstrating the ability of CNMF-E to demix overlapping signals). D) Calcium traces extracted by CNMF-E corresponding to ROIs in (C). Permission to publish miniscope drawing granted by Doric Lenses Inc.
5. Fluorescence microendoscopy systems
To visualize calcium dynamics in subsurface regions, implants must be used to relay excitation light to—and emitted signal from—GECI-expressing neurons. The widely used technique of fiber photometry is perhaps the most basic method to do this. A single optical fiber is implanted above a brain region of interest, and the measurements collected represent the overall, population-wide fluorescence of GECI-expressing cells. This technique allows access to neuronal populations deep in the brain, and animals can move freely during recording sessions. Moreover, the small footprint of the implant allows users to monitor multiple sites across the brain simultaneously, for example different hemispheres or a pair of connected neuronal populations (Sych et al., 2019) which requires more complex systems for imaging (Lecoq et al., 2014) at multiple sites (Barretto et al., 2011). This approach can enable photostimulation in one region through one optical fiber while recording the population activity of another region through a second optical fiber (Nieh et al., 2016). Since fiber photometry lacks single-cell resolution, it is generally best-suited for monitoring neuronal populations with predicted response properties that are not widely heterogeneous, otherwise changes in fluorescence may be diluted or undetectable. However, this will often be unknown a priori.
Functional dissection of neuronal populations is critical to understanding how the brain drives behavior. Even within a given region, a molecularly defined set of neurons will likely be comprised of a diverse collection of cell types (Mickelsen et al., 2019). Thus, while fiber photometry is a useful tool to capture the net activity of a population, analysis of cells as single units to understand the complexity and range of activity patterns has emerged as the key strategy for identifying nodes within a neuronal circuit. Interfacing a microscope with an implanted GRIN lens allows the visualization of the FOV in vivo with cellular and subcellular resolution (Broussard et al., 2018). By pairing this technology with GECIs, the calcium dynamics of single cells deep in the brain can be monitored in real time via one- or two-photon microscopy.
Two-photon microscopy was originally proposed in the dissertation of Maria Göppert-Mayer (Göppert-Mayer, 1931). This approach uses a pair of low energy photons generated by near infra-red light for excitation of the fluorophore of interest. The photons arrive at a fluorophore molecule nearly simultaneously, within 0.5 femtoseconds of each other, and the fluorophore emits a single photon of the wavelength corresponding to its profile (Helmchen and Denk, 2005). This results in low axial spread of the point function and enables optical sectioning along the z-axis with substantially reduced out-of-focus multiphoton absorption (Rubart, 2004; Helmchen and Denk, 2005). The primary advantages of two-photon microscopy are the use of long-wavelength light to excite fluorophores, which decreases the damage that occurs from short wavelengths (Chen et al., 2002), and the resolution, which enables visualization of calcium concentration dynamics in subcellular compartments (Meng et al., 2019) as transients driven by cellular events including action potentials (Yasuda et al., 2004). For excitable cell types like neurons, calcium concentration can vary widely between the soma and spines. Change in calcium concentration is dependent on net influx, extrusion, and buffering of calcium. Because buffering of calcium occurs very rapidly in the spines of the dendritic compartment, this calcium flux can have a local influence without changing the electrical potential of the entire dendrite. Therefore, high resolution imaging at a fast sampling rate is required to image these fast calcium events. Another major advantage of the two-photon system is the ability to simultaneously excite two fluorophores—one that is calcium-sensitive and one that is not. Use of a calcium-insensitive fluorophore that is visible at baseline can aid in identifying cellular structures and reduces sensitivity of the data to background fluctuations. In addition, the two-photon system reduces photobleaching and photodamage outside of the focal plane by focusing light onto the laser focal volume (Yang and Yuste, 2017). With proper habituation and careful task design, calcium dynamics can be monitored in awake head-fixed animals during behaviors of interest (Dombeck et al., 2010). For example, measurement of neuronal responses to environmental cues, rewards/punishments, or drugs of abuse lend themselves well to two-photon microscopy. However, two-photon microscopy has not yet been commercially optimized for use in freely moving animals because of challenges related to the beam scanning mechanism (Yang and Yuste, 2017).
Single-photon microscopy utilizes epifluorescence to illuminate a FOV under a miniaturized microendoscope (“miniscope”) in freely moving animals. The single-photon system is substantially lighter and smaller than commercial two-photon systems, enabling the head-mounted miniscope (Ferezou et al., 2006) to be tethered to an LED light source via a fiber optic cable (Fig. 1A). In addition, less motion artifact is observed in freely moving animals compared to head-fixed animals (Ghosh et al., 2011). The resolution is sufficient to visualize individual cells, and also dynamics in dense dendritic fields (Murayama et al., 2007). Acquisition can occur at rapid frame rates (>100 Hz) to allow for time-locking calcium events and behavioral events (Flusberg et al., 2008). Following habituation, neuronal activity during natural, learned, and complex behavioral paradigms in freely moving animals can be studied. Miniscopes are small enough for mice to conduct tasks such as the rotarod test, foot fault, and the Morris Water Maze (Lee et al., 2016). Single-photon microendoscopy combined with an elevated plus maze test identified vasoactive intestinal peptide neurons from the broader GABAergic circuitry in the prefrontal cortex by their ability to gate mouse activity on the open arms of the elevated plus maze (Lee et al., 2019), indicating their critical role in control of anxiety-like behavior. Taken together, these studies are examples of how in vivo calcium imaging can be used to enhance our understanding of neural mechanisms underlying behavior. Ready-to-use miniscope systems are commercially available through vendors such as Inscopix and Doric Lenses. They can also be assembled from an open-source parts list costing just over $1000 USD (http://miniscope.org).
