Abstract
In a thought-provoking study, Ocampo et al. show that the cyclic expression of stem cell reprogramming factors in vivo increases the lifespan of a mouse model of premature aging and provides health benefits to chronologically old, normal mice.
Reprogramming of somatic cells to induced pluripotent stem cells (iPSCs) not only holds great promise to model and treat many diseases but can also provide insights into aging and rejuvenation mechanisms (Studer et al., 2015). Indeed, reprogramming of aged somatic cells into iPSCs reverts some hallmarks of aging—including telomere length, mitochondrial fitness, abnormal nuclear morphologies, loss of heterochromatin markers, and gene expression—to a youthful embryonic state (Studer et al., 2015). In this issue of Cell, Ocampo et al. (2016) apply stem cell reprogramming factors in vivo to improve the health of an organism and to prolong its lifespan. They first find that transient in vitro expression of the four Yamanaka reprogramming factors—Oct4, Sox2, Klf4, and c-Myc (OSKM)—reverts age-associated features in mouse and human cells, without the loss of cellular identity. Remarkably, the authors show that at the organism level, short-term induction of the four reprogramming factors improves the physical state of a mouse model of premature aging and extends its lifespan. Importantly, they also provide evidence that transient expression of the four factors has beneficial effects in physiologically aged, normal mice.
An intriguing yet unexplored question in the aging and reprogramming field has been whether it is possible to uncouple the rejuvenation and the dedifferentiation process associated with iPSC reprogramming. Although previous studies have shown that transient expression of the four factors can induce a partially dedifferentiated state (without a complete loss of cellular identity) (Kurian et al., 2013), whether this partial state is sufficient to revert age-associated features remained unclear. The authors now show that short-term expression of the four Yamanaka factors is sufficient to revert some age-associated features in mouse and human cells without dedifferentiation. Using fibroblasts from a transgenic mouse model of premature aging (a model for Hutchinson Gilford Progeria Syndrome) that also carries an inducible OSKM polycistronic cassette, they show that short-term induction of OSKM (2 to 4 days) reduces markers of DNA damage (γH2AX), senescence, and mitochondrial impairment (reactive oxygen species production), without the loss of cellular identity. Moreover, expression of reprogramming factors restores the levels of two epigenetic marks of heterochromatin that have previously been associated with aging and progeria, namely trimethylated lysine 9 on histone H3 (H3K9me3) and H4K20me3 (Pal and Tyler, 2016). These effects are not observed upon induction of a single reprogramming factor, suggesting that a combination of the four factors is required. Interestingly, 8 days after OSKM induction, the progeria-associated features slowly come back, suggesting that OSKM expression provides a transient reversion of premature aging phenotypes. However, the re-establishment of age-associated features may be specific to the progeria model used, whose cells carry a genetic alteration. Additional experiments will be required to examine how well these findings recapitulate in cells from normal old mice.
What drives the reversion of these age-associated features? Ocampo et al. (2016) provide evidence that epigenetic changes may be at the core of this reversion. In time-course experiments, the epigenetic changes precede the reversion of DNA damage and senescence markers, and chemical inhibition of H3K9me3 methyl-transferases abrogates the rejuvenation effect. However, many questions remain; for example, it is unclear which precise histone mark is required. The authors show restored levels of H3K9me3 and H4K20me3, but the reprogramming factors induce a complete remodeling of the chromatin landscape (Takahashi and Yamanaka, 2016). Are alterations of other histone marks (e.g., H3K4me3 and H3K27me3, which also change which age) required, and to what extent? Interestingly, H3K9me3 marks mainly constitutive heterochromatin and is often found coating retrotransposal elements, thereby preventing their spurious reactivation. Expression of retrotransposal elements has been associated with cellular senescence and organismal aging, and suppression of these elements can reverse the senescence phenotype, at least in vitro (Pal and Tyler, 2016). These findings suggest a critical role of epigenetic remodeling in the rejuvenation process, perhaps by improving genomic stability. These results raise the exciting possibility that epigenetic changes supersede/precede other aging hallmarks in the physiological aging process, as well, and may thus constitute a key target for future rejuvenation strategies. It would be interesting to determine if transient pulses of OSKM are sufficient to erase other hallmarks of aging—including metabolism defects, increased inflammation, and increased protein aggregates—and determine their hierarchy compared to epigenetic events in this process.