Selection of an imaging system should be made when considering the experimental question to leverage the advantages of fiber photometry, single-photon microendoscopy, or two-photon microendoscopy. In addition, by understanding the disadvantages and challenges attributed to each method, experiments can be designed within the limitations of the selected imaging system. Careful consideration about the costs and skills required for the implementation of any in vivo imaging system can help users choose the setup that is most appropriate for their research (Table 1).
6. Data analysis
An important factor to consider when designing optical imaging experiments is the approach for data extraction and analysis following collection. Depending on acquisition parameters, functional imaging dataset file sizes approach or exceed 1 GB/min, which necessitates careful consideration of all subsequent data handling, processing, and storage. Moreover, these datasets require preprocessing before any downstream analysis can be performed. This can pose a significant barrier to new users without sophisticated programming skills. Here we summarize current approaches for analysis of optical imaging datasets.
Preprocessing pipelines are compatible with one or more time series file types, such as AVI or TIFF, which may differ from the file type collected by the recording software. Converting data formats with ImageJ/FIJI (Schindelin et al., 2012) is generally successful and recommended in such cases.
Typically, calcium imaging analysis begins with a motion correction step to remove artifacts evoked by natural brain movement, which can be exaggerated in freely moving animals (Fig. 1B). Subsequent neuronal activity extraction is critically dependent on a stable video, and thus removing movement artifacts is vitally important. For small FOVs, movement problems can be solved under assumptions of translational movement with standard rigid motion correction methods (Thevenaz et al., 1998), where for example all frames are matched to a standard template. However, as the size of the FOV grows, non-uniform motion artifacts arise across the FOV, requiring methods that correct for movement in a piecewise manner, which the non-rigid motion correction (NoRMCorre) approach sought to address (Pnevmatikakis and Giovannucci, 2017). However, by using block-based registration, assumptions and restrictions are made regarding the form and extent of movements that may not always be accurate to single-photon microendoscopic data. Moreover, since all frames from a given dataset are registered to one reference frame, movement correction errors can be prone to propagation. Recent pipelines for single-photon microendoscopic data typically employ NoRMCorre or combine rigid and non-rigid modules to increase processing efficiency and decrease registration time (Lu et al., 2018; Giovannucci et al., 2019).
Following motion correction, the main analysis task is “source extraction.” Calcium-imaging data are innately high-dimensional (npixels × nframes) and meaningful interpretation of the data requires the extraction of the spatial locations of putative neuronal components and their respective fluorescent signals across time. This was initially performed via region-of-interest (ROI) analysis, which involved manually circling clusters of pixels suggestive of GECI expression. While successful in some applications, ROI analysis is not well-suited to datasets containing a high density and large number of GCaMP-expressing cells, mainly due to labor intensity and difficulties associated with the manual separation of signal crosstalk (overlapping and/or out-of-focus regions). ROI analysis is also susceptible to background contamination. ROI analysis can be conducted with the ImageJ MosaicSuite plugins (Sbalzarini and Koumoutsakos, 2005). Python implementations of ROI analysis are also available in SIMA and SamuROI (Kaifosh et al., 2014; Rueckl et al., 2017).
Automated source extraction began with a combination of principal component analysis (PCA) to find and discard dimensions encoding noise, followed by independent component analysis (ICA) for extracting spatial footprints and fluorescent traces (Mukamel et al., 2009). While faster and less prone to crosstalk than ROI analysis, PCA/ICA has several pitfalls as well. First, the number of neuronal components must be manually entered following visual estimation. Second, as a stochastic algorithm, PCA/ICA results are likely to be different across repeated analyses of a dataset. Third, it is ill-suited to datasets from neuronal populations with sparse activity levels and low SNR. Finally, PCA/ICA is a linear demixing method and does not perform well when neural components substantially overlap (Pnevmatikakis et al., 2016; Lu et al., 2018).
Constrained nonnegative matrix factorization (CNMF) was introduced to simultaneously denoise, deconvolve, and demix calcium imaging data, accounting well for overlapping spatial footprints (Pnevmatikakis et al., 2016). Optimized for two-photon imaging data, CNMF works best in situations with high SNR, stable FOV, and simple spatiotemporal structure, similar to another two-photon data analysis package, Suite2p (Pachitariu et al., 2017), available in MATLAB and Python. However, CNMF does not perform well on single-photon microendoscopic datasets as the background is not modeled well. Thus, extended CNMF for microendoscopic data (CNMF-E) was developed to model a more accurate and flexible spatiotemporal background and incorporates better algorithms to initialize and fit neural components, producing better results in low SNR conditions and improving downstream analyses as compared to PCA/ICA extractions (Zhou et al., 2018) (Fig. 1C). As with PCA/ICA, CNMF-E can have problems with false-negative and false-positive ROI identification (although manual intervention steps have been successfully implemented), depends on parameter tuning, and will fail with a non-stable imaging field (Lu et al., 2018). Python and MATLAB each support CNMF in the Calcium Imaging Analysis (CAIMAN) pipeline (Giovannucci et al., 2019). CNMF-E is available in MATLAB, and core components of NoRMCorre and CNMF-E have been combined into a continuous analysis pipeline.
The EZCalcium pipeline was recently introduced as a graphical user interface (GUI)-based approach in MATLAB for users without prior programming experience (Cantu et al., 2020). This pipeline is currently optimized for two-photon imaging data, but its open source nature should allow a module such as CNMF-E to be easily integrated. Another source extraction module called A Technique for Extracting Neuronal Activity from Single Photon Neuronal Image Sequences (TENASPIS) is available in MATLAB (Kinsky et al., 2018; Mau et al., 2018), but has only been used for hippocampal populations and has not been benchmarked against other source extraction methods.
A recent end-to-end pipeline developed for microendoscopic data (MIN1PIPE) purported to solve both the movement correction problem (outperforming NoRMCorre) and source extraction problem (outperforming PCA/ICA and CNMF-E) (Lu et al., 2018). MIN1PIPE does not completely eliminate the need for some manual ROI pruning, but it is nearly automated, and the involved parameter-setting is straightforward and error-tolerant. MIN1PIPE’s precision may be improved with future advances, but it is likely the most easily implemented and accurate pipeline currently available for single-photon microendoscopic data. MIN1PIPE code is available for MATLAB.
Finally, one of the most powerful capabilities enabled by optical imaging is the ability to revisit the same FOV and neuronal populations across multiple sessions, and thus longitudinally monitor within-neuron changes in activity, which is crucial to research areas such as learning and long-term memory. Automated implementations of inter-session cell registration are available in CAIMAN and CellReg (Sheintuch et al., 2017; Giovannucci et al., 2019).
File sizes, hardware capabilities, and preprocessing methods chosen will all determine the time cost associated with optical imaging datasets. Processing speeds are substantially increased by spatial and/or temporal downsampling, which are often required as many analysis methods hold the video in random access memory (RAM) for analysis. Thus, computers with 32 GB RAM or more are generally desirable for analysis on local machines, and computers with up to 128 GB RAM have been used in published benchmark comparisons between analysis methods (Zhou et al., 2018). Cloud computing services have been used to significantly speed up processing, offering a promising avenue for future optimization (Namboodiri et al., 2019).
After obtaining the extracted data, the experimenter must overcome the next obstacle of figuring out what to do with it. This “post-processing analysis” will be determined by the experimental question. Simple baseline-corrected, time-locked fluorescence changes can be used to report how neurons from a dataset respond acutely to behavioral events such as foot shocks, social interaction, or food consumption (Jennings et al., 2015; Zhou et al., 2018; Jennings et al., 2019; Rossi et al., 2019) (Fig. 1D). More sophisticated analyses may seek to determine how hippocampal place cells encode spatial location (Ziv et al., 2013; Kinsky et al., 2018), how neuronal responses change over time (Namboodiri et al., 2019; Otis et al., 2019; Rossi et al., 2019), or how activity data from large populations of neurons can be used to decode behavioral states (Gründemann et al., 2019; Rubin et al., 2019). While the possibilities for data analyses and presentation are boundless, some consideration of post-processing analysis is required prior to experimentation to ensure all relevant collected data may be a factor in subsequent steps. Combining functional imaging methods with recent advances in sophisticated behavioral scoring such as SimBA and DeepLabCut (Nath et al., 2019; Nilsson et al., 2020) is likely to yield detailed insights into the relationship between neuronal activity and behavior in the near future. See Table 2 for a list of selected methods for functional imaging analysis.
Table 2.
Directory of selected methods for functional imaging analysis.
7. Conclusions
Over the past decade we have observed rapid development towards state-of-the-art techniques for imaging neuronal activity deep in the brain. Advancements in deep brain neuronal imaging have led to insights about how central inputs and outputs relate to hunger and thirst (Betley et al., 2015; Lutas et al., 2019), taste (Barretto et al., 2015; Patel et al., 2019), sleep (Weber et al., 2018; Blanco-Centurion et al., 2019), narcoleptic cataplexy (Sun et al., 2019), pain (Hua et al., 2020), anxiety (Shin et al., 2018), locomotion (Karnani et al., 2020), social interaction (Francis et al., 2017; Lu et al., 2017), threat responses (Gore et al., 2014), defensive strategies (Lecca et al., 2020), and drug abuse (Cameron et al., 2019; Heinsbroek et al., 2020). The increasing popularity of population-level analyses will continue to elucidate neuronal linkages to behavior (Gründemann et al., 2019; Rubin et al., 2019; Xu et al., 2020). Still, opportunities for major technical advancements remain, including optimization of surgical procedures, improvement of the endoscope-to-GRIN lens interface using screw-on microscope bodies, and miniaturization of the implant footprint for simultaneous multi-site recordings (de Groot et al., 2020). In addition, technical improvements to manipulate focus along the z-axis in single-photon systems and biological improvements for multi-channel imaging of cellular nuclei markers in combination with cytosolic markers to help distinguish cells will be beneficial. Once these challenges are resolved, versatile and low-cost imaging systems, potentially including in vivo two-photon imaging in freely moving animals (Helmchen et al., 2001; Engelbrecht et al., 2008; Helmchen et al., 2013), will take aim at the growing array of deep brain cell types defined by molecular characteristics (Saunders and Sabatini, 2015), anatomical connections (Chen et al., 2018; Li et al., 2020), and activity patterns (Guenthner et al., 2013; Grewe et al., 2017; Gründemann et al., 2019; Rubin et al., 2019).
Highlights.
Calcium imaging is a powerful tool for studying neuronal function
The many options for this technique make it flexible yet challenging
Deep brain imaging is possible with the use of GRIN lenses
We explain basic concepts and outline approaches to various experimental scenarios
Acknowledgements
The authors acknowledge with gratitude C. Lupica and I. Tzameret for comments on the manuscript and NIDA IRP Visual Media, particularly A. Russell and L. Brick, for brain slice drawings. This work is supported by the National Institute on Drug Abuse Intramural Research Program (NIDA IRP), U.S. National Institutes of Health (NIH).
Footnotes
CRediT Authorship Contribution Statement
Brenton T. Laing: Conceptualization, Investigation, Writing – Original Draft, Writing – Review & Editing, Visualization. Justin N. Siemian: Conceptualization, Formal analysis, Writing – Original Draft, Writing – Review & Editing, Visualization. Sarah Sarsfield: Resources, Writing – Review & Editing, Visualization. Yeka Aponte: Conceptualization, Funding acquisition, Resources, Supervision, Writing – Review & Editing.