Arguably, the most provocative question of this study is whether cellular reprogramming can also reset aging hallmarks at the organismal level (in vivo). Previous studies have been hampered by the fact that continuous expression of the four factors in vivo has resulted in extensive cancer development and high rate of mortality (Abad et al., 2013). The authors circumvent these issues by optimizing a protocol for in vivo cyclic induction of the reprogramming factors. When applied to a mouse model of premature aging that carries a mutation in the Lmna gene and produces a truncated Lamin A (LAKI mice), the mice exhibit improvements in features associated with their disease state, including physical appearance, histological changes in organs, as well as cardiovascular functions. Importantly, they also exhibit increased median and maximal lifespan (though the number of animals is relatively low [n < 30]). A central question is whether these findings can be extended to physiological aging. Interestingly, the authors show that cyclic expression of the four factors improves the regenerative capacity of pancreas and muscle following injury in physiologically old normal mice. A key remaining question is whether these health benefits produce lifespan extension in normal mice. Other key questions are concerning the stability of the health benefits and how late in life OSKM expression can be initiated to still provide a beneficial effect. Ultimately, it will be interesting to compare and contrast transient reprogramming with other interventions known to delay or reverse aging (Figure 1), including elimination of senescent cells (genetically or via senolytics) (Trabucco and Zhang, 2016), parabiosis (the fusion of young and old animals by blood circulation [Conboy et al., 2013]), rapamycin (Arriola Apelo and Lamming, 2016), and a fasting-mimicking diet (Brandhorst et al., 2015).
Figure 1. Comparison between Transient Reprogramming and Other Interventions for Tissue Rejuvenation and Lifespan.
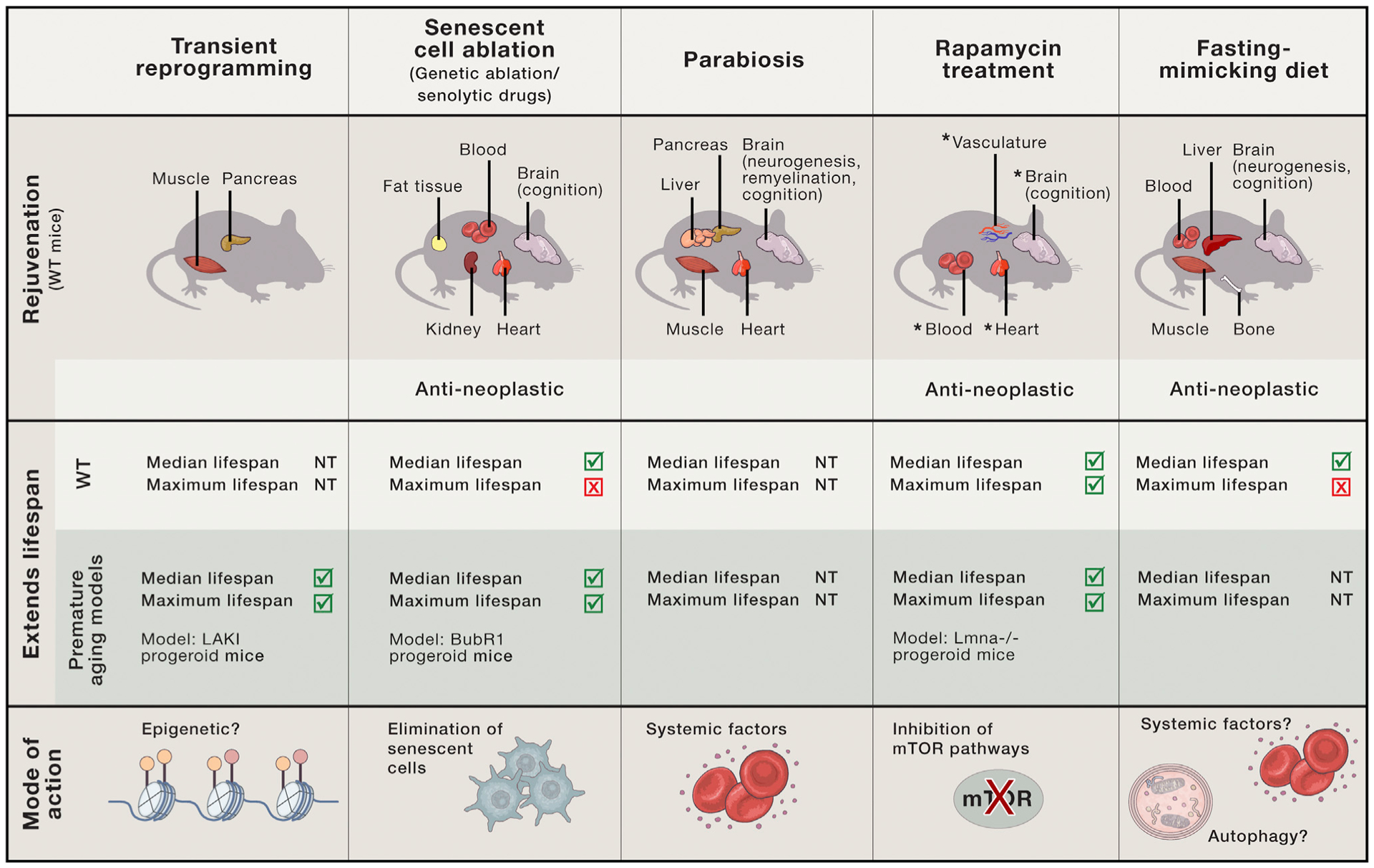
Comparison between interventions that have been shown to improve health span and lifespan in mice starting late in life, and their proposed mode of action. The intervention described in Ocampo et al. (2016), transient reprogramming, improves the regenerative capacity of pancreas and muscle following injury and increases lifespan in a premature aging mouse model, potentially via induced epigenetic changes. Only the organs/tissues that exhibit a rejuvenation/reverted phenotype in wild-type (WT) mice are indicated. “Blood” includes changes in immune system. NT, not tested. *, shown to also improve in young mice.