Ethics Statement
All experimental protocols were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and with the approval of the National Institute on Drug Abuse Animal Care and Use Committee.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akerboom J et al. (2013) Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Frontiers in Molecular Neuroscience 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andermann ML, Gilfoy NB, Goldey GJ, Sachdev RN, Wolfel M, McCormick DA, Reid RC, Levene MJ (2013) Chronic cellular imaging of entire cortical columns in awake mice using microprisms. Neuron 80:900–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aponte Y, Bischofberger J, Jonas P (2008) Efficient Ca2+ buffering in fast-spiking basket cells of rat hippocampus. J Physiol 586:2061–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy D, Aponte Y, Su HH, Sternson SM (2008) A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci 28:7025–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto-Chang OL, Dolmetsch RE (2009) Calcium Imaging of Cortical Neurons using Fura-2 AM. JoVE:e1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto RP, Ko TH, Jung JC, Wang TJ, Capps G, Waters AC, Ziv Y, Attardo A, Recht L, Schnitzer MJ (2011) Time-lapse imaging of disease progression in deep brain areas using fluorescence microendoscopy. Nature medicine 17:223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto RPJ, Gillis-Smith S, Chandrashekar J, Yarmolinsky DA, Schnitzer MJ, Ryba NJP, Zuker CS (2015) The neural representation of taste quality at the periphery. Nature 517:373–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betley JN, Xu S, Cao ZFH, Gong R, Magnus CJ, Yu Y, Sternson SM (2015) Neurons for hunger and thirst transmit a negative-valence teaching signal. Nature 521:180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Centurion C, Luo S, Spergel DJ, Vidal-Ortiz A, Oprisan SA, Van den Pol AN, Liu M, Shiromani PJ (2019) Dynamic Network Activation of Hypothalamic MCH Neurons in REM Sleep and Exploratory Behavior. The Journal of Neuroscience 39:4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocarsly ME, Jiang WC, Wang C, Dudman JT, Ji N, Aponte Y (2015) Minimally invasive microendoscopy system for in vivo functional imaging of deep nuclei in the mouse brain. Biomed Opt Express 6:4546–4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broussard GJ, Liang Y, Fridman M, Unger EK, Meng G, Xiao X, Ji N, Petreanu L, Tian L (2018) In vivo measurement of afferent activity with axon-specific calcium imaging. Nature Neuroscience 21:1272–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron CM, Murugan M, Choi JY, Engel EA, Witten IB (2019) Increased Cocaine Motivation Is Associated with Degraded Spatial and Temporal Representations in IL-NAc Neurons. Neuron 103:80–91.e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu DA, Wang B, Gongwer MW, He CX, Goel A, Suresh A, Kourdougli N, Arroyo ED, Zeiger W, Portera-Cailliau C (2020) EZcalcium: Open-Source Toolbox for Analysis of Calcium Imaging Data. Front Neural Circuits 14:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IH, Chu SW, Sun CK, Cheng PC, Lin BL (2002) Wavelength dependent damage in biological multi-photon confocal microscopy: A micro-spectroscopic comparison between femtosecond Ti:sapphire and Cr:forsterite laser sources. Optical and Quantum Electronics 34:1251–1266. [Google Scholar]
- Chen K-S, Xu M, Zhang Z, Chang W-C, Gaj T, Schaffer DV, Dan Y (2018) A Hypothalamic Switch for REM and Non-REM Sleep. Neuron 97:1168–1176.e1164. [DOI] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K, Kim DS (2013) Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder G, Ahanonu B, Grewe BF, Wang D, Schnitzer MJ, Scherrer G (2019) An amygdalar neural ensemble that encodes the unpleasantness of pain. Science 363:276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Pinto L, Dan Y (2016) Calcium imaging of sleep–wake related neuronal activity in the dorsal pons. Nature Communications 7:10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Sun Y, Mohar B, Hulse BK, Kerlin AM, Hasseman JP, Tsegaye G, Tsang A, Wong A, Patel R, Macklin JJ, Chen Y, Konnerth A, Jayaraman V, Looger LL, Schreiter ER, Svoboda K, Kim DS (2019) High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nature Methods 16:649–657. [DOI] [PubMed] [Google Scholar]
- Dana H, Mohar B, Sun Y, Narayan S, Gordus A, Hasseman JP, Tsegaye G, Holt GT, Hu A, Walpita D, Patel R, Macklin JJ, Bargmann CI, Ahrens MB, Schreiter ER, Jayaraman V, Looger LL, Svoboda K, Kim DS (2016) Sensitive red protein calcium indicators for imaging neural activity. eLife 5:e12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot A, van den Boom BJG, van Genderen RM, Coppens J, van Veldhuijzen J, Bos J, Hoedemaker H, Negrello M, Willuhn I, De Zeeuw CI, Hoogland TM (2020) NINscope, a versatile miniscope for multi-region circuit investigations. eLife 9:e49987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombeck DA, Harvey CD, Tian L, Looger LL, Tank DW (2010) Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat Neurosci 13:1433–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass AM, Kucukdereli H, Ponserre M, Markovic M, Gründemann J, Strobel C, Alcala Morales PL, Conzelmann K-K, Lüthi A, Klein R (2017) Central amygdala circuits modulate food consumption through a positive-valence mechanism. Nature Neuroscience 20:1384–1394. [DOI] [PubMed] [Google Scholar]
- Engelbrecht CJ, Johnston RS, Seibel EJ, Helmchen F (2008) Ultra-compact fiber-optic two-photon microscope for functional fluorescence imaging in vivo. Opt Express 16:5556–5564. [DOI] [PubMed] [Google Scholar]
- Ferezou I, Bolea S, Petersen CCH (2006) Visualizing the Cortical Representation of Whisker Touch: Voltage-Sensitive Dye Imaging in Freely Moving Mice. Neuron 50:617–629. [DOI] [PubMed] [Google Scholar]
- Flusberg BA, Nimmerjahn A, Cocker ED, Mukamel EA, Barretto RPJ, Ko TH, Burns LD, Jung JC, Schnitzer MJ (2008) High-speed, miniaturized fluorescence microscopy in freely moving mice. Nature Methods 5:935–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis TC, Chandra R, Gaynor A, Konkalmatt P, Metzbower SR, Evans B, Engeln M, Blanpied TA, Lobo MK (2017) Molecular basis of dendritic atrophy and activity in stress susceptibility. Molecular Psychiatry 22:1512–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaschuk O, Milos R-I, Konnerth A (2006) Targeted bulk-loading of fluorescent indicators for two-photon brain imaging in vivo. Nature Protocols 1:380–386. [DOI] [PubMed] [Google Scholar]
- Ghosh KK, Burns LD, Cocker ED, Nimmerjahn A, Ziv Y, Gamal AE, Schnitzer MJ (2011) Miniaturized integration of a fluorescence microscope. Nat Methods 8:871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci A, Friedrich J, Gunn P, Kalfon J, Brown BL, Koay SA, Taxidis J, Najafi F, Gauthier JL, Zhou P, Khakh BS, Tank DW, Chklovskii DB, Pnevmatikakis EA (2019) CaImAn an open source tool for scalable calcium imaging data analysis. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golshani P, Gonçalves JT, Khoshkhoo S, Mostany R, Smirnakis S, Portera-Cailliau C (2009) Internally Mediated Developmental Desynchronization of Neocortical Network Activity. The Journal of Neuroscience 29:10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, Gerfen CR (2007) Targeting Cre Recombinase to Specific Neuron Populations with Bacterial Artificial Chromosome Constructs. The Journal of Neuroscience 27:9817–9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göppert-Mayer M (1931) Über Elementarakte mit zwei Quantensprüngen. Annalen der Physik 401:273–294. [Google Scholar]
- Gore BB, Soden ME, Zweifel LS (2014) Visualization of plasticity in fear-evoked calcium signals in midbrain dopamine neurons. Learning & Memory 21:575–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe BF, Gründemann J, Kitch LJ, Lecoq JA, Parker JG, Marshall JD, Larkin MC, Jercog PE, Grenier F, Li JZ, Lüthi A, Schnitzer MJ (2017) Neural ensemble dynamics underlying a long-term associative memory. Nature 543:670–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründemann J, Bitterman Y, Lu T, Krabbe S, Grewe BF, Schnitzer MJ, Lüthi A (2019) Amygdala ensembles encode behavioral states. Science 364:eaav8736. [DOI] [PubMed] [Google Scholar]
- Guenthner Casey J, Miyamichi K, Yang Helen H, Heller HC, Luo L (2013) Permanent Genetic Access to Transiently Active Neurons via TRAP: Targeted Recombination in Active Populations. Neuron 78:773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinsbroek JA, Bobadilla A-C, Dereschewitz E, Assali A, Chalhoub RM, Cowan CW, Kalivas PW (2020) Opposing Regulation of Cocaine Seeking by Glutamate and GABA Neurons in the Ventral Pallidum. Cell reports 30:2018–2027.e2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmchen F, Denk W (2005) Deep tissue two-photon microscopy. Nature Methods 2:932–940. [DOI] [PubMed] [Google Scholar]
- Helmchen F, Denk W, Kerr JND (2013) Miniaturization of Two-Photon Microscopy for Imaging in Freely Moving Animals. Cold Spring Harbor Protocols 2013:pdb.top078147. [DOI] [PubMed] [Google Scholar]
- Helmchen F, Fee MS, Tank DW, Denk W (2001) A Miniature Head-Mounted Two-Photon Microscope: High-Resolution Brain Imaging in Freely Moving Animals. Neuron 31:903–912. [DOI] [PubMed] [Google Scholar]
- Hua T, Chen B, Lu D, Sakurai K, Zhao S, Han B-X, Kim J, Yin L, Chen Y, Lu J, Wang F (2020) General anesthetics activate a potent central pain-suppression circuit in the amygdala. Nature Neuroscience 23:854–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Takeuchi A, Horigane S-i, Ohkura M, Gengyo-Ando K, Fujii H, Kamijo S, Takemoto-Kimura S, Kano M, Nakai J, Kitamura K, Bito H (2015) Rational design of a high-affinity, fast, red calcium indicator R-CaMP2. Nature Methods 12:64–70. [DOI] [PubMed] [Google Scholar]
- Jennings JH, Kim CK, Marshel JH, Raffiee M, Ye L, Quirin S, Pak S, Ramakrishnan C, Deisseroth K (2019) Interacting neural ensembles in orbitofrontal cortex for social and feeding behaviour. Nature 565:645–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings JH, Ung RL, Resendez SL, Stamatakis AM, Taylor JG, Huang J, Veleta K, Kantak PA, Aita M, Shilling-Scrivo K, Ramakrishnan C, Deisseroth K, Otte S, Stuber GD (2015) Visualizing hypothalamic network dynamics for appetitive and consummatory behaviors. Cell 160:516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JC, Schnitzer MJ (2003) Multiphoton endoscopy. Opt Lett 28:902–904. [DOI] [PubMed] [Google Scholar]