How, mechanistically, does cyclic induction of reprogramming factors improve health span and lifespan of an organism? A key question is whether the extension of lifespan is solely due to the cellular rejuvenation provided by reprogramming or whether it is triggered by another mechanism. As observed in vitro, short-term induction of OSKM in vivo also leads to a decrease in DNA damage response genes and senescence-associated features and restored H3K9me3 and H4K20me3 levels in multiple organs. But an important question is whether all cells in a given organ experience this reversion or whether a subset of reprogrammed cells is enough to drive the increased functionality of the whole organ. At the organismal level, it would be interesting to know which organs are more susceptible to this induced rejuvenation and whether the rejuvenation of all organs is necessary for the lifespan effect. It is possible that rejuvenation of a central organ may help the entire organism, perhaps via systemic factors. Organ-specific induction of the four factors could expand our understanding of the organs that are critical for aging and that may drive the aging process of the whole organism. Also, as most age-associated features investigated in this study are related to DNA damage and cellular senescence, it would be interesting to determine how reprogramming could affect senescent cells in a tissue and if reprogramming potentially induces their selective killing. It would be interesting to combine this reprogramming approach with drugs that target senescent cells (senolytic) or with genetic models for elimination of senescent cells in vivo (Trabucco and Zhang, 2016). Testing the interaction between the two approaches could tease out whether the reprogramming factors mainly act by impacting the senescent cells and/or if it has additional beneficial effects.
The burning question is naturally whether this approach may also benefit human health and lifespan. The authors provide some initial observations that late-passage fibroblasts (redifferentiated from human iPSCs) exhibit a similar reversion in age-associated features upon short-term induction of the four factors. Although preliminary, these results suggest that these findings can be extended to human cells. A similar inducible whole-organism system in humans would be difficult to implement, yet future applications could involve more targeted approaches in key failing organs using non-invasive methods or the identification of compounds that can mimic it. A more extensive investigation of the risks associated with this approach would also be needed, especially in regard to tumor development in normal organisms. Because epigenetic changes appear to play a role early in the process, drugs that modulate specific epigenetic modifiers could be used, if targeted appropriately, as a way to mimic transient reprogramming. In addition, specific compounds or small RNAs that have been shown to enable reprogramming without exogenous gene expression (Takahashi and Yamanaka, 2016) could also be used to transiently ameliorate tissue functions without inducing dedifferentiation, thereby increasing the spectrum of thera pies for progeroid diseases and even physiological aging.
REFERENCES
- Abad M, Mosteiro L, Pantoja C, Cañamero M, Rayon T, Ors I, Graña O, Megías D, Domínguez O, Martínez D, et al. (2013). Nature 502, 340–345. [DOI] [PubMed] [Google Scholar]
- Arriola Apelo SI, and Lamming DW (2016). J. Gerontol. A Biol. Sci. Med. Sci 71, 841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandhorst S, Choi IY, Wei M, Cheng CW, Sedrakyan S, Navarrete G, Dubeau L, Yap LP, Park R, Vinciguerra M, et al. (2015). Cell Metab. 22, 86–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy MJ, Conboy IM, and Rando TA (2013). Aging Cell 12, 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian L, Sancho-Martinez I, Nivet E, Aguirre A, Moon K, Pendaries C, Volle-Challier C, Bono F, Herbert JM, Pulecio J, et al. (2013). Nat. Methods 10, 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo A, Reddy P, Martinez-Redondo P, Platero-Luengo A, Hatanaka F, Hishida T, Li M, Lam D, Kurita M, Beyret E, et al. (2016). Cell 167, this issue, 1719–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, and Tyler JK (2016). Sci. Adv 2, e1600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer L, Vera E, and Cornacchia D (2015). Cell Stem Cell 16, 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S (2016). Nat. Rev. Mol. Cell Biol 17, 183–193. [DOI] [PubMed] [Google Scholar]
- Trabucco SE, and Zhang H (2016). Cell Stem Cell 18, 305–306. [DOI] [PubMed] [Google Scholar]