- Jung JC, Mehta AD, Aksay E, Stepnoski R, Schnitzer MJ (2004) In vivo mammalian brain imaging using one- and two-photon fluorescence microendoscopy. J Neurophysiol 92:3121–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaifosh P, Zaremba JD, Danielson NB, Losonczy A (2014) SIMA: Python software for analysis of dynamic fluorescence imaging data. Frontiers in neuroinformatics 8:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamigaki T, Dan Y (2017) Delay activity of specific prefrontal interneuron subtypes modulates memory-guided behavior. Nature Neuroscience 20:854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani MM, Schöne C, Bracey EF, González JA, Viskaitis P, Li H-T, Adamantidis A, Burdakov D (2020) Role of spontaneous and sensory orexin network dynamics in rapid locomotion initiation. Progress in Neurobiology 187:101771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsky NR, Sullivan DW, Mau W, Hasselmo ME, Eichenbaum HB (2018) Hippocampal Place Fields Maintain a Coherent and Flexible Map across Long Timescales. Current Biology 28:3578–3588.e3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Ogawa SK, Roy DS, Okuyama T, Morrissey MD, Smith LM, Redondo RL, Tonegawa S (2017) Engrams and circuits crucial for systems consolidation of a memory. Science 356:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapoetke NC et al. (2014) Independent optical excitation of distinct neural populations. Nat Methods 11:338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RL, Hamby ME, Gong Y, Hirko AC, Wang S, Hughes JA, King MA, Meyer EM (2002) Dose and Promoter Effects of Adeno-Associated Viral Vector for Green Fluorescent Protein Expression in the Rat Brain. Experimental Neurology 176:66–74. [DOI] [PubMed] [Google Scholar]
- Lecca S, Namboodiri VMK, Restivo L, Gervasi N, Pillolla G, Stuber GD, Mameli M (2020) Heterogeneous Habenular Neuronal Ensembles during Selection of Defensive Behaviors. Cell reports 31:107752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoq J, Savall J, Vučinić D, Grewe BF, Kim H, Li JZ, Kitch LJ, Schnitzer MJ (2014) Visualizing mammalian brain area interactions by dual-axis two-photon calcium imaging. Nature Neuroscience 17:1825–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AT, Cunniff MM, See JZ, Wilke SA, Luongo FJ, Ellwood IT, Ponnavolu S, Sohal VS (2019) VIP Interneurons Contribute to Avoidance Behavior by Regulating Information Flow across Hippocampal-Prefrontal Networks. Neuron 102:1223–1234.e1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Han JH (2020) Successful In vivo Calcium Imaging with a Head-Mount Miniaturized Microscope in the Amygdala of Freely Behaving Mouse. J Vis Exp. [DOI] [PubMed] [Google Scholar]
- Lee SA, Holly KS, Voziyanov V, Villalba SL, Tong R, Grigsby HE, Glasscock E, Szele FG, Vlachos I, Murray TA (2016) Gradient Index Microlens Implanted in Prefrontal Cortex of Mouse Does Not Affect Behavioral Test Performance over Time. PLOS ONE 11:e0146533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene MJ, Dombeck DA, Kasischke KA, Molloy RP, Webb WW (2004) In vivo multiphoton microscopy of deep brain tissue. J Neurophysiol 91:1908–1912. [DOI] [PubMed] [Google Scholar]
- Li B, Nguyen TP, Ma C, Dan Y (2020) Inhibition of impulsive action by projection-defined prefrontal pyramidal neurons. Proceedings of the National Academy of Sciences 117:17278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, Zhang L, Moffitt C, Li Y, Lin D-T (2019) An open-source automated surgical instrument for microendoscope implantation. Journal of Neuroscience Methods 311:83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Li C, Singh-Alvarado J, Zhou ZC, Frohlich F, Mooney R, Wang F (2018) MIN1PIPE: A Miniscope 1-Photon-Based Calcium Imaging Signal Extraction Pipeline. Cell reports 23:3673–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Sun W, Liang Y, Kerlin A, Bierfeld J, Seelig JD, Wilson DE, Scholl B, Mohar B, Tanimoto M, Koyama M, Fitzpatrick D, Orger MB, Ji N (2017) Video-rate volumetric functional imaging of the brain at synaptic resolution. Nat Neurosci 20:620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutas A, Kucukdereli H, Alturkistani O, Carty C, Sugden AU, Fernando K, Diaz V, Flores-Maldonado V, Andermann ML (2019) State-specific gating of salient cues by midbrain dopaminergic input to basal amygdala. Nature Neuroscience 22:1820–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianova E, Aronson S, Proulx CD (2019) Multi-Fiber Photometry to Record Neural Activity in Freely-Moving Animals. J Vis Exp. [DOI] [PubMed] [Google Scholar]
- Marvin JS, Shimoda Y, Magloire V, Leite M, Kawashima T, Jensen TP, Kolb I, Knott EL, Novak O, Podgorski K, Leidenheimer NJ, Rusakov DA, Ahrens MB, Kullmann DM, Looger LL (2019) A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nature Methods 16:763–770. [DOI] [PubMed] [Google Scholar]
- Marvin JS, Borghuis BG, Tian L, Cichon J, Harnett MT, Akerboom J, Gordus A, Renninger SL, Chen T-W, Bargmann CI, Orger MB, Schreiter ER, Demb JB, Gan W-B, Hires SA, Looger LL (2013) An optimized fluorescent probe for visualizing glutamate neurotransmission. Nature Methods 10:162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mau W, Sullivan DW, Kinsky NR, Hasselmo ME, Howard MW, Eichenbaum H (2018) The Same Hippocampal CA1 Population Simultaneously Codes Temporal Information over Multiple Timescales. Current Biology 28:1499–1508.e1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng G, Liang Y, Sarsfield S, Jiang WC, Lu R, Dudman JT, Aponte Y, Ji N (2019) High-throughput synapse-resolving two-photon fluorescence microendoscopy for deep-brain volumetric imaging in vivo. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelsen LE, Bolisetty M, Chimileski BR, Fujita A, Beltrami EJ, Costanzo JT, Naparstek JR, Robson P, Jackson AC (2019) Single-cell transcriptomic analysis of the lateral hypothalamic area reveals molecularly distinct populations of inhibitory and excitatory neurons. Nat Neurosci 22:642–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamel EA, Nimmerjahn A, Schnitzer MJ (2009) Automated analysis of cellular signals from large-scale calcium imaging data. Neuron 63:747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama M, Pérez-Garci E, Lüscher H-R, Larkum ME (2007) Fiberoptic System for Recording Dendritic Calcium Signals in Layer 5 Neocortical Pyramidal Cells in Freely Moving Rats. Journal of Neurophysiology 98:1791–1805. [DOI] [PubMed] [Google Scholar]
- Nakai J, Ohkura M, Imoto K (2001) A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nature Biotechnology 19:137–141. [DOI] [PubMed] [Google Scholar]
- Namboodiri VMK, Otis JM, van Heeswijk K, Voets ES, Alghorazi RA, Rodriguez-Romaguera J, Mihalas S, Stuber GD (2019) Single-cell activity tracking reveals that orbitofrontal neurons acquire and maintain a long-term memory to guide behavioral adaptation. Nat Neurosci 22:1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath T, Mathis A, Chen AC, Patel A, Bethge M, Mathis MW (2019) Using DeepLabCut for 3D markerless pose estimation across species and behaviors. Nature Protocols 14:2152–2176. [DOI] [PubMed] [Google Scholar]
- Nieh EH, Vander Weele CM, Matthews GA, Presbrey KN, Wichmann R, Leppla CA, Izadmehr EM, Tye KM (2016) Inhibitory Input from the Lateral Hypothalamus to the Ventral Tegmental Area Disinhibits Dopamine Neurons and Promotes Behavioral Activation. Neuron 90:1286–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaos D, Randa K, Thomas W, Bakhos AT, Khalid S, Vasilis N (2008) Performance of the red-shifted fluorescent proteins in deep-tissue molecular imaging applications. Journal of Biomedical Optics 13:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson SR, Goodwin NL, Choong JJ, Hwang S, Wright HR, Norville ZC, Tong X, Lin D, Bentzley BS, Eshel N, McLaughlin RJ, Golden SA (2020) Simple Behavioral Analysis (SimBA) – an open source toolkit for computer classification of complex social behaviors in experimental animals. bioRxiv:2020.2004.2019.049452. [Google Scholar]
- Otis JM, Zhu M, Namboodiri VMK, Cook CA, Kosyk O, Matan AM, Ying R, Hashikawa Y, Hashikawa K, Trujillo-Pisanty I, Guo J, Ung RL, Rodriguez-Romaguera J, Anton ES, Stuber GD (2019) Paraventricular Thalamus Projection Neurons Integrate Cortical and Hypothalamic Signals for Cue-Reward Processing. Neuron 103:423–431.e424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachitariu M, Stringer C, Dipoppa M, Schröder S, Rossi LF, Dalgleish H, Carandini M, Harris KD (2017) Suite2p: beyond 10,000 neurons with standard two-photon microscopy. bioRxiv:061507. [Google Scholar]
- Paredes RM, Etzler JC, Watts LT, Zheng W, Lechleiter JD (2008) Chemical calcium indicators. Methods 46:143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JM, Swanson J, Ung K, Herman A, Hanson E, Ortiz-Guzman J, Selever J, Tong Q, Arenkiel BR (2019) Sensory perception drives food avoidance through excitatory basal forebrain circuits. eLife 8:e44548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriarchi T, Cho JR, Merten K, Howe MW, Marley A, Xiong W-H, Folk RW, Broussard GJ, Liang R, Jang MJ, Zhong H, Dombeck D, von Zastrow M, Nimmerjahn A, Gradinaru V, Williams JT, Tian L (2018) Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360:eaat4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnevmatikakis EA, Giovannucci A (2017) NoRMCorre: An online algorithm for piecewise rigid motion correction of calcium imaging data. J Neurosci Methods 291:83–94. [DOI] [PubMed] [Google Scholar]
- Pnevmatikakis Eftychios A, Soudry D, Gao Y, Machado TA, Merel J, Pfau D, Reardon T, Mu Y, Lacefield C, Yang W, Ahrens M, Bruno R, Jessell TM, Peterka Darcy S, Yuste R, Paninski L (2016) Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron 89:285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resendez SL, Jennings JH, Ung RL, Namboodiri VM, Zhou ZC, Otis JM, Nomura H, McHenry JA, Kosyk O, Stuber GD (2016) Visualization of cortical, subcortical and deep brain neural circuit dynamics during naturalistic mammalian behavior with head-mounted microscopes and chronically implanted lenses. Nat Protoc 11:566–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi MA, Basiri ML, McHenry JA, Kosyk O, Otis JM, van den Munkhof HE, Bryois J, Hubel C, Breen G, Guo W, Bulik CM, Sullivan PF, Stuber GD (2019) Obesity remodels activity and transcriptional state of a lateral hypothalamic brake on feeding. Science 364:1271–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubart M (2004) Two-Photon Microscopy of Cells and Tissue. Circulation Research 95:1154–1166. [DOI] [PubMed] [Google Scholar]
- Rubin A, Sheintuch L, Brande-Eilat N, Pinchasof O, Rechavi Y, Geva N, Ziv Y (2019) Revealing neural correlates of behavior without behavioral measurements. Nature Communications 10:4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueckl M, Lenzi SC, Moreno-Velasquez L, Parthier D, Schmitz D, Ruediger S, Johenning FW (2017) SamuROI, a Python-Based Software Tool for Visualization and Analysis of Dynamic Time Series Imaging at Multiple Spatial Scales. Frontiers in neuroinformatics 11:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Tsujino N, Hasegawa E, Akashi K, Abe M, Mieda M, Sakimura K, Sakurai T (2013) GABAergic neurons in the preoptic area send direct inhibitory projections to orexin neurons. Frontiers in Neural Circuits 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Sabatini BL (2015) Cre Activated and Inactivated Recombinant Adeno-Associated Viral Vectors for Neuronal Anatomical Tracing or Activity Manipulation. Current Protocols in Neuroscience 72:1.24.21–21.24.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Johnson CA, Sabatini BL (2012) Novel recombinant adeno-associated viruses for Cre activated and inactivated transgene expression in neurons. Front Neural Circuits 6:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbalzarini IF, Koumoutsakos P (2005) Feature point tracking and trajectory analysis for video imaging in cell biology. J Struct Biol 151:182–195. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnutgen F, Doerflinger N, Calleja C, Wendling O, Chambon P, Ghyselinck NB (2003) A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol 21:562–565. [DOI] [PubMed] [Google Scholar]
- Sheintuch L, Rubin A, Brande-Eilat N, Geva N, Sadeh N, Pinchasof O, Ziv Y (2017) Tracking the Same Neurons across Multiple Days in Ca(2+) Imaging Data. Cell reports 21:1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Pribiag H, Lilascharoen V, Knowland D, Wang X-Y, Lim BK (2018) Drd3 Signaling in the Lateral Septum Mediates Early Life Stress-Induced Social Dysfunction. Neuron 97:195–208.e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siciliano CA, Noamany H, Chang CJ, Brown AR, Chen X, Leible D, Lee JJ, Wang J, Vernon AN, Vander Weele CM, Kimchi EY, Heiman M, Tye KM (2019) A cortical-brainstem circuit predicts and governs compulsive alcohol drinking. Science 366:1008–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal VS, Zhang F, Yizhar O, Deisseroth K (2009) Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature 459:698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stosiek C, Garaschuk O, Holthoff K, Konnerth A (2003) In vivo two-photon calcium imaging of neuronal networks. Proc Natl Acad Sci U S A 100:7319–7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Blanco-Centurion C, Bendell E, Vidal-Ortiz A, Luo S, Liu M (2019) Activity dynamics of amygdala GABAergic neurons during cataplexy of narcolepsy. eLife 8:e48311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K, Block SM (1994) Biological Applications of Optical Forces. Annual Review of Biophysics and Biomolecular Structure 23:247–285. [DOI] [PubMed] [Google Scholar]
- Svoboda K, Denk W, Kleinfeld D, Tank DW (1997) In vivo dendritic calcium dynamics in neocortical pyramidal neurons. Nature 385:161–165. [DOI] [PubMed] [Google Scholar]
- Sych Y, Chernysheva M, Sumanovski LT, Helmchen F (2019) High-density multi-fiber photometry for studying large-scale brain circuit dynamics. Nature Methods 16:553–560. [DOI] [PubMed] [Google Scholar]
- Tada M, Takeuchi A, Hashizume M, Kitamura M, Kano M (2014) A highly sensitive fluorescent indicator dye for calcium imaging of neural activity in vitro and in vivo. European Journal of Neuroscience 39:1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenaz P, Ruttimann UE, Unser M (1998) A pyramid approach to subpixel registration based on intensity. IEEE Transactions on Image Processing 7:27–41. [DOI] [PubMed] [Google Scholar]
- Weber F, Dan Y (2016) Circuit-based interrogation of sleep control. Nature 538:51–59. [DOI] [PubMed] [Google Scholar]
- Weber F, Hoang Do JP, Chung S, Beier KT, Bikov M, Saffari Doost M, Dan Y (2018) Regulation of REM and Non-REM Sleep by Periaqueductal GABAergic Neurons. Nature Communications 9:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Abdelfattah AS, Miraucourt LS, Kutsarova E, Ruangkittisakul A, Zhou H, Ballanyi K, Wicks G, Drobizhev M, Rebane A, Ruthazer ES, Campbell RE (2014) A long Stokes shift red fluorescent Ca2+ indicator protein for two-photon and ratiometric imaging. Nat Commun 5:5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Yang H, Menon V, Lemire AL, Wang L, Henry FE, Turaga SC, Sternson SM (2020) Behavioral state coding by molecularly defined paraventricular hypothalamic cell type ensembles. Science 370:eabb2494. [DOI] [PubMed] [Google Scholar]
- Yang W, Yuste R (2017) In vivo imaging of neural activity. Nature Methods 14:349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda R, Nimchinsky EA, Scheuss V, Pologruto TA, Oertner TG, Sabatini BL, Svoboda K (2004) Imaging Calcium Concentration Dynamics in Small Neuronal Compartments. Science's STKE 2004:pl5. [DOI] [PubMed] [Google Scholar]
- Zhou P, Resendez SL, Rodriguez-Romaguera J, Jimenez JC, Neufeld SQ, Giovannucci A, Friedrich J, Pnevmatikakis EA, Stuber GD, Hen R, Kheirbek MA, Sabatini BL, Kass RE, Paninski L (2018) Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Burns LD, Cocker ED, Hamel EO, Ghosh KK, Kitch LJ, Gamal AE, Schnitzer MJ (2013) Long-term dynamics of CA1 hippocampal place codes. Nature Neuroscience 16:264–266. [DOI] [PMC free article] [PubMed] [Google Scholar]